Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis (original) (raw)
Abstract
We measured the expression pattern and analyzed codon usage in 8,133, 1,550, and 2,917 genes, respectively, from Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana. In those three species, we observed a clear correlation between codon usage and gene expression levels and showed that this correlation is not due to a mutational bias. This provides direct evidence for selection on silent sites in those three distantly related multicellular eukaryotes. Surprisingly, there is a strong negative correlation between codon usage and protein length. This effect is not due to a smaller size of highly expressed proteins. Thus, for a same-expression pattern, the selective pressure on codon usage appears to be lower in genes encoding long rather than short proteins. This puzzling observation is not predicted by any of the current models of selection on codon usage and thus raises the question of how translation efficiency affects fitness in multicellular organisms.
Nonrandom usage of synonymous codons is a widespread phenomenon, observed in genomes from many species in all domains of life. Such codon-usage biases may result from mutational biases, from natural selection acting on silent changes in DNA, or both. Selection on codon usage has been clearly demonstrated in several unicellular organisms (e.g., Escherichia coli, Saccharomyces cerevisiae) (for review, see ref. 1). Three characteristics of codon usage reflect such selective pressure in those organisms. First, codon usage is biased toward “preferred” codons that generally correspond to the most abundant tRNA species (2). Second, there is a positive correlation between codon-usage bias and the level of gene expression (3, 4). Finally, the rate of synonymous substitution between species is inversely correlated with codon usage bias, implying greater purifying selection on silent changes in highly biased genes (4, 5).
The selective differences between alternative synonymous codons are probably very small. Thus, in genes with low expression levels, or in species with small population sizes, selection is not sufficient to overcome genetic drift, and codon usage is essentially shaped by mutation patterns (for review, see ref. 6). The “selection–mutation–drift” model was proposed (4, 7, 8) to describe this balance between selection favoring optimal codons and mutation with drift allowing persistence of nonoptimal codons.
In multicellular eukaryotes, gene expression and tRNA abundance can be tissue- and developmental stage-specific and are difficult to quantify. However, the action of natural selection on codon usage has been established in Drosophila melanogaster: the limited data available show a relationship between codon preference and tRNA abundance (9, 10); negative correlations between codon usage bias and silent substitution rate have been observed in Drosophila (9, 11, 12); finally, anecdotal evidence suggests a relationship between codon-usage bias and gene expression level: genes known to be expressed at a high level, such as those encoding ribosomal proteins or glycolytic enzymes, show a greater-than-average codon bias (9, 12). Other studies, although less extensive, suggest that selection on codon usage may also occur in another invertebrate, Caenorhabditis elegans (13), and in the plant Arabidopsis thaliana (14).
Optimal codons probably confer fitness benefits by enhancing translation efficiency. However, it is not yet clear whether codon usage affects primarily the elongation rate, the cost of proofreading, or the accuracy of translation. Several studies suggested that in D. melanogaster, selection acts to increase translation accuracy (15, 16). However, in absence of expression data, it was not possible to directly test this hypothesis.
Recently, expressed sequence tag (EST) projects have been initiated in different species with the aim to make the inventory of all the mRNAs that they express. The thousands of obtained sequences are generally partial (typically, sequences are 300–500 nt long) and with a relatively high rate of sequencing errors (≈3%). However, these ESTs are accurate and long enough to unambiguously identify their corresponding genes. There is a high redundancy among those ESTs, which reflects the relative abundance of mRNAs in the tissue from which the cDNA library has been prepared. Thus, these data can be used to get rough estimates of gene expression patterns.
The purpose of the work presented here was to measure the expression levels of large sets of genes available for D. melanogaster, C. elegans, and A. thaliana to directly test whether there was selection on codon usage in those species. Our results demonstrate that selection acts on silent sites in those three distantly related multicellular eukaryotes. But surprisingly, we observed a strong negative correlation between codon-usage bias and protein length that is not due to a smaller size of highly expressed proteins. None of the current models of selection on silent site for translation efficiency accounts for this puzzling observation.
MATERIALS AND METHODS
Sequence Data.
C. elegans, D. melanogaster, and A. thaliana sequences were extracted from GenBank release 105 (February 1998) (17), by using the acnuc retrieval system (18). We selected complete protein-coding sequences (CDS) from nuclear genes, excluding pseudogenes and sequences described as ORFs or “unidentified reading frames”. Histone genes also were excluded (see below). Only genomic sequences were selected, except for D. melanogaster, for which we also included CDS from mRNA sequences to increase the sample size. All CDS were compared with each other with blastn2 (19) to remove redundant sequences. In case of alternative splicing, we retained only the longest CDS variant. The final data set included 8,133, 1,550, and 2,917 CDS, respectively from C. elegans, D. melanogaster, and A. thaliana, among which 7,891, 439, and 2,386 were interrupted by introns.
Expression Profiles.
Expression profiles were determined by counting the number of occurrence of each gene among EST sequences from different cDNA libraries that had been sampled with at least 9,000 ESTs. We selected in GenBank 67,987 C. elegans ESTs from whole-animal cDNA libraries at two developmental states (adult; embryo), 27,491 ESTs from D. melanogaster (adult ovary and head; embryo), and 36,207 ESTs from A. thaliana (one cDNA library pooled from four different tissues).
Selected CDS were first filtered with the xblast program (20) to mask repetitive elements. CDS were then compared with the species-specific EST data set by using blastn2 (19). blastn2 alignments showing at least 95% identity over 100 nt or more were counted as a sequence match. Because ESTs are derived from poly(A)+ selected cDNA libraries, they cannot be used to estimate the abundance of replication-dependent histone mRNAs (that are not polyadenylated).
RESULTS
Measuring Gene Expression with ESTs.
We selected from the databases 8,133, 1,550, and 2,917 complete protein-coding sequences from C. elegans, D. melanogaster, and A. thaliana, respectively. These large data sets represent 10–50% of the estimated number of genes in those three species, and are thus expected to be representative of whole genomes. EST sequences from different cDNA libraries were extracted from GenBank. For each species, and for each cDNA library, the expression level of selected genes was measured by counting the number of matching ESTs and dividing this number by the total number of ESTs sequenced in that cDNA library. Therefore, our measures reflect the relative mRNA abundance in those tissues where the cDNA libraries have been sampled. For C. elegans and D. melanogaster, libraries from two different stages (embryo, adult) were available. For genes expressed in both stages, we retained only their maximal relative abundance among these two cDNA libraries.
