Viral and Cellular Determinants of the Hepatitis C Virus Envelope-Heparan Sulfate Interaction (original) (raw)
Abstract
Cellular binding and entry of hepatitis C virus (HCV) are the first steps of viral infection and represent a major target for antiviral antibodies and novel therapeutic strategies. We have recently demonstrated that heparan sulfate (HS) plays a key role in the binding of HCV envelope glycoprotein E2 to target cells (Barth et al., J. Biol. Chem. **278:**41003-41012, 2003). In this study, we characterized the HCV-HS interaction and analyzed its inhibition by antiviral host immune responses. Using recombinant envelope glycoproteins, virus-like particles, and HCV pseudoparticles as model systems for the early steps of viral infection, we mapped viral and cellular determinants of HCV-HS interaction. HCV-HS binding required a specific HS structure that included N-sulfo groups and a minimum of 10 to 14 saccharide subunits. HCV envelope binding to HS was mediated by four viral epitopes overlapping the E2 hypervariable region 1 and E2-CD81 binding domains. In functional studies using HCV pseudoparticles, we demonstrate that HCV binding and entry are specifically inhibited by highly sulfated HS. Finally, HCV-HS binding was markedly inhibited by antiviral antibodies derived from HCV-infected individuals. In conclusion, our results demonstrate that binding of the viral envelope to a specific HS configuration represents an important step for the initiation of viral infection and is a target of antiviral host immune responses in vivo. Mapping of viral and cellular determinants of HCV-HS interaction sets the stage for the development of novel HS-based antiviral strategies targeting viral attachment and entry.
Hepatitis C virus (HCV) is a major cause of posttransfusion and community-acquired hepatitis in the world. The majority of HCV-infected individuals develop chronic hepatitis that may progress to liver cirrhosis and hepatocellular carcinoma (10). Treatment options are limited, and a vaccine to prevent HCV infection is not available (14).
HCV has been classified in a separate genus (Hepacivirus) of the Flaviviridae family. The virion contains a positive-strand RNA genome of approximately 9,600 nucleotides. The genome encodes a single polyprotein of 3,010 to 3,030 amino acids that is co- and posttranslationally processed by host and viral proteases into structural and nonstructural proteins. The HCV structural proteins comprise the core protein and the two envelope glycoproteins E1 and E2 (23). HCV preferentially replicates in the cytoplasm of hepatocytes, but distinct HCV sequences have also been isolated from B cells and dendritic cells (1). Several experimental systems have suggested that virus binding and entry are mediated by envelope glycoprotein E2. Using recombinant envelope glycoproteins (35), HCV-like particles (HCV-LPs) (5), retroviral HCV pseudotype particles (HCVpp) (3), and recombinant infectious virions (22, 48, 54) as model systems for the first steps of viral infection, CD81 (35), scavenger receptor class B type I (SR-BI) (38), dendritic cell-specific intercellular adhesion molecule 3 grabbing-nonintegrin (DC-SIGN) (25), and the glycosaminoglycan heparan sulfate (HS) (2) have been identified as HCV receptor candidates.
HS comprises a family of linear polysaccharides located at the surface of mammalian cells and in the extracellular matrix. HS varies with respect to composition and quantity among different species, cell types, tissues, and the stage of cellular development. HS consists mainly of repeating disaccharide units [GlcA-GlcNAc]n, where GlcA is glucuronic acid and GlcNAc is _N_-acetylglucosamine. However, these saccharides undergo N deacetylation and N sulfation of the GlcNAc residues, O sulfation at various positions, and epimerization of GlcA to iduronic acid (12). These secondary modifications give rise to an enormous structural diversity throughout the length of each chain. HS chains are attached to a core protein, forming a class of glycoproteins called proteoglycans. A number of studies have indicated the potential role of heparan sulfate proteoglycans (HSPGs) in the regulation of cell growth and transformation, differentiation processes, and cell adhesion. In hepatocytes, HSPGs are thought to bridge the extracellular matrix and the intracellular cytoskeleton (36).
Microorganisms may take advantage of the widespread distribution of proteoglycans on cell surfaces by using them as ligands for their attachment to the target cell. For several viruses, including members of the Flaviviridae family such as dengue virus (9), classical swine fever (18), and tick-borne encephalitis viruses (28), as well as herpes simplex virus 1 (40), human herpesvirus 8 (7), papillomavirus (39), and human immunodeficiency virus (46), HSPGs provide primary docking sites for the initiation of viral infection. Recently, we have demonstrated that HCV envelope glycoprotein E2 binds to highly sulfated HS expressed on the cell surface of human cell lines (2).
In this study, we mapped viral and cellular determinants of the HCV-HS interaction and demonstrate that binding of the viral envelope to a specific HS configuration represents an important step for the initiation of viral infection. Furthermore, we provide evidence that the HCV-HS interaction is targeted by antiviral host immune responses elicited during HCV infection in vivo.
MATERIALS AND METHODS
Reagents and cell lines.
Recombinant carboxy-terminal-truncated envelope glycoprotein E1 (comprising amino acid [aa] 192 to 326) and E2 (aa 384 to 673) were generated using recombinant vaccinia viruses containing HCV envelope cDNAs of European HCV 1b isolate (BE11) and purified as described previously (26, 45). HCV-LPs were synthesized and purified from insect cells infected with recombinant baculoviruses containing the cDNA of the HCV structural proteins of HCV strain H77c (genotype 1a) as described recently (49). Control preparations were derived from insect cells infected with a recombinant baculovirus containing the cDNA for β-glucuronidase (49). HCV-LP E2 concentration was determined by an E2-specific enzyme-linked immunosorbent assay (ELISA) (11). Heparin (bovine lung) was obtained from Merck Biosciences (Darmstadt, Germany). Kidney-derived normally sulfated HS and keratin sulfate were obtained from Sigma-Aldrich Corp. (Taufkirchen, Germany). De-2-O-sulfated, de-6-O-sulfated, de-N-sulfated, and fully N-sulfated heparin were purchased from Neoparin, Inc. (San Leandro, CA). Highly sulfated liver-derived HS was isolated as previously described (44). Heparin oligosaccharides of 2, 4, 6, 8, 10, 14, and 20 subunits and control saccharide (decasaccharide containing the pentasaccharide antithrombin III-binding site) were synthesized or isolated as previously described (34, 43). Mouse anti-E1 (11B7) and anti-E2 (16A6, 2F10, 917, and AP33) monoclonal antibodies (MAbs) have been described previously (11, 49). Rabbit anti-E1 (R852) polyclonal serum was raised in a New Zealand rabbit immunized with a secretory form of HCV E1 glycoprotein (genotype 1b strain J4L6S [51]; aa 197 to 334) as described previously (11). Chimpanzee anti-E2 MAbs 49F3, 11F11, and 22E1 were generated by fusing peripheral B lymphocytes from a chimpanzee immunized with recombinant E2 protein (aa 412 to 715) with the K6H6/B5 fusion partner (ATCC CRL-1832) for 11F11 and 22E1 and with SP2/0 (ATCC CRL-1581) as a fusion partner for 49F3 (immunoglobulin G1 [IgG1] isotype). Generation of rabbit anti-E2 (R646) polyclonal serum has been described previously (11). Human IgG was purchased from Binding Site (San Diego, CA). Sera from 12 patients with chronic HCV infection were obtained from the Department of Medicine II, University of Freiburg, Freiburg, Germany. All patients were serologically negative for hepatitis B virus and human immunodeficiency virus infection. Purification of human IgG from anti-HCV-positive sera and -HCV-negative control sera has been described previously (41). The origin and maintenance of human hepatoma cell lines HepG2 and Huh-7, mouse hepatoma cell line Hepa 1-6, and 293T human embryo kidney cells have been described previously (2, 3, 49). Human HeLa cells were kindly provided by M. Nassal (Department of Medicine II, University of Freiburg, Freiburg, Germany). The origin and maintenance of human immortalized OKF6 oral keratinocytes kindly provided by O. Opitz (Department of Medicine II, University of Freiburg) have been described previously (32).
