HPr Kinase/Phosphorylase, the Sensor Enzyme of Catabolite Repression in Gram-Positive Bacteria: Structural Aspects of the Enzyme and the Complex with Its Protein Substrate (original) (raw)
HPr kinase/phosphorylase (HprK/P), the sensor enzyme for catabolite repression in gram-positive bacteria, phosphorylates HPr, a phosphocarrier protein of a sugar transport and phosphorylation system (41), at a serine residue (10). To carry out its catalytic function in sugar transport and phosphorylation, HPr is phosphorylated at His15 by the phosphoenolpyruvate (PEP)-dependent protein kinase, enzyme I. For its regulatory function in catabolite repression, HprK/P phosphorylates HPr in an ATP-dependent reaction at Ser46 (9). The resulting serine-phosphorylated HPr (P-Ser-HPr) acts as a corepressor by binding to CcpA (catabolite control protein A) (8), a LacI-type repressor (22). This protein-protein interaction probably includes a strong electrostatic component, as unphosphorylated HPr has about 1,000-fold weaker affinity for CcpA than does P-Ser-HPr (27), although phosphorylation of HPr at Ser46 leads to only small structural changes (2, 54). The interaction with P-Ser-HPr allows CcpA to bind to specific operator sites (cre) (16) preceding a large number of catabolite-regulated genes (6). In Bacillus subtilis, the expression of about 400 genes, or 10% of the genome, is regulated by this mechanism, which enables the bacterium to respond efficiently to the availability of rapidly metabolizable carbon sources such as glucose (36, 39). P-Ser-HPr also plays a role in inducer exclusion of gram-positive bacteria, in which it probably binds to components of carbohydrate-specific ABC transporters and inhibits their activity (11, 37).
HprK/P is a bifunctional enzyme. It was first identified as a serine protein kinase, an activity that is enhanced in the presence of glycolytic intermediates such as fructose-1,6-bisphosphate (10). However, in the presence of inorganic phosphate, HprK/P also catalyzes the dephosphorylation of Ser46 (7). This reaction, for a long time assumed to be hydrolytic, has recently been demonstrated to be phosphorolytic and to produce pyrophosphate (35). X-ray studies played a major part in the identification of this novel reaction (14). Structures are available for both the enzyme and its complex with the substrate protein HPr (Table 1), showing that HprK/P is a homohexamer, with subunits of about 300 residues folding in two distinct structural domains. The C-terminal domain carries both catalytic activities, whereas the N-terminal domain has no defined function yet (15). The C-terminal domain contains a Walker A motif (17, 53), part of a phosphate-binding loop (P-loop) and of the nucleotide binding site (45). Its three-dimensional structure has no similarity to that of eukaryotic protein kinases, but it resembles adenylate kinase and other kinases that phosphorylate small molecules, especially PEP carboxykinase (18, 44). Thus, HprK/P defines a novel family of protein kinases.
TABLE 1.
X-ray structures of HprK/P
| PDB entry codea | Origin | Protein | Ligand(s) | mb | Resolu- tion (Å) | Refer- ence |
|---|---|---|---|---|---|---|
| 1JB1 | L. casei | C-domain | Pi | 1 | 2.8 | 15 |
| 1KKL | L. casei | C-domain | HPr | 3 | 2.8 | 14 |
| 1KKM | L. casei | C-domain | P-Ser-HPr, Pi, Ca2+ | 3 | 2.8 | 14 |
| 1KO7 | S. xylosus | Full length | Pi, Mg2+ | 2 | 1.95 | 32 |
| 1KNX | M. pneumoniae | Full length | 6 | 2.5 | 1 |
Five X-ray structures of HprK/P are available at present. After the C-terminal domain of Lactobacillus casei HprK/P had been reported (15), structures of the full-length protein were obtained for Staphylococcus xylosus (32) and Mycoplasma pneumoniae (1). The structures of the complexes between L. casei HprK/P and its protein substrates HPr and P-Ser-HPr were also determined (14). All are closely related, but minor differences can be observed, due to sequence changes, the presence of ligands, notably inorganic phosphate and metal ions, and the different crystal environments. We review the structure of the free enzyme and compare it to that of the L. casei C-terminal domain in complex with either unphosphorylated or phosphorylated B. subtilis HPr.
The mode of HPr and inorganic phosphate binding observed in the complexes suggests mechanisms for both the phosphorylation and phosphorolysis reactions (14). The unphosphorylated complex was used as a target for the CAPRI (critical assessment of predicted interactions) prediction experiment, where computational biologists were provided with the atomic coordinates of HPr and the L. casei C-terminal domain, and used docking simulations to derive models of the complex (24, 25). The models were then independently assessed by comparison to the X-ray structure, which had not been published at that time. We describe the results of this test and discuss features of the HPr-HprK/P interaction which prevented high-quality predictions in spite of the biochemical information that was available to the predictors (34).
THREE-DIMENSIONAL STRUCTURE OF HprK/P
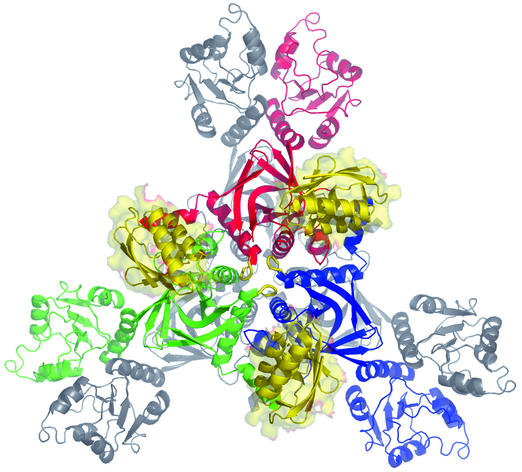
Hexamer. In the X-ray structures, HprK/P is a hexamer and has D3 symmetry, with a threefold axis orthogonal to three twofold axes (Fig. 1). The symmetry is crystallographic in the free L. casei C-terminal domain (Table 1) and local in all other structures, which allows for minor differences between the subunits. D3 symmetry is maintained in the complexes with HPr, where the hexamer binds six HPr molecules. Protein-protein contacts occur between the C-terminal catalytic domains, which form two trimers assembled in a hexameric core. Three pairs of N-terminal domains stick out, making little contact with the core. The peptide linking the two domains is flexible and allows each pair of N-terminal domains to adopt slightly different orientations relative to the core (1). In the X-ray structure of the L. casei C-terminal domains, the lack of N-terminal domain has little effect on either the fold or the mode of assembly of the core. In the two complexes with the C-terminal domains (14), HPr does not change conformation, and its binding induces no major change in the enzyme. Three HPr molecules bind on the top of the core, and three bind on the bottom (Fig. 1). They do not interact with each other, but each one makes contacts with two adjacent C-terminal domains. Their position makes it unlikely that additional contacts are made by HPr with the N-terminal domains in the full-length protein (14).
