Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression (original) (raw)
Abstract
Hox proteins are transcription factors involved in controlling axial patterning, leukaemias and hereditary malformations. Here, we show that HOXC10 oscillates in abundance during the cell cycle, being targeted for degradation early in mitosis by the ubiquitin-dependent proteasome pathway. Among abdominal-B subfamily members, the mitotic proteolysis of HOXC10 appears unique, since the levels of the paralogous HOXD10 and the related homeoprotein HOXC13 are constant throughout the cell cycle. When two destruction box motifs (D-box) are mutated, HOXC10 is stabilized and cells accumulate in metaphase. HOXC10 appears to be a new prometaphase target of the anaphase-promoting complex (APC), since its degradation coincides with cyclin A destruction and is suppressed by expression of a dominant-negative form of UbcH10, an APC-associated ubiquitin-conjugating enzyme. Moreover, HOXC10 co-immunoprecipitates the APC subunit CDC27, and its in vitro degradation is reduced in APC-depleted extracts or by competition with the APC substrate cyclin A. These data imply that HOXC10 is a homeoprotein with the potential to influence mitotic progression, and might provide a link between developmental regulation and cell cycle control.
Keywords: cell cycle/Hox/mitosis/origin of DNA replication/ubiquitin
Introduction
Hox proteins are transcription factors that assign positional identities along the embryonic body axis in animals ranging from arthropods to vertebrates (reviewed in Gehring et al., 1994). In mice and humans, there are 39 Hox genes arranged in four clusters (HoxA–D) on separate chromosomes. Based on sequence similarities and location within the clusters, Hox genes have been placed into 13 paralogous groups. The co-linearity of expression boundaries and position in the complex is conserved from flies to mammals (reviewed in Krumlauf, 1994). However, spatial co-linearity does not accurately describe Hox gene expression at later embryological stages, indicating that Hox proteins may also regulate structural protein synthesis (Godwin and Capecchi, 1998). As monomers, Hox proteins possess similar in vitro DNA-binding preferences. The diversity of Hox function in vivo is therefore crucially dependent on extradenticle/pre-B-cell leukaemia homeobox (PBX) and homothorax/MEIS cofactor families which target Hox proteins to a subset of their potential DNA-binding sites or modulate their transcriptional activation or repression (reviewed in Mann and Affolter, 1998). Depending on the cofactor, Hox proteins may also cause oncogenic transformation of haemopoietic cells. Mutations affecting Hox proteins have been involved in genetic malformations and spontaneous or experimentally induced leukaemias, suggesting that these diseases may result from defects of cell proliferation (reviewed in Boncinelli, 1997; Magli et al., 1997; van Oostveen et al., 1999; Abate-Shen, 2002). Homeotic transformation in vertebrates may be the consequence of differential growth and, similarly, modifications of local growth rates may account for Hox mutant limbs (reviewed in Duboule, 1995). Taken together, these observations suggest a direct role for Hox proteins in growth control and cell cycle progression, although the underlying mechanisms are still poorly understood.
The highly ordered progression of the eukaryotic cell cycle is achieved by a series of crucial events, which ensure faithful transmission of the genome. Irreversible directionality of cell cycle progression is achieved through proteolytic destruction of regulatory proteins via the ubiquitin–proteasome pathway (reviewed in Hochstrasser, 1996). Proteolysis of target proteins occurs within the 26S proteasome and is preceded by substrate polyubiquitylation through the sequential action of three enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2) and a ubiquitin ligase (E3). An important E3 enzyme in cell cycle control is the anaphase-promoting complex (APC) or cyclosome. APC triggers the metaphase to anaphase transition and exit from mitosis, and appears to play roles in G1 and in post-mitotic, differentiated cells (reviewed in Peters, 2002). In this process, APC is responsible for selective substrate recognition and positioning for transfer of activated ubiquitin by either one of its cognate E2 enzymes, called UbcH5 and UbcH10 in human cells. APC-mediated ubiquitylation depends on two rather poorly defined sequence elements in the substrate, the destruction box (D-box) and the KEN box that are found either singly or in combination in all APC substrates known to date (Glotzer et al., 1991; Pfleger and Kirschner, 2000). A complex regulatory network restricts APC activity towards diverse substrates to specific time windows, from cyclin A in prometaphase (den Elzen and Pines, 2001; Geley et al., 2001), cyclin B1 and securins in metaphase (Clute and Pines, 1999; Wakefield et al., 2000; Zur and Brandeis, 2001), to the spindle-associated protein Ase1 in anaphase (Juang et al., 1997). The most interesting feature here is that activation of the so-called mitotic spindle assembly checkpoint, which monitors the attachment of microtubules to kinetochores and delays the activation of the APC until all chromosomes are properly attached to the mitotic spindle, inhibits destruction of some APC substrates (namely securin and B-type cyclins), but not the degradation of others, such as cyclin A and Nek2A (den Elzen and Pines, 2001; Geley et al., 2001; Hames et al., 2001). This has been correlated to a so-called ‘A-type D-box’ that extends to residues C-terminal of the classical D-box and is found in these two proteins (den Elzen and Pines, 2001; Geley et al., 2001; Hames et al., 2001).
Using a yeast one-hybrid system, we previously have isolated HOXC10 and HOXC13 for their ability to bind to the human origin of DNA replication, mapped at the lamin B2 locus (lamin B2 origin) (De Stanchina et al., 2000). Since this origin shows a cell cycle-dependent in vivo DNA footprint (Abdurashidova et al., 1998), we have now investigated the levels of HOXC10 and other abdominal-B type Hox proteins during the cell cycle. Our data link HOXC10 with the destruction machinery controlling the cell cycle, and might therefore provide a connection between growth control and cell cycle progression.
Results
Cell cycle-dependent expression of HOXC10
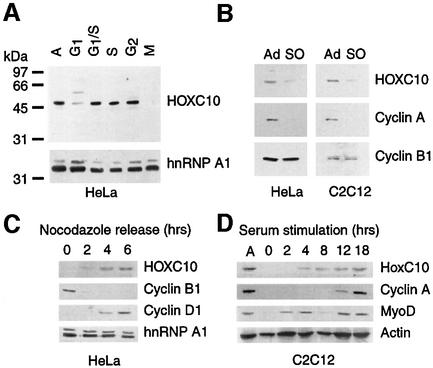
The levels of HOXC10 were analysed during the cell cycle by immunoblotting total extracts of chemically synchronized human HeLa cells with anti-HOXC10 antibodies (see figure S1, of the Supplementary data available at The EMBO Journal Online). As shown in Figure 1A and C, HOXC10 levels are reduced in early G1 phase, abundant from mid-G1 to G2 phases and become undetectable in mitosis. Interestingly, this mitotic disappearance is observed in the presence of nocodazole, which activates the spindle assembly checkpoint and thereby stabilizes proteins degraded at the metaphase to anaphase transition. We confirmed that the mitotic disappearance of HOXC10 is independent of cell line and artificial synchronization by culturing HeLa and C2C12 cells without nocodazole and comparing HOXC10 levels between adherent and shaken-off (mitotic) cells (Figure 1B). Upon nocodazole release of HeLa mitotic cells, HOXC10 arises ∼2 h later, slightly earlier than the appearance of cyclin D1 (Figure 1C). In quiescent mouse C2C12 cells (Figure 1D), Hoxc10 only becomes detectable 4 h after serum addition, slightly later than the mid-G1 MyoD peak (Kitzmann et al., 1998) and several hours earlier than cyclin A (reviewed in King et al., 1996a).