Genes were sorted according to their expression level and classified in four groups (Table 1). Genes without any EST made the first group. Expressed genes were classified in three other groups of low, moderate, and high expression. The limits between these classes were set for each species to obtain three samples of equal size (except for A. thaliana, where genes matching one single EST represent 53% of expressed genes).
Table 1.
Summary of relative mRNA abundance measures, based on EST sequence data
| Organism | Genes, no. | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| All | No Est | Expression | All | mRNA abundance × 105, expression | |||||
| Low | Moderate | High | Low | Moderate | High | ||||
| C. elegans | 8,133 | 4,495 | 1,150 | 1,242 | 1,246 | 9 (0–525) | 4 (3–5) | 10 (7–16) | 44 (17–525) |
| D. melanogaster | 1,550 | 675 | 214 | 330 | 331 | 16 (0–819) | 7 (5–10) | 13 (11–21) | 57 (22–819) |
| A. thaliana | 2,917 | 1,720 | 638 | 289 | 270 | 3 (0–146) | 3 (3–3) | 6 (6–6) | 16 (8–146) |
It should be noted that estimates of expression level derived from ESTs are imprecise. Notably, 17% of A. thaliana ESTs were obtained from a normalized cDNA collection (i.e., from which redundant clones have been removed) (21). As a consequence, the mRNA abundance of genes expressed at a high level is underestimated in A. thaliana. However, the relative order of genes sorted according their expression level should not be affected. Thus, even if ESTs are partly normalized, the classification in four broad groups of expression that we used remains correct.
Frequency of Favored Codons and Gene Expression.
Sharp and colleagues defined “optimal” codons as those showing a statistically significant increase in frequency between genes with low and high codon-usage bias (13, 22). Note that this definition does not necessarily imply any relationship with translation efficiency. In our opinion, the term optimal is equivocal here because it a priori suggests that there is a relationship with gene expression. Thus, codons found to occur significantly more often in highly than in lowly biased genes will hereafter be referred as favored codons. Here, optimal codons will refer only to those codons whose frequency has been shown to increase with gene expression.
Favored codons have been identified by multivariate analysis in D. melanogaster (22), C. elegans (13), and A. thaliana (14). We calculated for each gene the frequency of favored codons (Fav). Fav is a species-specific measure of codon-usage bias and is calculated as the number of occurrences of these favored codons divided by the total number of occurrences of the 18 amino acids having synonymous codons (13).
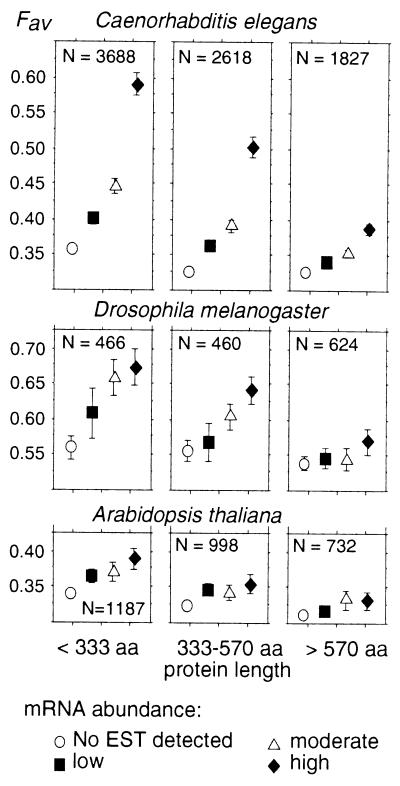
We computed average Fav in C. elegans, D. melanogaster, and A. thaliana for genes from the four groups of expression level. To investigate the effect of protein length on codon usage, the data set was split into three groups of equal sample size: short (<333 aa), intermediate, and long proteins (>570 aa) (Fig. 1).
Figure 1.
Frequency of favored codons and gene expression in C. elegans, D. melanogaster, and A. thaliana. Average Fav values have been computed for different expression levels and protein lengths. Error bars indicate the 95% confidence interval.
Three observations can made. First, Fig. 1 clearly shows that in the three species studied, there is an increase of Fav with expression level. Thus, the relationship between codon-usage bias and gene expression that had been suggested for those multicellular organisms is here directly demonstrated. Second, the range of variation of Fav with expression level is not the same in all species: the increase is very sharp in C. elegans and D. melanogaster and less pronounced in A. thaliana. It should be noted however, that the weaker correlation between Fav and expression in this latter species may be partly because of the fact that mRNA abundance of highly expressed genes is underestimated (see above). Finally, the increase of codon-usage bias with expression level is much stronger in genes coding for short than for long proteins.
Detailed Analysis for Each Amino Acid.
The relative synonymous codon usage (RSCU) is the observed frequency of a codon divided by the frequency expected if all synonyms for that amino acid were used equally. Thus, RSCU values close to 1.0 indicate a lack of bias. The RSCU is independent of amino acid composition and is thus useful for comparing different sets of genes (13). Table 2 reports average RSCU values of all codons from C. elegans, D. melanogaster, and A. thaliana genes, according to their relative mRNA abundance and the length of the protein they code. ΔRSCU corresponds to the difference of RSCU values between genes expressed at a high level and those for which we did not detect any EST. Favored codons (identified by multivariate analysis) and optimal codons (the ones with positive ΔRSCU values) are highlighted. There is a remarkable correspondence between optimal and favored codons: in C. elegans, D. melanogaster, and A. thaliana, respectively, 100% (21/21), 91% (20/22), and 86% (18/21) of favored codons are also optimal, whereas only 13% (5/38), 14% (5/37), and 11% (4/38) of nonfavored codons are optimal (and in all of these latter cases, ΔRSCU is weak).
Table 2.
