An actin–ribonucleoprotein interaction is involved in transcription by RNA polymerase II (original) (raw)
Abstract
To determine the function of actin in the cell nucleus, we sought to identify nuclear actin-binding proteins in the dipteran Chironomus tentans using DNase I-affinity chromatography. We identified the RNA-binding protein hrp65 as an actin-binding protein and showed that the C-terminal sequence of the hrp65-2 isoform is able to interact directly with actin in vitro. In vivo crosslinking and coimmunoprecipitation experiments indicated that hrp65 and actin are also associated in the living cell. Moreover, in vivo administration of a competing peptide corresponding to the C-terminal sequence of hrp65-2 disrupted the actin–hrp65-2 interaction and caused a specific and drastic reduction of transcription as judged by puff regression and diminished bromo-UTP incorporation. Our results indicate that an actin-based mechanism is implicated in the transcription of most if not all RNA polymerase II genes and suggest that an actin–hrp65-2 interaction is required to maintain the normal transcriptional activity of the cell. Furthermore, immunoelectron microscopy experiments and nuclear run-on assays suggest that the actin–hrp65-2 complex plays a role in transcription elongation.
Actin and myosin I are present not only in the cytoplasm but also in the cell nucleus (1, 2), where they have been implicated in transcription of protein-coding genes (3–5) and nuclear export (6). Many other cytoskeletal components have also been found in the cell nucleus, and some of them have been tentatively implicated in gene expression (reviewed in ref. 1).
Actin has been found associated with (pre)-messenger ribonucleoprotein (pre-mRNP) complexes in a variety of organisms, and we recently revealed that actin is a bona fide component of the Balbiani ring (BR) pre-mRNP particles in the salivary gland cells of the dipteran Chironomus tentans (7). The BR genes code for large secretory proteins that are expressed in the salivary gland cells of C. tentans (reviewed in ref. 8). The BR pre-mRNAs are assembled into large RNP particles, the BR pre-mRNPs, that can be visualized directly by transmission electron microscopy (reviewed in ref. 9). Actin is incorporated cotranscriptionally into the newly synthesized BR pre-mRNPs by binding to a subset of heterogeneous nuclear RNP (hnRNP) proteins such as the hnRNP A1-like protein hrp36 (7). Remarkably, the presence of actin in RNP complexes is not restricted to the BR particles of C. tentans, because actin has also been found associated with certain hnRNP proteins of the A/B group in mammalian pre-mRNPs (10).
The presence of actin in the cell nucleus is well documented, but the precise role of nuclear actin is still not understood. To obtain further insight into the function(s) of nuclear actin, we sought to identify additional proteins of C. tentans that bind to actin in the cell nucleus.
Materials and Methods
Detailed descriptions of the materials and methods can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
DNase I-Sepharose Pull-Down Assay. DNase I pull-down assays were performed as described (7). DNase I (Sigma) coupled to CNBr-activated Sepharose (Amersham Pharmacia) was pre-equilibrated in PBS and incubated with either nuclear or cytosolic extracts prepared from C. tentans tissue-culture cells. The bound proteins were analyzed by SDS/PAGE and detected by silver staining.
Peptides and Antibodies. All peptides used in this study were chemically synthesized by the solid-phase method and purified by reverse-phase HPLC to >85% homogeneity (Innovagen, Lund, Sweden). Peptide 65-2CTS was conjugated to keyhole limpet hemocyanin (KLH) and used to immunize rabbits according to standard procedures. The antibody against synaptonemal complex protein 3 (SCP3) used in negative controls was kindly provided by C. Höög (Karolinska Institute, Stockholm). Antibodies against hrp65 (11), hrp23 (12), and hrp45 (13) have been described in earlier studies.
Preparation of Proteins for Protein–Protein-Interaction Assays. The construction of plasmids containing the full-length ORF of hrp65-1, hrp65-2, and hrp65-3 with a T7 tag at the N terminus is described in detail in Supporting Materials and Methods. These plasmids were used as templates for TNT-coupled in vitro transcription/translation in the presence of [35S]methionine following manufacturer instructions (Promega). 35S-labeled native actin from nonmuscle cells was prepared by DNase I affinity chromatography essentially as described (14).
T7-Tag Antibody-Agarose Coimmunoprecipitations. T7-tagged 35S-labeled hrp65-1, hrp65-2, and hrp65-3 were immobilized onto T7-tag antibody agarose and incubated with saturating amounts of DNase I affinity-purified 35S-labeled native actin (molar ratio of actin/hrp65 ≈ 10-fold). For competition experiments, hrp65 beads were presaturated with either 65-2CTS peptide-KLH conjugate or KLH alone. Bound proteins were resolved by SDS/PAGE and detected by autoradiography.