Analysis of HCV envelope protein-heparin binding by SPR.
Surface plasmon resonance (SPR) analysis allowing a direct quantitative analysis of label-free molecular interactions in real time was used to characterize envelope protein E1- and E2-heparin interaction. SPR measurements were performed on a BIAcore 3000 system (BIAcore AB, Uppsala, Sweden) using the BIAcore 3000 version software. Immobilization of heparin-bovine serum albumin (BSA) on a flat carboxymethylated surface and binding of HCV envelope glycoproteins to heparin surface were performed as described previously (2). In brief, measurement of heparin-envelope protein interaction was performed as follows. Different dilutions of recombinant E1 and E2 in phosphate-buffered saline (PBS: 10 mM sodium phosphate, 137 mM NaCl, 2.7 mM KCl at pH 7.4) containing 0.05% betaine were injected (30 μl) at a flow rate of 10 μl/min. At the end of the sample injection, the same buffer flowed over the sensor surface to facilitate dissociation. After a 3-min dissociation time, the sensor surface was regenerated by injection first with 10 μl of 2 M NaCl and then with 10 μl of glycine-HCl, pH 2, and 10 μl of 50 mM NaOH (BIAcore) to get a fully regenerated surface.
Mapping of viral epitopes interacting with HS using an envelope-heparin binding assay.
Ninety-six-well ELISA plates pretreated by plasma polymerization (EpranEx; Plasso Technology Ltd., Portobello, Sheffield, United Kingdom) (27) were coated with 1 μg of heparin (Merck Biosciences) in PBS per well. After incubation overnight at room temperature, the plates were blocked with SAB buffer (100 mM NaCl, 50 mM NaAc, 0.2% Tween, pH 7.2) containing 1% BSA for 1 h at room temperature. HCV envelope glycoproteins E1 and E2 (1 μg/ml) were preincubated with anti-envelope MAbs (50 μg/ml), control IgG (50 μg/ml), purified IgG from HCV-infected patients (500 μg/ml), or IgG from healthy individuals (500 μg/ml) for 1 h at room temperature. Subsequently, envelope glycoprotein-antibody complexes were added to the well for 1 h at room temperature. Heparin-bound envelope glycoproteins were detected by the addition of rabbit anti-E2 (R646) or anti-E1 (R852) polyclonal antibodies and horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody. Protein-bound antibodies were detected by colorimetry as described previously (6).
Mapping of cellular determinants of HCV envelope-HS interaction using recombinant envelope glycoproteins and HCV-LPs.
Hepatoma cells (1.5 × 105 cells/100 μl) were incubated with recombinant envelope protein E1 (10 μg/ml) or E2 (2.5 μg/ml) or HCV-LPs (corresponding to 0.25 μg HCV-LP E2/ml) for 1 h at 4°C as described recently (2). Ligand concentrations used in the assay corresponded to the concentration required for half-maximal saturation of ligand binding to target cells (2). Following removal of nonbound viral proteins by washing with PBS, cell-bound ligands were detected by flow cytometry using mouse anti-E1 (11B7) or anti-E2 (16A6) and phycoerythrin (PE)-conjugated anti-mouse IgG as described previously (2). To assess whether binding of viral proteins was inhibited by soluble glycosaminoglycans (GAGs), viral proteins were incubated with GAGs, chemically modified heparins, or heparin oligosaccharides (10 μg/ml) for 30 min at room temperature prior to the addition to the cells as described previously (2).
Binding and entry of retroviral HCVpp.
HCVpp derived from H77C and HCV-J strains (genotypes 1a and 1b) were synthesized as described previously (3). To exclude that nonassembled envelope glycoproteins were detected in the HCVpp binding assay, HCVpp were purified by ultracentrifugation through a 20% sucrose cushion in an SW55 Ti Beckman rotor (30,000 rpm, 2.5 h, 4°C) prior to their use in binding studies (3). Cellular binding of sucrose gradient-purified HCVpp was analyzed by flow cytometry using anti-E2 (16A6) MAb as described above. To analyze whether infection of HCVpp was blocked by soluble GAGs, HCVpp were preincubated with GAGs (10 μg/ml) for 30 min. Then, Huh-7 cells were washed with ice-cold PBS and HCVpp-GAG complexes were added to cells for 1.5 h at 4°C, allowing binding of HCVpp-GAG complexes to cells. After washing of cells with PBS, Huh-7 cells were incubated for 72 h at 37°C and HCVpp entry as described previously (3). To study whether soluble GAGs interfered with early steps of HCVpp entry following viral attachment (postbinding events), Huh-7 cells first were incubated with HCVpp for 1 h at 4°C, washed with ice-cold PBS to remove unbound HCVpp, and then incubated with soluble GAGs (10 μg/ml) for another 1.5 h at 4°C. After washing of cells with PBS, temperature was shifted to 37°C, and HCVpp entry was measured 72 h later as described above.
To study neutralization of HCVpp infection by anti-HCV IgG antibodies, HCVpp were preincubated with purified human anti-HCV or control IgG (500 μg/ml), anti-E2 MAb (50 μg/ml), or control antibody (50 μg/ml) for 1 h at room temperature. HCVpp-antibody complexes were then added to the cells for 2 h at 37°C. After being washed with PBS to remove unbound HCVpp, Huh-7 cells were incubated for 72 h at 37°C and HCVpp entry was assessed as described previously (3).
RESULTS
HCV envelope-HS binding is mediated by both viral envelope glycoproteins E1 and E2.
Several lines of evidence have demonstrated that envelope glycoprotein E2 mediates binding of the virus to the host cell (13, 37). In line with these observations, we have previously demonstrated that HS represents a key molecule mediating binding of E2 to the cell surface. To study the role of HCV envelope glycoprotein E1 for interaction of the complete viral envelope with HS, we analyzed whether E1 binds heparin—a close structural homologue of highly sulfated HS expressed in various forms on the surface of defined cells and extracellular matrices, including hepatocytes (36).
To characterize the affinity of the E1 interaction with the HS homologue heparin, SPR analysis was performed. SPR allows a direct quantitative analysis of label-free molecular interactions in real time. As shown in Fig. 1A, E1 bound in a concentration-dependent manner to the heparin biosensor surface. E1-heparin association was demonstrated by the biosensor chip response following the initiation of sample injection. The resonance signal increased in a dose-dependent manner from approximately 32 resonance units (RU) at 50 nM to 236 RU at 400 nM. Assuming a 1:1 interaction (Langmuir model) between the immobilized ligand (heparin) and soluble analyte (E1 protein), the calculation of the kinetic parameters for E1-heparin binding revealed a dissociation constant (KD) of 5.3 × 10−8 (_k_on of 3.4 × 10−4 M−1 s−1, _k_off of 1.8 × 10−3 s−1). Compared to E2-heparin binding, this affinity was about 10-fold lower (Fig. 1B; KD of E2 5.2 × 10−9). It would be informative to have for comparative purposes functional E1-E2 complexes (20) analyzed in the same way. However, this approach is not easily feasible, since large quantities of highly purified E1-E2 heterodimers for SPR analysis are not readily available.
FIG. 1.