Codon usage (RSCU values), expression level, and protein length in C. elegans, D. melanogaster, and A. thaliana genes
| Codon | Favored | C. elegans | Favored | D. melanogaster | Favored | A. thaliana | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Short proteins | Long proteins | Short proteins | Long proteins | Short proteins | Long proteins | ||||||||||
| No EST | High | No EST | High | No EST | High | No EST | High | No EST | High | No EST | High | ||||
| Arg | 25,872 | 3,833 | 17,254 | 40,260 | 2,404 | 1,042 | 12,349 | 8,030 | 8,004 | 1,372 | 20,572 | 2,478 | |||
| AGA | 1.87 | 1.56 | 2.02 | 1.86 | 0.64 | 0.34 | 0.57 | 0.39 | 2.07 | 1.92 | 2.20 | 2.09 | |||
| AGG | 0.47 | 0.15 | 0.51 | 0.31 | 0.72 | 0.47 | 0.75 | 0.53 | ∗ | 1.17+ | 1.36 | 1.22 | 1.24 | ||
| CGA | 1.33 | 0.62 | 1.43 | 1.31 | 0.77 | 0.41 | 0.98 | 0.86 | 0.74 | 0.55 | 0.70 | 0.64 | |||
| CGC | ∗ | 0.58+++ | 1.34 | 0.45+ | 0.57 | ∗ | 1.92+++ | 2.80 | 1.83+++ | 2.29 | 0.43 | 0.41 | 0.40 | 0.37 | |
| CGG | 0.51 | 0.24 | 0.51 | 0.39 | 0.83 | 0.47 | 0.97 | 0.79 | 0.53 | 0.37 | 0.57 | 0.54 | |||
| CGU | ∗ | 1.24+++ | 2.09 | 1.08+++ | 1.56 | ∗ | 1.11+++ | 1.50 | 0.90++ | 1.15 | ∗ | 1.05+++ | 1.40 | 0.91++ | 1.11 |
| Leu | 46,057 | 4,576 | 32,074 | 61,200 | 3,540 | 1,433 | 20,782 | 13,530 | 12,438 | 2,191 | 38,506 | 4,980 | |||
| CUA | 0.58 | 0.19 | 0.63 | 0.43 | 0.46 | 0.23 | 0.56 | 0.51 | 0.62 | 0.52 | 0.63 | 0.59 | |||
| CUC | ∗ | 0.96+++ | 1.92 | 0.84++ | 1.10 | ∗ | 1.08 | 1.03 | 0.93 | 0.92 | ∗ | 1.12+++ | 1.50 | 0.92 | 0.96 |
| CUG | 0.76 | 0.69 | 0.79 | 0.73 | ∗ | 2.64+++ | 3.22 | 2.59 | 2.58 | 0.57 | 0.56 | 0.68 | 0.74 | ||
| CUU | ∗ | 1.45+++ | 1.76 | 1.41+++ | 1.78 | 0.60 | 0.41 | 0.56 | 0.62 | 1.54+ | 1.63 | 1.52 | 1.57 | ||
| UUA | 0.82 | 0.19 | 0.89 | 0.55 | 0.27 | 0.15 | 0.29 | 0.30 | 0.83 | 0.56 | 0.89 | 0.75 | |||
| UUG | 1.43 | 1.25 | 1.42 | 1.42 | 0.94 | 0.97 | 1.06 | 1.06 | 1.33 | 1.24 | 1.36 | 1.40 | |||
| Ser | 40,824 | 4,049 | 28,694 | 58,229 | 3,295 | 1,121 | 21,409 | 11,905 | 12,722 | 2,316 | 35,838 | 4,297 | |||
| AGC | 0.62+ | 0.78 | 0.54 | 0.54 | 1.53 | 1.24 | 1.52 | 1.44 | ∗ | 0.78 | 0.81 | 0.75 | 0.78 | ||
| AGU | 0.92 | 0.55 | 1.01 | 0.90 | 0.72 | 0.37 | 0.88 | 0.74 | 0.92 | 0.75 | 1.04 | 0.97 | |||
| UCA | 1.61 | 0.99 | 1.65 | 1.54 | 0.47 | 0.36 | 0.56 | 0.59 | 1.17 | 1.08 | 1.30 | 1.24 | |||
| UCC | ∗ | 0.76+++ | 1.39 | 0.73 | 0.73 | ∗ | 1.76+ | 1.92 | 1.37 | 1.43 | ∗ | 0.78+ | 0.89 | 0.67+ | 0.76 |
| UCG | 0.79+ | 0.98 | 0.76+ | 0.87 | ∗ | 1.03+++ | 1.64 | 1.23 | 1.21 | 0.68 | 0.59 | 0.55 | 0.59 | ||
| UCU | 1.30 | 1.31 | 1.32+ | 1.42 | 0.49 | 0.47 | 0.45+ | 0.59 | 1.67++ | 1.89 | 1.69 | 1.66 | |||
| Thr | 30,391 | 3,260 | 20,939 | 41,866 | 2,498 | 918 | 13,725 | 8,234 | 7,401 | 1,452 | 19,911 | 2,690 | |||
| ACA | 1.40 | 0.79 | 1.51 | 1.36 | 0.72 | 0.60 | 0.82 | 0.75 | 1.12 | 1.01 | 1.31 | 1.21 | |||
| ACC | ∗ | 0.69+++ | 1.50 | 0.59+ | 0.67 | ∗ | 1.74+++ | 2.22 | 1.48+ | 1.58 | ∗ | 0.85+ | 0.99 | 0.72+ | 0.80 |
| ACG | 0.57 | 0.42 | 0.57 | 0.53 | 0.79 | 0.67 | 1.08 | 0.97 | 0.73 | 0.57 | 0.54 | 0.57 | |||
| ACU | 1.34 | 1.29 | 1.33+ | 1.43 | 0.75 | 0.51 | 0.63 | 0.70 | 1.30+ | 1.43 | 1.43 | 1.42 | |||
| Pro | 23,312 | 2,961 | 15,553 | 34,714 | 2,442 | 809 | 13,932 | 7,782 | 6,716 | 1,373 | 18,010 | 2,424 | |||
| CCA | ∗ | 2.10+++ | 2.82 | 2.03++ | 2.31 | 0.93 | 0.78 | 1.01 | 0.97 | 1.27 | 1.25 | 1.35 | 1.38 | ||