Peptide-Binding Assays. Peptides shown in Fig. 1 were covalently bound to Sulfolink resin (Pierce) via terminal cysteine residues following manufacturer instructions. Peptide beads were incubated individually with saturating amounts of 35S-labeled native actin purified by DNase I affinity chromatography. Bound and unbound actin was quantified by phosphorimaging.
Fig. 1.
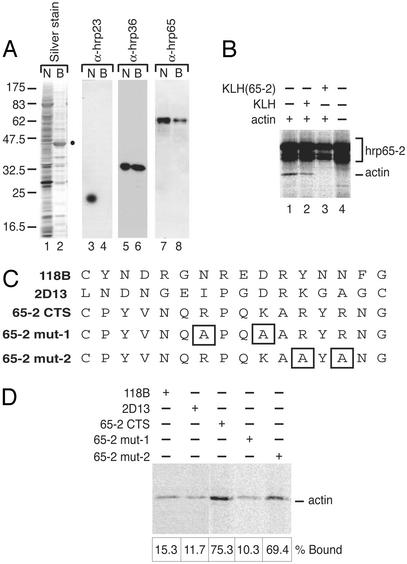
Actin binds directly to the C terminus of Hrp65-2. (A) Nuclear actin-associated proteins studied by DNase I affinity chromatography and Western blotting. Nuclear extracts from C. tentans tissue-culture cells were either fractionated by SDS/PAGE (N) or mixed with DNase I-Sepharose beads. The proteins in the DNase I-bound fraction (B) were eluted and separated by SDS/PAGE. Gels were silver-stained (lanes 1 and 2) or analyzed by Western blotting by using mAbs against three C. tentans proteins: hrp23 (lanes 3 and 4), hrp36 (lanes 5 and 6), and hrp65 (lanes 7 and 8). The actin band previously identified by mass spectrometry is marked with a dot. (B) Reconstitution experiments with purified [35S]hrp65-2 immobilized on T7 beads and saturating amounts of 35S-labeled G-actin. When indicated, binding was carried out in the presence of either nonconjugated KLH (lane 2) or 65-2CTS-conjugated KLH (lane 3) to compete binding of actin to hrp65-2. The bound proteins were separated by SDS/PAGE and analyzed by autoradiography. (C) Sequences of the peptides used in binding studies. (D) Specificity of the actin–hrp65-2 association assayed by affinity chromatography. The peptides shown in C were coupled to Sulfolink beads via their terminal cysteines and incubated with purified 35S-labeled G-actin. Bound actin was fractionated by SDS/PAGE and visualized by autoradiography. The bound actin was quantified by phosphorimaging, and the results are shown under each lane as percentages of input actin.
In Vivo Protein–Protein Crosslinking and Immunoprecipitation. In vivo crosslinking of proteins in C. tentans tissue-culture cells with dithiobis-succinimidylpropionate (DSP, Sigma) was done essentially as described (7). Nuclear and cytosolic extracts then were prepared as described (15) and where appropriate incubated with 8 M urea. The extracts were diluted 10 times to reduce the urea concentration and used immediately for immunoprecipitation. Immunoprecipitations were done following standard procedures.
Nuclear Run-on Transcription. Nuclear run-on transcription was done according to standard procedures (16) by using isolated C. tentans nuclei permeabilized with Triton X-100. When indicated, the permeabilized nuclei were incubated with peptides (final concentration 0.6 mg/ml) or antibodies (final concentration 0.12 mg/ml) before the run-on reaction. Control antibodies and control peptides were used at the same concentration. To estimate the run-on transcription corresponding to pre-mRNA, the radioactivity incorporated into RNA molecules in the 200- to 400-nt range was quantified by phosphorimaging.
Microinjection Experiments. C. tentans salivary glands were isolated from the fourth instar larvae. Microinjections were performed on isolated salivary glands with peptides at a concentration of 30–60 mg/ml in PBS. Immunofluorescence detection of hrp45 after microinjection was performed as described below. For transcription experiments, bromo-UTP (BrUTP) was injected at a concentration of 100 mg/ml 60 min after peptide injections.
Immunofluorescence of C. tentans Salivary Glands. Isolated salivary glands were fixed, permeabilized, and incubated with antibodies as indicated. Images of salivary gland nuclei were taken with an LSM 510 laser scanning microscope. The thickness of the optical section was 1 μm. When indicated, dissected glands were incubated before fixation in hemolymph containing actinomycin D at a final concentration of 4 μg/ml for 1 h.