FIG. 1.
The HprK/P hexamer. S. xylosus HprK/P is shown in ribbon representation, viewed along the threefold axis. Subunits of the top trimer are red, blue, and green; the bottom trimer is in gray. Three HPr molecules (yellow ribbons and surfaces) are drawn bound to the top trimer, at the location they take in the complex with the C-terminal domain of L. casei HprK/P. Drawn with PYMOL (W. L. DeLano, 2002, http://www.pymol.org).
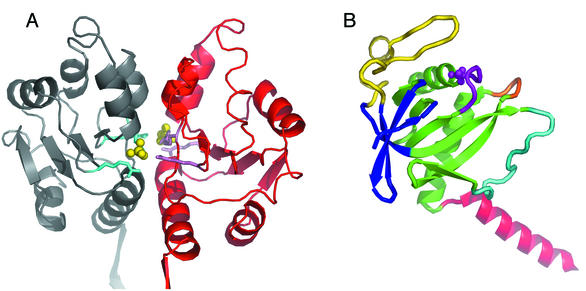
N-terminal domain. In the S. xylosus structure, two phosphate ions bind at the interface between pairs of N-terminal domains (Fig. 2A). They interact with three arginines (Arg33, Arg49, and Arg88) which are strictly conserved in the HprK/P sequences of gram-positive bacteria but not of gram-negative bacteria and mycoplasmas. The fold of the domain is similar to that of MurE (19), an enzyme implicated in the synthesis of cytoplasmic precursors for cell wall synthesis, which binds phosphorylated ligands in a similar way. This site, which may be designed to bind phosphorylated metabolites rather than just inorganic phosphate, very likely plays a part in the biological function of the N-terminal domain, whatever that function may be (32).
FIG. 2.
Subunit. (A) N-terminal domains. A pair of N-terminal domains of S. xylosus HprK/P are shown in ribbon representation. Two phosphate ions (yellow balls) bound at the dimer interface interact with arginine residues (cyan). The orientation is as in Fig. 1. (B) The C-terminal domain. The C-terminal domain of L. casei HprK/P is taken from the complex with P-Ser-HPr. The α/β unit is green; the P-loop and bound inorganic phosphate are purple; the central loop is orange. The second β-sheet in blue and the capping motif in yellow constitute a separate subdomain. The C-terminal α-helix is red, and the flexible loop, which is ordered in this structure, is cyan. The orientation is approximately that of the green subunit in Fig. 1.
C-terminal catalytic domain. The C-terminal domain (residues 127 to 319 in L. casei HprK/P) forms a globular structure, from which the C-terminal helix protrudes (Fig. 2B). It can be viewed as comprising two subdomains. One subdomain is an α/β unit made of a central five-stranded β-sheet with two α-helices on one face of it. These secondary-structure elements have the same particular topology as in the ATP-binding domain of PEP carboxykinase (33). The Walker A motif, G155DSGVGKS162 (L. casei numbering), forms a loop where an inorganic phosphate ion is found when the protein is crystallized in the presence of phosphate (15, 32). Thus, the Walker A motif is a bona fide P-loop (45), and it most likely binds the phosphate moiety of the nucleotide substrate. In the Mycoplasma pneumoniae X-ray structure (1), the inorganic phosphate site is empty and the loop takes an alternative conformation to fill the site.
In adenylate kinase and many other ATP-dependent kinases that act on small metabolites, the α/β unit of HprK/P and PEP carboxykinase exists with a different β-sheet topology. In the SCOP database (40), all these kinases belong to the superfamily of P-loop-containing proteins. The similarity of HprK/P to these proteins does not extend to the remainder of the C-terminal domain. It forms a subdomain specific to HprK/P, with a four-stranded antiparallel β-sheet that covers the face of the central β-sheet opposite the α-helices. The so-called capping motif (15), an unusual structure corresponding to the conserved HprK/P signature sequence (43), completes the globular core of the HprK/P subunit. Mutations affecting amino acids in the signature sequence of HprK/P from M. pneumoniae and B. subtilis abolished the dephosphorylation activity, and some also reduced the phosphorylation activity (21, 47).
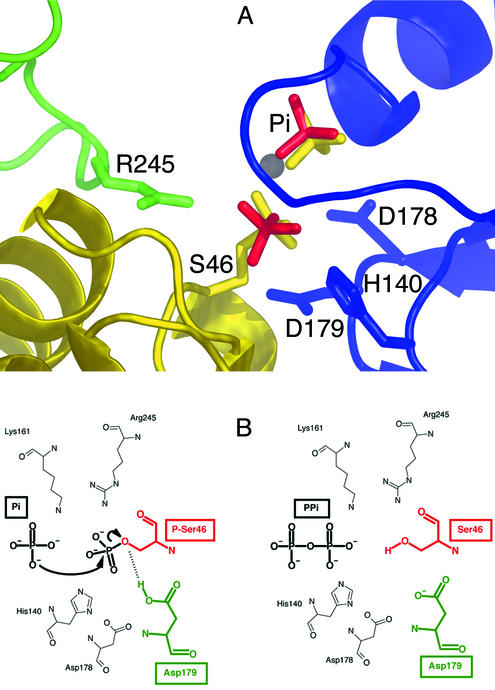
Flexible loop and C-terminal helix. A large polypeptide loop (residues 236 to 260 in L. casei HprK/P), connecting two β-strands of the α/β unit, is located on the surface of the protein above the active site. It is disordered in most HprK/P structures. A notable exception is the complex with phosphorylated HPr (Fig. 1), where a strictly conserved arginine of the loop (Arg245 in L. casei) interacts with the phosphoserine of HPr bound to an adjacent subunit (Fig. 3A). In some of the six subunits of the M. pneumoniae HprK/P asymmetric unit (1), the loop is partly ordered in a conformation similar to that in the complex, even though no ligand is present.