Fig. 1. HOXC10 oscillates during the cell cycle. (A) Asynchronous HeLa cells (A) were synchronized at the G1/S border by double thymidine block (G1/S) and released for 3 h into S phase (S) or synchronized by nocodazole and shaken to collect mitotic (M) and adherent (G2) cells or released in G1 phase for ∼3 h (G1). An anti-HOXC10 immunoblot is shown, whereas the anti-hnRNP A1 immunoblot is to control the loading. The Mr in kDa of pre-stained protein standards is shown alongside. (B) Asynchronous HeLa or C2C12 cells were harvested by shaking off (SO) mitotic or residual adherent (Ad) cells. The corresponding total lysates were immunoblotted with the indicated antibodies. (C) Kinetics (in hours) of HOXC10 during re-entry into G1 phase upon release of nocodazole block. On the same extracts, the levels of cyclin B1 and cyclin D1 were analysed to mark the M/G1 transition of the cell cycle. The hnRNP A1 immunoblot is to check the loading. (D) Time course (in hours) of HoxC10 in serum-stimulated quiescent C2C12 cells. MyoD and cyclin A mark the G1 progression and the beginning of S phase, respectively. The actin immunoblot is to control the loading.
Thus, HOXC10 accumulates from mid-G1 to G2 and disappears in M phase, presumably before the metaphase to anaphase transition, as well as in early G1 and in quiescent cells. This indicates that a member of the Hox family of developmental regulators can be subjected to cell cycle-regulated expression.
Mitotic HOXC10 disappearance parallels cyclin A degradation
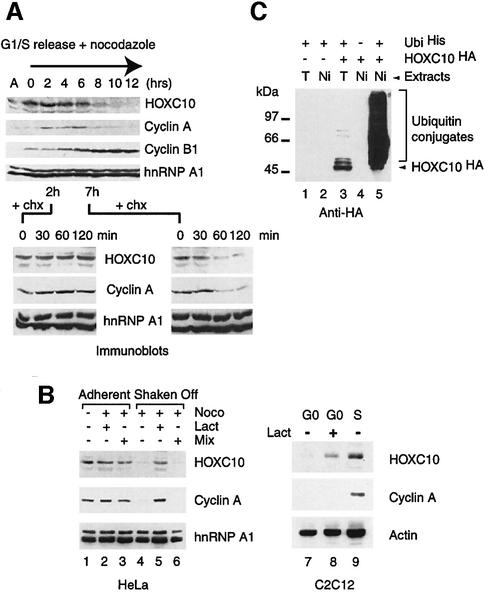
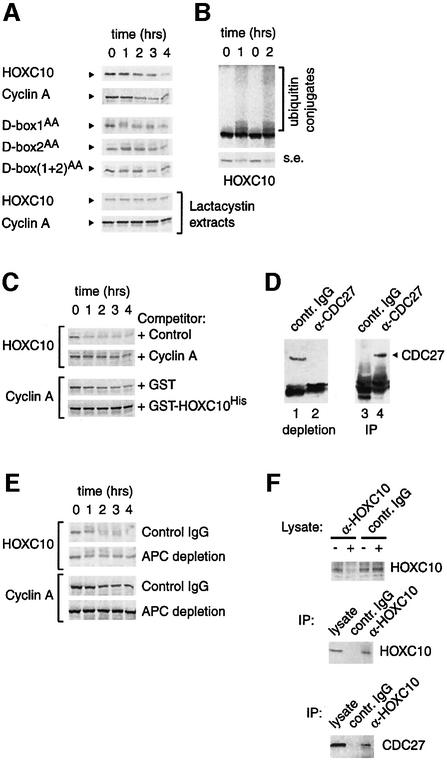
Northern blot analysis (see figure S2 of the Supplementary data) showed constant levels of Hoxc10 mRNA, suggesting that its mitotic disappearance is not caused by transcriptional downregulation. Therefore, mitotic proteolysis might be responsible for the drop in HOXC10 levels. To investigate this hypothesis further, HOXC10 oscillations in synchronized HeLa cells were compared with cyclin A and cyclin B1 (Figure 2A). Following the release of the G1/S block, HOXC10 is constantly detectable for up to 6 h and then progressively disappears, concomitant with an increase of the mitotic index. In this, HOXC10 disappearance parallels that of cyclin A (Figure 2A). In parallel experiments, aliquots of cells were collected 2 and 7 h after the release of the G1/S block and incubated in the presence of cycloheximide for up to 2 h to repress de novo synthesis of proteins. This revealed that HOXC10 becomes unstable in mitotic cells at the same time as cyclin A (Figure 2A). This observation was confirmed further by pulse–chase experiments on synchronized cells (not shown). We conclude from this that HOXC10 degradation is a post-translational event that occurs early in mitosis and, as shown in Figure 1A, is not controlled by the spindle checkpoint.
Fig. 2. HOXC10 becomes unstable in mitosis at the same time as cyclin A and in quiescent cells, and is targeted for degradation by the ubiquitin–proteasome pathway. (A) The stability of HOXC10 decreases in mitotic-enriched cell fractions. HeLa cells were synchronized at G1/S by a double thymidine block and released in nocodazole medium. At each time point, adherent and shaken off cells were harvested and lysed together. The kinetics of disappearance of cyclin A and the parallel appearance of cyclin B1 (8–12) mark the progressive accumulation of mitotic cells; nevertheless, a fraction of at least 10–20% residual adherent cells was always present in this synchronization procedure. Lower panels: aliquots of cells were collected 2 or 7 h after the release of the G1/S block, and incubated with cycloheximide (chx) for up to 120 min to inhibit neosynthesis of proteins. The indicated immunoblots show the stability of HOXC10, cyclin A and hnRNP A1 under these conditions. (B) The 26S proteasome inhibitor, lactacystin, impairs mitotic or G0 degradation of HOXC10. Asynchronous HeLa cells were cultured without any treatment (lane 1) or incubated with nocodazole for 14 h (lanes 2–6). Lactacystin (lanes 2 and 5) or a mixture of common lysosomal protease inhibitors (lanes 3 and 6) was added to the culture during the last 4 h of nocodazole treatment (Noco). Mitotic (Shaken Off) or residual adherent cells (Adherent) were lysed and analysed in immunoblots with the indicated antibodies. C2C12 quiescent cells (lane 7) were also incubated with lactacystin (lane 8) as above. An aliquot of cells (lane 9) was serum stimulated in S phase to check the expression of the indicated proteins. (C) In vivo ubiquitylation assay on human 293 cells transfected with different combinations of His6-tagged ubiquitin (UbiHis) and HA-tagged Hoxc10 (HOXC10HA) expression vectors. Ni-NTA affinity-purified ubiquitinHis conjugates (Ni) or SDS total lysates (T) of each transfection points were fractionated on an SDS–polyacrylamide gel and immunoblotted with mouse monoclonal anti-HA antibodies. Free HOXC10HA (arrowhead) and ubiquitinHis intermediates (Ubiquitin conjugates) are indicated. The Mr in kDa of pre-stained protein standards is shown next to the panel.