Isolation and Immunostaining of Polytene Chromosomes. Salivary glands were isolated from fourth instar C. tentans, and the polytene chromosomes were released from the glands by a pipetting procedure as described (17). Immunostaining was done according to standard procedures. For light microscopy, a 6-nm gold-conjugated secondary antibody (Jackson Immuno-Research) was used, and the gold labeling was silver-enhanced with IntenSE M (Amersham Pharmacia). For electron microscopy, the chromosomes were treated as described above, but the colloidal gold had a diameter of 12 nm and was visualized directly, and the chromosomes were embedded in Agar 100 resin after immunolabeling (18). The results were visualized in a Philips CM120 electron microscope at 60 kV.
Results
The RNA-Binding Protein Hrp65 Interacts with Actin in Vivo. To identify actin-binding partners in the cell nucleus, we made use of the specific binding of G-actin to DNase I (19) and carried out DNase I affinity chromatography experiments with nuclear extracts from C. tentans. Using SDS/PAGE and Western blotting, we analyzed the nuclear proteins that bound to the DNase I beads (Fig. 1 A). We confirmed the presence of actin and hrp36 in the DNase I-bound fraction, and we detected an additional band of ≈65 kDa that we identified as hrp65 (11) on the basis of its immunoreactivity with mAb 4F9 (Fig. 1 A, lanes 7 and 8). Hrp65 was not detectable in the DNase I-bound fraction when the experiment was repeated by using a cytoplasmic extract (data not shown). We concluded that hrp65 is associated with actin in the cell nucleus.
The Hrp65-2 Isoform Binds Directly to Actin in Vitro. We then carried out in vitro pull-down experiments with purified proteins to determine whether hrp65 was able to bind actin directly. Three hrp65 isoforms with different C-terminal sequences have been reported (20), and we assayed the ability of each isoform to bind actin. For this purpose, mammalian G-actin was labeled in vivo with [35S]methionine and purified by DNase I chromatography in low-salt G buffer (14). 35S-labeled hrp65 isoforms were prepared by in vitro transcription/translation and affinity-purified through an N-terminal T7 epitope. The purified hrp65 isoforms were immobilized individually on agarose beads coated with T7-tag antibody and incubated with saturating amounts of purified [35S]actin. Binding was performed in low salt, and the bound proteins were resolved by SDS/PAGE and detected by autoradiography. Hrp65-2 was the only hrp65 isoform able to interact specifically with actin (Fig. 1_B_, lane 1).
Because the hrp65 isoforms are identical from amino acid residue 1–499 (20), their differential behavior in the actin-binding assay suggested that the variable C-terminal sequences were involved in actin binding. To test this hypothesis, a peptide termed 65-2CTS containing the 15 C-terminal amino acid residues of hrp65-2 was chemically synthesized and conjugated to a carrier, KLH. The actin–hrp65-binding assays then were repeated in the presence of saturating amounts of either KLH-65-2 or KLH alone. As shown in Fig. 1_B_, KLH-65-2 was able to compete for binding of actin to hrp65-2, whereas KLH alone did not have any significant effect on the binding (Fig. 1_B_, compare lanes 2 and 3).
The hpr65-3 isoform failed to bind actin. Binding between actin and hrp65-1 was detectable to some extent but was not considered specific because it was abolished by both KLH-65-2 and KLH alone (data not shown). Thus our results show that actin binds specifically to hrp65-2, and they strongly suggest that the C terminus of hrp65-2 is implicated in this interaction.
The C Terminus of Hrp65-2 Is an Actin-Binding Site. To analyze further the actin–hrp65-2 binding specificity, we designed two peptides bearing double mutations likely to affect actin binding (Fig. 1_C_, 65-2mut-1 and 65-2mut-2). These two mutated peptides, the wild-type 65-2CTS, and two other synthetic peptides of unrelated sequence (118B and 2D13) were covalently coupled to Sulfolink beads via terminal cysteines and used in reconstitution experiments. The peptide-coupled beads were incubated with saturating amounts of [35S]actin. The bound actin was resolved by SDS/PAGE, detected by autoradiography, and quantified by phosphorimaging (Fig. 1_D_). As much as 75% of the input actin bound to the 65-2CTS beads, whereas the percentage of actin that bound to the control peptides was in all cases <16%. These results show that the 65-2CTS peptide contains a major determinant for actin-binding specificity, and that the C terminus of hrp65-2 constitutes an actin-binding site on the hrp65-2 molecule.