FIG. 3.
Catalytic mechanism. (A) The active site. Close-up of the L. casei C-terminal domain in complex with P-Ser-HPr, oriented as in Fig. 1, and comprising part of the blue subunit, the flexible loop of the adjacent green subunit, and part of HPr in yellow. The HPr phosphoserine (S46) and an inorganic phosphate ion (yellow) interact with the P-loop and residues H140, D178, and D179 of the blue subunit. The phosphoserine also interacts with Arg245 of the flexible loop of the green subunit. In S. xylosus, two phosphate ions (red) occupy the sites taken in L. casei by inorganic phosphate and the phosphoserine. (B) The mechanism of phosphorolysis. An oxygen atom of the inorganic phosphate substrate performs a nucleophilic attack on the phosphorus atom of the phosphoserine, yielding pyrophosphate and an oxyanion on Ser46 of HPr, which can accept a proton from Asp179. The same mechanism accounts for the reverse reaction, the phosphorylation of Ser46 by pyrophosphate.
Another mobile part of the C-terminal domain is the C-terminal helix of each subunit. It takes different orientations in the various HprK/P structures and rotates by about 30° when HPr binds to L. casei HprK/P. The orientation seen in the complex is the same as in the free S. xylosus protein, indicating that the helix can move under the influence of crystal contacts as well as HPr binding. The movement is of functional significance, for the C-terminal helix of each HprK/P subunit makes contacts with the HPr molecule bound to an adjacent subunit in the hexamer. It was one of the difficulties encountered when predicting the mode of HPr-HprK/P binding in the CAPRI experiment described below.
Central loop and quaternary structure. In each half of the hexamer, three C-terminal domains contain a loop that comes close to the threefold axis (Fig. 1). The loop (residues 266 to 271 in L. casei HprK/P), which we called the central loop because of its position, is in contact with the central loops of neighboring subunits and also with their P-loops. Although we do not have X-ray evidence yet, a nucleotide can be modeled at the active site of the HprK/P subunit on the basis of a comparison with other P-loop-containing kinases. In a model based on ADP bound to adenylate kinase (52), the phosphate moiety of the nucleotide fits nicely in the HprK/P active site, and the β-phosphate superimposes on the inorganic phosphate ion of the P-loop. However, the adenine base clashes with the central loop of an adjacent subunit (15), and a conformational change must occur in either the protein or the nucleotide. The change may affect subunit contacts within a trimer and, possibly, the quaternary structure of the protein. It is a likely source of the cooperativity seen when B. subtilis HprK/P binds ATP analogs and the activator fructose-1,6-bisphosphate (26). Interestingly, several mutations affecting the central loop specifically lowered the phosphorylase but not the kinase activity of HprK/Ps from L. casei and B. subtilis (38). These mutations led to the accumulation of P-Ser-HPr in the cells and to carbon catabolite repression even in the absence of a repressing sugar. Based on these results, it was assumed that the mutations in the central loop introduced structural changes which would propagate to the adjacent active sites.
Equilibrium centrifugation of the L. casei, B. subtilis, S. xylosus, Streptococcus salivarius, and Neisseria gonorrhoeae proteins showed HprK/P to be a hexamer in solution (S. Fieulaine and S. Nessler, unpublished data). Electrospray ionization mass spectrometry experiments suggested that, whereas the hexamer is the major form in solution at pH 6.8, it dissociates at pH 9.5 (42). Dynamic light scattering and surface plasmon resonance experiments confirmed the pH-dependent dissociation. Because the optimal pH for the kinase activity (pH 7.5) is higher than that for the phosphorylase activity (pH 6.2), it was suggested that the change in the oligomerization state may be relevant to the switch between the two activities. While the hexamer would be active as a phosphorylase, a dimeric or trimeric form of the protein was assumed to act as a kinase (42). It is worth noting that all HprK/P crystals were obtained near pH 7.5 except for the free catalytic domain of L. casei, which crystallized at pH 5.2. If dissociation does occur, HprK/P should be expected to yield trimers rather than dimers, as the subunit contacts are more extensive within the trimer than between trimers. Trimer contacts bury 2,000 Å2 out of a total interface area of 2,600 Å2 per monomer. Moreover, the HPr binding site is shared between two subunits of the same trimer.
CATALYTIC MECHANISM
Active site. In HprK/P, the active site comprises the P-loop and surrounding residues (Fig. 3A). As in other P-loop proteins (45), the primary role of the loop is to bind the phosphate moiety of the nucleotide substrate. This role is illustrated by the interaction between inorganic phosphate and main-chain NH groups of the loop (residues 155 to 162 in L. casei numbering) and between the phosphoserine of HPr and the conserved side chains of Lys161 and Ser157. Replacing the equivalent of Ser157 with an alanine in M. pneumoniae HprK/P had no effect on the kinase activity but increased the phosphorylase activity. By contrast, Lys160Ala and Lys160Arg substitutions in M. pneumoniae HprK/P led to a complete loss of the two activities (47). Several other mutations affecting amino acids in the P-loop of B. subtilis HprK/P reduced or abolished both catalytic activities (21). The replacement of Gly160 with a serine in L. casei HprK/P, while reducing the phosphorylase activity, had little effect on the kinase activity (38) and yielded a phenotype similar to that observed for mutations affecting the central loop. In the L. casei C-terminal domain, the Glu163 side chain also interacts with inorganic phosphate, which must therefore be present in the protonated PO4H2− species (15). This interaction is less likely to occur with a nucleotide, where the phosphate groups are generally unprotonated, unless the Glu163 side chain is protonated.