HOXC10 is a substrate of the ubiquitin–proteasome pathway
To investigate further if HOXC10 levels are controlled by proteolysis, HeLa cells were incubated with nocodazole in the presence of the 26S proteasome inhibitor lactacystin or a mix of common lysosomal protease inhibitors (Fenteany et al., 1995). Anti-HOXC10 immunoblots (Figure 2B) revealed that endogenous HOXC10 accumulates in nocodazole-treated mitotic cells after lactacystin treatment, whereas the protein is undetectable in control mitotic extracts or in mitotic extracts after treatment with the lysosomal protease inhibitor mix (Figure 2B). Importantly, the specific and effective inhibition of the 26S proteasome by lactacystin is demonstrated by the accumulation of cyclin A in extracts despite nocodazole treatment (Figure 2B). Similarly, proteasome-dependent proteolysis is also responsible for HOXC10 degradation in quiescent cells (Figure 2B). Short-lived regulatory proteins are often degraded through the proteasome pathway after being conjugated to polyubiquitin chains. To test if HOXC10 is ubiquitylated in the cell, HOXC10 and ubiquitin expression vectors were co-transfected in 293 cells and the ubiquitin intermediates were isolated by an in vivo ubiquitylation assay (Musti et al., 1996; Treier et al., 1994). By competing endogenous ubiquitin with transiently expressed histidine-tagged ubiquitin, the very unstable intermediates of ubiquitin chains covalently conjugated to the target protein can be affinity purified under denaturing conditions, resolved on an SDS–polyacrylamide gel and revealed with a specific antibody against the target protein. The typical ladder of polyubiquitin conjugates observed with anti-haemagglutinin (HA) antibody against the recombinant HA-tagged HOXC10 demonstrates that HOXC10 is polyubiquitylated in vivo (Figure 2C). These results show that the disappearance of HOXC10 in mitosis is indeed due to the ubiquitin–proteasome pathway.
Among members of the Hox family, HOXC10 is specifically targeted for degradation in mitosis of cycling cells
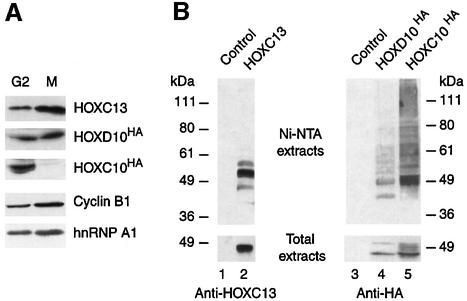
We asked whether other members of the Hox developmental regulator family might also undergo mitotic proteolysis. For this purpose, we chose to investigate the highly related HOXC13, and the paralogous HOXD10 that shows almost 100% identity to HOXC10 in the region of the homeodomain. In contrast to HOXC10, transiently expressed HOXD10 and HOXC13 proteins are detected not only in G2 but also in mitosis (Figure 3A) and other phases of the cell cycle (data not shown). Accordingly, we also observed constant levels of the endogenous HOXD10 and HOXC13 proteins throughout the cell cycle (data not shown). When subjected to the in vivo ubiquitylation assay used in Figure 2D, polyubiquitylated HOXD10 forms are however retained on the Ni-NTA beads (Figure 3B), whereas HOXC13 mainly gives rise to low molecular weight ubiquitylated species. Here, the pattern of HOXC13 and HOXD10 ubiquitylation was not investigated in more depth. It is possible that this ubiquitylation rather is involved in transcriptional activation by HOXC13 (Conaway et al., 2002). Although we cannot rule out the possibility that the ubiquitin–proteasome pathway might have a role in regulating the steady-state levels of HOXC13 and HOXD10 or in quality control through degradation of misfolded protein, none of these proteins fluctuates in abundance during the cell cycle. In conclusion, mitotic degradation, as observed for HOXC10, does not appear to be a common feature among abdominal-B Hox members.
Fig. 3. In cycling cells, HOXC10 is specifically targeted for degradation in mitosis, whereas other members of the Hox family show constant levels. (A) HeLa cells transiently transfected with Hoxc13, _Hoxd10_HA, _Hoxc10_HA or control expression vector were nocodazole synchronized. Lysates of mitotic (M) or adherent G2 phase (G2) cells were then immunoblotted with anti-HOXC13, anti-HA (HOXC10 and HOXD10), anti-cyclin B1 or anti-hnRNP A1 antibodies. (B) An in vivo ubiquitin assay (see legend of Figure 2D) was applied to members of the Hox family.
A dominant-negative form of UbcH10 suppresses HOXC10 ubiquitylation
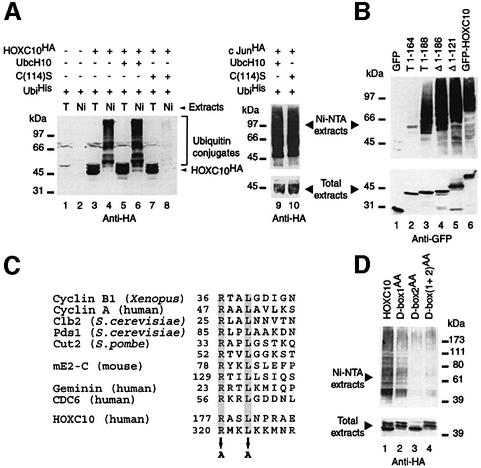
Mitotic proteolysis is largely mediated by the E3 APC in conjunction with the E2 enzymes UbcH5 and UbcH10. A dominant-negative form of UbcH10, UbcH10:C114S, inhibits destruction of cyclin A and B, arrests mammalian cells in mitosis and blocks onset of anaphase, presumably by blocking the ubiquitin-dependent proteolysis of proteins required for sister chromatid separation (Townsley et al., 1997). We thus asked whether this mutant might also interfere with HOXC10 in vivo ubiquitylation. As shown in Figure 4A, co-transfection of UbcH10:C114S cDNA strongly reduced the formation of HOXC10–ubiquitin conjugates, whereas, under the same experimental conditions, no reduction of c-Jun–ubiquitin conjugates was observed (Figure 4A, compare lanes 9 and 10). Ubiquitin-dependent proteolysis of HOXC10 may therefore involve ubiquitylation through the APC, as no other E3 has, so far, been shown to cooperate with UbcH10.
