Rapid Turnover of Extracellular Signal-Regulated Kinase 3 by the Ubiquitin-Proteasome Pathway Defines a Novel Paradigm of Mitogen-Activated Protein Kinase Regulation during Cellular Differentiation (original) (raw)
Abstract
Mitogen-activated protein (MAP) kinases are stable enzymes that are mainly regulated by phosphorylation and subcellular targeting. Here we report that extracellular signal-regulated kinase 3 (ERK3), unlike other MAP kinases, is an unstable protein that is constitutively degraded in proliferating cells with a half-life of 30 min. The proteolysis of ERK3 is executed by the proteasome and requires ubiquitination of the protein. Contrary to other protein kinases, the catalytic activity of ERK3 is not responsible for its short half-life. Instead, analysis of ERK1/ERK3 chimeras revealed the presence of two destabilization regions (NDR1 and -2) in the N-terminal lobe of the ERK3 kinase domain that are both necessary and sufficient to target ERK3 and heterologous proteins for proteasomal degradation. To assess the physiological relevance of the rapid turnover of ERK3, we monitored the expression of the kinase in different cellular models of differentiation. We observed that ERK3 markedly accumulates during differentiation of PC12 and C2C12 cells into the neuronal and muscle lineage, respectively. The accumulation of ERK3 during myogenic differentiation is associated with the time-dependent stabilization of the protein. Terminal skeletal muscle differentiation is accompanied by cell cycle withdrawal. Interestingly, we found that expression of stabilized forms of ERK3 causes G1 arrest in NIH 3T3 cells. We propose that ERK3 biological activity is regulated by its cellular abundance through the control of protein stability.
Protein kinases are signaling molecules whose activity must be tightly regulated. Indeed, deregulation of protein kinase expression and/or activity has been implicated in various human diseases such as cancer (11), insulin resistance syndromes (19), inflammatory disorders (21, 22), and congenital malformations (53). The activity of protein kinases is controlled by multiple mechanisms that include extracellular ligands, second messengers, phosphorylation, subcellular localization, protein-protein interactions, and regulated proteolysis.
Mitogen-activated protein (MAP) kinases are among the most intensively studied families of protein kinases. These evolutionarily conserved enzymes have been shown to participate in pathways controlling embryogenesis, cell proliferation, differentiation, cell survival, and adaptation (4, 23, 38, 55). All of the MAP kinases characterized thus far are stable proteins whose expression is generally not limiting for activity. The biological activity of MAP kinases is mainly regulated by their phosphorylation state and subcellular localization (8, 47). MAP kinases are enzymatically activated by phosphorylation of two residues within the activation loop motif Thr-Xaa-Tyr, catalyzed by a family of dual-specificity protein kinases (37). The amplitude and duration of MAP kinase activation, which are controlled by the activity of MAP kinase kinases and phosphatases, play an important role in determining the final biological response (25, 29). The targeting of MAP kinases to specific subcellular locations is also a key determinant of their biological activity (3, 25, 43).
Extracellular signal-regulated kinase 3 (ERK3) was cloned originally by virtue of its homology to the MAP kinase ERK1 (2). The ERK3 protein is nearly 50% identical to ERK1/2 in the kinase domain and has similar lengths of inserts between conserved subdomains, indicating that ERK3 belongs to the MAP kinase family of protein kinases (2, 27, 57). However, ERK3 presents structural features that are distinct from other MAP kinases. Notably, ERK3 displays the sequence Ser-Glu-Gly instead of the highly conserved Thr-Xaa-Tyr motif in the activation loop. Moreover, ERK3 has a unique C-terminal extension that is absent in classical MAP kinases. The impact of these features on ERK3 function is unknown.
The physiological functions of ERK3 remain to be established. ERK3 mRNA is detected in every adult tissue examined, although the relative levels of expression vary considerably between tissues (2, 52). Interestingly, expression of ERK3 mRNA is acutely regulated during mouse development, suggesting a possible role of the kinase during embryogenesis (52). Another study reported that ERK3 mRNA is upregulated during in vitro differentiation of P19 embryonal carcinoma cells toward the neuronal or muscle lineage (2). However, nothing is known about the regulation of ERK3 protein and kinase activity. Here we report that, unlike other MAP kinases, ERK3 is an unstable protein that is rapidly and constitutively degraded by the ubiquitin-proteasome pathway in exponentially proliferating cells. Neither kinase activity nor the C-terminal extension of ERK3 is required for the high turnover of the enzyme. Instead, we have identified two degradation domains in the N-terminal lobe of the kinase that are both necessary and sufficient to target ERK3 for ubiquitination and degradation by the proteasome. Importantly, we show that the ERK3 half-life increases during muscle differentiation, leading to accumulation of the protein. We also show that expression of stabilized forms of ERK3 inhibits S-phase entry in fibroblasts. We propose that ERK3 biological activity is mainly controlled by regulated protein turnover.
MATERIALS AND METHODS
Reagents and antibodies.
MG-132 and lactacystin were obtained from Biomol and Boston Biochem, respectively. E-64 and Z-VAD-FMK were supplied by Calbiochem. Leupeptin was from ICN. 5-Bromo-2-deoxyuridine (BrdU) was from Roche Diagnostics. Nerve growth factor 2.5S (NGF) was from Invitrogen. Commercial antibodies were obtained from the following suppliers: polyclonal anti-ERK3 (sc-156), anti-Cdk2 (sc-163), and polyclonal anti-Myc (Santa-Cruz Biotechnology); monoclonal antibody (MAb) anti-p21 (Pharmingen); polyclonal anti-green fluorescent protein (GFP; Affinity BioReagents); biotinylated goat anti-rabbit immunoglobulin G (IgG) and horseradish peroxidase (HRP)-conjugated streptavidin (Vector Laboratories); HRP-conjugated goat anti-mouse and anti-rabbit IgG (Bio-Rad); monoclonal anti-BrdU (Calbiochem); fluorescein isothiocyanate-conjugated donkey anti-rabbit and rhodamine-conjugated donkey anti-rabbit (Jackson Immunoresearch). MAb 12CA5 to influenza virus hemagglutinin (HA) was a gift from M. Dennis (SignalGene). Anti-Myc MAb was prepared in-house from 9E10 hybridoma-producing cells. Polyclonal antibody to the C terminus of ERK3 (E3-CTE4) was obtained by immunization of rabbits with purified His6-E3365-721-glutathione _S_-transferase (GST) fusion protein expressed in Escherichia coli (7). The ERK3 phospho-Ser189-specific antibody was generated in collaboration with Cell Signaling Technology by immunizing rabbits with the synthetic phospho-Ser189 peptide CSHKGHL(p)SEGLVTKW coupled to keyhole limpet hemocyanin. The antibody was purified by protein A and peptide affinity chromatography.
Plasmid constructs and mutagenesis.
All recombinant ERK3 DNA constructs were derived from the human cDNA sequence (27). Point mutations were introduced into ERK3 cDNA by using the Altered Sites in vitro mutagenesis system (Promega). pcDNA3-HA was constructed by inserting the HA sequence from pcDNA I NEO (28) into the _Hin_dIII/_Eco_RI sites of pcDNA3 (Invitrogen Life Technologies). pcDNA-HA-ERK1 was constructed by subcloning the coding sequence of hamster ERK1 (28) into _Eco_RI-digested pcDNA3-HA. pcDNA-HA-ERK3 was constructed by first inserting an _Eco_RI site at the initiator ATG codon of ERK3 cDNA by site-directed mutagenesis, followed by subcloning of the ERK3 coding sequence into the _Eco_RI site of pcDNA3-HA. Expression vectors for HA-tagged p38β2 (J. Han, Scripps Research Institute) and JNK2 (J. Woodgett, Ontario Cancer Institute) were constructed by amplifying the respective coding sequences by PCR, followed by subcloning into the _Eco_RI/_Xba_I (p38β2) or _Xho_I/_Xba_I (JNK2) sites of pcDNA3-HA. pREP10-HA-ERK5 was kindly provided by C. Prody (University of Toronto). The K49A/K50A (KD) and S189A mutants of ERK3 were obtained by site-directed mutagenesis and subcloned into pcDNA3-HA vector. HA-ERK31-365Δ was generated by PCR by using pcDNA3-HA-ERK3 as a template and subcloned into the _Eco_RI/_Xba_I sites of pcDNA3. To construct the GST-tagged ERK3 expression vector, the GST coding sequence from pGEX-KG (13) was amplified by PCR and inserted into the _Eco_RI/_Xba_I sites of pcDNA3-HA to generate pcDNA3-HA-GST; the coding sequence of ERK3 wild-type and S189A was then amplified by PCR to remove the stop codon and subcloned into the _Eco_RI site of pcDNA3-HA-GST. pcDNA3-HA-ERK1-GST was constructed similarly by subcloning the PCR-amplified ERK1 coding sequence into the _Eco_RI site of pcDNA3-HA-GST. Retroviral expression vectors for HA-ERK1 and HA-ERK3 were constructed by subcloning the inserts into the _Bam_HI/_Eco_RI sites of pBabe-Puro (30). pMT123 vector encoding HA-tagged ubiquitin was kindly provided by Dirk Bohmann (51). To generate chimeras between ERK1 and ERK3, specific regions of the two proteins were amplified by PCR while introducing restriction sites that allow conservation of the reading frame. The two inserts were then ligated simultaneously into pcDNA3-HA vector. To construct expression vectors for enhanced GFP (EGFP) fusion proteins, the EGFP coding sequence from pEGFP-C2 (Clontech) was amplified by PCR and inserted into the _Eco_RI/_Xba_I sites of pcDNA3-HA to generate pcDNA3-HA-EGFP; the coding region of ERK31-365 or ERK31-112 was then amplified by PCR and subcloned into the _Eco_RI site of pcDNA3-HA-EGFP. pcDNA3-HA-ERK1-EGFP and pcDNA3-HA-ERK11-116-EGFP were constructed by using a similar strategy. Myc6-EGFP was constructed by inserting EGFP into _Eco_RI/_Xba_I of pCS3MT (gift of Andrew Jan Waskiewicz). To construct the bicistronic expression vector, poliovirus internal ribosome entry site (IRES) (40) and Myc6-EGFP were amplified by PCR and cloned into _Xba_I/_Apa_I-digested pcDNA3, yielding pcDNA3-IRES-EGFP. The indicated coding sequences were then subcloned into the _Hin_dIII/_Eco_RI sites of pcDNA3-IRES-EGFP. All mutations were confirmed by DNA sequencing. The sequences of the primers used for PCR and details about the cloning strategies are available upon request.