| CCC | 0.38 | 0.19 | 0.39 | 0.29 | ∗ | 1.52+++ | 1.88 | 1.26 | 1.31 | ∗ | 0.41+ | 0.52 | 0.43 | 0.46 | |
| CCG | 0.72 | 0.61 | 0.73 | 0.65 | 1.02 | 0.92 | 1.28 | 1.16 | 0.82 | 0.76 | 0.63 | 0.67 | |||
| CCU | 0.79 | 0.38 | 0.86 | 0.76 | 0.53 | 0.42 | 0.45+ | 0.56 | 1.50 | 1.48 | 1.58 | 1.48 | |||
| Ala | 29,935 | 5,149 | 19,697 | 49,193 | 3,432 | 1,494 | 18,160 | 11,932 | 8,775 | 2,171 | 23,348 | 3,535 | |||
| GCA | 1.35 | 0.66 | 1.48 | 1.21 | 0.57 | 0.43 | 0.74 | 0.72 | 1.00 | 0.82 | 1.19 | 1.11 | |||
| GCC | ∗ | 0.74+++ | 1.40 | 0.66+ | 0.74 | ∗ | 2.01+++ | 2.32 | 1.74 | 1.77 | ∗ | 0.68+ | 0.78 | 0.58 | 0.60 |
| GCG | 0.48 | 0.31 | 0.51 | 0.42 | 0.59 | 0.49 | 0.81 | 0.66 | 0.64 | 0.58 | 0.50 | 0.52 | |||
| GCU | ∗ | 1.42++ | 1.63 | 1.35++ | 1.62 | 0.83 | 0.77 | 0.71+ | 0.86 | 1.68+ | 1.82 | 1.73 | 1.77 | ||
| Gly | 25,766 | 4,491 | 16,959 | 39,020 | 3,281 | 1,284 | 15,131 | 10,315 | 9,656 | 2,068 | 23,343 | 3,579 | |||
| GGA | ∗ | 2.26+++ | 2.77 | 2.28+++ | 2.59 | 1.23 | 0.94 | 1.13 | 1.12 | 1.53 | 1.48 | 1.43 | 1.49 | ||
| GGC | 0.50 | 0.40 | 0.48 | 0.36 | ∗ | 1.66++ | 1.94 | 1.70 | 1.69 | ∗ | 0.60 | 0.59 | 0.52 | 0.53 | |
| GGG | 0.36 | 0.15 | 0.37 | 0.22 | 0.25 | 0.16 | 0.32 | 0.23 | 0.54 | 0.47 | 0.69 | 0.61 | |||
| GGU | 0.87 | 0.69 | 0.87 | 0.83 | 0.86+ | 0.96 | 0.84+ | 0.96 | ∗ | 1.34+ | 1.46 | 1.36 | 1.37 | ||
| Val | 31,831 | 3,934 | 21,386 | 45,812 | 2,662 | 1,123 | 13,116 | 8,981 | 9,361 | 1,784 | 25,453 | 3,497 | |||
| GUA | 0.67 | 0.25 | 0.77 | 0.59 | 0.30 | 0.28 | 0.43 | 0.44 | 0.56 | 0.47 | 0.67 | 0.60 | |||
| GUC | ∗ | 0.83+++ | 1.56 | 0.74+ | 0.88 | ∗ | 1.13+ | 1.29 | 0.93 | 0.94 | ∗ | 0.81 | 0.87 | 0.69 | 0.69 |
| GUG | 0.90 | 0.71 | 0.88 | 0.79 | ∗ | 1.79 | 1.84 | 1.92 | 1.81 | 1.07 | 1.11 | 1.02 | 1.06 | ||
| GUU | 1.60 | 1.47 | 1.61+ | 1.74 | 0.78 | 0.58 | 0.72+ | 0.80 | 1.56 | 1.56 | 1.63 | 1.64 | |||
| Lys | 34,361 | 4,512 | 22,049 | 47,882 | 2,529 | 1,441 | 11,653 | 9,450 | 9,257 | 1,930 | 24,600 | 3,135 | |||
| AAA | 1.27 | 0.59 | 1.31 | 1.11 | 0.59 | 0.35 | 0.60 | 0.54 | 0.96 | 0.80 | 1.00 | 0.94 | |||
| AAG | ∗ | 0.73+++ | 1.41 | 0.69++ | 0.89 | ∗ | 1.41++ | 1.65 | 1.40 | 1.46 | ∗ | 1.04+ | 1.20 | 1.00 | 1.06 |
| Asn | 25,983 | 2,605 | 18,131 | 34,502 | 1,936 | 718 | 11,562 | 7,439 | 6,174 | 998 | 17,808 | 2,166 | |||
| AAC | ∗ | 0.75+++ | 1.22 | 0.70 | 0.76 | ∗ | 1.23++ | 1.47 | 1.09+ | 1.17 | ∗ | 1.03+ | 1.15 | 0.90+ | 0.99 |
| AAU | 1.25 | 0.78 | 1.30 | 1.24 | 0.77 | 0.53 | 0.91 | 0.83 | 0.97 | 0.85 | 1.10 | 1.01 | |||
| Gln | 19,766 | 2,424 | 13,462 | 32,392 | 2,071 | 665 | 14,445 | 8,767 | 4,845 | 875 | 14,059 | 1,783 | |||
| CAA | 1.37 | 1.19 | 1.39 | 1.37 | 0.55 | 0.41 | 0.60 | 0.58 | 1.18 | 1.07 | 1.12 | 1.07 | |||
| CAG | ∗ | 0.63+ | 0.81 | 0.61 | 0.63 | ∗ | 1.45+ | 1.59 | 1.40 | 1.42 | ∗ | 0.82+ | 0.93 | 0.88 | 0.93 |
| His | 11,626 | 1,270 | 7,872 | 17,198 | 1,136 | 342 | 7,300 | 3,789 | 3,222 | 606 | 8,933 | 1,100 | |||
| CAC | ∗ | 0.75+++ | 1.23 | 0.72 | 0.73 | ∗ | 1.25++ | 1.46 | 1.21 | 1.22 | ∗ | 0.81+ | 0.90 | 0.69+ | 0.82 |
| CAU | 1.25 | 0.77 | 1.28 | 1.27 | 0.75 | 0.54 | 0.79 | 0.78 | 1.19 | 1.10 | 1.31 | 1.18 | |||
| Glu | 30,622 | 3,979 | 21,650 | 55,173 | 2,508 | 1,231 | 13,586 | 11,132 | 9,708 | 1,623 | 26,319 | 3,529 | |||
| GAA | 1.30 | 0.92 | 1.36 | 1.30 | 0.58 | 0.35 | 0.62 | 0.61 | 1.02 | 1.00 | 1.08 | 1.00 | |||
| GAG | ∗ | 0.70+++ | 1.08 | 0.64 | 0.70 | ∗ | 1.42++ | 1.65 | 1.38 | 1.39 | ∗ | 0.98 | 1.00 | 0.92+ | 1.00 |