Fig. 4. In vivo ubiquitylation of HOXC10 requires two functional destruction boxes and is suppressed by a dominant-negative form of UbcH10. (A) An in vivo ubiquitylation assay was carried out as in Figure 2. Note that the overexpression of the UbcH10:C(114)S mutant abolishes the accumulation of ubiquitinHis conjugates of HOXC10 (lane 8), but not that of c-Jun (lane 10). (B) Vectors expressing GFP alone (GFP), GFP-HOXC10 wt (GFP-HOXC10), or a set of GFP–HOXC10 fusion proteins containing the first 164 (T1-164), 188 (T1-188) or lacking the first 121 (Δ1–121) or 186 (Δ1–186) N-terminal residues were subjected to an in vivo ubiquitylation assay. (C) Comparison of D-box sequences identified in APC substrates. The conserved arginine at position 1 and leucine at position 4 are highlighted by grey boxes. Potential HOXC10 D-boxes are located at positions 177–185 (D-box 1) and 320–328 (D-box 2). Alanine replacements in the N-terminal (D-box1AA), C-terminal (D-box2AA) or in both [D-box(1+2)AA] D-box mutants are indicated by arrows. (D) D-box mutants of HOXC10 were subjected to in vivo ubiquitylation assays.
HOXC10 contains two functional D-boxes
All APC substrates identified to date contain a D-box motif, initially identified in the N-terminal region of mitotic cyclins (Glotzer et al., 1991; King et al., 1996b), and/or a KEN box (Pfleger and Kirschner, 2000). Since the aforementioned results suggested that HOXC10 might be targeted for destruction by APC, we tried to identify degradation signals in HOXC10. We found two putative D-boxes, one in the N-terminal region (between residues 177 and 185, D-box 1) and one in the homeodomain (between residues 320 and 328, D-box 2). To investigate whether these putative D-boxes are functional, a set of different deletion mutants of HOXC10 fused to green fluorescent protein (GFP) was subjected to in vivo ubiquitylation assays. As shown in Figure 4B, ubiquitin conjugates were detected with the control protein GFP–HOXC10, with the truncated T1-188 mutant containing only D-box 1 and with the N-terminal deletion mutants, Δ1–121 or Δ1–186, carrying both D-boxes or D-box 2 alone, respectively. Conversely, no ubiquitin conjugates were detected with the GFP moiety alone or GFP fused to the N-terminal 164 amino acids of HOXC10. We also investigated missense mutations of the HOXC10 D-boxes, since the substitution of the two most conserved D-box residues with alanine (Figure 4C) impairs ubiquitin-dependent degradation of target proteins (King et al., 1996b). D-box 1 alanine substitutions (D-box1AA) caused a significant reduction of ubiquitin conjugates, and an even more dramatic decrease was observed with D-box 2 (D-box2AA) and the double D-box [D-box(1+2)AA] mutants (Figure 4D). In summary, these results indicate that both D-boxes of HOXC10 are functional and required for an efficient ubiquitylation, and provide further evidence for APC-mediated destruction of HOXC10.
HOXC10 appears to be a substrate of the APC
We employed a cell-free degradation assay to explore the hypothesis that APC may target HOXC10 for degradation. In this assay, extracts of mitotic HeLa cells are incubated with in vitro translated substrate. In this system, HOXC10 is efficiently degraded (Figure 5A), although with slightly slower kinetics than cyclin A, which we included to compare our experimental conditions with those previously reported (Bastians et al., 1999). Moreover, HOXC10 ubiquitin conjugates are also clearly observed when the crude extracts are supplemented with His6-tagged ubiquitin (Figure 5B). As expected from the in vivo observations (Figure 2B), HOXC10 degradation was dependent on the 26S proteasome, since extracts of mitotic HeLa cells pre-incubated with the proteasome inhibitor, lactacystin, were unable to degrade either protein (Figure 5A). Similar results were obtained when we added lactacystin directly to mitotic extracts instead of pre-incubating the cells with it (data not shown). In contrast to wild-type HOXC10, the double D-box mutant of HOXC10 and, to a lesser extent, the single D-box mutants were significantly stabilized (Figure 5A). These results indicate that D-boxes of HOXC10 are required for its mitotic degradation. Moreover, a 100-fold excess of cyclin A strongly delayed the kinetics of HOXC10 proteolysis, suggesting that cyclin A, a known APC substrate, can quench HOXC10 degradation, probably by competing for the same ubiquitin ligase. The competition for degradation was reciprocal, as an excess of HOXC10 inhibited cyclin A proteolysis almost completely (Figure 5C). The involvement of APC in targeting HOXC10 for degradation was supported further by the observations that anti-HOXC10 antibodies co-immunoprecipitated endogenous CDC27, a core subunit of the APC, in extracts from cells transiently overexpressing HOXC10 (Figure 5F) and that HOXC10 proteolysis was reduced in CDC27-depleted extracts (Figure 5D and E). In the latter assay, we noticed that cyclin A was stabilized more efficiently than HOXC10 by CDC27 depletion. HOXC10 was also subjected to an in vitro ubiquitylation assay system containing only purified components (E1, ubiquitin, ATP, UbcH10 or UbcH5 and activated APC), but we failed to observe significant polyubiquitylation (data not shown). Additional cofactors might be needed to reconstitute the APC-mediated ubiquitylation of this specific protein in this minimal system.