Cell culture, transfections, and infections.
Human embryonic kidney 293 cells were cultured in minimal essential medium supplemented with 10% fetal bovine serum plus antibiotics. COS-7, BALB/c 3T3, and NIH 3T3 cells were cultured in Dulbecco modified Eagle medium (DMEM) containing 10% calf serum and antibiotics. Rat1 cells were cultured and transfected as described previously (46). PC12 cells were maintained on poly-l-lysine-coated dishes in DMEM containing 10% heat-inactivated horse serum, 5% fetal bovine serum, and antibiotics. To induce neuronal differentiation, PC12 cells were plated on collagen type VII (Sigma) and stimulated with NGF (100 ng/ml) for the times indicated. C2C12 myoblasts were cultured in DMEM supplemented with 20% fetal bovine serum and antibiotics (growth medium [GM]). Muscle differentiation of C2C12 cells was induced by replacing GM with differentiation medium (DM; DMEM, 10 mM HEPES [pH 7.4], 2% calf serum) when the cells reached confluence.
HEK 293 cells were transiently transfected by the calcium phosphate precipitation method with 6 to 9 μg of plasmid DNA per 60-mm dish. Cells were harvested at 48 h posttransfection unless otherwise indicated. COS-7 and NIH 3T3 cells were transfected by using Lipofectamine reagent according to the manufacturer's specifications (Invitrogen). Retroviral infection of A31 and ts20 cells was performed by using the pCL-Ampho packaging vector (Imgenex Corporation). Briefly, HEK 293 cells were cotransfected with pBabe-Puro constructs and pCL-Ampho. After 48 h, the virus-containing medium was collected, supplemented with 10 μg of Polybrene (Sigma)/ml, and incubated with subconfluent A31 or ts20 cells for 24 h at 34°C (permissive temperature). The medium was changed and, after an additional period of 24 h, infected ts20 and A31 cells were selected for 2 days in complete medium containing 2 and 4 μg of puromycin/ml, respectively. The cells were either kept at 34°C or shifted to 39°C for the indicated times.
Immunoblot analysis, immunoprecipitation, and immunofluorescence.
Cell lysis, immunoprecipitation, and immunoblot analysis was performed as described previously (49). The concentrations of primary antibodies used for immunoblotting were as follows: anti-HA MAb 12CA5 (1:2,500), commercial anti-ERK3 antibody (1:1,000), E3-CTE4 serum (1:1,000), anti-GFP antibody (1:1,000), and anti-phospho-ERK3(S189) (1:1,000). For detection of ectopically expressed ERK3 constructs, cell lysates (750 μg of protein) were subjected to immunoprecipitation with 2 μl of anti-HA MAb preadsorbed on protein A-Sepharose beads. Immunoprecipitated proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. The blocked membrane was first incubated for 1 h with anti-ERK3 or anti-GFP primary antibody. After extensive washing, the membrane was incubated for 1 h with biotinylated anti-rabbit IgG (1:700) followed by a third incubation of 1 h with HRP-conjugated streptavidin (1:20,000). Immunoreactive bands were visualized by using enhanced chemiluminescence. For the detection of Ser189-phosphorylated ERK3, GST-tagged or endogenous ERK3 protein was isolated from cell extracts (750 μg of protein) by glutathione-agarose pull-down or immunoprecipitation with antibody E3-CTD4, respectively. Precipitated proteins were resolved by SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting.
For BrdU incorporation studies, NIH 3T3 were seeded at 40,000 cells/well onto poly-l-lysine-coated glass coverslips placed in six-well-plates, and transfected with expression plasmids (1 μg of DNA) by using Lipofectamine reagent. After 24 h, the cells were serum starved for 24 h and stimulated with 10% serum for 20 h in the presence of 10 μM BrdU. The cells were fixed with 3.7% paraformaldehyde-phosphate-buffered saline for 20 min at 37°C. After a quenching step in 0.1 M glycine for 5 min, the cells were permeabilized by incubation in 0.1% Triton X-100 for 5 min at room temperature. Nonspecific sites were blocked in phosphate-buffered saline-1% bovine serum albumin for 60 min at 37°C. The transfected proteins were detected by staining with anti-Myc as primary antibody and fluorescein isothiocyanate-conjugated anti-rabbit IgG as a secondary reagent. Then, DNA was denatured with 2 N HCl for 10 min at room temperature. Staining for BrdU incorporation was performed by incubating the cells for 1 h at 37°C with anti-BrdU antibody, followed by incubation with rhodamine-conjugated anti-mouse IgG as secondary reagent. DAPI (4′,6′-diamidino-2-phenylindole) staining was performed to visualize the nuclei. Cell samples were analyzed by fluorescence microscopy (Leica DM RB). The results are expressed as the percentage of transfected cells showing nuclear labeling for BrdU.
In vivo ubiquitination assay.
HEK 293 cells were cotransfected with expression vectors for HA-ERK3-GST (4 μg) and HA-ubiquitin (2 μg). After 48 h, the cells were treated with 25 μM MG-132 for 12 h. The cells were lysed in Triton X-100 lysis buffer supplemented with 10 mM _N_-ethylmaleimide (NEM; Sigma), and the clarified lysates were further incubated for 10 min with 2.5 mM dithiothreitol to quench any free NEM. Cell lysates (750 μg of protein) were then incubated for 2 h at 4°C with 20 μl of glutathione-agarose resin. The bound proteins were washed four times in lysis buffer and resolved by SDS-PAGE. Ubiquitin-containing conjugates were detected by immunoblotting with MAb 12CA5 as described above.
Biosynthetic labeling experiments and in vitro translation.
Biosynthetic labeling experiments were carried out as described previously (49). To assess the rate of biosynthesis of HA-ERK1 and HA-E31-73-E187-369, transfected HEK 293 cells were starved of methionine and cysteine for 30 min. 35S-labeled methionine-cysteine (300 μCi/ml) was then added to the medium, and the cells were incubated for 5 min at 37°C. The cells were lysed in Triton X-100 lysis buffer, and HA-tagged proteins were immunoprecipitated as described above. The labeled proteins were separated by SDS-PAGE and detected by fluorography. To determine the turnover rate of HA-ERK1 and HA-E31-73-E187-369 proteins, transfected HEK 293 cells were pulse-labeled for 2 h at 37°C with 100 μCi of 35S-labeled methionine-cysteine/ml and then chased for the indicated times in complete medium containing 100 μg of methionine/ml and 150 μg of cysteine/ml. Labeled HA-tagged proteins were detected as described. In vitro transcription-translation was performed by using TNT coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions. The reaction products were detected by fluorography with Amplify (Amersham).
Statistical analysis.
The results are expressed as the mean ± the standard error of the mean. Statistical significance between experimental groups was determined by one-way analysis of variance, followed by Bonferroni analysis by using GraphPad software (version 2.01). P values of <5% were considered significant.
Modeling of ERK3 structure.
The ternary structure of ERK3 was modeled by using the SWISS-MODEL server (14, 39), based on the structure of unphosphorylated rat ERK2 protein (pdb number 1ERK_) (56). The model contains amino acids 3 to 346 of human ERK3.
RESULTS
ERK3 is a highly unstable protein.
To get insights into the regulation of ERK3, we constructed several expression vectors encoding N-terminal HA-tagged human ERK3 cDNA. The constructs were transiently transfected into HEK 293 cells, and the expression of ERK3 was analyzed by immunoblotting with anti-HA antibody. Surprisingly, the expression of ERK3 (apparent molecular mass of 105 kDa) was undetectable in HEK 293 cells, whereas similar constructions of the other MAP kinase family members ERK1, p38β2, JNK2, and ERK5 yielded high protein levels (Fig. 1A). The failure to ectopically express ERK3 was also observed in other cell types, such as Rat1, COS-7, NIH 3T3, or BHK cells (data not shown). Identical results were obtained by using the mouse ERK3 cDNA (data not shown). In vitro transcription-translation of ERK3 expression vector in the presence of [35S]methionine confirmed the integrity of the plasmid and showed that ERK3 and ERK1 constructs are translated with comparable efficiency (Fig. 1B). We also tested the possibility that ERK3 might be present in the insoluble fraction obtained from the cell lysis procedure. However, no expression of ERK3 could be detected in either the soluble or the insoluble fraction of transfected HEK 293 cells (data not shown). Notably, the overexpressed ERK1 protein was found in both fractions.