SPR analysis of envelope glycoprotein E1-heparin binding. (A) Heparin-BSA or BSA was covalently immobilized onto the surface of a biosensor chip as described recently (2). Subsequently, different concentrations of recombinant highly purified envelope glycoprotein E1 were injected onto the biosensor surface. The biosensor chip response is indicated on the y axis (measured in RU) as a function of time (x axis) at 25°C. The sensorgram shows the difference in the BSA-heparin coated chip response compared to the BSA-coated control chip response following E1 injection. (B) Side-by-side analysis of E1 and E2 (both at 100 nM).
Next, we analyzed whether cellular binding of E1 was mediated by cell surface HS. As shown in Fig. 2, E1 exhibited a dose-dependent and saturable binding to human hepatoma cell lines HepG2 and Huh-7 (Fig. 2A). Cellular binding of E1 to HepG2 and Huh-7 was significantly lower when compared to binding of E2 to target cells (Fig. 2A). To address the specificity of HCV envelope glycoprotein binding to human hepatoma cells, we studied the ability of HCV envelope glycoproteins to bind to human nonliver cell lines (HeLa, OKF6, and 293T) and non-human liver cells (mouse hepatoma Hepa 1-6) in a side-by-side analysis. Whereas human hepatoma cell lines were characterized by high-level binding of HCV envelope glycoproteins, human nonliver and non-human liver cell lines were characterized by absent or low-level binding of E1 and E2 glycoproteins (Fig. 2B). Binding of envelope glycoproteins appeared to be more pronounced to Huh-7 than to HepG2 cells (Fig. 2B). Since human liver-derived cell lines as well as human hepatocytes are characterized by cell surface expression of highly sulfated HS (2, 47), this comparative analysis of envelope glycoprotein binding provides further evidence that highly sulfated HS expressed on human hepatoma cell lines may contribute to cellular HCV binding.
FIG. 2.
Cellular binding of envelope glycoproteins E1 and E2 to human hepatoma cells is HS dependent. (A) Dose-dependent binding of recombinant E1 and E2 to HepG2 and Huh-7 cells. Cells were incubated with increasing concentrations of E1 or E2. Binding of E1 and E2 was analyzed by flow cytometry using a mouse anti-E1 (11B7) or anti-E2 (16A6) MAb, respectively, and PE-conjugated anti-mouse IgG. On the y axis, net mean fluorescence intensity (ΔMFI) values for each protein concentration were calculated by subtracting the MFI of negative controls (cells incubated in PBS without envelope protein and the addition of anti-envelope MAb and PE-conjugated anti-mouse IgG antibodies) from that obtained with cells incubated with envelope proteins at the concentration indicated on the x axis. Data are shown as mean ΔMFI ± standard deviation (SD) of three (E1 and E2 at 50 and 100 μg/ml) or four (all other envelope protein concentrations) experiments. Significant differences in ΔMFIs obtained for E2 versus E1 binding are indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001 [determined by two-tailed t test]). (B) Binding of E1 and E2 to human hepatoma (Huh-7 and HepG2), human nonhepatoma (293T, HeLa, and OKF6), and mouse hepatoma (Hepa 1-6) cell lines. Cells were incubated with recombinant envelope glycoproteins (2.5 μg/ml), and cellular binding of envelope glycoproteins was quantified by flow cytometry in side-by-side experiments as described in panel A. ΔMFI ± SD of a representative experiment performed in triplicate is shown. (C) Flow cytometry histograms of E1 binding to Huh-7 cells (black line, unshaded peak) in the presence of soluble GAGs. Recombinant E1 protein was preincubated with PBS, chondroitin sulfate A (CSA), normally sulfated HS (HS), highly sulfated HS (hsHS), or heparin (each at 10 μg/ml) for 30 min at room temperature. E1-GAG complexes were added to cells, and cellular binding of E1 was quantified by flow cytometry as described for panel A. Background fluorescence (gray-shaded peak) corresponds to cells incubated without envelope protein. (D) Percent cellular binding of E1 protein to HepG2 and Huh-7 in the presence of soluble GAGs relative to binding of E1 without GAGs (100%). Mean ± SD of a representative experiment performed in triplicate is shown.
HS homologue heparin as well as liver-derived highly sulfated HS—but not kidney-derived normally sulfated HS or chondroitin sulfate A—markedly inhibited cellular binding of recombinant E1 to HepG2 (Fig. 2C and D) and Huh-7 cells (Fig. 2D), respectively. Interestingly, the magnitude of cellular binding of envelope glycoproteins E1 and E2 correlated with their affinity to the HS homologue heparin. These results suggest that cellular binding of the viral envelope is mediated by an interaction of HS with both glycoproteins E1 and E2.
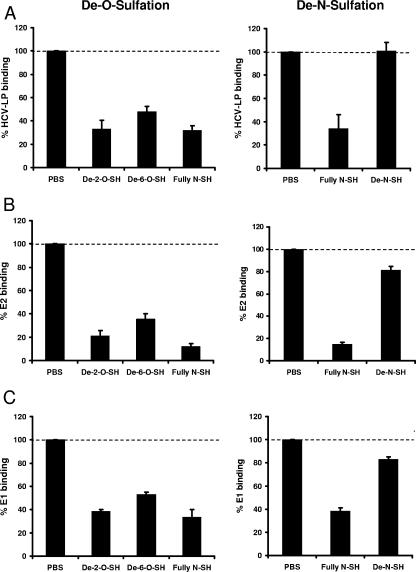
Envelope-HS interaction is mediated by a specific HS configuration that includes N-sulfo groups and a minimum of 10 to 14 saccharide subunits.
The structural complexity of HS in various tissues and species arises from an extensive series of modifications during HS biosynthesis. These modifications include the introduction of N-sulfo groups into glucosamine residues and O-sulfo groups into glucosamine or iduronic or glucuronic acid residues (12). To identify the relevance of these modifications for the envelope-HS interaction, we performed envelope glycoprotein binding experiments using chemically modified heparins as competitors. This approach has been successfully applied to map cellular determinants of HS for respiratory syncytial virus infection (16). As shown in Fig. 3, preincubation of recombinant envelope glycoproteins E1 and E2 with fully N-sulfated heparin or de-2-O-sulfated heparin and de-6-O-sulfated heparin markedly inhibited binding of viral envelope proteins to hepatoma cells. In contrast, de-N-sulfated heparin almost completely lost its ability to inhibit binding of recombinant envelope glycoproteins to hepatoma cells. Since the use of C-terminally-truncated recombinant E1 and E2 protein as a surrogate ligand for virus binding may be limited by the fact that the proper conformation of the envelope proteins requires coexpression of both proteins, we studied HCV envelope-HS interaction using HCV-LPs. In contrast to individually expressed recombinant E1 and E2 proteins, HCV-LP envelope glycoproteins are presented as an E1/E2 heterodimer expressed from a full-length E1/E2 cDNA (49). Using HCV-LPs as an HCV surrogate ligand for cellular binding, similar results were obtained (Fig. 3). These results strongly suggest that the presence of N-sulfo groups in HS is required for efficient HCV envelope-HS binding.
FIG. 3.
Cellular binding of HCV-LPs and recombinant envelope glycoprotein requires N sulfation of cell surface HS. Sucrose gradient-purified HCV-LPs (A) and recombinant E2 (B) and E1 (C) proteins were incubated with chemically modified heparins (10 μg/ml) for 30 min at room temperature. Envelope protein-GAG complexes were added to human hepatoma cells for 1 h at 4°C, and cellular binding of HCV-LPs and recombinant proteins was quantified by flow cytometry as described for Fig. 2A. Data are shown as percent binding of ligands (mean ± standard deviation [SD] of a representative experiment performed in triplicate) relative to binding of ligands in the absence of modified heparins (100%). De-2-O- and de-6-O-sulfated heparin (SH) lacks O-sulfo groups at position 2 or 6; de-N-SH lacks N-sulfo groups.