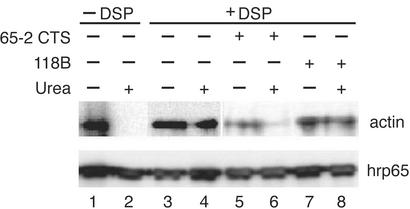
Actin and Hrp65-2 Are also Directly Associated in Vivo. We performed in vivo crosslinking experiments to establish whether actin and hrp65 are directly associated in living cells. C. tentans tissue-culture cells were treated in vivo with DSP, a reversible short-range (≈11-Å) crosslinker that reacts with primary amino groups (21). After crosslinking, nuclear protein extracts were assayed by immunoprecipitation with the mAb 4E9 against hrp65. The bound fractions were studied by Western blotting with polyclonal antibodies against either hrp65 or actin. As shown in Fig. 2 (lanes 1–4), actin was coimmunoprecipitated from DSP-treated extracts even under denaturing conditions (e.g., in the presence of 8 M urea), which shows that hrp65 is crosslinked directly to actin and hence that actin and hrp65 interact directly in vivo.
Fig. 2.
Actin binds directly to Hrp65 in vivo: coimmunoprecipitation after in vivo crosslinking. C. tentans cells were treated with the cell-permeable crosslinker DSP. Nuclear extracts were prepared and split into two equal portions; one was treated with urea. Nuclear extracts were also prepared from DSP-untreated cells (lanes 1 and 2). All samples were subjected to immunoprecipitation with mAb 4E9 against hrp65. Bound proteins were analyzed by SDS/PAGE and Western blotting with rabbit polyclonal antibodies against hrp65 and actin. In some experiments, the cells were incubated in the presence of peptides, either 65-2CTS (lanes 5 and 6) or 118B (lanes 7 and 8), and then treated as described above.
The 65-2CTS Peptide Disrupts the Actin–Hrp65-2 Complex in Vivo. We next determined whether the 65-2CTS peptide could disrupt the actin–hrp65 interaction in vivo. It is well known that short hydrophilic peptides can be taken up by cells from the culture medium (reviewed in ref. 22). To test whether 65-2CTS was a cell-permeable peptide useful for in vivo studies, we labeled the 65-2CTS peptide, as well as a control peptide of unrelated sequence (peptide 118B), with fluorescein, added the fluorescein-labeled peptides to the culture medium, and assessed by confocal microscopy whether C. tentans tissue-culture cells could incorporate the peptides. Peptides 65-2CTS and 118B could be internalized efficiently from the culture medium in a dose-dependent manner (see Fig. 5, which is published as supporting information on the PNAS web site). To test whether 65-2CTS could disrupt the binding between actin and hrp65 in the living cell, the coimmunoprecipitation experiment was repeated with the addition of 65-2CTS to the medium before the DSP treatment (Fig. 2, lanes 5–8). The presence of 65-2CTS caused a drastic reduction in the amount of actin bound to hrp65 (Fig. 2, compare lanes 3 and 4 with lanes 5 and 6). From this result we conclude that the peptide 65-2CTS efficiently disrupts the actin–hrp65-2 interaction in vivo.
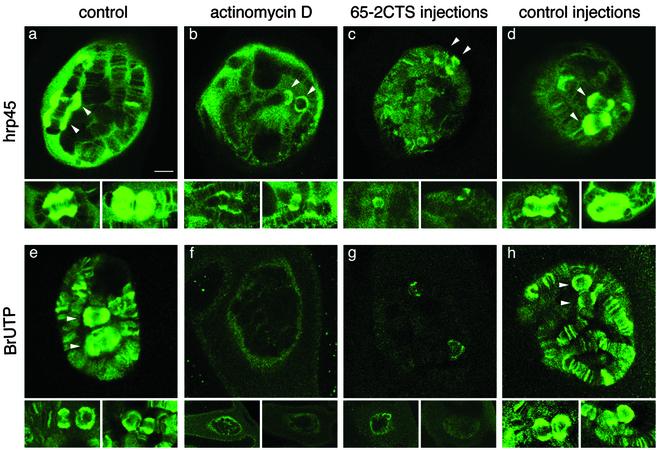
Disruption of the Actin–Hrp65-2 Complex Inhibits Transcription in Vivo. To investigate the functional significance of the actin–hrp65-2 interaction, we injected the 65-2CTS peptide into the nucleus of salivary gland cells and analyzed the effects of the injection on the expression of the BR genes. The BR genes display all the typical features of protein-coding genes, and their expression can be monitored easily at the cytological level by using immunofluorescence microscopy. Two giant BR puffs, BR1 and BR2, are observed under normal growth conditions in the nuclei of salivary gland cells immunostained with antibodies against the abundant SR protein hrp45 (13) as shown in Fig. 3 a_–_d. Microinjection of the 65-2CTS peptide into the salivary gland cell resulted in a pronounced regression of the BR puffs and caused a general decrease of immunostaining in the polytene chromosomes (Fig. 3_c_). This effect was comparable to that of actinomycin D (Fig. 3_b_), a transcription inhibitor known to cause a drastic reduction in the size of the BR puffs (23). Injection of control peptides (either 65-2mut-1 or 118B) did not have any visible effect (Fig. 3_d_). These observations strongly suggest that the 65-2CTS peptide specifically inhibits transcription. To confirm this hypothesis we combined the peptide injections with BrUTP incorporation under conditions that reveal transcription by RNA polymerase II (24), and we detected the transcription sites by immunofluorescence with an antibody against BrUTP (Fig. 3 e_–_h). As expected, the BrUTP incorporation was abolished by actinomycin D (Fig. 3_f_). Administration of the 65-2CTS peptide followed by 60 min of incubation and subsequent BrUTP injection drastically reduced BrUTP incorporation (Fig. 3_g_). The effect of 65-2CTS was highly specific as judged by the normal levels of BrUTP incorporation observed when control peptides were injected under the same experimental conditions (Fig. 3_h_). These results show that the overall RNA synthesis by RNA polymerase II is drastically down-regulated by the 65-2CTS peptide. Furthermore, the effect of 65-2CTS is global and not restricted to a small subset of genes.