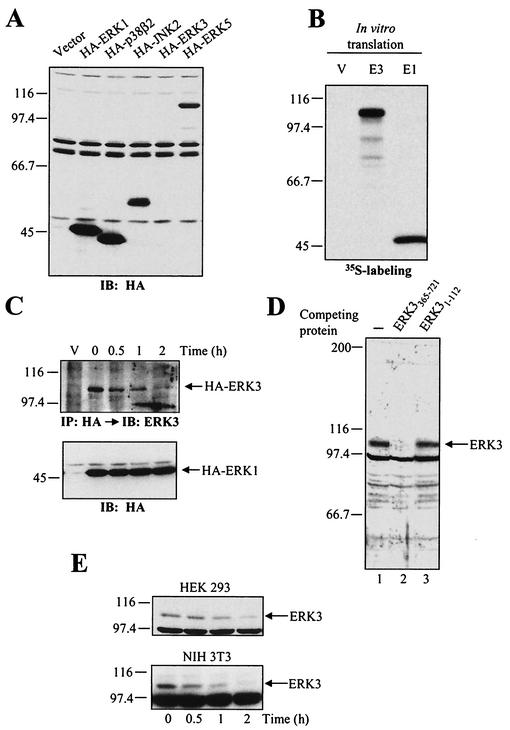
FIG. 1.
ERK3 is an unstable protein. (A) HEK 293 cells were transiently transfected with the indicated constructs, and cell lysates were analyzed by immunoblotting with anti-HA MAb. (B) Expression vectors for HA-ERK3 (E3) and HA-ERK1 (E1) were translated in vitro in the presence of [35S]methionine-cysteine and analyzed by fluorography. (C) HEK 293 cells were transfected with HA-ERK3 and HA-ERK1. After 48 h, the cells were treated with cycloheximide (100 μg/ml) for the indicated times. HA-ERK3 protein was immunoprecipitated with anti-HA MAb and detected by immunoblotting with a commercial anti-ERK3 antibody coupled to a biotin-streptavidin amplification system (upper panel). HA-ERK1 was analyzed as in panel A (lower panel). (D) Specificity of anti-ERK3 antibody. The polyclonal anti-ERK3 antibody E3-CTE4 was incubated in the absence (lane 1) or presence of the immunogen His6ERK3365-721GST (lane 2) or ERK3 N terminus (lane 3) prior to immunoblotting analysis of HEK 293 cell lysate. (E) HEK 293 (upper panel) and NIH 3T3 (lower panel) cells were treated with cycloheximide (100 μg/ml) for the indicated times. Endogenous ERK3 was detected by immunoblotting with a polyclonal antibody to the C terminus of ERK3 (E3-CTE4).
One possible explanation for the above findings is that ectopically expressed ERK3 is rapidly degraded in cells. To test this hypothesis, we measured the half-life of ectopic ERK3 after shutting off protein synthesis with cycloheximide. In order to detect the expression of HA-tagged ERK3, the protein was enriched by immunoprecipitation with anti-HA MAb, followed by immunoblotting analysis with a commercial polyclonal antibody to ERK3 coupled to a biotin-streptavidin amplification system. Using this sensitive approach, HA-ERK3 was detected in transfected HEK 293 cells as an ∼105-kDa band (Fig. 1C). Cycloheximide-chase experiments indicated that HA-ERK3 is a highly unstable protein with a half-life of ca. 30 min in exponentially growing HEK 293 cells (Fig. 1C, upper panel). In contrast, HA-ERK1 has an apparent half-life of >2 h (Fig. 1C, lower panel). We also tried to determine the half-life of HA-ERK3 by pulse-chase analysis, but the level of expression was too low to yield a detectable signal (data not shown).
To confirm that the observed instability of HA-ERK3 is an intrinsic property of the protein, we generated a rabbit polyclonal antibody against the C terminus of ERK3 (E3-CTE4). This antibody detects an endogenous protein of 105 kDa in HEK 293, NIH 3T3, Rat1, HeLa, and BALB/c 3T3 cell lysates (Fig. 1D and data not shown). Incubation of the antiserum with the immunogen ERK3 C-terminus fusion protein, but not with ERK3 N terminus, completely abolishes the immunoreactivity, thus confirming the specificity of the antibody (Fig. 1D). By cycloheximide-chase, we found that endogenous ERK3 is also unstable, with a half-life comparable to that measured for transfected HA-ERK3 (Fig. 1E).
ERK3 is ubiquitinated in vivo and is degraded by the 26S proteasome.
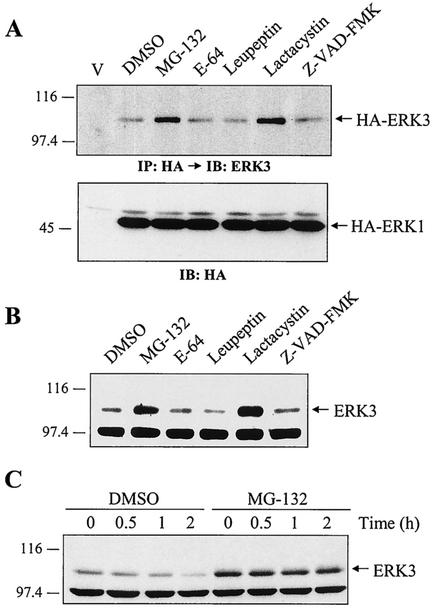
We used various pharmacological inhibitors to address the role of known proteolytic systems in mediating the observed rapid ERK3 degradation. Two inhibitors of the proteasome, the peptide-aldehyde MG-132 (45) and the Streptomyces metabolite lactacystin (9), were found to significantly increase the steady-state level of transfected ERK3 (Fig. 2A, upper panel). In contrast, inhibitors of cathepsins (E-64 and leupeptin), calpains (E-64), or caspases (Z-VAD-FMK) had no effect on HA-ERK3 expression. No change in the abundance of HA-ERK1 protein was observed under these conditions (Fig. 2A, lower panel). Similarly, treatment of HEK 293 cells with MG-132 or lactacystin resulted in the accumulation of endogenous ERK3 protein, as revealed by immunoblot analysis (Fig. 2B). Cycloheximide-chase experiments confirmed that proteasome inhibitors act by increasing ERK3 stability. The half-life of ERK3 increased from 30 min to >2 h upon MG-132 treatment (Fig. 2C). These results indicate that ERK3 is actively degraded by the proteasome.
FIG. 2.
ERK3 is degraded by the proteasome. (A) HEK 293 cells were transfected with empty vector or with expression vectors for HA-ERK3 and HA-ERK1. After 48 h, the cells were treated for a period of 12 h with various protease inhibitors: MG-132 (25 μM), E-64 (25 μM), leupeptin (0.1 mM), lactacystin (20 μM), and Z-VAD-FMK (25 μM). Expression of HA-ERK3 protein was detected by immunoprecipitation with anti-HA MAb, followed by immunoblotting with anti-ERK3 antibody coupled to a biotin-streptavidin amplification system. Expression of ERK1 was analyzed by immunoblotting with anti-HA MAb. (B) HEK 293 cells were treated for 12 h with the indicated protease inhibitors. Endogenous ERK3 expression was detected by immunoblotting with E3-CTE4 antibody. (C) HEK 293 cells were treated with dimethyl sulfoxide (0.1%) or MG-132 (25 μM). After 30 min, cycloheximide was added to the medium for the times indicated. Expression of endogenous ERK3 was detected as in panel B.
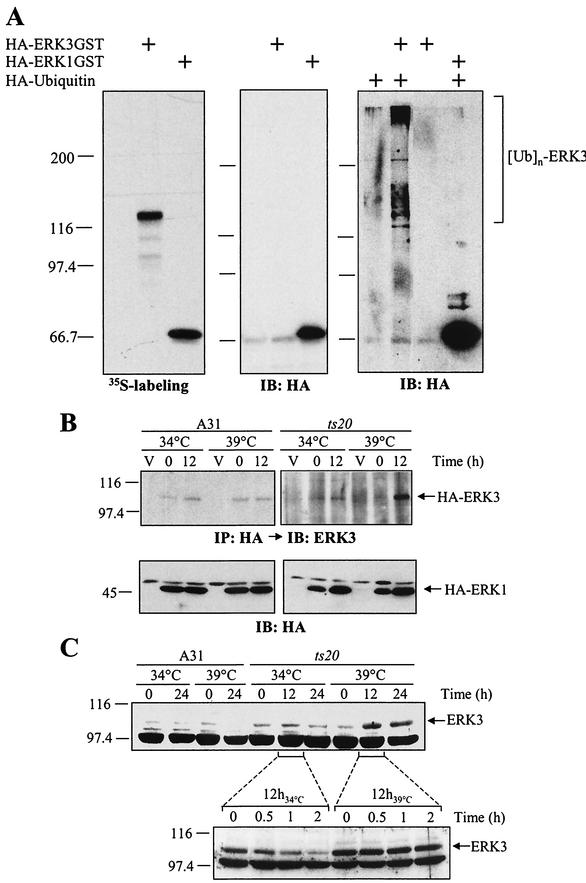
Efficient degradation of proteins by the proteasome generally requires the conjugation of multiple molecules of ubiquitin to the substrate (17, 18). We thus sought to determine whether ERK3 is ubiquitinated in vivo. To this end, we engineered C-terminal GST-tagged versions of HA-ERK3 and HA-ERK1 as a means to specifically purify the kinases from cell extracts. In vitro translation experiments confirmed that the GST-tagged ERK3 and ERK1 constructs are translated with similar efficiency (Fig. 3A, left panel). However, when the two constructs were transfected into HEK 293 cells, no detectable expression of ERK3-GST could be observed under standard immunoblotting conditions, whereas ERK1-GST was expressed to high levels (Fig. 3A, middle panel). The weak expression of ERK3-GST was detected by using a biotin-streptavidin amplification system (data not shown). To verify the in vivo ubiquitination status of ERK3, HEK 293 cells were cotransfected with ERK3-GST and HA-tagged ubiquitin, followed by treatment with MG-132. The GST fusion kinases were then purified from cellular extracts and analyzed by anti-HA immunoblotting. High-molecular-weight immunoreactive species were detected only in cells transfected with both ERK3-GST and HA-ubiquitin and not in cells transfected with ERK1-GST (Fig. 3A, right panel). These results strongly suggest that ERK3 is polyubiquitinated in vivo.