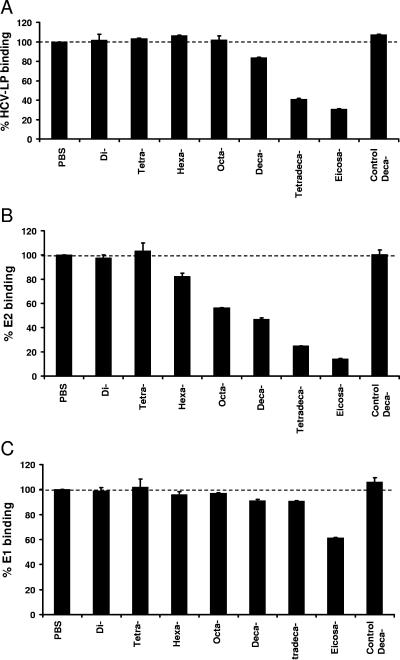
To study whether the HCV-HS interaction requires a defined HS oligosaccharide length, we analyzed the effect of defined heparin oligosaccharides with different chain lengths (di- to eicosasaccharide) on the inhibition of envelope glycoprotein binding to hepatoma cells. For purified recombinant E2 protein, a marked inhibition of binding required a minimum oligosaccharide chain length of 10 subunits (Fig. 4B). In contrast, E1 binding to hepatoma cells was only partially inhibited by a 20-subunit oligosaccharide (40% inhibition) (Fig. 4C). The minimum heparin oligosaccharide chain length that resulted in more than 50% inhibition of cellular HCV-LP binding (containing both envelope glycoproteins E1/E2 on the particle surface) was a 14-subunit oligosaccharide (Fig. 4A).
FIG. 4.
Cellular binding of HCV-LPs and recombinant envelope E1 and E2 in the presence of heparin oligosaccharides. HCV-LPs (A), recombinant E2 (B), and E1 protein (C) were preincubated withheparin-derived oligosaccharides ranging in size from di- to eicosasaccharides or a control decasaccharide containing the pentasaccharide antithrombin-III binding site (each at 50 μg/ml) for 30 min at room temperature. Viral protein-oligosaccharide complexes were added to human hepatoma cells, and cellular binding of viral proteins was quantified by flow cytometry using anti-envelope MAb as described for Fig. 2A. Data are shown as percent cellular viral protein binding relative to binding of viral proteins without oligosaccharides (100%).
Mapping of viral epitopes mediating HCV-HS interaction.
To identify key viral epitopes mediating HCV envelope-HS interaction, we developed an envelope-HS binding assay using immobilized HS homologue heparin as a capture antigen based on a previously established ELISA (2). In this assay, heparin is noncovalently immobilized at the bottom of the ELISA plate (27), serving as a surrogate molecule for highly sulfated HS on the cell surface. Using this envelope-heparin binding assay, we quantified binding of recombinant envelope glycoproteins to heparin in the presence of monoclonal antienvelope antibodies directed against defined viral epitopes. In contrast to model systems using cell lines for cellular binding—expressing several different envelope binding molecules such as CD81, SR-BI, and HS—the envelope-heparin binding assay allows us to specifically study the interaction of the HCV envelope glycoproteins with HS homologue heparin. Since we had identified E2 as the major envelope glycoprotein mediating envelope-HS binding (Fig. 1 and 2), we focused on the identification of E2 epitopes required for the envelope-HS interaction. Preincubation of E2 with defined anti-E2 monoclonal antibodies resulted in a marked inhibition of E2 binding to immobilized heparin, allowing us to map viral epitopes involved in the binding of the virus to cell surface HS (Fig. 5A). Antibodies inhibiting E2-heparin interaction targeted the following regions on the E2 protein: E2398-403 corresponding to HVR1 region (2F10) as well as E2480-487 (11F11) and E2516-530 (49F3) corresponding to CD81 binding regions 1 and 2(50). Interestingly, epitope E2412-423 targeted by MAb AP33 has been shown to represent a neutralization epitope in HCV-infected patients (41). In contrast, anti-E2 antibodies 22E1 (E2544-555) and 917 (E2460-479), human IgG (Fig. 5A), or anti-E1 antibody 11B7 (data not shown) did not inhibit E2 binding to immobilized heparin. Inhibition of E2-heparin interaction by anti-E2 antibodies was concentration dependent, exhibiting a 50% inhibition of E2-heparin binding at remarkably low antibody concentrations (5 μg/ml; Fig. 5B). These results identify at least four viral epitopes mediating envelope-HS interaction and confirm the specificity of E2-HS binding.
FIG. 5.
Mapping of viral epitopes interacting with HS using an E2-heparin binding assay. (A) Inhibition of E2-heparin binding by monoclonal anti-E2 antibodies. E2 (1 μg/ml) was preincubated with the anti-E2 MAbs or IgG (50 μg/ml) for 1 h at 37°C and then added to ELISA plates coated with heparin (10 μg/ml). Heparin-bound E2 was detected using a polyclonal anti-E2 rabbit serum and colorimetric reaction as described in Materials and Methods. Results are shown as percent inhibition of E2-heparin interaction. Data are shown as mean percent inhibition of E2-heparin binding ± standard deviation (SD) obtained from three independent experiments. (B) Concentration-dependent inhibition of E2-heparin binding by anti-E2 antibodies. Recombinant E2 protein (1 μg/ml) was incubated with anti-E2 monoclonal antibody 2F10 (squares) or 49F3 (diamonds) or with control IgG (open triangles) at various concentrations for 1 h at 37°C. E2-antibody complexes were added to heparin immobilized on plates, and the E2-heparin interaction was analyzed as described above. The OD of the colorimetric reaction is proportional to heparin-bound E2. Results are presented as mean OD ± SD of a representative experiment performed in triplicate.
Since the ability of an anti-E2 antibody to inhibit heparin-E2 binding did not correlate with the affinity of the same antibody to recombinant E2 protein, as studied by E2 ELISA (data not shown), it is unlikely that the observed inhibition of binding is predominantly due to sterical hindrance. Sterical hindrance is also very unlikely following the observation that antibodies targeting epitopes in close proximity exhibited a very different profile in the inhibition of E2-heparin binding. For example, antibody 49F3 targeting aa 516 to 530 exhibited a marked inhibition of E2-heparin binding, whereas antibody 22E1 targeting aa 544 to 555 did not inhibit E2-heparin binding (Fig. 5).
To assess whether binding of the identified E2 epitopes to HS is relevant for the initiation of HCV infection, we studied whether the identified antibodies capable of blocking E2-heparin binding inhibited viral entry of HCVpp. Interestingly, three anti-E2 MAbs inhibiting E2-HS binding also inhibited HCVpp entry into human hepatoma cells (Fig. 5A). These data suggest that binding of HCV to heparan sulfate mediated by viral epitopes E2 398-403, E2412-423, and E2 516-530 plays an important role in the initiation of HCV infection.
HCV-HS binding is targeted by anti-envelope antibodies from patients with chronic HCV infection.
To study the potential impact of HCV-HS binding on HCV infection in vivo, we studied whether HCV-HS interaction is targeted by human antiviral humoral immune responses. Using the envelope-HS binding assay, we quantified binding of recombinant envelope glycoproteins to heparin in the presence of anti-HCV antibodies from sera from HCV-infected individuals.