| Asp | 25,098 | 3,551 | 17,514 | 43,357 | 2,166 | 1,052 | 11,557 | 8,516 | 7,676 | 1,366 | 20,700 | 2,743 | |||
| GAC | ∗ | 0.66++ | 0.92 | 0.60 | 0.58 | ∗ | 0.98+ | 1.08 | 0.95 | 0.96 | ∗ | 0.64+ | 0.72 | 0.60 | 0.63 |
| GAU | 1.34 | 1.08 | 1.40 | 1.42 | 1.02 | 0.92 | 1.05 | 1.04 | 1.36 | 1.28 | 1.40 | 1.37 | |||
| Tyr | 18,124 | 1,765 | 11,538 | 20,250 | 1,407 | 500 | 6,456 | 3,871 | 4,061 | 774 | 11,149 | 1,403 | |||
| UAC | ∗ | 0.84+++ | 1.35 | 0.80+ | 0.89 | ∗ | 1.35++ | 1.57 | 1.24 | 1.29 | ∗ | 0.97++ | 1.19 | 0.86 | 0.93 |
| UAU | 1.16 | 0.65 | 1.20 | 1.11 | 0.65 | 0.43 | 0.76 | 0.71 | 1.03 | 0.81 | 1.14 | 1.07 | |||
| Cys | 12,135 | 1,016 | 7,540 | 12,899 | 1,174 | 244 | 4,433 | 2,242 | 2,768 | 439 | 7,481 | 755 | |||
| UGC | ∗ | 0.87+++ | 1.28 | 0.82 | 0.84 | ∗ | 1.44++ | 1.68 | 1.44 | 1.42 | ∗ | 0.80+ | 0.89 | 0.77+ | 0.86 |
| UGU | 1.13 | 0.72 | 1.18 | 1.16 | 0.56 | 0.32 | 0.56 | 0.58 | 1.20 | 1.11 | 1.23 | 1.14 | |||
| Phe | 29,190 | 2,470 | 17,908 | 28,498 | 1,536 | 599 | 7,614 | 4,898 | 6,274 | 1,039 | 16,537 | 2,103 | |||
| UUC | ∗ | 0.90+++ | 1.50 | 0.88++ | 1.17 | ∗ | 1.34++ | 1.59 | 1.27 | 1.31 | ∗ | 0.98++ | 1.18 | 0.87 | 0.93 |
| UUU | 1.10 | 0.50 | 1.12 | 0.83 | 0.66 | 0.41 | 0.73 | 0.69 | 1.02 | 0.82 | 1.13 | 1.07 | |||
| Ile | 34,721 | 3,057 | 23,018 | 40,126 | 2,136 | 937 | 10,904 | 7,260 | 7,243 | 1,288 | 20,984 | 2,753 | |||
| AUA | 0.54 | 0.10 | 0.57 | 0.32 | 0.51 | 0.22 | 0.60 | 0.49 | 0.72 | 0.50 | 0.79 | 0.62 | |||
| AUC | ∗ | 0.84+++ | 1.75 | 0.80++ | 1.01 | ∗ | 1.59+++ | 1.95 | 1.41 | 1.48 | ∗ | 1.09++ | 1.33 | 0.94+ | 1.08 |
| AUU | 1.62 | 1.16 | 1.63 | 1.66 | 0.90 | 0.83 | 0.99 | 1.03 | 1.20 | 1.17 | 1.28 | 1.29 | |||
| Codon | Favored | C. elegans | Favored | D. melanogaster | Favored | A. thaliana | |||||||||
| Short proteins | Long proteins | Short proteins | Long proteins | Short proteins | Long proteins | ||||||||||
| No EST | High | No EST | High | No EST | High | No EST | High | No EST | High | No EST | High | ||||
| Ter | 2,543 | 298 | 429 | 663 | 255 | 80 | 232 | 140 | 680 | 122 | 458 | 59 | |||
| UAA | 1.43+++ | 2.00 | 1.37+++ | 1.75 | 1.53+++ | 1.95 | 1.14+++ | 1.59 | 1.29+ | 1.40 | 1.19 | 0.86 | |||
| UAG | 0.51 | 0.53 | 0.58 | 0.52 | 0.73 | 0.79 | 1.00 | 0.69 | 0.64 | 0.39 | 0.50+ | 0.66 | |||
| UGA | 1.06 | 0.46 | 1.05 | 0.73 | 0.74 | 0.26 | 0.87 | 0.73 | 1.08+ | 1.20 | 1.31+ | 1.47 |
For all codons, ΔRSCU is strongest in C. elegans and weakest in A. thaliana, in agreement with the global measure of Fav (Fig. 1).
Generally, ΔRSCU values are higher in short than in long proteins. In these latter, many amino acids appear to have no optimal codons. Overall, long proteins do not have a particular set of optimal codons, but simply show a weakest codon-usage bias.
The usage of UAA terminator clearly increases with gene expression in C. elegans and D. melanogaster (Table 2). In A. thaliana, the terminator usage is less biased.
Mutational Bias or Selection?
Biased codon usage may be explained either by selection on silent sites or by directional mutation pressure. The observed relationship between codon usage and mRNA abundance argues in favor of the selection model. However, it does not allow definitive rejection of the mutational bias hypothesis, because there may be a relationship between gene expression and mutation pattern. For example, it has been shown that the frequency of C-to-T mutations increases with the expression level of E. coli genes (23, 24).