Fig. 5. APC may be involved in HOXC10 destruction. (A) HOXC10 is degraded in vitro by mitotic extracts, whereas D-box mutations enhance its stability. In vitro translated 35S-labelled cyclin A and D-box or wild-type HOXC10 proteins were incubated with HeLa mitotic extracts. At the indicated times, in hours, equal aliquots of each reaction were removed, resolved by SDS–PAGE and autoradiographed. Representative kinetics of at least four independent experiments are shown. Mitotic extracts were also prepared from shaken off HeLa cells incubated with lactacystin during the last 4 h of nocodazole treatment. Note that under these conditions, the in vitro degradation of HOXC10 and cyclin A is impaired. (B) HOXC10 is ubiquitylated in mitotic extracts. 35S-labelled HOXC10 was incubated in mitotic extracts supplemented with in vitro translated His6-tagged ubiquitin and processed as above. Longer (upper panel) and shorter (s.e.) exposures are shown. (C) In the same degradation assay, an ∼100-fold excess of in vitro translated cyclin A or bacterial Ni-NTA-purified GST-HOXC10His (0.5 µg) blocks HOXC10 or cyclin A degradation, respectively. Unprogrammed reticulocyte lysate (Control) and GST were assayed as controls. (D) APC immunodepletion of HeLa mitotic extracts. Shaken-off cells were lysed in low salt buffer and incubated for 45 min at room temperature with irrelevant IgG or with anti-CDC27 antibody. The anti-CDC27 immunoblot shows a significant depletion of the APC subunit CDC27 from the supernatant (compare lanes 1 and 2). The corresponding control IgG and anti-CDC27 immunoprecipitations are also shown (lanes 3 and 4). (E) Mitotic lysates depleted as in (D) were incubated with the in vitro translated HOXC10 or cyclin A. Equal aliquots were analysed as above, and exposure time was the same for all gels. (F) Upon transfection of HeLa cells, the recombinant HOXC10 was immunoprecipitated with anti-HOXC10 or irrelevant IgG antibodies. The immunoprecipitations were resolved by SDS–PAGE and analysed with anti-CDC27 antibody (lower panel) or anti-HOXC10 antibody (middle panel). The level of HOXC10 was checked in the supernatant before and after immunoprecipitation (upper panel).
A stable form of HOXC10 accumulates cells in mitosis by delaying the metaphase to anaphase transition
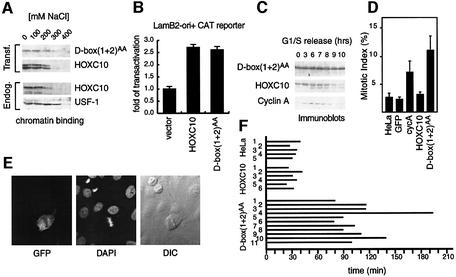
To investigate the biological relevance of mitotic proteolysis of HOXC10, the transcriptional activity of the double D-box mutant, D-box(1+2)AA, was initially investigated and compared with that of HOXC10. Chromatin binding assays show that the mutant is eluted from the chromatin with ionic strength similar to endogenous or transfected HOXC10 (Figure 6A). In addition, D-box(1+2)AA and HOXC10 were able to transactivate with comparable efficiency a Hox-binding site-dependent CAT reporter gene (Figure 6B). Thus, HOXC10 transcriptional activity appears to be unaffected by amino acid substitutions in the consensus D-box residues.
Fig. 6. A stable mutant of HOXC10 can delay the metaphase–anaphase transition. (A) Chromatin binding of the D-box(1+2)AA mutant, endogenous or transfected wild-type HOXC10 and the control USF1. Nuclear pellet was extracted in buffer containing the indicated concentration of salt, and the solubilized nuclear proteins were separated from the chromatin-bound proteins by centrifugation. The chromatin nuclear matrix-bound fraction is shown. (B) CAT assay for analysing the transactivation activity of D-box (1+2)AA or wild-type HOXC10 on the LamB2-ori+ CAT reporter gene. (C) HeLa cells were transiently transfected with the GFP-based bicistronic vectors for D-box(1+2)AA or wild-type Hoxc10, synchronized as described in Figure 2B. Anti-HOXC10 immunoblots show that D-box(1+2)AA is more stable than the wild-type in mitotic-enriched extracts. (D) HeLa cells were transiently transfected with the indicated GFP-based bicistronic vectors. The percentage of mitosis (mitotic index) of untransfected HeLa or GFP-positive cells was measured 42–48 h after transfection. The mitotic index (+SD) was calculated from five independent experiments in which at least 350 cells were counted in each experiment. (E) Confocal and transmitted light image of a D-box(1+2)AA-expressing GFP cell. Left panel: GFP fluorescence. Middle panel: DAPI nuclear staining. Right panel: phase contrast. (F) Wild-type HOXC10- or D-box(1+2)AA-expressing GFP-positive cells transfected as above or untransfected HeLa cells were analysed by time-lapse and fluorescence microscopy as shown in Figure 7. A horizontal bar indicates the period of time (in minutes) from a visible chromosome condensation to metaphase–anaphase transition of each cell indicated by a number.
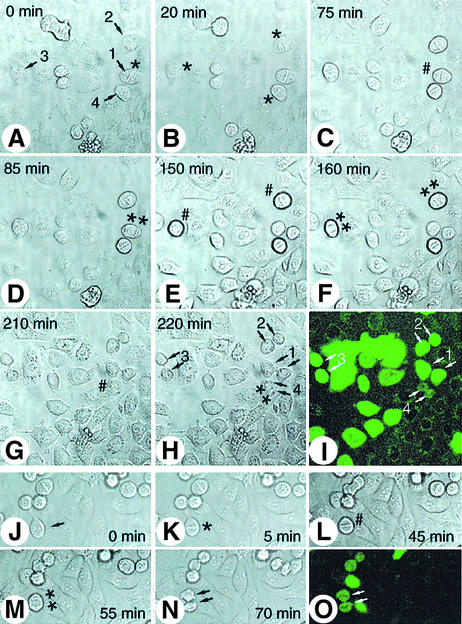
We then asked whether the increased stability of D-box(1+2)AA observed in vitro and in vivo (Figures 5 and 6C) would perturb cell cycle progression. With this aim, D-box(1+2)AA or wild-type Hoxc10 cDNAs were cloned into a GFP-based bicistronic vector and transiently transfected into HeLa cells. Since this vector allows the expression of both genes in a single transcription unit in which the gene of interest precedes the GFP gene, it is predictable that each GFP-expressing cell is also positive for the protein of interest. Additionally, the presence of the recombinant proteins together with GFP was confirmed by indirect immunofluorescence with anti-HOXC10 antibodies (not shown). As such, putative perturbation of the cell cycle caused by the protein of interest should be directly observed by scoring GFP-positive cells. Thus, the mitotic index was evaluated 42–48 h after transfection, among GFP cells. D-box(1+2)AA-expressing GFP-positive cells showed a 3- to 5-fold higher mitotic index than untransfected cells or HOXC10-expressing GFP or GFP control cells (Figure 6D). The mitotic index of the double D-box mutant was even higher than that observed with cyclin A-expressing GFP cells (Figure 6D) (Geley et al., 2001). After nuclear dye staining, the condensation status and chromosome alignment indicated that most of the D-box(1+2)AA-positive cells were accumulated in a pre-anaphase status, morphologically indistinguishable from a typical metaphase (Figure 6E). To investigate whether the mitotic accumulation by D-box(1+2)AA resulted from a mitotic block or a delay of mitotic progression, the aforementioned living cells were subjected to time-lapse microscopy analysis. With this procedure, the interval between initial chromosome condensation and chromosome separation was evaluated over a range of 75–190 min for D-box(1+2)AA-expressing GFP cells, and ∼25–40 min for control or HOXC10-expressing GFP cells (Figures 6D and 7). In contrast, the time from anaphase onset to re-entry into G1 phase in D-box(1+2)AA-positive cells was comparable with control cells. Thus, the D-box(1+2)AA-dependent mitotic accumulation resulted from a delay rather than an irreversible block of events occurring earlier or at the time of the metaphase to anaphase transition. These observations suggest that HOXC10 might be able to play a direct role in regulating mitotic progression.