Outside the P-loop, and in addition to the arginine of the flexible loop (Arg245) mentioned above, a histidine (His140 in L. casei HprK/P) and two consecutive aspartates (Asp178 and Asp179) play major parts in catalysis. Although no homology is detected in the overall sequence, HprK/P shares these active-site residues with PEP carboxykinase (18, 44). His140 and Asp179 of L. casei HprK/P interact with Ser46 of HPr, but they are mobile in the absence of a substrate. In the S. xylosus structure, two inorganic phosphate ions occupy the P-loop (Fig. 3A), and two histidine conformations coexist. In the complex with unphosphorylated HPr, the carboxylate of Asp179 forms a hydrogen bond with Ser46, which suggests that it is the catalytic base that removes the serine OH proton upon phosphorylation (14). Asp178 does not directly contact HPr or the phosphates, but it is nevertheless essential to the catalytic activity of the enzyme (18). In the complex with HPr, Asp178 interacts with Ser162, which is part of the divalent cation coordination sphere, as in other P-loop-containing proteins. Replacing one of the two equivalent aspartates of B. subtilis HprK/P (Asp176 and Asp177) with an alanine led to a complete loss of the phosphorylase activity, although the Asp176Ala mutant retained weak kinase activity (18).
Phosphorolysis reaction. With P-Ser-HPr as the substrate, HprK/P catalyzes the dephosphorylation of Ser46 in the presence of inorganic phosphate (28). The reaction was originally assumed to be a hydrolysis (and the enzyme a phosphatase) and inorganic phosphate an activator until the X-ray structure of the L. casei C-terminal domain showed that inorganic phosphate binds in the P-loop and only there. This location was compatible with inorganic phosphate being a competitive inhibitor of the phosphorylation reaction, but it could not explain the activating effect of inorganic phosphate on P-Ser-HPr dephosphorylation. This prompted us to analyze the products of the reaction, which proved to be free HPr and pyrophosphate (35). Thus, dephosphorylation of Ser46 proceeds by phosphorolysis rather than hydrolysis of the phosphoester. HprK/P catalyzes the two phosphate transfer reactions: HO-Ser-HPr + ATP ↔ P-Ser-HPr + ADP, and P-Ser-HPr + inorganic phosphate ↔ HO-Ser-HPr + inorganic pyrophosphate. Thermodynamics should drive the first reaction to the right and the second to the left. As a result, HprK/P can phosphorylate HPr with pyrophosphate as the phosphate donor (35). When performed in the crystal of B. subtilis HPr complexed to the L. casei C-terminal domain, this reaction produced P-Ser-HPr and an inorganic phosphate ion which remained bound (14). To be efficient, HPr dephosphorylation actually depends on coupling phosphorolysis with a third reaction, catalyzed by pyrophosphatases: inorganic pyrophosphate + H2O ↔ 2 inorganic phosphate.
In B. subtilis, the hprK gene is part of an operon also containing the gene coding for YvoE, which proved to have pyrophosphatase activity (35). In the cell, the equilibrium ratio between the free and phosphorylated forms of HPr therefore depends on the relative concentrations not only of ATP and ADP, but also of phosphate and pyrophosphate.
Mechanism of phosphate transfer. The X-ray structure of the complex with P-Ser-HPr was obtained in the presence of pyrophosphate (14). The resulting phosphorylation of HPr Ser46 suggests a detailed mechanism for the phosphate transfer during pyrophosphate-dependent HPr phosphorylation and by microscopic reversibility during phosphorolysis (Fig. 3B). The inorganic phosphate ion, product of the reaction, occupies the same location as in the free L. casei C-terminal domain and the S. xylosus full-length HprK/P. One of the inorganic phosphate oxygen atoms is at the correct distance (about 3.2 Å) for performing in-line nucleophilic attack on the P atom of the phosphoserine, forming the P-O-P bond of pyrophosphate while the Ser-O-P bond breaks. The serine may then receive a proton from Asp179, which acts as an acid catalyst in this reaction. The Asp179 side chain is within hydrogen bonding distance of the phosphoserine and therefore likely to be protonated in the presence of its negative charge (14). The metal ion that bridges inorganic phosphate and the phosphoserine (Ca2+ in this particular X-ray structure) should assist phosphate transfer by reducing the electrostatic repulsion between the two phosphates. Cations have been shown to play an essential role in the HprK/P mechanism (30), as in many other phosphate transfer enzymes (49).
The present set of HprK/P structures provides less information about the mechanism of ATP-dependent phosphorylation, as we still lack one with a bound nucleotide. When the C-terminal domain of L. casei HprK/P is superimposed with the corresponding domain of the PEP carboxykinase (18), the phosphate ion in the P-loop of HprK/P superimposes with the β-phosphate of ATP in PEP carboxykinase (50). Thus, nucleotide-dependent phosphate transfer is likely to involve the same catalytic groups as in the pyrophosphate-dependent reaction. However, we already mentioned that ATP cannot bind to the form of HprK/P observed in the X-ray structures, at least not in the conformation observed in other P-loop-containing proteins such as adenylate kinase. This is also true of the ATP conformation observed in PEP carboxykinase. A proper description of the reaction will require new X-ray structures including the nucleotide and transition state analogs (48), detailing the conformational changes that must accompany ATP binding and may explain the activity regulation by fructose-1,6-bisphosphate.
DIVERSITY OF HprK/P
Functional and structural changes in HprK/P can be related to sequence changes and to physiological differences between bacteria. The three proteins for which the structure has been determined come from low-G+C gram-positive bacteria, and they are very similar. Among these, M. pneumoniae HprK/P has the more divergent sequence. This is most obvious in its N-terminal domain, which lacks two of the three arginines that bind inorganic phosphate in S. xylosus. No homologue of CcpA, the catabolite repressor protein of gram-positive bacteria, has been found in sequenced mycoplasmal genomes (1), suggesting that HprK/P and P-Ser-HPr play a different role. Compared to that of B. subtilis, the HprK/P of M. pneumoniae is active as a kinase at much lower ATP concentrations and is stimulated by lower concentrations of fructose-1,6-bisphosphate (47).