Fig. 3.
Disruption of the actin–Hrp65-2 complex down-regulates mRNA transcription. (a_–_d) Salivary glands were isolated from C. tentans fourth instar larvae, immunostained with mAb 2E4 against hrp45, and visualized by confocal microscopy. Individual salivary gland cells untreated (a), treated with the transcription inhibitor actinomycin D before immunostaining (b), injected with peptide 65-2CTS before immunostaining (c), or injected with control peptides (either 118B or 65-2mut1) before immunostaining (d) are shown. After peptide injections, the glands were incubated in hemolymph at 18°C for 90 min before fixation and immunostaining. For each treatment, three examples are presented: one showing a full nucleus and two additional ones showing only the BR puffs. (e_–_h) Detection of nascent transcripts by incorporation of BrUTP. BrUTP was injected into the cytoplasm of salivary gland cells, and the incorporation of BrUTP into nascent RNA was visualized by immunofluorescence after 20 min of incubation in hemolymph at 18°C. (e) Incorporation of BrUTP in untreated cells. (f) Glands were incubated with hemolymph containing actinomycin D before BrUTP injection and immunostaining. (g and h) Either peptide 65-2CTS or a control peptide (65-2mut-1 or 118B) was injected into the nucleus of the salivary gland cells, the glands were incubated in hemolymph for 60 min at 18°C, and BrUTP was administered by cytoplasmic injection. The glands then were incubated for an additional 20 min before fixation and immunostaining as described above. (Bar in a, 10 μm.) All images are at the same magnification. The arrowheads point at the BR1 and BR2 puffs in each nucleus.
We confirmed the inhibitory effect of the 65-2CTS peptide in diploid tissue-culture cells of C. tentans by measuring incorporation of [32P]ATP into poly(A)+ RNA in vivo in the presence or absence of the 65-2CTS peptide. The [32P]ATP incorporation was reduced by 70% when the 65-2CTS peptide was added to the culture medium at a final concentration of 25 μM (data not shown).
Hrp65-2 Is Associated with Nascent Transcripts. We determined the location of hrp65-2 inside the nucleus using an antibody raised against the 65-2CTS peptide that recognized the native hrp65-2. This antibody stained the BR puffs and other smaller puffs in isolated polytene chromosomes, which indicated that hrp65-2 is located at transcriptionally active sites. The specificity of the anti-hrp65-2 antibody in Western blot and immunostaining of isolated chromosomes is shown in Fig. 6, which is published as supporting information on the PNAS web site.
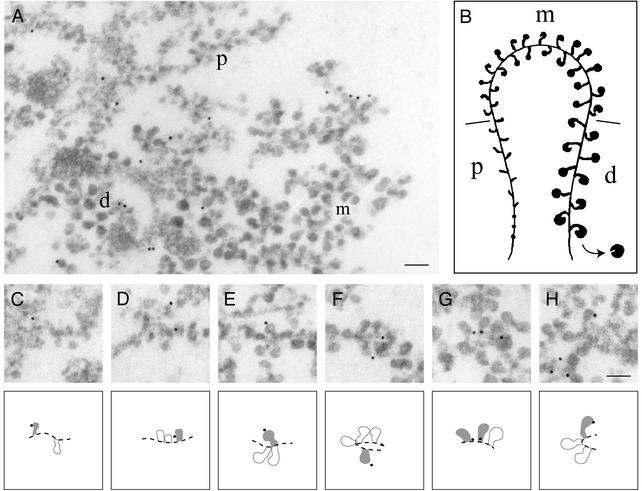
We analyzed the location of hrp65-2 in the active BR transcription unit by immunoelectron microscopy (Fig. 4). The active BR genes, represented schematically in Fig. 4_B_, show a distinct polarity defined by the size and structure of the nascent pre-mRNPs along the gene (25). Full-length transcription units are not found in the sections used for electron microscopy, but partial segments can be assigned to defined positions along the gene based on their morphology. The possibility of identifying the different parts of the BR transcription unit allowed us to map the binding of hrp65-2 to the proximal (p), middle (m), and distal (d) segments of the BR gene by immunoelectron microscopy with the anti-hrp65-2 antibody. As shown in Fig. 4 C_–_H, we found hrp65-2 in all three segments of the active BR transcription unit (distribution of 400 consecutive gold markers: P = 50%, m = 31%, d = 19%). From this result we conclude that hrp65-2 is located along the entire BR gene.