Preincubation of E2 protein with anti-HCV antibodies from HCV-infected individuals resulted in a marked inhibition of E2-heparin binding in 5 out of 12 HCV-infected patients (Fig. 6A,pt2, pt5, pt8, pt11, and pt12). The inhibition was concentration dependent and specific for sera containing anti-HCV antibodies since IgG from anti-HCV-negative individuals did not inhibit E2-heparin binding (Fig. 6B) (data not shown). To further address the specificity of the ELISA for antibody-mediated inhibition of heparin-E2 binding, we performed a series of control experiments. Since the addition of purified anti-HCV IgG derived from HCV-infected patients or control IgG derived from healthy individuals to immobilized E2 did not result in an alteration of the optical density (OD) of the colorimetric reaction when compared to the detection of E2 without the addition of supplementary IgG (data not shown), it is unlikely that serum-derived IgG interfered with binding of the primary (polyclonal rabbit anti-E2) or secondary (anti-rabbit IgG) detection antibody to E2 or anti-E2, respectively. To study whether the E1-heparin interaction was also affected by anti-HCV antibodies from human sera, we studied binding of E1 in the presence of anti-HCV IgG (Fig. 6A). In contrast to findings for antibody-mediated inhibition of the E2-heparin binding, only a modest antibody-mediated inhibition of E1-heparin binding could be demonstrated for one patient (Fig. 6A, pt10). Similar to the findings observed for inhibition of E1/E2-heparin binding by monoclonal antibodies, inhibition of antibody-mediated envelope glycoprotein-heparin binding did not correlate with the affinity of the purified IgG to recombinant envelope glycoproteins (data not shown). Thus, it is unlikely that the observed inhibition of binding is predominantly due to sterical hindrance by binding of the antibody to the envelope protein. More likely, anti-HCV antibodies specifically interfere with the HCV envelope-HS interaction.
FIG. 6.
Inhibition of envelope glycoprotein binding to immobilized heparin by purified human anti-HCV IgG. (A) Envelope glycoproteins E1 and E2 were incubated with IgG purified from sera from anti-HCV-positive patients (pt1 to pt12) or healthy individuals (control [C]). Envelope glycoprotein-antibody complexes were added to immobilized heparin, and bound envelope glycoproteins were detected by rabbit anti-E1 or anti-E2 polyclonal serum as described in the legend to Fig. 5. Data are shown as mean percent inhibition of E2-heparin binding ± standard deviation (SD) obtained from three independent experiments. To study inhibition of HCVpp entry in the presence of anti-HCV IgG, HCVpp (HCV-J strain) were incubated with purified IgG from HCV-infected patients and human control IgG before the addition to Huh-7 cells. HCVpp entry was analyzed by flow cytometry as described in Materials and Methods. (B) Concentration-dependent inhibition of E2-heparin binding by purified anti-HCV IgG (pt12; solid diamonds) and control IgG (open squares). Analysis of inhibition was performed as described in the legend to Fig. 5. Results are presented as mean OD ± SD of a representative experiment performed in triplicate.
To address the relevance of this finding for antibody-mediated virus neutralization, we studied whether the ability of antibodies to inhibit envelope-heparin binding correlated with the ability of the antibody to neutralize HCVpp infection. Interestingly, all polyclonal anti-HCV antibodies markedly inhibiting HCV envelope-heparin binding strongly inhibited HCVpp entry into Huh-7 cells (inhibition of HCVpp entry of >25%; Fig. 6A). Although this correlation was not present for all anti-HCV antibodies, these findings suggest that HCV infection results in the induction of neutralizing antiviral antibodies recognizing viral epitopes required for envelope-HS binding.
HCV-HS binding is required for the initiation of viral infection.
To study the functional relevance of HCV envelope-HS interaction for the initiation of viral infection, we performed competition experiments using retroviral HCVpp displaying functional envelope glycoproteins on their surface (3). This experimental system has been shown to be a convenient and powerful model for the study of the first steps of HCV infection. Similar to previously published results (3, 4, 17), HCVpp only infected Huh-7 cells (Fig. 2B). As described previously, the nonpermissiveness of HepG2 cells to HCVpp infection is due to a lack of CD81 expression required for postbinding events (4, 17). As shown in Fig. 7A, HCVpp entry into Huh-7 cells was markedly inhibited by heparin and highly sulfated HS but not normally sulfated HS and chondroitin sulfate A, suggesting that the interaction of HCV glycoproteins with cellular HS contributes to the initiation of HCV infection.
FIG. 7.
Inhibition of HCVpp infection of Huh-7 cells by heparin and highly sulfated HS. (A) For HCVpp infection, sucrose gradient-purified HCVpp (HCV-J strain) were preincubated with PBS, heparin, highly sulfated HS (hsHS), normally sulfated HS (HS), or chondroitin sulfate A (CSA) (each at 10 μg/ml) for 30 min at room temperature. HCVpp-GAG complexes were added to Huh-7 cells and incubated for 1.5 h at 4°C. HCVpp entry was determined by GFP reporter gene expression using flow cytometry. Data are shown as mean percent cells positive for GFP relative to infection of HCVpp without GAG (100%). (B) To study whether soluble GAGs interfere with the first step of HCVpp infection, HCVpp binding, HCVpp-GAG complexes were added to Huh-7 cells for 1 h at 4°C and cellular binding of sucrose gradient-purified HCVpp was quantified by anti-E2 MAb (16A6) and flow cytometry. Data are shown as percent binding (mean ± standard deviation [SD] of a representative experiment performed in triplicate) relative to binding of HCVpp without GAG (100%). (C) To study whether soluble GAGs interfere with viral entry mechanisms following viral attachment (temperature-dependent postbinding events), Huh-7 cells were first incubated with HCVpp for 1 h at 4°C. Following binding of HCVpp and washing of cells with PBS to remove unbound HCVpp, soluble GAGs were added to cells (1.5 h at 4°C). Following a shift of incubation temperature to 37°C, HCVpp entry was assessed after 72 h by flow cytometric analysis of GFP reporter gene expression. Data are shown as mean percent cells positive for GFP relative to infection of HCVpp without GAG (100%).
To assess whether highly sulfated HS acts predominantly as a cell surface molecule for viral binding or represents a coreceptor for viral entry following attachment of the viral envelope, we studied the impact of soluble GAGs on HCVpp binding and postbinding steps. As shown in Fig. 7B, HCVpp binding to Huh-7 cells was markedly inhibited by heparin and highly sulfated HS. In contrast, envelope-dependent HCVpp binding was not affected by preincubation of HCVpp with normally sulfated HS and chondroitin sulfate A (Fig. 7B). To exclude that the HCVpp preparation contained nonassembled envelope glycoproteins interfering with HCVpp-GAG interaction, HCVpp had been partially purified by sucrose gradient centrifugation prior to their use in entry and binding studies. Next, we studied whether soluble GAGs interfered with viral entry mechanisms following viral attachment (temperature-dependent postbinding events). Therefore, we incubated Huh-7 cells with HCVpp at 4°C and then added soluble GAGs to bound HCVpp and assessed temperature-dependent HCVpp entry following a shift of the temperature to 37°C. As shown in Fig. 7C, addition of highly sulfated HS following cellular HCVpp binding resulted in a partial inhibition of HCVpp entry.
Taken together, these data suggest that highly sulfated HS is an important molecule for binding of the viral envelope to the cell surface, as shown in Fig. 7B, and may contribute to viral entry in concert with other cell surface molecules such as CD81 and SR-BI, as shown in Fig. 7C.
