NCBI Reference Sequence Project: update and current status (original) (raw)
Abstract
The goal of the NCBI Reference Sequence (RefSeq) project is to provide the single best non-redundant and comprehensive collection of naturally occurring biological molecules, representing the central dogma. Nucleotide and protein sequences are explicitly linked on a residue-by-residue basis in this collection. Ideally all molecule types will be available for each well-studied organism, but the initial database collection pragmatically includes only those molecules and organisms that are most readily identified. Thus different amounts of information are available for different organisms at any given time. Furthermore, for some organisms additional intermediate records are provided when the genome sequence is not yet finished. The collection is supplied by NCBI through three distinct pipelines in addition to collaborations with community groups. The collection is curated on an ongoing basis. Additional information about the NCBI RefSeq project is available at http://www.ncbi.nih.gov/RefSeq/.
BACKGROUND
RefSeq is unique in providing a large multi-species curated sequence database that explicitly links chromosomal, transcript and protein information; this establishes a critical baseline for integrating sequence, genetic, expression and functional information into a single consistent framework. All RefSeq records include attribution to the original sequence data. When a molecule is represented by multiple GenBank sequences, an effort is made to select the ‘best’ sequence to instantiate as a RefSeq. The goal is to avoid mutations, sequencing errors and cloning artifacts. Should a RefSeq be identified with a problem of this type, it is corrected. Sequences are computationally validated to confirm that the genomic sequence corresponding to an annotated mRNA feature does match the mRNA sequence record and that coding region features really can be translated into the corresponding protein sequence.
RefSeq offers a significant advantage for gene characterization, database searching and sequence identification, whether by BLAST, text or accession queries, or inclusion in a local customized database. The RefSeq collection may include alternatively spliced transcripts that share some identical exons, or identical proteins expressed from these alternatively spliced transcripts, or close paralogs or homologs. It has the advantage of a representing an objective and experimentally verifiable definition of ‘non-redundant’ in providing one example of each natural biomolecule per organism.
Thus the RefSeq collection establishes a consistent baseline and clear model of the central dogma. The RefSeq collection supports:
- Facile identification of the sequence standard for a genome, transcript, or protein
- Genome annotation
- Comparative genomics
- Reduction of redundancy in clustering approaches
- Analysis and comparison
- Unambiguous association of functional information
- Navigation to additional sources of information
Distinct processing pipelines are used to provide the RefSeq collection:
- Computed annotation pipeline
- Entrez genomes pipeline
- LocusLink supported pipeline
- Collaboration
These pipelines are overlapping and the RefSeq dataset for some genomes may be provided by more than one pipeline.
Computed Annotation Pipeline
The Computed Annotation Pipeline relies on automated computation to provide scaffolds, transcripts and proteins. Transcript and protein records may represent an ab initio prediction with varying levels of transcript or protein homology support, or they may be fully supported by GenBank transcript data. These records are not subject to incremental individual updates or to direct curation. They are sometimes more predicted in nature and represent an approach that complements manual curation in providing comprehensive genome annotation.
Entrez Genomes Pipeline
NCBI's Entrez Genomes database represents a collection of complete, or nearly complete, genomes and chromosomes (2). It is divided into six large taxonomic groups: Archaea, Eubacteria, Eukaryotae, Viroids, Viruses and Plasmids. Entrez Genomes RefSeq records include genomic, transcript and protein records and are provided by collaboration, in-house automatic processing and in-house curation. The Entrez Genomes web site includes custom displays, analysis and tools for some genomes.
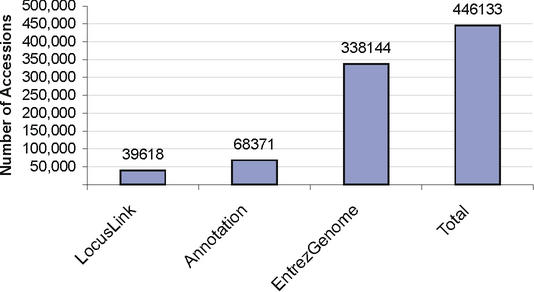
In general, these RefSeq records undergo an initial automated validation step before being released. The resulting record is a copy of a GenBank entry, but validation may make some corrections and provides more consistent feature annotation. Records provided via collaboration have a status of ‘Reviewed’ and attribution to the collaborating group. Records provided by in-house processing have a ‘Provisional’ or ‘Reviewed’ status. This pipeline has provided RefSeq records for over 1780 distinct organisms and makes significant contribution to the NCBI RefSeq collection (Fig. 1).
Figure 1.
Number of RefSeq proteins contributed by the three source pipelines (as of August, 2002).
LocusLink pipeline—new features and current statistics
The Entrez Genomes and LocusLink supported pipelines are similar in that they both make use of automatic computation to validate records and are also subject to ongoing in-house curation of individual records. Both of these pipelines welcome opportunities to collaborate. These two pipelines differ in infrastructure, targeted scope and processing details. The curated RefSeq set is used as one of several reagents in the annotation pipeline; the human RefSeq collection is targeted for focused in-house curation.