Fig. 7. The time-lapse and fluorescence microscopy show the delay of metaphase–anaphase transition by the D-box(1+2)AA mutant. Living D-box(1+2)AA (A–I) or wild-type HOXC10 (J–O) GFP-positive cells (numbered arrows) were photographed for GFP fluorescence (I and O) and in phase contrast at an interval of 5 min. The photograms show the indicated cells during entry into mitosis, when chromosome condensation becomes visible (*); at the metaphase to anaphase transition (#); and in late anaphase/telophase (**). The most representative photograms are shown.
Discussion
In the present report, we provide evidence that HOXC10, a member of the Hox family of developmental selectors, oscillates during the cell cycle and is targeted for degradation early in mitosis by the ubiquitin–proteasome pathway. Mitotic proteolysis of HOXC10 appears to be a feature not shared with other Hox proteins, and it seems to be regulated by the APC. We also show that failure of HOXC10 destruction causes mitotic accumulation by delaying the metaphase to anaphase transition, unveiling a putative, yet unpredicted, novel function of a Hox family member in cell cycle progression.
Our data are compatible with previous observations involving HOXC10 in cell proliferation control. In mouse embryos, Hoxc10, a member of the abdominal-B gene subfamily of the Hoxc cluster (from Hoxc9 to Hoxc13), is expressed in the mesenchyme of the developing hindlimb, and in distal epidermal regions of developing fore- and hindlimb (Peterson et al., 1994). It is thought to have some functions specifically required in hindlimb morphogenesis, probably in controlling the proliferation rate (Duboule, 1995). Malformations of the hindlimb probably due to a defect of proliferation or apoptosis are described for Hoxc10 gene targeting (Suemori et al., 1995). In amphibians, Hoxc10 expression is strongly induced in regenerating mesenchymal cells upon amputation of a limb or tail, and it is the only posterior Hox that is specifically induced during regeneration of anterior limb (Simon and Tabin, 1993; Carlson et al., 2001). In mammalian cultured cells, it is associated with a proliferative status (De Stanchina et al., 2000) and its expressed sequence tags are frequently isolated from cDNA libraries of tumours, whereas it is absent in the corresponding normal tissue (http://cgap.nci.nih.gov).
Several transcription factors involved in proliferation control, namely E2F, MyoD, Myc and c-Jun (Ciechanover et al., 1991; Treier et al., 1994; Hateboer et al., 1996; Breitschopf et al., 1998; Salghetti et al., 1999), are known to be substrates of the ubiquitin–proteasome pathway. HOXC10 now joins them as a novel substrate, but at the same time stands out, since its mitotic degradation seems rather unique (Figures 1 and 2).
The timing of HOXC10 proteolysis coincides with the activity of UbcH10 (Yamanaka et al., 2000), and is strongly reduced in the presence of dominant-negative UbcH10. These observations support a role for APC, known to cooperate with UbcH10, in degradation of HOXC10; they do not, however, rule out the possibility that another APC-associated E2, such as UbcH5, may also target HOXC10. Furthermore, the timing of HOXC10 proteolysis is compatible with the activity of APC. A major hallmark for APC-dependent proteolysis is the presence of D-box and/or KEN-box motifs in the target protein. HOXC10 mutagenesis demonstrates that two D-box motifs, conserved between mammalian and amphibian HOXC10, are indeed required for its ubiquitin-dependent degradation (Figures 4 and 5). It was shown that substitutions of the conserved R and L residues by alanines strikingly reduce or even suppress ubiquitylation and enhance protein stability (King et al., 1996b). Consistently, the stability of D-box mutants of HOXC10 in vitro and in vivo (Figures 5 and 6) leads to the conclusion that mitotic degradation of HOXC10 is mainly dependent on the presence of functional D-boxes. Interestingly, although the paralogous HOXD10 and the cognate HOXC13 contain three and one putative D-box core sequence (RXXL), respectively, the levels of these proteins do not oscillate throughout the cell cycle (Figure 3 and data not shown). This observation confers a specific peculiarity among Hox members to HOXC10, which is even more surprising considering the high degree of similarity to HOXD10. It implies that HOXC10 might have a specific function during development, possibly in regulating the amount of cells that proliferate in a regional-specific fashion. Despite the high overall identity with HOXD10, the central portion of HOXC10, containing D-box 1, is less conserved between them. In this region, we noticed what could be an extended D-box of the cyclin A type, since the nearby C-terminal amino acids share some similarity with the A-type D-boxes of cyclin A and Nek2A (not shown), two proteins that are destroyed in early mitosis and not stabilized by activation of the spindle checkpoint (den Elzen and Pines, 2001; Geley et al., 2001; Hames et al., 2001). Consistently, the kinetics of HOXC10 proteolysis in mitosis parallels cyclin A degradation and precedes cyclin B1 proteolysis. Cyclin A overexpression in mammalian cells delays progression through mitosis, transiently accumulating prometaphase cells, probably by competition for the APC (Sigrist et al., 1995; den Elzen and Pines, 2001; Geley et al., 2001). Although a mitotic delay by Nek2 stabilization has not been described so far, a stable mutant of the Nek2-related NIMA kinase does cause a mitotic arrest in Aspergillus nidulans (Pu and Osmani, 1995). Thus, down-regulation of cyclin A or NIMA activity appears to be necessary to allow a physiological mitotic progression. Similarly, stable forms of HOXC10 accumulate mitotic cells by delaying the metaphase to anaphase transition (Figures 6 and 7), implying a need to reduce the levels of HOXC10 for mitotic progression. Since DNA binding and transcriptional activation of D-box(1+2)AA were not affected, we can exclude altered transcriptional activity as the cause for this accumulation. We cannot, however, rule out the possibility that non-degradable HOXC10 causes the delay by interfering with the destruction of other mitotic regulators. It is tempting to speculate that HOXC10 and cyclin A may be involved in the same regulatory network or common factor(s) may be required for their targeted degradation. The reciprocal interference of HOXC10 and cyclin A in in vitro mitotic degradation, the delayed degradation of HOXC10 in APC-depleted mitotic extracts and the association of HOXC10 with the APC core subunit, CDC27, altogether support the hypothesis that HOXC10 may be a novel APC substrate (Figure 5). Thus, the failure of HOXC10 ubiquitylation in in vitro APC-dependent ubiquitylation assays implies that the in vivo reaction might be somewhat more complex. In the case of the APC substrate SnoN, a negative regulator of transforming growth factor-β (TGF-β) signalling, moderate in vitro ubiquitylation could only be reconstituted if the purified system also contained a Smad cofactor to help SnoN recruitment to the APC (Stroschein et al., 2001; Wan et al., 2001). Moreover, to study complex degradation pathways, extract systems might prove to be superior to simplified in vitro ubiquitylation systems, since they more closely reflect the physiological conditions and differential influences, e.g. of TGF-β signalling on SnoN degradation (Stroschein et al., 2001; Wan et al., 2001). In this respect, it is interesting to note that Smad family members can form complexes with Hox proteins (Shi et al., 1999). It will therefore be important to investigate whether these or other known HOX-associated proteins are involved in targeting HOXC10 for destruction.