If such expression-linked mutational bias was responsible for the biased nucleotide content at silent sites of highly expressed genes, it should affect not only exons but also introns. In the three species considered here, most optimal codons end in G or C, and thus G + C content at third codon position increases with expression level. But we did not detect any significant increase in introns G + C content with expression. On the contrary, intron G + C content decreases slightly (but significantly) with expression in C. elegans (data not shown). Therefore, the correlation between codon usage and expression is not due to a mutational bias, but to selection.
Codon Usage and Development Stages.
We compared codon usage in C. elegans genes expressed only in embryo, only in adult or in both (respectively 898, 1,366, and 1,374 genes). For each of these subsets, we computed RSCU values for different gene expression levels and protein length. Optimal codons in embryo-specific and adult-specific genes are exactly the same ones as in genes expressed in both stages. Thus, there is no evidence of development stage-specific codon usage in C. elegans.
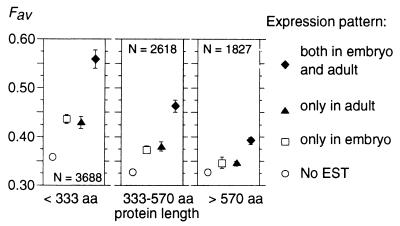
In multicellular organisms, selective pressure on codon usage is expected to depend not only on expression level of genes but also on the number of tissues or development stages where they are expressed. Indeed, we observed that Fav is in average much stronger in genes expressed both in embryo and adult than in stage-specific genes (Fig. 2). As noticed previously with data on mRNA abundance, the impact of expression pattern on codon usage is stronger in genes encoding short than long proteins.
Figure 2.
Frequency of favored codons and development stage expression in C. elegans. Average Fav values have been computed for different patterns of expression and protein lengths. Error bars indicate the 95% confidence interval.
Protein Length and Codon Usage.
In all species, Fav decreases with the length of encoded proteins (Fig. 1). In genes of moderate or high expression (where the Fav variability is most pronounced), there is a significant negative correlation between Fav and the logarithm of protein length (Table 3).
Table 3.
Linear correlations between Fav and protein length (logarithmic scale)
| Organism | Expression | |
|---|---|---|
| Moderate | High | |
| C. elegans | R = −0.45 | R = −0.60 |
| D. melanogaster | R = −0.48 | R = −0.41 |
| A. thaliana | R = −0.33 | R = −0.37 |
We observed the same phenomenon for terminator usage. Among moderately or highly expressed genes, the optimal terminator (UAA) is used more frequently in genes encoding short than long proteins, both in D. melanogaster (χ2 = 5.8, P = 0.016) and C. elegans (χ2 = 6.9, P = 0.009).
In some species, codon usage has been shown to vary along the length of genes (25–27). In D. melanogaster, there seems to be an increase of G + C content at the start of genes followed by an overall decline (28). This decline affects not only exons, but also introns, and may be caused by a within-gene variation of mutational bias. Such effect could potentially be responsible for differences of G + C content between short and long genes. However, intron G + C content does not decrease with the length of the encoded protein (data not shown).
The observed decrease of the global Fav with protein length could be explained if the functional constraints responsible for selection against nonoptimal codons were restricted to a limited portion of the gene. To test this hypothesis, we measured the maximum Fav value (_Fav_max) along coding sequences, by using a sliding window of 150 codons, moved by steps of three codons. Where the selective pressure on codon usage is weak (in A. thaliana and in genes expressed at a low level in C. elegans and D. melanogaster), we observed a slight increase of _Fav_max with protein length (data not shown). Stochastic effects can probably explain this: the longer the sequence, the higher the chance of finding a segment of high frequency of favored codons. But in genes of moderate or high expression, _Fav_max is significantly higher in short than in long proteins, both in D. melanogaster (Student’s t test = 2.7, P = 0.0067) and C. elegans (Student’s t test = 12.7, P < 0.0001). In C. elegans genes expressed at a high level, where the strongest effect was observed (Table 3), there is a significant negative correlation between _Fav_max and protein length (r = −0.42, P < 0.0001).
Therefore, the negative correlation between Fav and protein length cannot be explained simply by localization of constraints on codon usage.
DISCUSSION
Selection for translation efficiency has been proposed for several years to explain codon usage bias in some multicellular eukaryotes (9, 13, 14). The large data set analyzed here shows a clear correlation between codon-usage bias and gene expression levels in D. melanogaster, C. elegans, and A. thaliana and thus provides direct evidence for selection on silent sites in those three distantly related organisms. It should be stressed that ESTs give only a rough picture of gene expression. Thus, we believe that in reality the correlation between codon-usage bias and expression may be even stronger than what we observed. In C. elegans, where expression data from adult and from embryo is available, we did not find any evidence of development stage-specific codon usage. However, the codon-usage bias is higher in genes expressed both in embryo and adult than in stage-specific genes. This is consistent with a selective pressure on codon usage depending not only on the expression level of genes but also on the number of tissues or development stages where they are expressed.
Surprisingly, we found that in the three species studied, the frequency of optimal codons decreases with the length of the encoded protein. A similar tendency has already been described in D. melanogaster and yeast (12, 16). But because the authors did not have expression data, they could not determine whether this correlation was a direct relationship between protein length and Fav or if it was caused by a tendency of genes expressed at a high level to encode short proteins. The authors retained this latter explanation and proposed that selection acts to reduce the length of proteins expressed at a high level (16). However, we did not find any evidence for such a selection (Table 4). In A. thaliana, there is no significant variation of protein length with expression level; in D. melanogaster, the only significant difference is that genes with no ESTs encode shorter proteins than expressed genes; and in C. elegans, there is an increase of average protein length with expression level. Indeed, for a same expression pattern, codon usage clearly decreases with increasing protein length (Fig. 1, 2). Thus, the selective pressure on codon usage is lower in genes encoding long than short proteins.
Table 4.