DISCUSSION
HS is a common cell surface receptor for numerous viruses, and defined sequences are believed to determine host cell specificity (31). In this study, we demonstrate that (i) binding of the HCV envelope glycoproteins to a specific HS structure on host cells is an important step for the initiation of viral infection and (ii) this interaction represents an important target of antiviral host immune responses in HCV infection in vivo.
Viral determinants of the HCV-HS interaction.
Extending our previous findings that HCV envelope glycoprotein binding to target cells requires cell surface HS, we now demonstrate that the interaction of the viral envelope is mediated by both envelope glycoproteins E1 and E2. Similar to findings for envelope glycoprotein E2, highly sulfated HS played an important role in mediating binding of E1. Although recombinant soluble E1 had a lower affinity for immobilized heparin in SPR assays than E2 (Fig. 1) and E1 exhibited a significantly different cellular binding profile from E2 (Fig. 2A), the functional differences of E1 and E2 envelope glycoproteins may not be directly extrapolated to the in vivo situation, where E1 is likely to be in a different conformation as a complex with E2.
Through mapping studies using a panel of monoclonalantienvelope antibodies and a biochemical assay for HCV-HS binding, we have demonstrated that HCV envelope binding to HS was predominantly mediated by four viral epitopes overlapping the E2 hypervariable region 1 (HVR1) and E2-CD81 binding domains. A number of studies have been undertaken to define binding sites of proteins to the HS homologue heparin (for review, see reference 31). Heparin binding motifs frequently contain clusters of one, two, or three basic amino acids, such as arginine and lysine. However, the absence of heparin binding motifs in many other heparin binding proteins suggests that linear patterns of amino acids may not be necessary. Instead, proteins could use a similar spatial structural motif to bind heparin efficiently, in which the basic residues are close in space but not necessary close in amino acid sequence (29). The E2 sequence contains a number of arginine, lysine, and histidine residues scattered across the protein but not clustered to suggest a conventional binding motif to heparin or highly sulfated HS. Our mapping studies identified two additional heparin binding sites (E2480-487 and E2516-530) within the putative E2 CD81 binding regions, suggesting that this central E2 domain may interact with both HS and CD81. Furthermore, we identified two epitopes in the N-terminal E2 region mediating HCV-HS binding, confirming experimental and computational data showing that HVR1 represents a candidate region for HS (33). Antibody 2F10 targeting the middle of HVR1 (E2398-403) markedly blocked the interaction of E2 with immobilized heparin (up to 90% inhibition), suggesting an important role of E2398-403 as a heparan sulfate binding epitope. Moreover, antibody AP33 targeting amino acid position E2412-423 close to HVR1 (outside the E2-CD81 binding domains) demonstrated a marked inhibition of the E2-heparin interaction. Since these antibodies inhibit HCVpp infection (Fig. 5) (42), it is conceivable that binding of the N-terminal E2 glycoprotein (including epitopes E2398-403 and E2412-423) to cell surface HS plays an important role in the initiation of HCV infection.
Interestingly, we observed a strong correlation between antibody inhibition of E2-HS binding and antibody inhibition of HCVpp entry for all monoclonal anti-envelope antibodies with the exception of anti-E2 antibody 11F11 (Fig. 5A). It is possible that the epitope (E2480-487)) targeted by this antibody is not sufficient for binding of the virion during HCV infection in vivo. Alternatively, steric hinderance occurring in the E2-heparin ELISA may explain the lack of functional correlation within the two assays.
HCV virions from chronically infected patients (15, 53) as well as tissue cell culture-derived recombinant HCV (HCVcc) have been shown to bind efficiently to heparan sulfate homologue heparin (T. Wakita, K. Morikawa, T. Date, M. Miyamoto, A. Murayama, S. Sone, and N. Tanabe, 12th Int. Meet. Hepatitis C Virus Related Viruses, presentation P-146, 2005). Furthermore, this high-affinity interaction can be used to purify HCV virions from plasma or tissue culture medium using heparin-Sepharose columns (15, 53; Wakita et al., 12th Int. Meet. Hepatitis C Virus Related Viruses). These observations clearly underline the relevance of our findings for the authentic HCV virion.
In this context, it is of interest to note that Callens et al. (8) observed an interaction only of intracellular but not extracellular detergent-solubilized HCVpp envelope glycoproteins with heparin coupled to Sepharose beads. Whether this lack of interaction of extracellular HCVpp envelope glycoprotein is due to technical aspects of the assay system (e.g., alteration of envelope glycoprotein conformation by the use of detergent) is currently under investigation.
Cellular determinants of the HCV-HS interaction.
Binding of viral or cellular ligands to HS depends on defined patterns and orientations of the sulfo and carboxyl groups along the polysaccharide chain (31). Heparin is distinguished from HS by much higher levels of N- and O-sulfo groups containing an average of 2.4 sulfo groups/disaccharide unit. Detailed analysis of the composition and organization of liver HS has revealed a remarkable proportion of heparin-like structures. A combination of high N- and O-sulfo group contents in liver HS is translated to a total sulfate content of 1.3 sulfo groups/disaccharide, which is 50% higher than those for most HS species (47). In recent years, numerous HS biosynthetic enzymes have been identified which are involved in the modification of the HS chain (12). These modifications include 2-O sulfation of iduronic and glucuronic acid, N sulfation of glucosamine, as well as 6-O sulfation and 3-O sulfation of glucosamine (12). Here we provide evidence that N sulfation, but not 2-O and 6-O sulfation, is required for HCV envelope-HS interaction. This conclusion is supported by two key observations: (i) chemically modified, de-N-sulfated heparin lost its ability to block cellular binding of recombinant envelope glycoproteins; and (ii) both de-2-O- and de-6-O-sulfated heparins retain their ability to block cellular binding of HCV envelope glycoproteins, indicating that these sulfate groups are not important for binding of the HCV envelope. These findings support the conclusion that specific sulfate groups on cellular HS rather than the total level of sulfation may be important for mediating HCV-host cell interaction. Besides the involvement of N-sulfate groups, the size of the saccharide chain appears to play an important role in efficient viral protein-HS binding. Heparin oligosaccharides of less than 10 subunits failed to inhibit binding of envelope glycoprotein E2 to target cells. For HCV-LPs, marked inhibition of cellular binding (>50%) was observed for a 14-subunit oligosaccharide. These findings indicate that the interaction of envelope glycoproteins with highly sulfated HS on target cells is not simply the result of charge interactions but requires a specific HS structure. Interestingly, similar findings have been reported for the interaction of HS with other viruses (9, 16, 24).
Functional relevance of HCV-HS binding.
To study the functional relevance of HCV envelope-HS interaction for the initiation of HCV infection, we performed competition experiments using HCVpp (3). Infectious HCVpp that are assembled by displaying unmodified HCV envelope glycoproteins on retroviral core particles have been successfully used for studies of HCV internalization. Similar to HCVcc (22, 48, 54), this model closely mimics the first steps of natural HCV infection (3, 17). Entry of HCVpp is dependent on the presence of at least two cell surface molecules, the tetraspanin CD81 and scavenger receptor SR-BI. However, the failure of HCVpp or HCVcc to infect human cell lines of nonhepatic origin expressing both CD81 and SR-BI on their cell surface (4) suggests that additional cell surface molecules determine viral tropism in natural HCV infection. In this study, we provide evidence that highly sulfated HS may play an important role in the initiation of HCV infection in the HCVpp model of HCV infection. The marked and specific inhibition of cellular HCVpp binding and partial inhibition of HCVpp entry by highly sulfated HS and heparin suggest that highly sulfated HS (i) is an important molecule for binding of the viral envelope to the cell surface and (ii) may contribute to viral entry in concert with other cell surface molecules such as CD81 and SR-BI.