FIG.3.
A functional ubiquitin-conjugating system is required for ERK3 proteolysis. (A) ERK3 is ubiquitinated in vivo. In the left panel is shown in vitro translation of expression vectors encoding HA-ERK3GST and HA-ERK1GST in the presence of [35S]methionine-cysteine. The products were detected by fluorography. In the central panel, HEK 293 cells were transfected with HA-ERK3GST and HA-ERK1GST expression constructs. After 48 h, GST-tagged proteins were precipitated with glutathione-agarose beads and analyzed by immunoblotting with anti-HA MAb. In the right panel, HEK 293 cells were cotransfected with GST-tagged expression vectors together with HA-ubiquitin. After 48 h, the cells were treated with MG-132 (25 μM) for an additional 12 h, and GST-tagged proteins were precipitated with glutathione-agarose beads. Immunoreactive ubiquitin conjugates were detected by immunoblotting with anti-HA MAb. (B) Parental BALB/c 3T3 (A31) and E1 mutant ts20 cells were infected with retroviruses encoding HA-ERK3 or HA-ERK1. At 3 days after selection in puromycin-containing medium, the infected cells were shifted or not at the restrictive temperature (39°C) for the times indicated. Expression of ectopic ERK3 and ERK1 proteins was detected as in Fig. 1E. (C) In the upper panel, A31 and ts20 cells were incubated at the permissive or restrictive temperature for the indicated times. Endogenous ERK3 expression was detected by immunoblotting with E3-CTE4 antibody. In the lower panel, ts20 cells were incubated at the permissive or restrictive temperature for 12 h. The cells were then treated with cycloheximide for the indicated times. Expression of endogenous ERK3 protein was monitored with E3-CTE4 antibody.
In rare cases, such as p21Cip1 (50), ubiquitination is not necessary for proteasomal degradation. To test the importance of ubiquitin conjugation in targeting ERK3 to the proteasome, we used the ts20 cell line that harbors a temperature-sensitive allele of the ubiquitin-activating enzyme E1 (6). For these experiments, ts20 and parental A31 cells were infected with retroviruses encoding HA-ERK3 or HA-ERK1. As shown in Fig. 3B (upper panel), the ectopic HA-ERK3 protein significantly accumulated when ts20 cells were shifted to the nonpermissive temperature (39°C), whereas no change in expression was observed in A31 cells expressing a normal E1. In contrast, the expression of HA-ERK1 was unaffected by inactivation of E1 at the nonpermissive temperature (Fig. 3B, lower panel). The steady-state level of endogenous ERK3 expression also markedly increased when ts20 cells were incubated at 39°C (Fig. 3C, upper panel). No temperature-dependent upregulation of ERK3 was observed in A31 cells. To unambiguously demonstrate that the increase of endogenous ERK3 expression observed at the restrictive temperature results from stabilization of the protein, we measured its half-life by cycloheximide-chase experiments. The half-life of ERK3 increased from ca. 30 min at 34°C to more than 2 h at 39°C (Fig. 3C, lower panel). We conclude from these results that a functional ubiquitin-conjugating system is required for efficient degradation of ERK3 by the proteasome.
Ser189 phosphorylation, kinase activity, and C-terminal extension of ERK3 are not involved in its constitutive degradation.
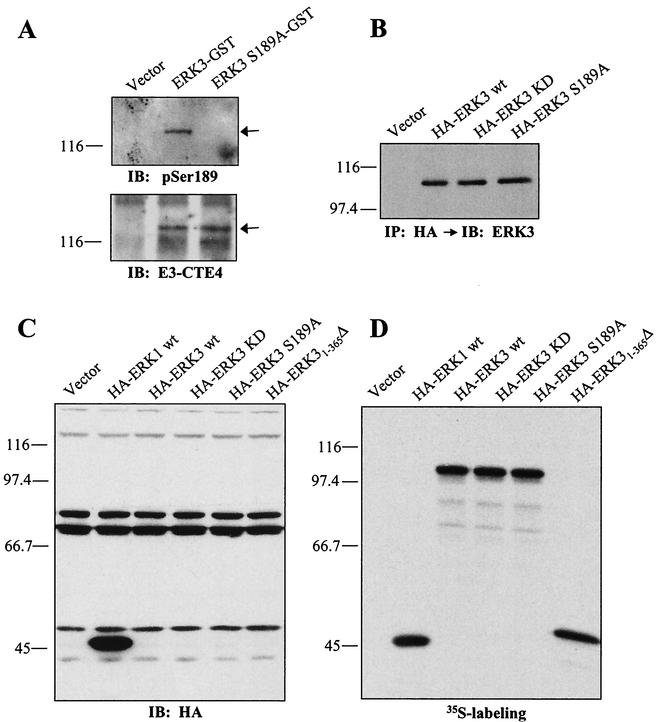
We next carried out experiments to identify signals in ERK3 that are important for degradation. ERK3 displays the sequence S189EG in its activation loop instead of the conserved TXY motif. In agreement with previous findings (5), we observed by in vivo labeling experiments that ERK3 is phosphorylated on Ser189 in living cells (P. Coulombe and S. Meloche, unpublished data). To further study the regulation of ERK3 phosphorylation on Ser189, we developed a phospho-specific antibody to this residue. The specificity of the antibody is illustrated in Fig. 4A, which confirms that ERK3 is phosphorylated on Ser189 in proliferating HEK 293 cells. We next evaluated the impact of Ser189 phosphorylation on ERK3 degradation. No change in the steady-state level of ERK3 expression was observed for the S189A mutant, suggesting that activation loop phosphorylation does not influence ERK3 stability (Fig. 4A to C). To investigate the contribution of ERK3 kinase activity on its stability, we generated a catalytically inactive mutant by changing two lysine residues present in subdomain II to alanine (KD; kinase dead). Transient transfections into HEK 293 cells showed that ERK3 KD is expressed at levels similar those for the wild-type protein (Fig. 4B and C). This suggests that the kinase activity of ERK3 is not required for the high turnover rate of the protein.
FIG. 4.
Role of Ser189 phosphorylation, kinase activity, and C-terminal extension on the stability of ERK3. (A) ERK3 is phosphorylated on Ser189. HEK 293 cells were transfected with GST fusion constructs of ERK3 wild-type or S189A mutant. GST-tagged proteins were purified from lysates by using glutathione-agarose beads and analyzed by immunoblotting with anti-phospho-ERK3(S189) (upper panel) or E3-CTD4 antibody (lower panel). (B) HEK 293 cells were transfected with the indicated expression constructs. Ectopically expressed ERK3 proteins were detected by immunoprecipitation with anti-HA MAb, followed by immunoblotting with anti-ERK3 antibody coupled to a biotin-streptavidin amplification system. (C) HEK 293 cells were transfected with the indicated expression constructs and cell lysates were analyzed by immunoblotting with anti-HA MAb. (D) The same expression constructs were translated in vitro in the presence of [35S]methionine-cysteine and analyzed by fluorography.
ERK3 possesses a unique C-terminal extension that is not found in ERK1 or other classical MAP kinases. We therefore tested the hypothesis that sequences contained within this region might have a role in regulating ERK3 stability. As shown in Fig. 4C, deletion of the C-terminal extension of ERK3 (ERK31-365Δ) did not increase its expression to detectable levels. In vitro assays confirmed that the deletion mutant of ERK3 was translated efficiently (Fig. 4D). These results argue that the major determinants of ERK3 stability are contained within the first 365 amino acids of the kinase.
Mapping of the regions conferring ERK3 instability.
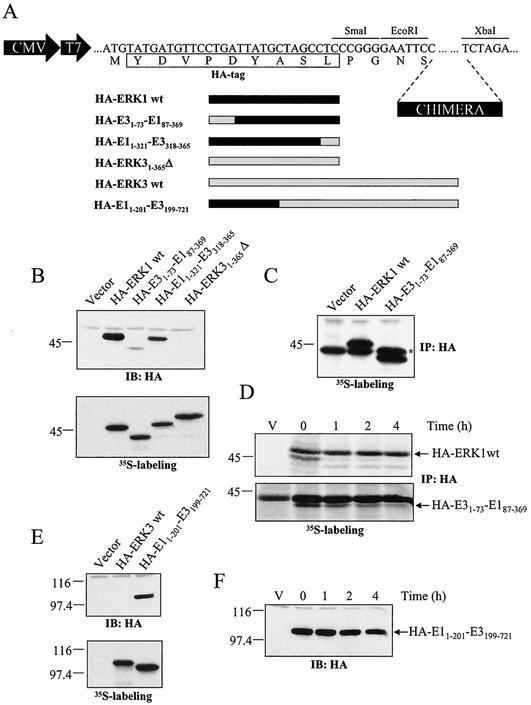
To identify the regions that target ERK3 for rapid degradation, we generated a series of chimeras between ERK1 (E1) and ERK3 (E3) (Fig. 5A). These chimeric constructs were subcloned into pcDNA3-HA vector and transiently expressed in HEK 293 cells. We found that a chimera containing the first 73 amino acids of ERK3 was expressed at low level compared to ERK1 or to a chimera containing amino acids 318 to 365 of ERK3 (Fig. 5B, upper panel). Although no difference was observed in the in vitro translation rates of the constructs (Fig. 5B, lower panel), it is still possible that the weak expression of the chimera containing the N-terminal region of ERK3 is due to a reduction in de novo biosynthesis. To exclude this possibility, pulse-labeling experiments were performed in transfected cells. The results clearly indicated that the rate of synthesis of HA-E31-73-E187-369 was similar to that of HA-ERK1 (Fig. 5C). Thus, the N-terminal region of ERK3 must regulate the turnover of the protein. Indeed, pulse-chase experiments confirmed that the half-life of HA-E31-73-E187-369 (1.2 h) is more than 15-fold less than that of HA-ERK1 (>20 h) (Fig. 5D). This difference is likely responsible for the marked differences in steady-state levels of the proteins observed by immunoblotting. In a complementary experiment, we replaced the first 199 amino acids of ERK3 by the equivalent residues of ERK1. Although wild-type HA-ERK3 and HA-E11-201-E3199-721 were translated with comparable efficiency in vitro, the HA-E11-201-E3199-721chimera is expressed to much higher levels than HA-ERK3 when transiently transfected into HEK 293 cells (Fig. 5E). This enhanced expression results from the stabilization of the protein. As revealed by cycloheximide-chase experiments, the HA-E11-201-E3199-721 chimera has a half-life of 4 to 6 h, which is at least eight times longer than wild-type ERK3 (Fig. 5F). These data indicate the presence of a functional degradation domain in the N-terminal extremity of ERK3.