The differences are greater between HprK/Ps from gram-positive and gram-negative bacteria. Enteric bacteria such as E. coli and S. enterica serovar Typhimurium have no HprK/P, but homologues are found in other gram-negative bacteria, including neisseriae, bordetellae, and xylellae, as well as in spirochetes, green sulfur bacteria, fusobacteria, and others (4, 17, 23). These bacteria do not have CcpA, and most lack a functional phosphotransferase system, yet they have HPr and HprK/P (4). Based on the gene arrangement in several of these organisms, HprK/P and P-Ser-HPr were suggested to control the activity of sigma factors resembling E. coli RpoN (4) or of transcription regulators belonging to the EnvZ/OmpR family of two-component systems (4, 23). Like its gram-positive homologue, the HprK/P of gram-negative bacteria probably catalyzes both the kinase and the phosphorylase reactions, although this has so far been demonstrated only for neisseriae (4).
HprK/P-containing gram-negative organisms have an HPr sequence that strongly resembles that of gram-positive bacteria near Ser46 and differs from that of E. coli. Their HprK/P contains all the elements of the active site (P-loop, catalytic His and Asp residues, signature sequence), but the similarity to gram-positive HprK/Ps is otherwise weak. The divergence is largest in the N-terminal domain. The arginine residues binding inorganic phosphate in S. xylosus are absent in gram-negative HprK/Ps. Those of α-proteobacteria, including Agrobacterium tumefaciens and Sinorhizobium meliloti, lack the domain altogether (4, 23). Even more surprising, the genome of Fusobacterium nucleatum encodes a protein composed of two complete HprK/P domains fused in tandem (4). The two domains exhibit 22% sequence identity compared to each other and about 30% identity compared to L. casei HprK/P. The N-terminal part lacks most of the active-site elements, including a proper Walker motif A, but it carries the equivalent of Arg245, the arginine of the flexible loop which, in L. casei, contributes to the active site of the neighboring subunit by interacting with the phosphoserine on HPr. The P-loop lysine, which is essential to the phosphorylase function, and the first glycine are also conserved in the N-terminal copy.
This unusual organization may lead to a separation of the two antagonistic activities in F. nucleatum, with the C-terminal copy carrying the kinase and the N-terminal copy carrying the phosphorylase. Alternatively, F. nucleatum HprK/P may have a different quaternary structure, where the N-terminal copy can contribute to HPr binding and provide the equivalent of Arg245 to catalytic activities carried by the C-terminal copy. These hypotheses are presently being tested in our laboratories. Given these differences, it will be interesting to determine the structure of HprK/Ps from gram-negative bacteria, especially from α-proteobacteria and F. nucleatum.
PREDICTING PROTEIN-PROTEIN INTERACTIONS: HprK/P AND THE CAPRI EXPERIMENT
Assessing protein-protein docking predictions. Having structures for HPr and HprK/P, could we have predicted their mode of interaction by computational methods instead of crystallizing them together? Recent reviews present docking procedures that have been developed for this purpose (20, 55). The algorithms rotate and translate ligand molecules to generate a large number of candidate complexes and retain those that score favorably in terms of shape complementarity, hydrogen bonding, and electrostatic and hydrophobic interactions.
The CAPRI (critical assessment of predicted interactions; http://capri.ebi.ac.uk) experiment was designed to test predictions made in this way. In CAPRI, the prediction is made blindly and assessed by comparison to an unpublished experimental structure. Round 1 of CAPRI began in the summer of 2001, with the HPr-HprK/P complex as its first target. Participating groups were given the atomic coordinates of the L. casei HprK/P C-terminal domain (15) and of B. subtilis HPr (31). Two other targets, two antibody Fab fragments bound to large viral antigens, were also submitted for prediction. A few weeks later, a total of 69 atomic models of the HPr-HprK/P complex and equivalent numbers of models of the other two targets were returned.
The predictions were evaluated by comparison to the X-ray structures of the complexes, which had just been completed (34). The receptor component (HprK/P or the viral antigen) of the models was superimposed onto the target X-ray structures, and the positions of the ligand component (HPr or the Fab) were compared. A ligand misorientation angle, θ, and the root-mean-square distance, _I_rmsd, of interface atoms in the model and the target were calculated. We also evaluated the capacity of the procedures to predict which residues make the contacts (epitope prediction) and which interactions they make (pair prediction). This was done by comparing lists of residues and residue pairs forming the interface in the models and the X-ray structures and calculating the fraction of the contact residues (fR for the receptor, fL for the ligand) and residue pairs (_f_pair) that was correctly predicted in the models.
Prediction of the HPr-HprK/P complex. Table 2 summarizes results obtained on the HPr-HprK/P complex (T01) and the two other targets. A more complete analysis, including results of a later round of CAPRI, is given in reference 34. A “high-quality” model had to have an _I_rms of less than 1 Å. No predictor achieved this, but several submitted “medium-quality” models of T02 and T03, where most of the viral epitope and over 30% of the contacts with antibody residues were correctly predicted. In contrast, the best models of the HPr-HprK/P complex were just “acceptable”: the ligand (HPr) was misplaced by 2 to 4 Å and grossly misoriented relative to the receptor (HprK/P), but about half of the contact residues were correctly predicted on both proteins and also 20 to 30% of the pairs.
TABLE 2.