This hypothesis is supported by results obtained at the same time in the infectious tissue culture system using recombinant HCV. Koutsoudakis and colleagues reported a dose-dependent inhibition of infection of HCVcc by heparin—a homologue of highly sulfated HS—but not by normally sulfated HS and other GAGs (19). Furthermore, selective enzymatic degradation of highly sulfated HS domains present on cell surface HS by specific heparinases markedly reduced HCVcc infectivity (19). Similar to our results obtained in the HCVpp model system, heparin inhibited HCVcc infection only when administrated during virus binding and not afterwards. The authors of this study concluded that the presence of defined glycosaminoglycans such as highly sulfated HS is required for efficient viral particle attachment and consequently contributes to virus invasion (19). Taken together, these findings suggest that highly sulfated HS may play a key role in the initiation of HCV infection. Interaction of HCV envelope glycoproteins with cellular highly sulfated HS seems to mediate the first step of binding the viral envelope to the cell surface. In a second step, the virus may then be transferred to a second high-affinity receptor such as CD81 or SR-BI, triggering viral entry.
Interference with the HCV-HS binding by anti-HCV antibodies strongly suggests that this interaction plays an important role in HCV infection in vivo. Interestingly, several anti-HCV-positive human sera markedly inhibiting HCV envelope-heparin interaction also demonstrated a marked inhibition of HCVpp entry into Huh-7 cells, suggesting that HCV infection results in the induction of antiviral antibodies targeting envelope-HS binding. Antibodies without correlation between inhibition of E2-HS binding and antibody inhibition of HCVpp entry (e.g., anti-HCV IgG derived from pt9 in Fig. 6) may target the interaction of the HCV envelope with other cell surface molecules (e.g., SR-BI, CD81) or interfere with virus-host interactions on steps occurring postbinding.
Finally, mapping of viral and cellular determinants of HCV-HS interaction sets the stage for the development of novel HS-based antiviral strategies targeting viral attachment and entry. The systematic generation and screening of small anionic drugs, heparin-like molecules, and semisynthetic derivatives (30) are currently being explored for the inhibition of dengue virus infectivity. In mouse models for dengue virus and the encephalitis flaviviruses, the HS mimetic PI-88 (52) showed a significant benefit with respect to disease outcome (21), supporting the evaluation of glycosaminoglycan derivatives as antiviral drugs in vivo.
Acknowledgments
The authors thank Bettina Gissler and Anita Haberstroh for excellent technical assistance.
This work was supported by grants from the European Union, Brussels, Belgium (QLK-2-2002-01329 and ViRgiL NoE); the Deutsche Forschungsgemeinschaft (Ba1417/11-1); the Bundesministerium für Forschung und Technologie (01KI9951) Bonn, Germany; INSERM, Paris; the ANR Chair of Excellence Program (R06026MM) Paris, and ANRS, Paris, France; and the National Institutes of Health (HL62244, HL52622, and GM 38060).
Footnotes
▿
Published ahead of print on 23 August 2006.
REFERENCES
- 1.Bain, C., A. Fatmi, F. Zoulim, J. P. Zarski, C. Trepo, and G. Inchauspe. 2001. Impaired allostimulatory function of dendritic cells in chronic hepatitis C infection. Gastroenterology 120**:**512-524. [DOI] [PubMed] [Google Scholar]
- 2.Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278**:**41003-41012. [DOI] [PubMed] [Google Scholar]
- 3.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197**:**633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278**:**41624-41630. [DOI] [PubMed] [Google Scholar]
- 5.Baumert, T. F., S. Ito, D. T. Wong, and T. J. Liang. 1998. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72**:**3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumert, T. F., S. Wellnitz, S. Aono, J. Satoi, D. Herion, J. Tilman Gerlach, G. R. Pape, J. Y. Lau, J. H. Hoofnagle, H. E. Blum, and T. J. Liang. 2000. Antibodies against hepatitis C virus-like particles and viral clearance in acute and chronic hepatitis C. Hepatology 32**:**610-617. [DOI] [PubMed] [Google Scholar]
- 7.Birkmann, A., K. Mahr, A. Ensser, S. Ya ǧ ubo ǧ lu, F. Titgemeyer, B. Fleckenstein, and F. Neipel. 2001. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75**:**11583-11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callens, N., Y. Ciczora, B. Bartosch, N. Vu-Dac, F. L. Cosset, J.-M. Pawlotsky, F. Penin, and J. Dubuisson. 2005. Basic residues in hypervariable region 1 of hepatitis C virus envelope glycoprotein E2 contribute to virus entry. J. Virol. 79**:**15331-15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y., T. Maguire, R. E. Hileman, J. R. Fromm, J. D. Esko, R. J. Linhardt, and R. M. Marks. 1997. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 3**:**866-871. [DOI] [PubMed] [Google Scholar]
- 10.Chisari, F. V. 2005. Unscrambling hepatitis C virus-host interactions. Nature 436**:**930-932. [DOI] [PubMed] [Google Scholar]
- 11.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76**:**7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esko, J. D., and U. Lindahl. 2001. Molecular diversity of heparan sulfate. J. Clin. Investig. 108**:**169-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farci, P., A. Shimoda, D. Wong, T. Cabezon, D. De Gioannis, A. Strazzera, Y. Shimizu, M. Shapiro, H. J. Alter, and R. H. Purcell. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 93**:**15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feld, J. J., and J. H. Hoofnagle. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436**:**967-972. [DOI] [PubMed] [Google Scholar]
- 15.Germi, R., J. M. Crance, D. Garin, J. Guimet, H. Lortat-Jacob, R. W. Ruigrok, J. P. Zarski, and E. Drouet. 2002. Cellular glycosaminoglycans and low density lipoprotein receptor are involved in hepatitis C virus adsorption. J. Med. Virol. 68**:**206-215. [DOI] [PubMed] [Google Scholar]
- 16.Hallak, L. K., D. Spillmann, P. L. Collins, and M. E. Peeples. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 74**:**10508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100**:**7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulst, M. M., H. G. van Gennip, A. C. Vlot, E. Schooten, A. J. de Smit, and R. J. M. Moormann. 2001. Interaction of classical swine fever virus with membrane-associated heparan sulfate: role for virus replication in vivo and virulence. J. Virol. 75**:**9585-9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80**:**5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambot, M., S. Fretier, A. Op De Beeck, B. Quatannens, S. Lestavel, V. Clavey, and J. Dubuisson. 2002. Reconstitution of hepatitis C virus envelope glycoproteins into liposomes as a surrogate model to study virus attachment. J. Biol. Chem. 277**:**20625-20630. [DOI] [PubMed] [Google Scholar]