HOXC10 and HOXC13 were isolated for the binding to lamin B2 origin of DNA replication in a yeast one-hybrid screening. They also bind the lamin B2 origin sequence in in vitro assays, and transactivate the lamin B2 origin- based reporter in mammalian cells (De Stanchina et al., 2000). The lamin B2 origin is an early firing origin that shows a cell cycle-regulated in vivo DNA footprint (Abdurashidova et al., 1998) and contains consensus binding sites for abdominal-B-type Hox proteins (Shen et al., 1997; Bromleigh and Freedman, 2000), some of which also overlap the starting point of bi-directional DNA replication (Abdurashidova et al., 2000; De Stanchina et al., 2000). It is known that transcription factors may participate in regulating replication origin firing, probably by driving the chromatin into a configuration that allows or hinders the assembly of replication complexes at the origin (Bosco et al., 2001; Cayirlioglu and Duronio, 2001). Further investigations are, however, required to understand whether HOXC10, and perhaps other members of the Hox family, may be part of a mechanism that ensures the timely licensing of replication origins, as described for other APC substrates (McGarry and Kirschner, 1998; Petersen et al., 2000; Tada et al., 2001).
Finally, several examples have been reported in which molecules that pattern organs also promote organ growth and cell proliferation during metazoan development (reviewed in Vidwans and Su, 2001). In some cases, progression through specific stages of the cell cycle may also have a more direct and essential role in development. Elucidation of molecular pathways coupling cell cycle regulators, i.e. cyclins, cyclin-dependent kinases (CDKs) and their inhibitors (CKIs), which are conserved in different cell types among eukaryotes, with patterning molecules, i.e. Hox, to meet the specialized needs of cells during development, represents a challenging task for the future.
Materials and methods
Miscellaneous methods
Cell lines, cell culture, transfection, cell synchronization, immunoprecipitation, immunoblotting, RNA analysis, plasmids and site-directed mutagenesis were performed according to standard procedures and are described in the Supplementary data.
Bacterial recombinant proteins and antibody production
GST–HOXC10His and GST–HOXC10: 1–231 were purified under native condition as described (Peverali et al., 1994, 1996). Rabbit anti-HOXC10 antibody was generated against GST–HOXC10: 1–231 that lacks the conserved homeodomain and affinity purified as described (Peverali et al., 1994).
Purification of ubiquitin conjugates
Purification of His6-ubiquitin conjugates was performed 36 h after transfection as described (Treier et al., 1994; Musti et al., 1996).
Production of in vitro translated proteins and in vitro degradation assay
A coupled transcription and translation system (TNT; Promega) with or without [35S]methionine (1000 Ci/mmol) was used to produce in vitro translated proteins for the degradation assay as described (Bastians et al., 1999). To assay the effect of APC depletion on HOXC10, mitotic extracts were prepared in low salt buffer followed by three serial incubations at room temperature for 15 min with mouse anti-CDC27 (AF3.1, Santa Cruz Biotechnology) (0.5 µg/µl extract) or irrelevant IgG antibodies pre-conjugated to (0.5 µl beads/µl extract) protein A/G Plus-Agarose (Santa Cruz Biotechnologies).
Chromatin binding and CAT assay
Endogenous HOXC10, the control USF1 and the transiently expressed wild-type or double D-box mutant of HOXC10 were assayed for binding to chromatin as described (Mendez and Stillman, 2000). To release the chromatin-bound proteins, cell nuclei (P1) were incubated for 30 min on ice in buffer B containing increasing concentrations (0–400 mM) of sodium chloride; the final chromatin pellet (P3) was then analysed by immunoblot.
To verify the transactivation efficiency of D-box(1+2)AA, HeLa cells (70% confluency) were co-transfected by the Effectene procedure (Qiagen) with LamB2-ori+ CAT reporter vector (De Stanchina et al., 2000), pSV-β-galactosidase and either pCMV-Hoxc10, pCMV-D-box(1+2)AA or pCMV vector. CAT activity was assayed 40 h later by the Quan-T-CAT assay system (Amersham).
Immunofluorescence and time-lapse microscopy
Wild-type or the D-box(1+2)AA mutant of Hoxc10 and cyclin A cDNAs were cloned into pIRES-hrGFP-2a bicistronic expression vector (Stratagene), transfected into HeLa cells and 42–48 h later were processed for immunofluorescence analysis (Peverali et al., 1994) or analysed by time-lapse DIC and fluorescence microscopy. To evaluate the percentage of mitosis (mitotic index) among GFP-positive cells or untransfected cells, HeLa cells were stained with Hoechst 33258 or 4′,6-diamidino-2-phenylindole (DAPI) nuclear dye (Sigma) and analysed for the expression of hrGFP. Interphase or mitotic cells were morphologically scored by fluorescence microscopy. Time-lapse DIC and fluorescence microscopy were carried out with a Leitz confocal microscope at 488 nm by incubating the transfected cells at 37°C with a home-made heating system in Dulbecco’s modified Eagle’s medium–HEPES (Invitrogen-Life Tecnologies) plus 10% fetal calf serum.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The authors are grateful to Drs D.Bohmann, G.Dreyfuss, K.Helin, J.M.Ruderman and V.Zappavigna for providing reagents, Drs J.M.Peters and D.Orioli for helpful discussion and reading the manuscript, Dr E.Prosperi for cytofluorimetric analysis of DNA content, Dr P.Vaghi, CGS, University of Pavia for the expertise in confocal microscopy, and Mr D.Arena for technical support. This work was partially supported by an EU (HPRN-CT-2000-00089) contract to S.R. and by the ‘Programma di Settore-Biomolecole per la Salute Umana’ CNR/MURST grant to G.B.