Targets and predictions in (CAPRI) round 1
| Target complex | Ligand | Receptor | No. of predictor groups | No. of models sub- mitteda | No. of models of qualityb: | Geometry of best modelc | Fraction of correctly predicted interactionsd | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Medium | Accept- able | Ligand mis- orientation θ (°) | Interface RMSD (Å) | Ligand residues (fL) | Receptor residues (fR) | Residue pairs (_f_pair) | |||||
| T01 | HPr | HprK/P | 16 | 69 | 0 | 0 | 8 | 34 | 2.5 | 0.6 | 0.4 | 0.2 |
| T02 | Fab | Rotavirus VP6 | 15 | 70 | 0 | 1 | 6 | 5 | 1.2 | 1.0 | 1.0 | 1.0 |
| T03 | Fab | Influenza virus hemagglutinin | 13 | 62 | 0 | 2 | 0 | 12 | 1.7 | 0.5 | 0.8 | 0.7 |
In the past, antigen-antibody complexes have proved difficult to predict even with the help of biochemical data. Such data were available for T02 but not T03, and the success on that target validated the docking procedures. In comparison, the HPr-HprK/P complex should have been an easy one, as Ser46 of HPr, the site of phosphorylation, had to be close to the P-loop of HprK/P. We used this information (15) to draw a tentative model of the complex, but the orientation of HPr was incorrect, as in the CAPRI predictions. This appears to be due to the movement of the C-terminal HprK/P helix. Docking procedures which could handle the side chain rotations and small main chain movements that occur in HPr (or in T02 and T03) failed with a larger conformation change. For HprK/P, the predictors expected changes in the flexible loop and dealt with it either by deleting the loop or by modeling its conformation, but the loop was still disordered in T01, and it did not affect the predictions. In contrast, the C-terminal helix cannot interact with HPr in the position seen in the free L. casei structure, and important contacts were missed. After the X-ray structure of the complex was disclosed, its movement was modeled by one of the predictor groups, who obtained a significantly better prediction by docking HPr in HprK/P while leaving the C-terminal helix free to rotate (46).
The HPr-HprK/P complex has been teaching important lessons to computational biologists interested in protein-protein interaction. Predicting simple movements like C-terminal helix rotation is now on their agenda, but they must complement the docking approach with others that are less sensitive to conformation changes. One approach uses residue conservation in sequence families. If a conserved site is on the protein surface, it is likely to interact with ligands (51). Two CAPRI predictors tested this approach in conjunction with docking. With HPr, which has many partners, the conservation of surface residues did not say much beyond what was known from site-directed mutagenesis (56), but HprK/P showed a patch of conserved residues that surrounded its active site. The two groups of CAPRI predictors who used this information identified up to 20 of the 25 residues in contact with HPr, and they did significantly better than other groups, although HPr was still misoriented in their models (3).
CONCLUSION
As in eukaryotes, protein phosphorylation in bacteria plays an important role in the regulation of many different cellular functions. Nevertheless, HprK/P, the first bacterial serine protein kinase for which the structure has been determined, is unrelated to the eukaryotic kinases. HprK/P belongs to another large structural family, the P-loop-containing kinases, which has many members in all phyla, about 350 in Saccharomyces cerevisiae and 900 in humans (29). P-loop-containing kinases have been assumed to phosphorylate only small molecules, but the example of HprK/P suggests that some may have proteins as substrates, potentially defining novel cellular signal transduction pathways. Another interesting feature of HprK/P is its capacity to use both pyrophosphate and ATP as phosphate donors. It shares this property with a few other kinases (12), but in the case of HprK/P, it is the reverse reaction that seems to be of physiological importance, the phosphorolysis of the phosphoserine yielding pyrophosphate. As a consequence, the P-Ser-HPr-dependent catabolite repression regulatory pathway does not require a protein phosphatase, although a pyrophosphatase seems to be needed. The two antagonistic catalytic activities are carried out at the same active site by a rather small protein (less than 200 residues in the C-terminal domain). Their association may constitute a mode of regulation which developed earlier in evolution than those presently known in eukaryotic cells. It is well worth determining whether it has been retained only in bacteria, or whether eukaryotic cells have equivalents to the kinase/phosphorylase double function of HprK/P.
Acknowledgments
This work was supported by the Association pour la Recherche Contre le Cancer and by the CNRS. S.F. is grateful to the Ligue Nationale de Lutte Contre le Cancer for a personal fellowship.
REFERENCES
- 1.Allen, G. S., K. Steinhauer, W. Hillen, J. Stulke, and R. G. Brennan. 2003. Crystal structure of HPr kinase/phosphatase from Mycoplasma pneumoniae. J. Mol. Biol. 326**:**1203-1217. [DOI] [PubMed] [Google Scholar]
- 2.Audette, G. F., R. Engelmann, W. Hengstenberg, J. Deutscher, K. Hayakawa, J. W. Quail, and L. T. Delbaere. 2000. The 1.9 Å resolution structure of phospho-serine 46 HPr from Enterococcus faecalis. J. Mol. Biol. 303**:**545-553. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Zeev, E., A. Berchanski, A. Heifetz, B. Shapira, and M. Eisenstein. 2003. Prediction of the unknown: Inspiring experience with the CAPRI experiment. Proteins **52:**41-46. [DOI] [PubMed]
- 4.Boël, G., I. Mijakovic, A. Mazé, S. Poncet, M.-K. Taha, M. Larribe, E. Darbon, A. Khemiri, A. Galinier, and J. Deutscher. 2003. Transcriptional regulators potentially controlled by HPr kinase/phosphorylase in gram-negative bacteria. J. Mol. Microbiol. Biotechnol. 5**:**206-215. [DOI] [PubMed]