- 21.Lee, E., M. Pavy, N. Young, C. Freeman, and M. Lobigs. 2006. Antiviral effect of the heparan sulfate mimetic, PI-88, against dengue and encephalitic flaviviruses. Antivir. Res. 69**:**31-38. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309**:**623-626. [DOI] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., and C. M. Rice. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436**:**933-938. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., Z. Shriver, M. Pope, S. C. Thorp, M. B. Duncan, R. J. Copeland, C. S. Raska, K. Yoshida, R. J. Eisenberg, G. Cohen, R. J. Linhardt, and R. Sasisekharan. 2002. Characterization of a heparan sulfate octasaccharide that binds to herpes simplex viral type 1 glycoprotein D. J. Biol. Chem. 277**:**33456-33467. [DOI] [PubMed] [Google Scholar]
- 25.Lozach, P. Y., A. Amara, B. Bartosch, J. L. Virelizier, F. Arenzana-Seisdedos, F. L. Cosset, and R. Altmeyer. 2004. C-type lectins L-SIGN and DC-SIGN capture and transmit infectious hepatitis C virus pseudotype particles. J. Biol. Chem. 279**:**32035-32045. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig, I. S., A. N. Lekkerkerker, E. Depla, F. Bosman, R. J. P. Musters, S. Depraetere, Y. van Kooyk, and T. B. Geijtenbeek. 2004. Hepatitis C virus targets DC-SIGN and L-SIGN to escape lysosomal degradation. J. Virol. 78**:**8322-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahoney, D. J., J. D. Whittle, C. M. Milner, S. J. Clark, B. Mulloy, D. J. Buttle, G. C. Jones, A. J. Day, and R. D. Short. 2004. A method for the non-covalent immobilization of heparin to surfaces. Anal. Biochem. 330**:**123-129. [DOI] [PubMed] [Google Scholar]
- 28.Mandl, C. W., H. Kroschewski, S. L. Allison, R. Kofler, H. Holzmann, T. Meixner, and F. X. Heinz. 2001. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J. Virol. 75**:**5627-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margalit, H., N. Fischer, and S. A. Ben-Sasson. 1993. Comparative analysis of structurally defined heparin binding sequences reveals a distinct spatial distribution of basic residues. J. Biol. Chem. 268**:**19228-19231. [PubMed] [Google Scholar]
- 30.Marks, R. M., H. Lu, R. Sundaresan, T. Toida, A. Suzuki, T. Imanari, M. J. Hernaiz, and R. J. Linhardt. 2001. Probing the interaction of dengue virus envelope protein with heparin: assessment of glycosaminoglycan-derived inhibitors. J. Med. Chem. 44**:**2178-2187. [DOI] [PubMed] [Google Scholar]
- 31.Munoz, E. M., and R. J. Linhardt. 2004. Heparin-binding domains in vascular biology. Arterioscler. Thromb. Vasc. Biol. 24**:**1549-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opitz, O. G., Y. Suliman, W. C. Hahn, H. Harada, H. E. Blum, and A. K. Rustgi. 2001. Cyclin D1 overexpression and p53 inactivation immortalize primary oral keratinocytes by a telomerase-independent mechanism. J. Clin. Investig. 108**:**725-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penin, F., C. Combet, G. Germanidis, P.-O. Frainais, G. Deléage, and J.-M. Pawlotsky. 2001. Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J. Virol. 75**:**5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pervin, A., C. Gallo, K. A. Jandik, X. J. Han, and R. J. Linhardt. 1995. Preparation and structural characterization of large heparin-derived oligosaccharides. Glycobiology 5**:**83-95. [DOI] [PubMed] [Google Scholar]
- 35.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282**:**938-941. [DOI] [PubMed] [Google Scholar]
- 36.Rabenstein, D. L. 2002. Heparin and heparan sulfate: structure and function. Nat. Prod. Rep. 19**:**312-331. [DOI] [PubMed] [Google Scholar]
- 37.Rosa, D., S. Campagnoli, C. Moretto, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. Weiner, J. Y. N. Lau, Q.-L. Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 93**:**1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21**:**5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shafti-Keramat, S., A. Handisurya, E. Kriehuber, G. Meneguzzi, K. Slupetzky, and R. Kirnbauer. 2003. Different heparan sulfate proteoglycans serve as cellular receptors for human papillomaviruses. J. Virol. 77**:**13125-13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99**:**13-22. [DOI] [PubMed] [Google Scholar]
- 41.Steinmann, D., H. Barth, B. Gissler, P. Schürmann, M. I. Adah, J. T. Gerlach, G. R. Pape, E. Depla, D. Jacobs, G. Maertens, A. H. Patel, G. Inchauspé, T. J. Liang, H. E. Blum, and T. F. Baumert. 2004. Inhibition of hepatitis C virus-like particle binding to target cells by antiviral antibodies in acute and chronic hepatitis C. J. Virol. 78**:**9030-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarr, A. W., A. M. Owsianka, J. M. Timms, C. P. McClure, R. J. Brown, T. P. Hickling, T. Pietschmann, R. Bartenschlager, A. H. Patel, J. K. Ball, A. Owsianka, V. S. Juttla, D. Lavillette, B. Bartosch, and F. L. Cosset. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43**:**592-601. [DOI] [PubMed] [Google Scholar]
- 43.Toida, T., R. E. Hileman, A. E. Smith, P. I. Vlahova, and R. J. Linhardt. 1996. Enzymatic preparation of heparin oligosaccharides containing antithrombin III binding sites. J. Biol. Chem. 271**:**32040-32047. [DOI] [PubMed] [Google Scholar]
- 44.Toida, T., H. Yoshida, H. Toyoda, I. Koshiishi, T. Imanari, R. E. Hileman, J. R. Fromm, and R. J. Linhardt. 1997. Structural differences and the presence of unsubstituted amino groups in heparan sulphates from different tissues and species. Biochem. J. 322**:**499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Doorn, L. J., K. van Hoek, G. de Martinoff, F. Bosman, L. Stuyver, T. Kos, I. Frantzen, P. Sillekens, G. Maertens, and W. Quint. 1997. Serological and molecular analysis of hepatitis C virus envelope regions 1 and 2 during acute and chronic infections in chimpanzees. J. Med. Virol. 52**:**441-450. [DOI] [PubMed] [Google Scholar]
- 46.Vives, R. R., A. Imberty, Q. J. Sattentau, and H. Lortat-Jacob. 2005. Heparan sulfate targets the HIV-1 envelope glycoprotein gp120 coreceptor binding site. J. Biol. Chem. 280**:**21353-21357. [DOI] [PubMed] [Google Scholar]
- 47.Vongchan, P., M. Warda, H. Toyoda, T. Toida, R. M. Marks, and R. J. Linhardt. 2005. Structural characterization of human liver heparan sulfate. Biochim. Biophys. Acta 1721**:**1-3. [DOI] [PubMed] [Google Scholar]
- 48.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11**:**791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wellnitz, S., B. Klumpp, H. Barth, S. Ito, E. Depla, J. Dubuisson, H. E. Blum, and T. F. Baumert. 2002. Binding of hepatitis C virus-like particles derived from infectious clone H77C to defined human cell lines. J. Virol. 76**:**1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yagnik, A. T., A. Lahm, A. Meola, R. M. Roccasecca, B. B. Ercole, A. Nicosia, and A. Tramontano. 2000. A model for the hepatitis C virus envelope glycoprotein E2. Proteins 40**:**355-366. [DOI] [PubMed] [Google Scholar]
- 51.Yanagi, M., M. St. Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244**:**161-172. [DOI] [PubMed] [Google Scholar]
- 52.Yu, G., N. S. Gunay, R. J. Linhardt, T. Toida, J. Fareed, D. A. Hoppensteadt, H. Shadid, V. Ferro, C. Li, K. Fewings, M. C. Palermo, and D. Podger. 2002. Preparation and anticoagulant activity of the phosphosulfomannan PI-88. Eur. J. Med. Chem. 37**:**783-791. [DOI] [PubMed] [Google Scholar]
- 53.Zahn, A., and J. P. Allain. 2005. Hepatitis C virus and hepatitis B virus bind to heparin: purification of largely IgG-free virions from infected plasma by heparin chromatography. J. Gen. Virol. 86**:**677-685. [DOI] [PubMed] [Google Scholar]
- 54.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102**:**9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]