Protein length and gene expression
| Organism | Protein length, amino acids | ||||
|---|---|---|---|---|---|
| All genes | Without EST | Expression | |||
| Low | Moderate | High | |||
| C. elegans | 445 ± 388 | 336 ± 221 | 452 ± 369 | 542 ± 408 | 734 ± 617 |
| D. melanogaster | 624 ± 546 | 541 ± 481 | 731 ± 591 | 686 ± 583 | 662 ± 577 |
| A. thaliana | 452 ± 311 | 459 ± 316 | 456 ± 322 | 429 ± 273 | 425 ± 285 |
It is likely that in the three species studied here, selection on codon usage acts to increase translation efficiency, as in other organisms such as yeast or E. coli (2–4). It is thought that the use of codons that match the most abundant tRNA reduces the time to find and bind the correct tRNA. Hence, optimal codons not only increase the elongation rate but also decrease the likelihood of binding a noncognate tRNA. Thus, different models have been proposed to account for the effect of translation efficiency on fitness: selection may act to maximize elongation rate, minimize the cost of proofreading, or maximize the accuracy of translation (8). All three models predict that codon-usage bias should be higher in genes expressed at high levels (8, 15, 29). But interestingly, none of these models accounts for the protein length effect on codon usage observed here. These different models are discussed below.
Powell and Moriyama (12) hypothesized that this length effect could be explained by selection for translation rate. Assume for example that a nonoptimal codon requires twice as long to incorporate an amino acid as does the optimal codon. Mutations to nonoptimal codons will have a greater relative effect in smaller genes than in longer ones: in a short gene with 100 codons, such mutation would increase translation time by 1%, whereas the same mutation in a gene with 1,000 codons would increase translation time by only 0.1%. Thus, such mutations are more likely to be counterselected in short genes than in long ones.
However, several arguments suggest that this model is not realistic. First, it is unlikely that selection acts to increase the rate of synthesis of a particular protein (except in specialized tissues that are devoted to the production of a few extremely abundant proteins such as silk glands in Bombyx mori) (30). Rather, selection is thought to act to increase the cell growth rate. Therefore, selection should act to increase the production of all cell constituents, and not of a particular protein species. Moreover, it seems that initiation is the limiting step in protein translation (reviewed in refs. 1 and 8). Thus, the rate of production of a given protein is determined by the initiation rate and not by the elongation rate. Indeed, the effect of codon usage on protein synthesis is thought to be indirect: the use of optimal codons in a given gene will increase the elongation rate and thus reduce the time the ribosome is bound to the mRNA; this leads to an increase in the pool of free ribosomes and hence to an increase in the translation initiation rate of all mRNA species (1, 8). Hence, the use of optimal codon in a given gene is thought to increase the production of all proteins in a cell, not only its own protein product. The delay caused by nonoptimal codons—and the resulting decrease in concentration of free ribosomes—will be the same in long and short mRNAs. Thus, the strength of selection on codon usage for an increased rate of protein production should be independent of protein length.
Selection to minimize the cost of proofreading is also expected to be independent of protein length. The process of rejecting noncognate tRNAs decreases elongation rate and consumes energy. The energy waste caused by a nonoptimal codon should be the same in a mRNA encoding a long or a short protein. And, as explained above, the strength of selection on elongation rate does not depend on protein length.
The model of selection on translation accuracy predicts that codon-usage bias should be higher in long than in short proteins: because the cost of producing a protein is proportional to its length, selection in favor of codons that increase accuracy should be higher in longer genes, as has been observed in E. coli (16, 29). This model is thus in total contradiction with our observations. Therefore, even if there is evidence for selection on translation accuracy in D. melanogaster (15), this is not the major factor that shapes codon usage in the three organisms studied here.
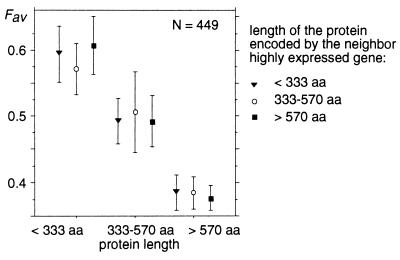
Another possible explanation for this length effect comes from population genetics theory. In a simulation study, Li (7) noticed that if the selective advantages conferred by optimal codons are additive (which is the case for the three models mentioned above) and if all sites within a gene are completely linked (i.e., recombination within a gene is rare relatively to mutation), then the efficacy of selection on optimal codons decreases with increasing protein length. This interference between linked sites is analogous to Muller’s ratchet effect (31, 32): the efficacy of natural selection is reduced when genetic linkage exists among multiple sites affected by selection. A prediction of that model is that two tightly linked genes affected by selection on codon usage (i.e., highly expressed) should also interfere and behave as if they were a single gene encoding a longer protein. Thus, a long gene having a short neighbor should have the same codon usage as a short gene having a long neighbor. In the C. elegans data set, 449 genes expressed at high level have a neighbor that is expressed at a high level, <5 kb from their 5′ or 3′ end. Fig. 3 clearly shows that their codon usage is affected by their own length but not by the length of their neighbor. Therefore, neighbor genes do not seem to interfere. It is unlikely that recombination is frequent enough so that two genes within 5 kb are not genetically linked. If this were the case, we would expect that recombination should also occur within genes (that are 2.4 kb long on average), and hence the Li effect should not be detected. Moreover, even when considering closer genes, we do not find any evidence of interference: genes expressed at high level that have a very close neighbor with genes expressed at high level (<2 kb, n = 229) do not have a lower codon usage than genes expressed at high level that have no neighbor with genes expressed at a high or moderate level within 5 kb (n = 409) (t test = 0.4 P = 0.69).
Figure 3.
Frequency of favored codons in C. elegans genes expressed at high level having a neighbor expressed at high level less than 5 kb from their 5′ or 3′ end. Average Fav values have been computed for different protein lengths. Error bars indicate the 95% confidence interval.
In conclusion, none of the current models of selection on codon usage is consistent with the observed decrease of codon-usage bias in genes encoding long proteins. This length effect has been observed both in plant and metazoan species. It seems to occur also in yeast but not in E. coli, where codon-usage bias increases with protein length (16, 29). Thus, the selective pressure acting on codon usage may be different in eukaryotes and eubacteria.