FIG. 5.
A signal located in the N terminus of ERK3 regulates its stability. (A) Schematic representation of the constructs used in these experiments. (B) In the upper panel, HEK 293 cells were transfected with the indicated expression constructs and cell lysates were analyzed by immunoblotting with anti-HA MAb. In the lower panel, in vitro translation of the same constructs was visualized by fluorography. (C) Synthesis of ERK1 and E31-73-E187-369 proteins. HEK 293 cells were transfected with the indicated constructs and pulse-labeled with [35S]methionine-cysteine for 5 min. The HA-tagged proteins were immunoprecipitated with anti-HA MAb and analyzed by fluorograply. (D) Turnover of ERK1 and E31-73-E187-369 proteins. Transfected HEK 293 cells were pulse-labeled for 2 h with [35S]methionine-cysteine and then chased for the indicated times in complete medium. The HA-tagged proteins were immunoprecipitated with anti-HA MAb and visualized with a PhosphorImager apparatus. (E) In the upper panel, HEK 293 cells were transfected with the indicated expression constructs, and lysates were analyzed by immunoblotting with anti-HA MAb. In the lower panel, in vitro translation of the same constructs was visualized by fluorography. (F) HEK 293 cells were transfected with an expression vector encoding HA-ERK11-201-ERK3201-721 chimera. After 48 h, the cells were treated with cycloheximide for the indicated times, and expression of the protein was detected by immunoblotting with anti-HA MAb.
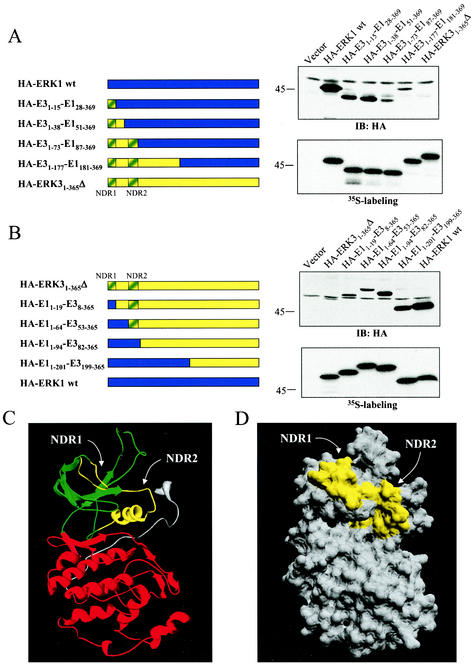
To define more precisely this degradation domain, we constructed additional chimeras between ERK1 and ERK3. We first engineered ERK1 chimeras containing N-terminal ERK3 sequences of increasing length (Fig. 6A). Substitution of the first 15 or 38 amino acids of ERK1 by the equivalent residues of ERK3 resulted in a two- to threefold reduction in protein expression. This finding suggests the presence of a destabilization signal in the first 15 amino acids of ERK3. A chimeric kinase containing the first 73 amino acids of ERK3 was expressed four- to sixfold less than the HA-E31-38-E151-369 chimera, thereby indicating the presence of an additional destabilization signal located between residues 38 and 73 of ERK3. A chimeric protein containing the N-terminal 177 amino acids of ERK3 was expressed at levels comparable to HA-E31-73-E187-369 chimera. Expression of HA-ERK31-365Δ was barely detectable under these conditions. In the complementary approach, we generated ERK3 chimeras containing N-terminal sequences of ERK1 (Fig. 6B). Chimeras containing the first 19 or 64 amino acids of ERK1 were expressed at similar levels and were slightly stabilized compared to HA-ERK31-365Δ. This is consistent with the loss of a destabilization signal present in the very first amino acids of ERK3. Substitution of the first 81 or 198 amino acids of ERK3 by the corresponding sequences of ERK1 further stabilized the protein, yielding expression levels that were four to six times higher than HA-E11-64-E353-365 chimera. These last results confirm the existence of a second destabilization motif present between residues 53 and 82 of ERK3. Notably, the destabilization regions identified by these two independent series of chimeras were found to overlap (Fig. 6A and B). Because of their critical role in regulating ERK3 protein stability, these two regions were named N-terminal degradation regions 1 and 2 (NDR1 and -2).
FIG.6.
The N-terminal lobe of ERK3 kinase domain contains two regions that control protein stability. (A) Left panel, schematic representation of the constructs. In the upper right panel, HEK 293 cells were transfected with the indicated expression constructs, and cell lysates were analyzed by immunoblotting with anti-HA MAb. In the lower right panel is shown in vitro translation of the same constructs as visualized by fluorography. (B) Same as in panel A. (C) Ribbon diagram of ERK3 structure (amino acids 3 to 346) modeled from the X-ray structure of unphosphorylated ERK2. The N-terminal lobe is green, and the C-terminal lobe is red. NDR1 and NDR2 are yellow. (D) Surface representation of ERK3. NDR1 and NDR2 are yellow.
To further analyze the NDR1 and NDR2, we generated a structural model of ERK31-365Δ based on the X-ray crystal structure of unphosphorylated ERK2 (see Materials and Methods). In this theoretical model comprising amino acids 3 to 346 of ERK3, the enzyme adopts a structure highly similar to that of ERK2, with an N-terminal lobe rich in beta strands and a C-terminal lobe rich in alpha helices (Fig. 6C). The predicted structure of ERK3 reveals that the two degradation regions identified by chimera analysis are both found within ERK3 N-terminal lobe. Interestingly, the NDR1 and NDR2 are located on the same side of the molecule (Fig. 6C, highlighted in yellow). A surface representation of ERK3 structure clearly shows that NDR1 and NDR2 are juxtaposed and could define a putative interaction domain (Fig. 6D).
The N-terminal lobe of ERK3 is sufficient to target proteins for proteasomal degradation.
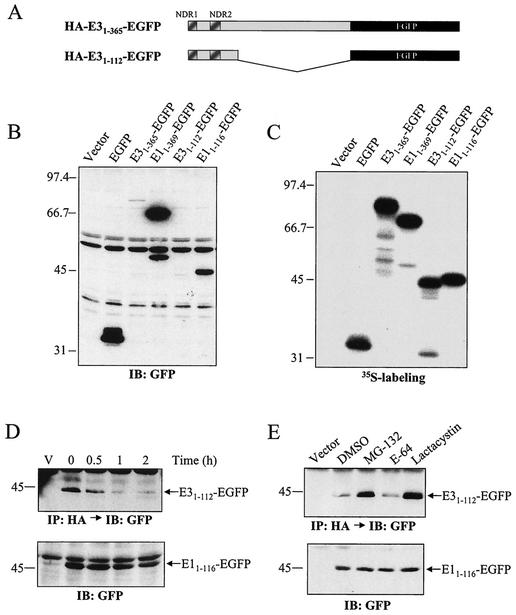
The results presented above suggest that NDR1 and NDR2 form a transferable degradation signal that is independent of the kinase C-terminal lobe. To test this hypothesis, we constructed fusion proteins between ERK3 and the stable heterologous protein EGFP (Fig. 7A). Immunoblot analysis indicated that EGFP alone is expressed at high levels in transfected HEK 293 cells (Fig. 7B). Addition of the first 365 amino acids of ERK3 to EGFP almost completely abolished expression of the fusion protein, whereas the equivalent fusion of ERK1 had no significant effect. Fusion of only the N-terminal lobe of ERK3 (residues 1 to 112) was sufficient to downregulate EGFP expression, whereas the equivalent sequence in ERK1 had little effect. A cycloheximide-chase experiment confirmed that the N-terminal lobe of ERK3 increases the degradation rate of EGFP. The E31-112-EGFP fusion protein has a half-life of less than 30 min in transfected cells compared to a half-life of more than 2 h for E11-116-EGFP hybrid protein (Fig. 7D). Results with pharmacological inhibitors indicated that the N-terminal lobe of ERK3 targets proteins for degradation by the proteasome. Indeed, treatment with MG-132 and lactacystin increased the steady-state level of E31-112-EGFP, whereas E-64 had no effect (Fig. 7E). Under these conditions, the expression of E11-116-EGFP was not significantly modified by these protease inhibitors. These results demonstrate that the N-terminal lobe of ERK3 is a transferable degradation signal that is sufficient to efficiently target proteins to the proteasome.
FIG. 7.