Fig. 4.
Hrp65-2 is associated with the nascent BR pre-mRNP particles: Immunoelectron microscopic localization of hrp65-2 in the active BR transcription unit. Polytene chromosomes were isolated, incubated with the anti-hrp65-2 antibody, and detected with a secondary antibody conjugated to colloidal gold markers. The immunolabeled chromosomes were fixed, dehydrated, embedded in agar resin, and sectioned. (A) Distribution of gold markers in the proximal (p), middle (m), and distal (d) portions of the BR gene. (B) Schematic representation of the active BR transcription unit. (C_–_H) Examples of immunolabeling in nascent BR pre-mRNP particles. Interpretations of the images are provided under each micrograph. (Bar, 100 nm.)
To establish whether hrp65-2 was located on the DNA axis or harbored in the growing pre-mRNP, we studied the location of the gold markers in the distal region of the BR gene. The markers were classified as close to (within 25 nm) or distant from (>25 nm) the axis. This latter group only contains gold markers associated with BR pre-mRNP. The markers close to the axis could represent hrp65-2 molecules bound to the stalk of the growing pre-mRNP particles and/or molecules bound to the RNA polymerase or the chromatin axis. We examined 13 well defined distal segments of the BR gene and found 17 gold markers close to the axis and 13 markers distant from the axis. From these results we cannot rule out that a fraction of hrp65-2 is bound to the RNA polymerase or the chromatin axis, but we can conclude that hrp65-2 is incorporated cotranscriptionally into the nascent BR pre-mRNP particles.
Perturbation of the Actin–Hrp65-2 Complex Diminishes Transcription Elongation. The immunoelectron microscopy results reported above together with data from a previous study (7) show that both actin and hrp65-2 are associated with the nascent BR pre-mRNP along the gene, and therefore it is plausible that these proteins play a role in transcription elongation. To test this possibility, we carried out in vitro run-on transcription assays in the presence or absence of either anti-actin antibody or 65-2CTS peptide. The nuclear run-on assays, devised to analyze transcription rates on natural chromatin templates, do not support new initiation events and are based on measurements of transcription elongation (reviewed in ref. 26). Nuclei from C. tentans cells were isolated, permeabilized, and submitted to run-on analysis in the presence of radiolabeled RNA precursors. As summarized in Table 1, the addition of anti-actin antibody or 65-2CTS peptide in the run-on reaction resulted in a significant and specific reduction of pre-mRNA synthesis. These run-on assays confirm the results from our in vivo experiments, where disruption of the actin–hrp65-2 interaction results in transcription inhibition. Moreover, from the run-on experiments we can conclude that the inhibitory effects of the anti-actin antibody and the 65-2CTS peptide are exerted, at least partially, at the level of transcription elongation.
Table 1. Effect of anti-actin antibody and 65-2CTS peptide on nuclear run-on transcription.
| Reaction conditions | Relative run-on transcription* |
|---|---|
| Standard | 100 |
| Anti-actin antibody | 54 |
| 65-2CTS peptide | 62 |
| Control antibody (SCP3) | 82 |
| Control peptide (65-2mut-1) | 83 |
Discussion
We have identified hrp65-2 as a nuclear actin-binding protein and shown that the synthetic peptide 65-2CTS corresponding to the C terminus of hrp65-2 binds directly to actin in vitro, disrupts the actin–hrp65-2 interaction both in vivo and in vitro, and causes a global transcription inhibition in living cells. These results raise the possibility that an actin-based mechanism is involved in transcription of most if not all RNA polymerase II genes. Both globular and filamentous actin might be present in the cell nucleus, as discussed previously (ref. 10 and references therein), and the conformational state of the actin involved in transcription remains to be investigated.
Hrp65 belongs to a family of evolutionarily conserved proteins characterized by the presence of a DBHS domain (27). The DBHS group includes the mammalian proteins PSF, p54nrb/NonO, and PSP1 and the Drosophila protein NonA/Bj6 (reviewed in ref. 28). Although the molecular function of the DBHS proteins is not understood fully, they have been implicated in several gene-expression events from splicing (29) to nuclear retention of edited RNAs of viral origin (30) to transcription regulation (ref. 31 and references therein). Whether the functions of these other DBHS proteins also require actin remains to be elucidated.