- 5.Chen, R., W. Tong, J. Mintseris, L. Li, and Z. Weng. 2003. ZDOCK predictions for the CAPRI challenge. Proteins **52:**68-73. [DOI] [PubMed]
- 6.Deutscher, J., A. Galinier, and I. Martin-Verstraete. 2001. Carbohydrate transporters and regulation of carbohydrate uptake and metabolism, p. 129-150. In A. L. Sonenschein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 7.Deutscher, J., U. Kessler, and W. Hengstenberg. 1985. Streptococcal phosphoenolpyruvate:sugar phosphotransferase system: purification and characterization of a phosphoprotein phosphatase which hydrolyzes the phosphoryl bond in seryl-phosphorylated histidine-containing protein. J. Bacteriol. 163**:**1203-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutscher, J., E. Küster, U. Bergstedt, V. Charrier, and W. Hillen. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol. Microbiol. 15**:**1049-1053. [DOI] [PubMed] [Google Scholar]
- 9.Deutscher, J., B. Pevec, K. Beyreuther, H. H. Kiltz, and W. Hengstenberg. 1986. Streptococcal phosphoenolpyruvate-sugar phosphotransferase system: amino acid sequence and site of ATP-dependent phosphorylation of HPr. Biochemistry 25**:**6543-6551. [DOI] [PubMed] [Google Scholar]
- 10.Deutscher, J., and M. H. Saier, Jr. 1983. ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 80**:**6790-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dossonnet, V., V. Monedero, M. Zagorec, A. Galinier, G. Perez-Martinez, and J. Deutscher. 2000. Phosphorylation of HPr by the bifunctional HPr Kinase/P-ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion. J. Bacteriol. 182**:**2582-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duclos, B., E. Vaganay, M. Dadssi, and A. J. Cozzone. 1996. Pyrophosphate is a source of phosphoryl groups for Escherichia coli protein phosphorylation. FEMS Microbiol. Lett. 145**:**49-54. [DOI] [PubMed] [Google Scholar]
- 13.Fernández-Recio, J., M. Totrov, and R. Abagyan. 2003. ICM-DISCO docking by global energy optimization with fully flexible side-chains. Proteins **52:**113-117. [DOI] [PubMed]
- 14.Fieulaine, S., S. Morera, S. Poncet, I. Mijakovic, A. Galinier, J. Janin, J. Deutscher, and S. Nessler. 2002. X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr. Proc. Natl. Acad. Sci. USA 99**:**13437-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fieulaine, S., S. Morera, S. Poncet, V. Monedero, V. Gueguen-Chaignon, A. Galinier, J. Janin, J. Deutscher, and S. Nessler. 2001. X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain. EMBO J. 20**:**3917-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita, Y., Y. Miwa, A. Galinier, and J. Deutscher. 1995. Specific recognition of the _Bacillus subtilis gnt cis_-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol. Microbiol. 17**:**953-960. [DOI] [PubMed] [Google Scholar]
- 17.Galinier, A., M. Kravanja, R. Engelmann, W. Hengstenberg, M. C. Kilhoffer, J. Deutscher, and J. Haiech. 1998. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc. Natl. Acad. Sci. USA 95**:**1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galinier, A., J. P. Lavergne, C. Geourjon, S. Fieulaine, S. Nessler, and J. M. Jault. 2002. A new family of phosphotransferases with a P-loop motif. J. Biol. Chem. 277**:**11362-11367. [DOI] [PubMed] [Google Scholar]
- 19.Gordon, E., B. Flouret, L. Chantalat, J. van Heijenoort, D. Mengin-Lecreulx, and O. Dideberg. 2001. Crystal structure of UDP-N-acetylmuramoyl-l-alanyl-d-glutamate: meso-diaminopimelate ligase from Escherichia coli. J. Biol. Chem. 276**:**10999-11006. [DOI] [PubMed] [Google Scholar]
- 20.Halperin, I., B. Ma, H. Wolfson, and R. Nussinov. 2002. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins 47**:**409-443. [DOI] [PubMed] [Google Scholar]
- 21.Hanson, K. G., K. Steinhauer, J. Reizer, W. Hillen, and J. Stulke. 2002. HPr kinase/phosphatase of Bacillus subtilis: expression of the gene and effects of mutations on enzyme activity, growth and carbon catabolite repression. Microbiology 148**:**1805-1811. [DOI] [PubMed] [Google Scholar]
- 22.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a _trans_-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol. Microbiol. 5**:**575-584. [DOI] [PubMed] [Google Scholar]
- 23.Hu, K. Y., and M. H. J. Saier. 2002. Phylogeny of phosphoryl transfer proteins of the phosphoenolpyruvate-dependent sugar-transporting phosphotransferase system. Res. Microbiol. 153**:**405-415. [DOI] [PubMed] [Google Scholar]
- 24.Janin, J. 2002. Welcome to CAPRI: a critical assessment of predicted interactions. Proteins **47:**257. [DOI] [PubMed]
- 25.Janin, J., K. Henrick, J. Moult, L. Ten Eyck, M. Sternberg, S. Vajda, I. Vakser, and S. J. Wodak. 2003. CAPRI: a critical assessment of predicted interactions. Proteins **52:**2-9. [DOI] [PubMed]
- 26.Jault, J. M., S. Fieulaine, S. Nessler, P. Gonzalo, A. Di Pietro, J. Deutscher, and A. Galinier. 2000. The HPr kinase from Bacillus subtilis is a homo-oligomeric enzyme which exhibits strong positive cooperativity for nucleotide and fructose-1,6-bisphosphate binding. J. Biol. Chem. 275**:**1773-1780. [DOI] [PubMed] [Google Scholar]
- 27.Jones, B. E., V. Dossonet, E. Küster, W. Hillen, J. Deutscher, and R. E. Klevit. 1997. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J. Biol. Chem. 272**:**26530-26535. [DOI] [PubMed] [Google Scholar]
- 28.Kravanja, M., R. Engelmann, V. Dossonnet, M. Blüggel, H. E. Meyer, R. Frank, A. Galinier, J. Deutscher, N. Schnell, and G. Hengstenberg. 1999. The hprK gene of Enterococcus faecalis encodes a novel bifunctional enzyme: the HPr kinase/phosphatase. Mol. Microbiol. 31**:**59-66. [DOI] [PubMed] [Google Scholar]
- 29.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409**:**860-921. [DOI] [PubMed] [Google Scholar]