The N-terminal lobe of ERK3 kinase domain is sufficient to target proteins for rapid proteasomal degradation. (A) Schematic representation of the constructs used. (B) HEK 293 cells were transfected with the indicated expression constructs, and cell lysates were analyzed by immunoblotting with anti-GFP antibody. (C) The same expression constructs were translated in vitro in the presence of [35S]methionine-cysteine and analyzed by fluorography. (D) HEK 293 cells were transfected with expression vectors encoding HA-ERK31-112-EGFP and HA-ERK11-116-EGFP. After 48 h, the cells with treated with cycloheximide for the indicated times. The HA-ERK31-112-EGFP protein was detected by immunoprecipitation with anti-HA MAb, followed by immunoblotting with anti-GFP antibody coupled to a biotin-streptavidin amplification system (upper panel). HA-ERK11-116-EGFP was detected by immunoblotting with anti-GFP antibody (lower panel). (E) Transfected HEK 293 cells were treated for 12 h with the following protease inhibitors: MG-132 (25 μM), E-64 (25 μM), and lactacystin (20 μM). Expression of EGFP-tagged proteins was detected as in panel D.
ERK3 is stabilized and accumulates to high levels during muscle differentiation.
To establish the physiological relevance of the short half-life of ERK3, it is important to identify stimuli that cause the accumulation of the kinase in cells. To address this question, we first examined the expression of ERK3 in growth-arrested and proliferating cells. No change in the steady-state levels of endogenous ERK3 protein could be observed during cell cycle progression (data not shown). Treatment with DNA-damaging agents, CoCl2 (hypoxia-mimetic), or tumor necrosis factor alpha had no effect either on ERK3 turnover.
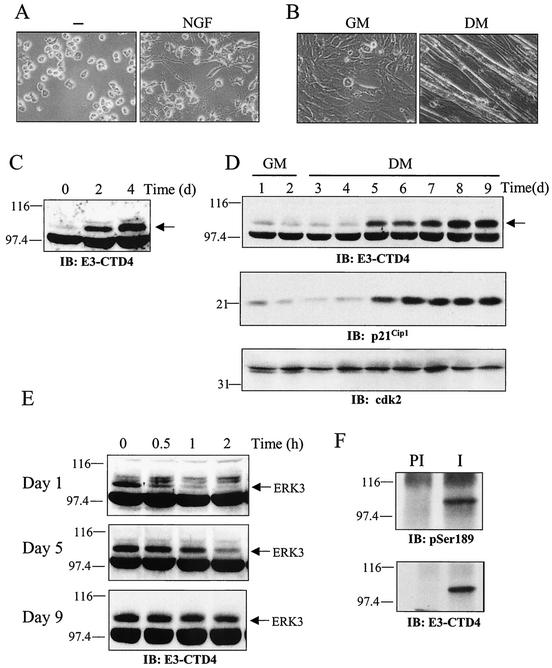
Because of the importance of MAP kinase pathways in cell differentiation and lineage commitment (23, 38, 54), we also monitored the expression of ERK3 in different cellular models undergoing differentiation. The phaeochromocytoma cell line PC12 differentiates into a sympathetic neuron-like phenotype, characterized by neurite outgrowth and other cellular changes (12), in response to neurotrophic factors (Fig. 8A). C2C12 is a myogenic cell line derived from muscle satellite cells (1) that form postmitotic, multinucleated myotubes when grown in low serum (Fig. 8B). In both of these cellular models, differentiation was accompanied by a dramatic and time-dependent accumulation of ERK3 protein (Fig. 8C and D). In C2C12 cells, ERK3 upregulation paralleled that of the cell cycle inhibitor p21Cip1, a marker of skeletal muscle differentiation (15, 36) (Fig. 8D). Most importantly, cycloheximide-chase experiments revealed that ERK3 accumulation results in large part from its time-dependent stabilization during the differentiation process. The half-life of ERK3 changed from ∼15 min in undifferentiated C2C12 myoblasts to 1 h by the fifth day of differentiation to more than 2 h in fully differentiated cells (Fig. 8E). Immunoblot analysis with anti-phospho-ERK3(S189)-specific antibody indicated that ERK3 is phosphorylated in the activation loop in differentiated C2C12 cells, suggesting that the kinase is active.
FIG. 8.
Induction of ERK3 expression during cellular differentiation. (A) PC12 cells were stimulated or not with NGF for 4 days. Phase-contrast micrographs show the outgrowth of neurites in NGF-treated cells. (B) C2C12 cells were grown in GM for 2 days or in DM for 7 days. Phase-contrast micrographs show the formation of myotubes in DM. (C) PC12 cells were treated with NGF for the indicated times. The expression of endogenous ERK3 was analyzed by immunoblotting with E3-CTE4 antibody. (D) C2C12 cells were incubated in GM or DM for the indicated times. The expression of endogenous ERK3, p21Cip1 and Cdk2 was analyzed by immunoblotting. (E) The half-life of ERK3 was evaluated by cycloheximide-chase experiments in proliferating (day 1) and differentiating (day 5 and 9) C2C12 myoblasts. (F) ERK3 is phosphorylated on Ser189 in differentiated muscle cells. Endogenous ERK3 was immunoprecipitated from differentiated C2C12 cells (day 9) with either preimmune serum (lane PI) or E3-CTE4 antibody (lane I). Phosphorylation of ERK3 was monitored by immunoblotting with anti-phospho-Ser189-specific antibody.
Expression of stabilized forms of ERK3 inhibits S-phase entry.
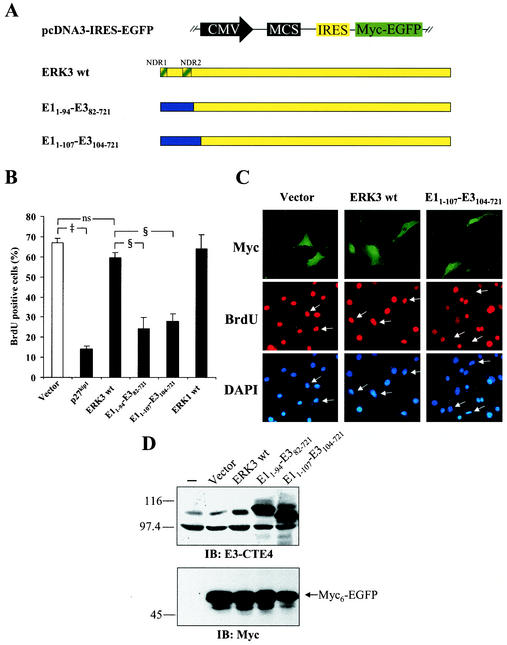
Cellular differentiation is accompanied by cell cycle arrest in the G1 phase. We thus evaluated the impact of ERK3 expression on cell cycle progression. For these experiments, we constructed a bicistronic vector expressing Myc-tagged EGFP downstream of the poliovirus IRES to enable detection of transfected cells (Fig. 9A). Transfected NIH 3T3 fibroblasts were synchronized in G0 by serum starvation and then stimulated with serum in the presence of BrdU to monitor entry into S phase (S-phase entry). As expected, overexpression of p27Kip1, used as control, strongly inhibited the entry of cells into S phase (Fig. 9B) (42). Under these conditions, ectopically expressed wild-type ERK3 had little effect on cell cycle progression (Fig. 9B and C). Using the information gained from our chimera analysis, we next tested the effect of stabilized forms of ERK3 (see Fig. 6B). Two chimeric constructs were made in which part of ERK3 N-terminal lobe is replaced by the corresponding ERK1-derived sequences (Fig. 9A). Immunoblot analysis confirmed that the two chimeric proteins are expressed at high levels compared to wild-type ERK3 (Fig. 9D). Ectopic expression of the stabilized forms of ERK3 markedly decreased the number of BrdU-positive cells (Fig. 9B and C). No morphological change typical of apoptosis or cell death was observed under these conditions. These results suggest that ERK3 can act as a negative regulator of G1-phase progression.
FIG.9.
Stable forms of ERK3 inhibit S-phase entry in NIH 3T3 fibroblasts. (A) Schematic representation of chimeric constructs used in these experiments. The NDR1 and NDR2 regions are highlighted. (B) NIH 3T3 cells were transfected with the indicated constructs. The cells were serum starved and then stimulated with 10% serum for 20 h in the presence of BrdU. The percentage of transfected cells (Myc-positive) that incorporated BrdU was evaluated by fluorescence microscopy. ns, not significantly different; ‡, P < 0.01 compared to vector; §, P < 0.01 compared to wild-type ERK3. (C) Representative example of the results shown in panel B. (D) NIH 3T3 cells were transfected or not with the indicated constructs. After 44 h, cell lysates were prepared and analyzed by immunoblotting with E3-CTE4 antibody (upper panel) and 9E10 MAb (lower panel).
DISCUSSION
Regulated intracellular proteolysis is now recognized as a versatile and efficient mechanism to control gene expression. The regulation of protein turnover can have a significant impact on the activity of the corresponding target gene and is associated with either a decrease or an increase in protein stability. For example, the inhibitors of NF-κB (IκB) and the cyclin-dependent kinase inhibitor p27Kip1 are expressed to high levels in resting cells but are rapidly degraded in response to cytokines or cell cycle progression, respectively (34, 35). In contrast, the tumor suppressor gene product p53 and the transcription factor hypoxia inducible factor 1α (HIF-1α) are both constitutively degraded in unstimulated cells. However, in response to specific stimuli, i.e., genotoxic stress for p53 (33) or hypoxia for HIF-1 (48), the proteins accumulate as a result of the inhibition of protein degradation, thereby allowing them to exert their biological functions. Here we report that the atypical MAP kinase homologue ERK3 is a highly unstable protein, with a half-life of ca. 30 min, that is constitutively degraded in exponentially proliferating cells. This is the first documented example of a MAP kinase family member whose activity is acutely regulated by protein turnover. Interestingly, Lu et al. (24) recently reported that prolonged exposure to sorbitol induces polyubiquitination of the MAP kinases ERK1/2 and promotes their degradation by a mechanism dependent on the plant homeobox domain (PHD) of MEKK1. In agreement with the results presented here, no ubiquitination of ERK1/2 was observed in response to serum or other stress stimuli, such as UV radiation or anisomycin. In contrast to ERK3, which is rapidly degraded in proliferating cells, ubiquitination of ERK1/2 (and possibly other MAP kinases) may represent an alternative feedback mechanism for downregulating kinase activity upon a persistent stress stimulus.