In light of our results, the most interesting question is the nature of the mechanistic coupling between the actin–hrp65 complex and transcription. In mammalian cells, changes in the G-actin pool are responsible for the regulation of a specific subset of SRF target genes through cofactors that bind G-actin and sense G-actin levels (5, 32). In this scenario, the binding of hrp65-2 (or 65-2CTS) to actin might regulate interactions between actin and other cofactors with crucial roles in transcription. Such a mechanism would not require the presence of the actin–hrp65 complex at the transcription site. However, both actin and hrp65-2 are located at the active BR transcription unit (ref. 7, and our present results), and thus we favor a model in which the actin–hrp65 complex acts at the transcription site. Moreover, two types of observations give support to the idea that the actin–hrp65 complex plays a role in transcription elongation. First, perturbation of the actin–hrp65-2 complex causes a significant reduction in the elongation of nascent transcripts in run-on assays. Second, using immunoelectron microscopy we have shown that actin (7) and hrp65-2 (our present results) are associated with the nascent pre-mRNP along the entire transcription unit.
What is the function of the actin–hrp65 complex along the active gene? The fact that actin and actin-related proteins are components of chromatin-remodeling complexes (reviewed in ref. 33) and the observation that PSF and p54nrb/NonO participate in the regulation of the CYP17 gene by recruiting histone deacetylases (31, 34) suggest that the actin–hrp65 interaction may participate in the recruitment of complexes with chromatin-remodeling activities required for effective elongation. Alternatively, the actin–hrp65 interaction may be required to stimulate the processivity of the elongating polymerase.
It is well accepted that the large subunit of RNA polymerase II facilitates cotranscriptional pre-mRNA processing by recruiting pre-mRNA processing factors to the nascent pre-mRNA (reviewed in ref. 35). Our present observations suggest a reciprocal mechanism by which factors associated with the pre-mRNA would stimulate transcription. A recent report by Fong and Zhou (36) has provided another example of such a reciprocal mechanism: The spliceosomal U small nuclear RNPs interact with the transcription elongation factor TAT-SF1 and stimulate polymerase elongation. Thus, these reports reveal the importance of bidirectional crosstalk between the nascent pre-mRNP and the elongating polymerase for productive gene expression in vivo.
Supplementary Material
Supporting Information
Acknowledgments
We thank C. Höög for the gift of the anti-synaptonemal complex protein 3 antibody and L. Wieslander for the 118B peptide. This work was supported by grants from the Swedish Research Council, the Human Frontier Science Program Organization, the Åke Wiberg Foundation, the Carl Trygger Foundation, the Lars Hiertas Minne Foundation, and the Jeansson Foundation. P.P., N.F., and F.M. were supported by fellowships from the Blanceflor-Ludovici Foundation, the European Community, and the European Molecular Biology Organization, respectively.
Abbreviations: RNP, ribonucleoprotein; mRNP, messenger RNP; BR, Balbiani ring; KLH, keyhole limpet hemocyanin; DSP, dithiobis-succinimidylpropionate; BrUTP, bromo-UTP.
References
- 1.Rando, O. J., Zhao, K. & Crabtree, G. R. (2000) Trends Cell Biol. 10**,** 92–97. [DOI] [PubMed] [Google Scholar]
- 2.Pestic-Dragovich, L., Stojiljkovic, L., Philimonenko, A. A., Nowak, G., Ke, Y., Settlage, R. E., Shabanowitz, J., Hunt, D. F., Hozak, P. & deLanerolle, P. (2000) Science 290**,** 337–341. [DOI] [PubMed] [Google Scholar]
- 3.Scheer, U., Hinssen, H., Franke, W. W. & Jockusch, B. M. (1984) Cell 39**,** 111–122. [DOI] [PubMed] [Google Scholar]
- 4.Zhao, K., Wang, W., Rando, O. J., Xue, Y., Swiderek, K., Kuo, A. & Crabtree, G. R. (1998) Cell 95**,** 625–636. [DOI] [PubMed] [Google Scholar]