- 30.Lavergne, J. P., J. M. Jault, and A. Galinier. 2002. Insights into the functioning of Bacillus subtilis HPr kinase/phosphatase: affinity for its protein substrates and role of cations and phosphate. Biochemistry 41**:**6218-6225. [DOI] [PubMed] [Google Scholar]
- 31.Liao, D. I., and O. Herzberg. 1994. Refined structures of the active Ser83→Cys and impaired Ser46→Asp histidine-containing phosphocarrier proteins. Structure 2**:**1203-1216. [DOI] [PubMed] [Google Scholar]
- 32.Marquez, J. A., S. Hasenbein, B. Koch, S. Fieulaine, S. Nessler, R. B. Russell, W. Hengstenberg, and K. Scheffzek. 2002. Structure of the full-length HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution: Mimicking the product/substrate of the phospho transfer reactions. Proc. Natl. Acad. Sci. USA 99**:**3458-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matte, A., H. Goldie, R. M. Sweet, and L. T. Delbaere. 1996. Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase: a new structural family with the P-loop nucleoside triphosphate hydrolase fold. J. Mol. Biol. 256**:**126-143. [DOI] [PubMed] [Google Scholar]
- 34.Mendez, R., R. Leplae, L. de Maria, and S. J. Wodak. 2003. Assessment of blind predictions of protein-protein interactions: current status of docking methods. Proteins **52:**51-67. [DOI] [PubMed]
- 35.Mijakovic, I., S. Poncet, A. Galinier, V. Monedero, S. Fieulaine, J. Janin, S. Nessler, J. A. Marquez, K. Scheffzek, S. Hasenbein, W. Hengstenberg, and J. Deutscher. 2002. Pyrophosphate-producing protein dephosphorylation by HPr kinase/phosphorylase: a relic of early life? Proc. Natl. Acad. Sci. USA 99**:**13442-13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miwa, Y., A. Nakata, A. Ogiwara, M. Yamamoto, and Y. Fujita. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 28**:**1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monedero, V., O. P. Kuipers, E. Jamet, and J. Deutscher. 2001. Regulatory functions of serine-46-phosphorylated HPr in Lactococcus lactis. J. Bacteriol. 183**:**3391-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monedero, V., S. Poncet, I. Mijakovic, S. Fieulaine, V. Dossonnet, I. Martin-Verstraete, S. Nessler, and J. Deutscher. 2001. Mutations lowering the phosphatase activity of HPr kinase/phosphatase switch off carbon metabolism. EMBO J. 20**:**3928-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39**:**1366-1381. [DOI] [PubMed] [Google Scholar]
- 40.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247**:**536-540. [DOI] [PubMed] [Google Scholar]
- 41.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57**:**543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramström, H., S. Sanglier, E. Leize-Wagner, C. Philippe, A. Van Dorsselaer, and J. Haiech. 2003. Properties and regulation of the bifunctional enzyme HPr kinase/phosphatase in Bacillus subtilis. J. Biol. Chem. 278**:**1174-1185. [DOI] [PubMed] [Google Scholar]
- 43.Reizer, J., C. Hoischen, F. Titgemeyer, C. Rivolta, R. Rabus, J. Stulke, D. Karamata, M. H. Saier, and W. Hillen. 1998. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol. Microbiol. 27**:**1157-1169. [DOI] [PubMed] [Google Scholar]
- 44.Russell, R. B., J. A. Marquez, W. Hengstenberg, and K. Scheffzek. 2002. Evolutionary relationship between the bacterial HPr kinase and the ubiquitous PEP-carboxykinase: expanding the P-loop nucleotidyl transferase superfamily. FEBS Lett. 517**:**1-6. [DOI] [PubMed] [Google Scholar]
- 45.Saraste, M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop-a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 15**:**430-434. [DOI] [PubMed] [Google Scholar]
- 46.Schneidman-Duhovny, D., Y. Inbar, V. Polak, M. Shatsky, I. Halperin, H. Benyamini, A. Barzilai, O. Dror, N. Haspel, R. Nussinov, and H. J. Wolfson. 2003. Taking geometry to its edge: fast unbound rigid (and hinge-bent) docking. Proteins **52:**107-112. [DOI] [PubMed]
- 47.Steinhauer, K., T. Jepp, W. Hillen, and J. Stulke. 2002. A novel mode of control of Mycoplasma pneumoniae HPr kinase/phosphatase activity reflects its parasitic lifestyle. Microbiology 148**:**3277-3284. [DOI] [PubMed] [Google Scholar]
- 48.Sudom, A. M., L. Prasad, H. Goldie, and L. T. Delbaere. 2001. The phosphoryl-transfer mechanism of Escherichia coli phosphoenolpyruvate carboxykinase from the use of AlF(3). J. Mol. Biol. 314**:**83-92. [DOI] [PubMed] [Google Scholar]
- 49.Tari, L., A. Matte, H. Goldie, and L. T. J. Delbaere. 1997. Mg2+-Mn2+ clusters in enzyme-catalyzed phosphoryl-transfer reactions. Nat. Struct. Biol. 4**:**990-994. [DOI] [PubMed] [Google Scholar]
- 50.Tari, L. W., A. Matte, U. Pugazhenthi, H. Goldie, and L. T. Delbaere. 1996. Snapshot of an enzyme reaction intermediate in the structure of the ATP-Mg2+-oxalate ternary complex of Escherichia coli PEP carboxykinase. Nat. Struct. Biol. 3**:**355-363. [DOI] [PubMed] [Google Scholar]
- 51.Valencia, A., and F. Pazos. 2002. Computational methods for the prediction of protein interactions. Curr. Opin. Struct. Biol. 12**:**368-373. [DOI] [PubMed] [Google Scholar]
- 52.Vonrhein, C., H. Bönisch, G. Schäfer, and G. E. Schulz. 1998. The structure of a trimeric archaeal adenylate kinase. J. Mol. Biol. 282**:**167-179. [DOI] [PubMed] [Google Scholar]
- 53.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1**:**945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wittekind, M., J. Reizer, J. Deutscher, M. H. Saier, and R. E. Klevit. 1989. Common structural changes accompany the functional inactivation of HPr by seryl phosphorylation or by serine to aspartate substitution. Biochemistry 28**:**9908-9912. [DOI] [PubMed] [Google Scholar]
- 55.Wodak, S. J., and J. Janin. 2002. Structural basis of macromolecular recognition. Adv. Protein Chem. 61**:**9-73. [DOI] [PubMed] [Google Scholar]
- 56.Zhu, P. P., O. Herzberg, and A. Peterkofsky. 1998. Topography of the interaction of HPr(Ser) kinase with HPr. Biochemistry 37**:**11762-11770. [DOI] [PubMed] [Google Scholar]