The finding of selection on codon usage in very distantly related taxa, such as plants and animals, suggests that this is a widespread phenomenon. However, we did not observe any correlation between codon usage and expression level in human genes (33) (unpublished data). As noticed by others (6, 9), this absence of selection may be explained by population genetics: a mutation that is advantageous in a species with large effective population size may be neutral in a small population, where random drift overcomes selection. In mammals, effective population sizes have been estimated to be ≈104, i.e., 102 to 103 smaller than in Drosophila species (34). Therefore, it is likely that in most human genes, fitness differences among synonymous codons is not sufficient to overcome drift.
A prediction of our observation is that the genes on which selection for optimal codons could operate in mammals should be short and expressed at very high levels in many different tissues and development stages. Indeed, to our knowledge, the only example of selection on silent site in mammals was described in H3 histones, that are short genes (137 codons) and expressed at extremely high level during S phase of the cell cycle in every cell of the animal (35). How translation efficiency affects fitness in eukaryotes, however, remains unexplained.
Acknowledgments
We are grateful to M. Gouy, C. Gautier, and A. Eyre-Walker for many helpful discussions. This work is supported by the CNRS (Centre National de la Recherche Scientifique).
ABBREVIATIONS
CDS
protein-coding sequence
EST
expressed sequence tag
Fav
frequency of favored codons
RSCU
relative synonymous codon usage
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
*
To whom reprint requests should be addressed. e-mail: duret@biomserv.univ-lyon1.fr.
References
- 1.Kurland C G. FEBS Lett. 1991;285:165–169. doi: 10.1016/0014-5793(91)80797-7. [DOI] [PubMed] [Google Scholar]
- 2.Ikemura T. In: Transfer RNA in Protein Synthesis. Hatfield D L, Lee B J, Pirtle R M, editors. Boca Raton, FL: CRC; 1992. pp. 87–111. [Google Scholar]
- 3.Gouy M, Gautier C. Nucleic Acids Res. 1982;10:7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharp P M, Li W H. J Mol Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 5.Sharp P M, Li W H. Mol Biol Evol. 1987;4:222–230. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- 6.Sharp P M, Averof M, Lloyd A T, Matassi G, Peden J F. Philos Trans R Soc London B. 1995;349:241–247. doi: 10.1098/rstb.1995.0108. [DOI] [PubMed] [Google Scholar]
- 7.Li W H. J Mol Evol. 1987;24:337–345. doi: 10.1007/BF02134132. [DOI] [PubMed] [Google Scholar]
- 8.Bulmer M. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shields D C, Sharp P M, Higgins D G, Wright F. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- 10.Moriyama E N, Powell J R. J Mol Evol. 1997;45:514–523. doi: 10.1007/pl00006256. [DOI] [PubMed] [Google Scholar]
- 11.Sharp P M, Li W H. J Mol Evol. 1989;28:398–402. doi: 10.1007/BF02603075. [DOI] [PubMed] [Google Scholar]
- 12.Powell J R, Moriyama E N. Proc Natl Acad Sci USA. 1997;94:7784–7790. doi: 10.1073/pnas.94.15.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenico M, Lloyd A T, Sharp P M. Nucleic Acids Res. 1994;22:2437–2446. doi: 10.1093/nar/22.13.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiapello H, Lisacek F, Caboche M, Henaut A. Gene. 1998;209:GC1–GC38. doi: 10.1016/s0378-1119(97)00671-9. [DOI] [PubMed] [Google Scholar]
- 15.Akashi H. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriyama E N, Powell J R. Nucleic Acids Res. 1998;26:3188–3193. doi: 10.1093/nar/26.13.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F F. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gouy M, Gautier C, Attimonelli M, Lanave C, Di-Paola G. Comp Appl Biosci. 1985;1:167–172. doi: 10.1093/bioinformatics/1.3.167. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claverie J-M, States D J. Comput Chem. 1993;17:191–201. [Google Scholar]
- 21.Cooke R, Raynal M, Laudie M, Grellet F, Delseny M, Morris P C, Guerrier D, Giraudat J, Quigley F, Clabault G, et al. Plant J. 1996;9:101–124. doi: 10.1046/j.1365-313x.1996.09010101.x. [DOI] [PubMed] [Google Scholar]
- 22.Sharp P M, Lloyd A T. In: An Atlas of Drosophila Genes: Sequences and Molecular Features. Maroni G, editor. New York: Oxford Univ. Press; 1993. pp. 378–397. [Google Scholar]
- 23.Beletskii A, Bhagwat A S. Proc Natl Acad Sci USA. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beletskii A, Bhagwat A S. Biol Chem. 1998;379:549–551. [PubMed] [Google Scholar]
- 25.Bulmer M. J Theor Biol. 1988;133:67–71. doi: 10.1016/s0022-5193(88)80024-9. [DOI] [PubMed] [Google Scholar]
- 26.Chen G F, Inouye M. Nucleic Acids Res. 1990;18:1465–1473. doi: 10.1093/nar/18.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eyre-Walker A, Bulmer M. Nucleic Acids Res. 1993;21:4599–4603. doi: 10.1093/nar/21.19.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kliman R M, Eyre-Walker A. J Mol Evol. 1998;46:534–541. doi: 10.1007/pl00006334. [DOI] [PubMed] [Google Scholar]
- 29.Eyre-Walker A. Mol Biol Evol. 1996;13:864–872. doi: 10.1093/oxfordjournals.molbev.a025646. [DOI] [PubMed] [Google Scholar]
- 30.Garel J P, Hentzen D, Daillie J. FEBS Lett. 1974;39:359–363. doi: 10.1016/0014-5793(74)80149-3. [DOI] [PubMed] [Google Scholar]
- 31.Muller H J. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 32.Felsenstein J. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlin S, Mrázek J. J Mol Biol. 1996;262:459–472. doi: 10.1006/jmbi.1996.0528. [DOI] [PubMed] [Google Scholar]
- 34.Nei M, Graur D. Evol Biol. 1984;17:73–118. [Google Scholar]
- 35.Debry R W, Marzluff W F. Genetics. 1994;138:191–202. doi: 10.1093/genetics/138.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]