Protein degradation by the ubiquitin-proteasome system is involved in the regulation of many important cellular processes, including signal transduction, transcription, cell cycle progression, and class I major histocompatibility complex antigen presentation (10, 26). Using a combination of pharmacological, biochemical, and genetic approaches, we found that the ubiquitin-proteasome pathway is responsible for the rapid degradation of ERK3. Treatment with MG-132 and lactacystin, two structurally unrelated proteasome inhibitors, markedly increased the steady-state levels of ERK3 protein without changing the levels of ERK1. Importantly, we showed that MG-132 treatment markedly increases the half-life of endogenous ERK3, clearly demonstrating that ERK3 is a direct substrate of the proteasome.
Targeting of proteins to the proteasome generally requires the attachment of a multiubiquitin chain to the substrate (17, 18). However, there are examples, such as ornithine decarboxylase (32) and p21Cip1 (50), for which it has been demonstrated that ubiquitination is not necessary for degradation by the proteasome. Our results clearly indicate that ERK3 is ubiquitinated in vivo and that ubiquitination is a required step for efficient degradation by the proteasome. This conclusion is supported by the following observations. First, cotransfection experiments of ERK3 with HA-tagged ubiquitin revealed the accumulation of high-molecular-weight HA-ubiquitin conjugates in MG-132-treated cells. These slowly migrating HA-immunoreactive species were not observed in cells transfected with ERK1. Second, the half-life of endogenous ERK3 protein was markedly increased at the nonpermissive temperature in cells bearing a thermolabile allele of the ubiquitin-activating enzyme E1. Third, fusion of the degradation domain of ERK3 to the heterologous protein EGFP was sufficient to promote its ubiquitination (data not shown) and to target it for proteasomal degradation (Fig. 7D).
A role for the proteasome in the regulation of ERK3 expression was recently reported by Zimmermann et al., who showed, by using a microarray-based differential cDNA hybridization technology, that ERK3 mRNA is upregulated by various proteasome inhibitors (58). Proteasome inhibition also resulted in the accumulation of ERK3 protein. It was concluded that proteasome inhibitors influence ERK3 expression mainly at the transcriptional level. However, these authors did not evaluate the half-life or the ubiquitination status of ERK3. Whereas our study does not exclude a role of the proteasome in the transcriptional activation of ERK3 gene, the experiments presented here clearly demonstrate the importance of this proteolytic system in directly controlling ERK3 protein stability in proliferating cells.
It is interesting that for most protein kinases shown to be unstable, destabilization of the kinase is linked to enzymatic activation. For example, activation of the serine/threonine kinases PKCα (20) and GRK2 (41) by their specific activators bryostatin and isoproterenol, respectively, leads to their proteasomal degradation. Similarly, the turnover of many receptor and soluble tyrosine kinases is also regulated by their phosphotransferase activity. Ligand-activated platelet-derived growth factor β receptor (31) or constitutively activated v-Src (16) are both ubiquitinated and targeted to the 26S proteasome. The activation-dependent destabilization of the enzyme seems to be part of a negative feedback regulatory loop aimed at decreasing overall protein kinase activity. In contrast, our findings indicate that the control of ERK3 stability is independent of its kinase activity and of activation loop phosphorylation.
From the results of mutagenesis analysis, we concluded that the destabilizing signal is contained within the first 365 amino acids of ERK3. To further define the degradation domain of ERK3, we examined the stability of a series of chimeric kinases made between the stable ERK1 kinase and ERK3. We were able to delimit two regions in the N-terminal lobe of ERK3 kinase domain that contribute to protein instability. These two regions correspond to the first 15 amino acids of ERK3 (NDR1) and parts of kinase subdomains II and III (NDR2, residues 53 to 73). As one would expect, primary sequence analysis revealed that these two regions display low identity to the corresponding ERK1 sequences. Close inspection of the sequences did not reveal any obvious resemblance with known degradation motifs. Interestingly, in a recent study aimed at defining structural determinants controlling subcellular localization and MEK recognition, Robinson et al. observed that a chimera containing the N-terminal lobe of ERK3 and the C-terminal lobe of ERK2 is expressed at a low level compared to wild-type ERK2 (44). This observation is in agreement with the results presented here. Despite the fact that NDR1/2 are not contiguous in the primary sequence, a theoretical model of the structure of ERK33-346 revealed that the two regions lie on the same side of the molecule in close proximity. We postulate that the NDR1/2 form a docking site for assembly of an E3 ubiquitin-protein ligase. In support of this hypothesis, we showed that ERK3 N-terminal lobe is sufficient to target heterologous proteins for degradation by the proteasome. It remains to be determined whether NDR1/2 activity is regulated by posttranslational modifications or, alternatively, if the assembly of the ERK3-E3 complex is regulated by interacting proteins. The ERK3-specific E3 ligase may also be subject to direct regulation.
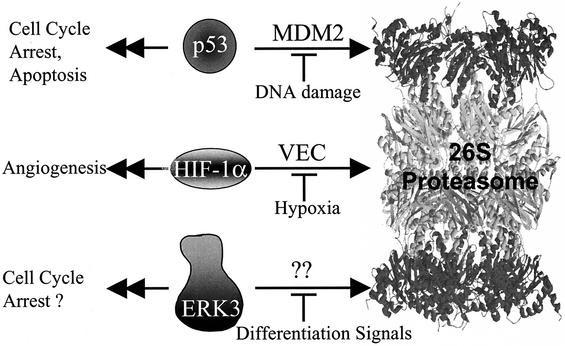
Our results provide strong evidence for the physiological relevance of ERK3 rapid turnover. We found that the stability of ERK3 increases with time during in vitro myogenic differentiation of C2C12 cells, resulting in a marked upregulation of the protein. The accumulation of ERK3 is concomitant with the induction of 21Cip1 protein and exit from the cell cycle, suggesting that ERK3 may contribute in some way to cell cycle withdrawal. In agreement with this idea, BrdU incorporation studies revealed that expression of stabilized forms of ERK3, but not unstable wild-type ERK3, inhibits the induction of S phase in fibroblasts. This is the first biological effect of ERK3 described to date. All of these observations suggest that unidentified cellular ERK3-specific E3 ligase(s) is able to repress the cell cycle inhibitory activity of ERK3 by promoting its ubiquitination and degradation by the proteasome. We thus propose that the biological activity of ERK3 is mainly regulated by its cellular abundance through the control of protein degradation by the ubiquitin-proteasome pathway (Fig. 10). This model is reminiscent of the regulation mode of the transcription factors p53 and HIF-1α.
FIG. 10.
Model of ERK3 regulation by proteasome-mediated degradation.
Acknowledgments
We thank D. Bohmann, R. Davis, J. Han, H. Ozer, C. Prody, J. Woodgett, and P. Jolicoeur for reagents.
This work was supported by a grant (MOP-38010) to S.M. from the Canadian Institutes for Health Research (CIHR). P.C. and S.P. are recipients of a studentship from the CIHR and Heart and Stroke Foundation of Canada, respectively. G.R. is the recipient of a fellowship from the National Cancer Institute of Canada. S.M. is an Investigator of the CIHR.