Molecule types
The collection has been expanded to include non-coding transcripts such as those derived from structural RNA genes and transcribed pseudogenes. See Table 1 for a complete listing of the available molecule types, corresponding accession prefix and pipeline source.
Table 1. RefSeq accession prefixes, molecule types, originating pipeline and annotated status categories.
| Accession prefix | Molecule | Pipeline | Status category |
|---|---|---|---|
| NT_ | Genomic | Computed annotation | Model |
| NW_ | Genomic | Computed annotation | Model |
| XM_ | mRNA | Computed annotation | Model |
| XR_ | RNA | Computed annotation | Model |
| XP_ | Protein | Computed annotation | Model |
| NC_ | Genomic | Entrez genomes | Provisional, Reviewed |
| NG_ | Genomic | LocusLink | Provisional, Reviewed |
| NM_ | mRNA | LocusLink Entrez genomes | Provisional, Predicted, Reviewed |
| NR_ | RNA | LocusLink | Provisional, Reviewed |
| NP_ | Protein | LocusLink Entrez genomes | Provisional, Predicted, Reviewed |
Status categories
RefSeq records include annotation that provides a general indication of their curation status and reliability. Records generated via the automated annotation pipeline are annotated as ‘Model RefSeq’ whereas those contributed by the curation pipelines may be annotated as provisional (not yet reviewed), predicted (transcript or protein is not fully supported), or reviewed. Reviewed records are the most highly curated and effort has been made to ensure the quality and comprehensive coverage of the sequence itself as well as to apply descriptive information that leads the user to functionally relevant data (publications, names, summaries). RefSeq records annotated as predicted do have some level of support; they do not merely instantiate ab initio predictions (see Table 1).
Organisms
The LocusLink pipeline now provides RefSeq records for Danio rerio (zebrafish) and Drosophila melanogaster in addition to records for human, mouse and rat. Gene-to-sequence associations for human, mouse and rat are curated both in-house and in collaboration with the Human Gene Nomenclature Committee (3), OMIM (4), the Mouse Genome Informatics group (5), and the Rat Genome Database (6). Records for Drosophila are provided in collaboration with FlyBase (7). Collaboration with the zebrafish community ZFIN database (via LocusLink) supports RefSeq annotation (8).
Growth
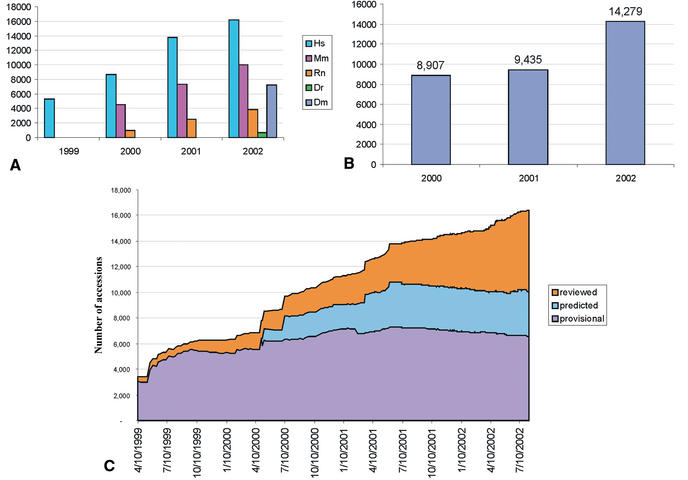
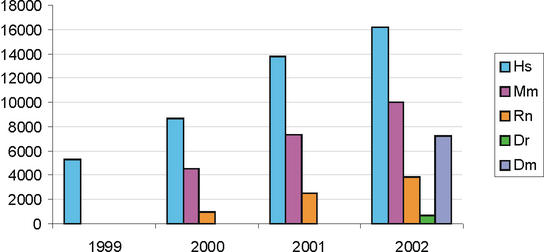
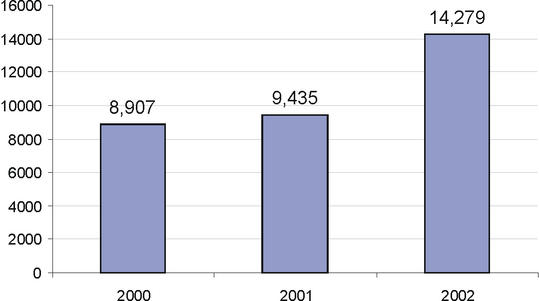
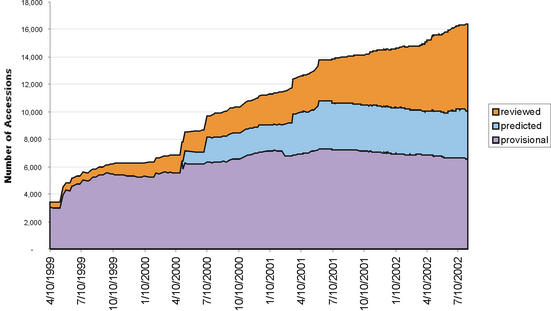
The LocusLink pipeline provided over 14 000 new RefSeq records between July 2001 and July 2002 (Fig. 2). Much of this growth was realized by the addition of zebrafish and Drosophila records (692 and 7243 respectively). The annual growth trend by organism and by new accessions added is presented in Figure 2A and B; the annual growth trend of the manually curated records (with annotated status of Reviewed) is presented in Figure 2C.