- 5.Sotiropoulos, A., Gineitis, D., Copeland, J. & Treisman, R. (1999) Cell 98**,** 159–169. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann, W., Reichart, B., Ewald, A., Muller, E., Schmitt, I., Stauber, R. H., Lottspeich, F., Jockusch, B. M., Scheer, U., Hauber, J. & Dabauvalle, M. C. (2001) J. Cell Biol. 152**,** 895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Percipalle, P., Zhao, J., Pope, B., Weeds, A., Lindberg, U. & Daneholt, B. (2001) J. Cell Biol. 153**,** 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wieslander, L. (1994) Prog. Nucleic Acid Res. Mol. Biol. 48**,** 275–313. [DOI] [PubMed] [Google Scholar]
- 9.Daneholt, B. (2001) Proc. Natl. Acad. Sci. USA 98**,** 7012–7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Percipalle, P., Jonsson, A., Nashchekin, D., Karlsson, C., Bergman, T., Guialis, A. & Daneholt B. (2002) Nucleic Acids Res. 30**,** 1725–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miralles, F., Öfverstedt, L. G., Sabri, N., Aissouni, Y., Hellman, U., Skoglund, U. & Visa, N. (2000) J. Cell Biol. 148**,** 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun, X., Alzhanova-Ericsson, A. T., Visa, N., Aissouni, Y., Zhao, J. & Daneholt, B. (1998) J. Cell Biol. 142**,** 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alzhanova-Ericsson, A. T., Sun, X., Visa, N., Kiseleva, E., Wurtz, T. & Daneholt, B. (1996) Genes Dev. 10**,** 2881–2893. [DOI] [PubMed] [Google Scholar]
- 14.Zechel, K. (1980) Eur. J. Biochem. 110**,** 343–348. [DOI] [PubMed] [Google Scholar]
- 15.Wurtz, T., Kiseleva, E., Nacheva, G., Alzhanova-Ericcson, A., Rosen, A. & Daneholt, B. (1996) Mol. Cell. Biol. 16**,** 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spector, D. L., Goldman, R. D. & Leinwand L. A. (1998) Cells: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. 1.
- 17.Björkroth, B., Ericsson, C., Lamb, M. M. & Daneholt, B. (1988) Chromosoma 96**,** 333–340. [Google Scholar]
- 18.Kiseleva, E., Wurtz, T., Visa, N. & Daneholt, B. (1994) EMBO J. 13**,** 6052–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazarides, E. & Lindberg, U. (1974) Proc. Natl. Acad. Sci. USA 71**,** 4742–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miralles, F. & Visa, N. (2001) Exp. Cell Res. 264**,** 284–295. [DOI] [PubMed] [Google Scholar]
- 21.Wang, K. & Richards, F. M. (1974) J. Biol. Chem. 249**,** 8005–8018. [PubMed] [Google Scholar]
- 22.Fischer, P. M., Krausz, E. & Lane, D. P. (2001) Bioconjugate Chem. 12**,** 825–841. [DOI] [PubMed] [Google Scholar]
- 23.Daneholt, B., Edström, J.-E., Egyházi, E., Lambert, B. & Ringborg, U. (1969) Chromosoma 28**,** 399–417. [DOI] [PubMed] [Google Scholar]
- 24.Masson, C., Bouniol, C., Fomproix, N., Szollosi, M. S., Debey, P. & Hernandez-Verdun, D. (1996) Exp. Cell Res. 226**,** 114–125. [DOI] [PubMed] [Google Scholar]
- 25.Skoglund, U., Andersson, K., Björkroth, B., Lamb, M. M. & Daneholt, B. (1983) Cell 34**,** 847–855. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava, R. A. & Schonfeld, G. (1994) Methods Mol. Biol. 31**,** 281–288. [DOI] [PubMed] [Google Scholar]
- 27.Dong, B., Horowitz, D. S., Kobayashi, R. & Krainer, A. R. (1993) Nucleic Acids Res. 21**,** 4085–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shav-Tal, Y. & Zipori, D. (2002) FEBS Lett. 531**,** 109–114. [DOI] [PubMed] [Google Scholar]
- 29.Gozani, O., Patton, J. G. & Reed, R. (1994) EMBO J. 13**,** 3356–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, Z. & Carmichael, G. G. (2001) Cell 106**,** 465–475. [DOI] [PubMed] [Google Scholar]
- 31.Sewer, M. B., Nguyen, V. Q., Huang, C. J., Tucker, P. W., Kagawa, N. & Waterman, M. R. (2002) Endocrinology 143**,** 1280–1290. [DOI] [PubMed] [Google Scholar]
- 32.Gineitis, D. & Treisman, R. (2001) J. Biol. Chem. 276**,** 24531–24539. [DOI] [PubMed] [Google Scholar]
- 33.Olave, I. A., Reck-Peterson, S. L. & Crabtree, G. R. (2002) Annu. Rev. Biochem. 71**,** 755–781. [DOI] [PubMed] [Google Scholar]
- 34.Mathur, M., Tucker, P. W. & Samuels, H. H. (2001) Mol. Cell. Biol. 21**,** 2298–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maniatis, T. & Reed, R. (2002) Nature 416**,** 499–506. [DOI] [PubMed] [Google Scholar]
- 36.Fong, Y. W. & Zhou, Q. (2001) Nature 414**,** 929–933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information