REFERENCES
- 1.Blau, H. M., G. K. Pavlath, E. C. Hardeman, C. P. Chiu, L. Silberstein, S. G. Webster, S. C. Miller, and C. Webster. 1985. Plasticity of the differentiated state. Science 230**:**758-766. [DOI] [PubMed] [Google Scholar]
- 2.Boulton, T. G., S. H. Nye, D. J. Robbins, N. Y. Ip, E. Radziejewska, S. D. Morgenbesser, R. A. DePinho, N. Panayotatos, M. H. Cobb, and G. D. Yancopoulos. 1991. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65**:**663-675. [DOI] [PubMed] [Google Scholar]
- 3.Brunet, A., D. Roux, P. Lenormand, S. Dowd, S. Keyse, and J. Pouyssegur. 1999. Nuclear translocation of p42/p44 mitogen-activated protein kinase is required for growth factor-induced gene expression and cell cycle entry. EMBO J. 18**:**664-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410**:**37-40. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, M., T. G. Boulton, and M. H. Cobb. 1996. ERK3 is a constitutively nuclear protein kinase. J. Biol. Chem. 271**:**8951-8958. [DOI] [PubMed] [Google Scholar]
- 6.Chowdary, D. R., J. J. Dermody, K. K. Jha, and H. L. Ozer. 1994. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 14**:**1997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulombe, P., and S. Meloche. 2002. Dual-tag prokaryotic vectors for enhanced expression of full-length recombinant proteins. Anal. Biochem. 310**:**219-222. [DOI] [PubMed] [Google Scholar]
- 8.English, J., G. Pearson, J. Wilsbacher, J. Swantek, M. Karandikar, S. Xu, and M. H. Cobb. 1999. New insights into the control of MAP kinase pathways. Exp. Cell Res. 253**:**255-270. [DOI] [PubMed] [Google Scholar]
- 9.Fenteany, G., R. F. Standaert, W. S. Lane, S. Choi, E. J. Corey, and S. L. Schreiber. 1995. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268**:**726-731. [DOI] [PubMed] [Google Scholar]
- 10.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82**:**373-428. [DOI] [PubMed] [Google Scholar]
- 11.Goodfellow, P. J. 1994. Inherited cancers associated with the RET proto-oncogene. Curr. Opin. Genet. Dev. 4**:**446-452. [DOI] [PubMed] [Google Scholar]
- 12.Greene, L. A., and A. S. Tischler. 1976. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. USA 73**:**2424-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione _S_-transferase. Anal. Biochem. 192**:**262-267. [DOI] [PubMed] [Google Scholar]
- 14.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18**:**2714-2723. [DOI] [PubMed] [Google Scholar]
- 15.Halevy, O., B. G. Novitch, D. B. Spicer, S. X. Skapek, J. Rhee, G. J. Hannon, D. Beach, and A. B. Lassar. 1995. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 267**:**1018-1021. [DOI] [PubMed] [Google Scholar]
- 16.Harris, K. F., I. Shoji, E. M. Cooper, S. Kumar, H. Oda, and P. M. Howley. 1999. Ubiquitin-mediated degradation of active Src tyrosine kinase. Proc. Natl. Acad. Sci. USA 96**:**13738-13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67**:**425-479. [DOI] [PubMed] [Google Scholar]
- 18.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30**:**405-439. [DOI] [PubMed] [Google Scholar]
- 19.Kahn, C. R., D. Vicent, and A. Doria. 1996. Genetics of non-insulin-dependent (type-II) diabetes mellitus. Annu. Rev. Med. 47**:**509-531. [DOI] [PubMed] [Google Scholar]
- 20.Lee, H. W., L. Smith, G. R. Pettit, A. Vinitsky, and J. B. Smith. 1996. Ubiquitination of protein kinase C-alpha and degradation by the proteasome. J. Biol. Chem. 271**:**20973-20976. [PubMed] [Google Scholar]
- 21.Lee, J. C., S. Kumar, D. E. Griswold, D. C. Underwood, B. J. Votta, and J. L. Adams. 2000. Inhibition of p38 MAP kinase as a therapeutic strategy. Immunopharmacology 47**:**185-201. [DOI] [PubMed] [Google Scholar]
- 22.Levitzki, A. 1999. Protein tyrosine kinase inhibitors as novel therapeutic agents. Pharmacol. Ther. 82**:**231-239. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, T. S., P. S. Shapiro, and N. G. Ahn. 1998. Signal transduction through MAP kinase cascades. Adv. Cancer Res. 74**:**49-139. [DOI] [PubMed] [Google Scholar]
- 24.Lu, Z., S. Xu, C. Joazeiro, M. H. Cobb, and T. Hunter. 2002. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9**:**945-956. [DOI] [PubMed] [Google Scholar]
- 25.Marshall, C. J. 1995. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80**:**179-185. [DOI] [PubMed] [Google Scholar]
- 26.Mayer, R. J. 2000. The meteoric rise of regulated intracellular proteolysis. Nat. Rev. Mol. Cell. Biol. 1**:**145-148. [DOI] [PubMed] [Google Scholar]
- 27.Meloche, S., B. G. Beatty, and J. Pellerin. 1996. Primary structure, expression and chromosomal locus of a human homolog of rat ERK3. Oncogene 13**:**1575-1579. [PubMed] [Google Scholar]
- 28.Meloche, S., G. Pages, and J. Pouyssegur. 1992. Functional expression and growth factor activation of an epitope-tagged p44 mitogen-activated protein kinase, p44mapk. Mol. Biol. Cell 3**:**63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meloche, S., K. Seuwen, G. Pages, and J. Pouyssegur. 1992. Biphasic and synergistic activation of p44MAPK (ERK1) by growth factors: correlation between late phase activation and mitogenicity. Mol. Endocrinol. 6**:**845-854. [DOI] [PubMed] [Google Scholar]
- 30.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18**:**3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori, S., C. H. Heldin, and L. Claesson-Welsh. 1992. Ligand-induced polyubiquitination of the platelet-derived growth factor beta-receptor. J. Biol. Chem. 267**:**6429-6434. [PubMed] [Google Scholar]
- 32.Murakami, Y., S. Matsufuji, T. Kameji, S. Hayashi, K. Igarashi, T. Tamura, K. Tanaka, and A. Ichihara. 1992. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360**:**597-599. [DOI] [PubMed] [Google Scholar]
- 33.Oren, M. 1999. Regulation of the p53 tumor suppressor protein. J. Biol. Chem. 274**:**36031-36034. [DOI] [PubMed] [Google Scholar]
- 34.Pagano, M., S. W. Tam, A. M. Theodoras, P. Beer-Romero, G. Del Sal, V. Chau, P. R. Yew, G. F. Draetta, and M. Rolfe. 1995. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269**:**682-685. [DOI] [PubMed] [Google Scholar]
- 35.Palombella, V. J., O. J. Rando, A. L. Goldberg, and T. Maniatis. 1994. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell 78**:**773-785. [DOI] [PubMed] [Google Scholar]
- 36.Parker, S. B., G. Eichele, P. Zhang, A. Rawls, A. T. Sands, A. Bradley, E. N. Olson, J. W. Harper, and S. J. Elledge. 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267**:**1024-1027. [DOI] [PubMed] [Google Scholar]
- 37.Payne, D. M., A. J. Rossomando, P. Martino, A. K. Erickson, J. H. Her, J. Shabanowitz, D. F. Hunt, M. J. Weber, and T. W. Sturgill. 1991. Identification of the regulatory phosphorylation sites in pp42/mitogen-activated protein kinase (MAP kinase). EMBO J. 10**:**885-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson, G., F. Robinson, G. T. Beers, B. E. Xu, M. Karandikar, K. Berman, and M. H. Cobb. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocrinol. Rev. 22**:**153-183. [DOI] [PubMed] [Google Scholar]
- 39.Peitsch, M. C. 1996. ProMod and Swiss-Model: internet-based tools for automated comparative protein modeling. Biochem. Soc. Trans. 24**:**274-279. [DOI] [PubMed] [Google Scholar]
- 40.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334**:**320-325. [DOI] [PubMed] [Google Scholar]
- 41.Penela, P., A. Ruiz-Gomez, J. G. Castano, and F. Mayor, Jr. 1998. Degradation of the G protein-coupled receptor kinase 2 by the proteasome pathway. J. Biol. Chem. 273**:**35238-35244. [DOI] [PubMed] [Google Scholar]
- 42.Polyak, K., M. H. Lee, H. Erdjument-Bromage, A. Koff, J. M. Roberts, P. Tempst, and J. Massague. 1994. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78**:**59-66. [DOI] [PubMed] [Google Scholar]
- 43.Robinson, M. J., S. A. Stippec, E. Goldsmith, M. A. White, and M. H. Cobb. 1998. A constitutively active and nuclear form of the MAP kinase ERK2 is sufficient for neurite outgrowth and cell transformation. Curr. Biol. 8**:**1141-1150. [DOI] [PubMed] [Google Scholar]
- 44.Robinson, M. J., B. E. Xu Be, S. Stippec, and M. H. Cobb. 2002. Different domains of the mitogen-activated protein kinases ERK3 and ERK2 direct subcellular localization and upstream specificity in vivo. J. Biol. Chem. 277**:**5094-5100. [DOI] [PubMed] [Google Scholar]
- 45.Rock, K. L., C. Gramm, L. Rothstein, K. Clark, R. Stein, L. Dick, D. Hwang, and A. L. Goldberg. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78**:**761-771. [DOI] [PubMed] [Google Scholar]
- 46.Rodier, G., A. Montagnoli, L. Di Marcotullio, P. Coulombe, G. F. Draetta, M. Pagano, and S. Meloche. 2001. p27 cytoplasmic localization is regulated by phosphorylation on Ser10 and is not a prerequisite for its proteolysis. EMBO J. 20**:**6672-6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaeffer, H. J., and M. J. Weber. 1999. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell. Biol. 19**:**2435-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semenza, G. L. 2001. HIF-1, O(2), and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell 107**:**1-3. [DOI] [PubMed] [Google Scholar]
- 49.Servant, M. J., P. Coulombe, B. Turgeon, and S. Meloche. 2000. Differential regulation of p27Kip1 expression by mitogenic and hypertrophic factors: involvement of transcriptional and posttranscriptional mechanisms. J. Cell Biol. 148**:**543-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheaff, R. J., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5**:**403-410. [DOI] [PubMed] [Google Scholar]
- 51.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78**:**787-798. [DOI] [PubMed] [Google Scholar]
- 52.Turgeon, B., M. K. Saba-El-Leil, and S. Meloche. 2000. Cloning and characterization of mouse extracellular-signal-regulated protein kinase 3 as a unique gene product of 100 kDa. Biochem. J. 346(Pt. 1)**:**169-175. [PMC free article] [PubMed] [Google Scholar]
- 53.Webster, M. K., and D. J. Donoghue. 1997. FGFR activation in skeletal disorders: too much of a good thing. Trends Genet. 13**:**178-182. [DOI] [PubMed] [Google Scholar]
- 54.Weston, C. R., and R. J. Davis. 2002. The JNK signal transduction pathway. Curr. Opin. Genet. Dev. 12**:**14-21. [DOI] [PubMed] [Google Scholar]
- 55.Widmann, C., S. Gibson, M. B. Jarpe, and G. L. Johnson. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79**:**143-180. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, F., A. Strand, D. Robbins, M. H. Cobb, and E. J. Goldsmith. 1994. Atomic structure of the MAP kinase ERK2 at 2.3 Å resolution. Nature 367**:**704-711. [DOI] [PubMed] [Google Scholar]
- 57.Zhu, A. X., Y. Zhao, D. E. Moller, and J. S. Flier. 1994. Cloning and characterization of p97MAPK, a novel human homolog of rat ERK-3. Mol. Cell. Biol. 14**:**8202-8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimmermann, J., N. Lamerant, R. Grossenbacher, and P. Furst. 2001. Proteasome- and p38-dependent regulation of ERK3 expression. J. Biol. Chem. 276**:**10759-10766. [DOI] [PubMed] [Google Scholar]