Figure 2.
Annual growth trends for the LocusLink supported RefSeq collection. (A) The number of RefSeq records (NM accessions) publicly available on July 1 of 1999 through 2002 for human (Hs), mouse (Mm), rat (Rn), zebrafish (Dr) and fly (Dm). These numbers include genes for which multiple reference sequences are provided to represent splice variants and their products. As of July 2002, 37919 RefSeqs were available; this includes 16142 human, 10004 mouse, 3838 rat, 692 zebrafish and 7243 Drosophila RefSeq records. (B) New growth. The number of new accessions (NM) added to the collection per 12 month window for the sum of human, mouse, rat, zebrafish and fly. (C) Distribution of human RefSeq records in different status categories over the same time window.
Manual curation improves the RefSeq collection in several ways including:
- Extension of the transcript
- Correction of sequence artifacts (vector, linker sequence)
- Provision of additional records representing alternative splice products
- Functional (descriptive) annotation of the nucleotide and protein record
- Representing the correct gene name-to-sequence association
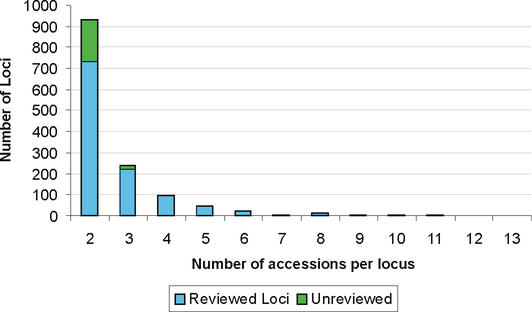
Alternate transcript records are provided for over 1368 genes, of these the vast majority (930) instantiate a single additional transcript variant (see Fig. 3). Two loci have instantiated a larger number of alternate splice forms; 18 variants are provided for the DMD gene, in collaboration with Dr den Dunnen and 19 variants are provided for the collagen gene COL13A1.
Figure 3.
Number of splice variant records provided per locus. The manual curation effort instantiates additional splice variants as a new accession number when the CDS is full-length and when there is some level of support either in available sequence data or in the literature for the existence of the new splice form. Because of these conservative criteria, the RefSeq collection under-represents the number of alternate splice forms that may occur. Additional records are provided primarily through the manual curation process, however, a smaller number are made available prior to the full review process following preliminary sequence review.
REFERENCES
- 1.Pruitt K.D., Katz,K.S., Sicotte,H. and Maglott,D.R. (2000) Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet., 16, 44–47. [DOI] [PubMed] [Google Scholar]
- 2.Tatusova T.A., Karsch-Mizrachi,I. and Ostell,J.A. (1999) Complete genomes in WWW Entrez: data representation and analysis. Bioinformatics, 15, 536–543. [DOI] [PubMed] [Google Scholar]
- 3.White J.A., McAlpine,P.J., Antonarakis,S., Cann,H., Eppig,J.T., Frazer,K., Frezal,J., Lancet,D., Nahmias,J., Pearson,P., Peters,J., Scott,A., Scott,H., Spurr,N., Talbot,C. Jr and Povey,S. (1997) Guidelines for human gene nomenclature (1997). HUGO Nomenclature Committee. Genomics, 45, 468–471. [DOI] [PubMed] [Google Scholar]
- 4.Hamosh A., Scott,A.F., Amberger,J., Valle.D. and McKusick,V.A. (2000) Online Mendelian Inheritance in Man (OMIM). Hum. Mutat., 15, 57–61. [DOI] [PubMed] [Google Scholar]
- 5.Blake J.A., Eppig,J.T., Richardson,J.E. and Davisson,M.T. (2000) The Mouse Genome Database (MGD): expanding genetic and genomic resources for the laboratory mouse. The Mouse Genome Database Group. Nucleic Acids Res., 28, 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Twigger S., Lu,J., Shimoyama,M., Chen,D., Pasko,D., Long,H., Ginster.J., Chen,C.F., Nigam,R., Kwitek,A., Eppig,J., Maltais,L., Maglott,D., Schuler,G., Jacob,H. and Tonellato,P.J. (2002) Rat Genome Database (RGD): mapping disease onto the genome. Nucleic Acids Res., 30, 125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.FlyBase Consortium (1999) The FlyBase database of the Drosophila Genome Projects and community literature. The FlyBase Consortium. Nucleic Acids Res., 27, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westerfield M., Doerry,E., Kirkpatrick,A.E. and Douglas,S.A. (1999) Zebrafish informatics and the ZFIN database. Methods Cell Biol., 60, 339–355. [DOI] [PubMed] [Google Scholar]