References
- Abate-Shen C. (2002) Deregulated homeobox gene expression in cancer: cause or consequence? Nat. Rev. Cancer, 2, 777–785. [DOI] [PubMed] [Google Scholar]
- Abdurashidova G., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (1998) Cell cycle modulation of protein–DNA interactions at a human replication origin. EMBO J., 17, 2961–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdurashidova G., Deganuto,M., Klima,R., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science, 287, 2023–2026. [DOI] [PubMed] [Google Scholar]
- Bastians H., Topper,L.M., Gorbsky,G.L. and Ruderman,J.V. (1999) Cell cycle-regulated proteolysis of mitotic target proteins. Mol. Biol. Cell, 10, 3927–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncinelli E. (1997) Homeobox genes and disease. Curr. Opin. Genet. Dev., 7, 331–337. [DOI] [PubMed] [Google Scholar]
- Bosco G., Du,W. and Orr-Weaver,T.L. (2001) DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol., 3, 289–295. [DOI] [PubMed] [Google Scholar]
- Breitschopf K., Bengal,E., Ziv,T., Admon,A. and Ciechanover,A. (1998) A novel site for ubiquitination: the N-terminal residue and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J., 17, 5964–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromleigh V.C. and Freedman,L.P. (2000) p21 is a transcriptional target of HOXA10 in differentiating myelomonocytic cells. Genes Dev., 14, 2581–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M.R., Komine,Y., Bryant,S.V. and Gardiner,D.M. (2001) Expression of Hoxb13 and Hoxc10 in developing and regenerating axolotl limbs and tails. Dev. Biol., 229, 396–406. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu P. and Duronio,R.J. (2001) Cell cycle: flies teach an old dogma new tricks. Curr. Biol., 11, R178–R181. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., DiGiuseppe,J.A., Bercovich,B., Orian,A., Richter,J.D., Schwartz,A.L. and Brodeur,G.M. (1991) Degradation of nuclear oncoproteins by the ubiquitin system in vitro. Proc. Natl Acad. Sci. USA, 88, 139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute P. and Pines,J. (1999) Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol., 1, 82–87. [DOI] [PubMed] [Google Scholar]
- Conaway R.C., Brower,C.S. and Conaway,J.W. (2002) Emerging roles of ubiquitin in transcription regulation. Science, 296, 1254–1258. [DOI] [PubMed] [Google Scholar]
- De Stanchina E., Gabellini,D., Norio,P., Giacca,M., Peverali,F.A., Riva,S., Falaschi,A. and Biamonti,G. (2000) Selection of homeotic proteins for binding to a human DNA replication origin. J. Mol. Biol., 299, 667–680. [DOI] [PubMed] [Google Scholar]
- den Elzen N. and Pines,J. (2001) Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol., 153, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D. (1995) Vertebrate Hox genes and proliferation: an alternative pathway to homeosis? Curr. Opin. Genet. Dev., 5, 525–528. [DOI] [PubMed] [Google Scholar]
- Fenteany G., Standaert,R.F., Lane,W.S., Choi,S., Corey,E.J. and Schreiber,S.L. (1995) Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science, 268, 726–731. [DOI] [PubMed] [Google Scholar]
- Gehring W.J., Affolter,M. and Burglin,T. (1994) Homeodomain proteins. Annu. Rev. Biochem., 63, 487–526. [DOI] [PubMed] [Google Scholar]
- Geley S., Kramer,E., Gieffers,C., Gannon,J., Peters,J.M. and Hunt,T. (2001) Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol., 153, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M., Murray,A.W. and Kirschner,M.W. (1991) Cyclin is degraded by the ubiquitin pathway. Nature, 349, 132–138. [DOI] [PubMed] [Google Scholar]
- Godwin A.R. and Capecchi,M.R. (1998) Hoxc13 mutant mice lack external hair. Genes Dev., 12, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames R.S., Wattam,S.L., Yamano,H., Bacchieri,R. and Fry,A.M. (2001) APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J., 20, 7117–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hateboer G., Kerkhoven,R.M., Shvarts,A., Bernards,R. and Beijersbergen,R.L. (1996) Degradation of E2F by the ubiquitin–proteasome pathway: regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev., 10, 2960–2970. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (1996) Ubiquitin-dependent protein degradation. Annu. Rev. Genet., 30, 405–439. [DOI] [PubMed] [Google Scholar]
- Juang Y.L., Huang,J., Peters,J.M., McLaughlin,M.E., Tai,C.Y. and Pellman,D. (1997) APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science, 275, 1311–1314. [DOI] [PubMed] [Google Scholar]
- King R.W., Deshaies,R.J., Peters,J.M. and Kirschner,M.W. (1996a) How proteolysis drives the cell cycle. Science, 274, 1652–1659. [DOI] [PubMed] [Google Scholar]
- King R.W., Glotzer,M. and Kirschner,M.W. (1996b) Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol. Biol. Cell, 7, 1343–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann M., Carnac,G., Vandromme,M., Primig,M., Lamb,N.J. and Fernandez,A. (1998) The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J. Cell Biol., 142, 1447–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. (1994) Hox genes in vertebrate development. Cell, 78, 191–201. [DOI] [PubMed] [Google Scholar]
- Mendez J. and Stillman,B. (2000) Chromatin association of human origin recognition complex, cdc6 and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol., 20, 8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magli M.C., Largman,C. and Lawrence,H.J. (1997) Effects of HOX homeobox genes in blood cell differentiation. J. Cell. Physiol., 173, 168–177. [DOI] [PubMed] [Google Scholar]
- Mann R.S. and Affolter,M. (1998) Hox proteins meet more partners. Curr. Opin. Genet. Dev., 8, 423–429. [DOI] [PubMed] [Google Scholar]
- McGarry T.J. and Kirschner,M.W. (1998) Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell, 93, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Musti A.M., Treier,M., Peverali,F.A. and Bohmann,D. (1996) Differential regulation of c-Jun and JunD by ubiquitin-dependent protein degradation. Biol. Chem., 377, 619–624. [DOI] [PubMed] [Google Scholar]
- Peters J.M. (2002) The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell, 9, 931–943. [DOI] [PubMed] [Google Scholar]
- Petersen B.O. et5 al. (2000) Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev., 14, 2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R.L., Papenbrock,T., Davda,M.M. and Awgulewitsch,A. (1994) The murine Hoxc cluster contains five neighboring AbdB-related Hox genes that show unique spatially coordinated expression in posterior embryonic subregions. Mech. Dev., 47, 253–260. [DOI] [PubMed] [Google Scholar]
- Peverali F.A., Ramqvist,T., Saffrich,R., Pepperkok,R., Barone,M.V. and Philipson,L. (1994) Regulation of G1 progression by E2A and Id helix–loop–helix proteins. EMBO J., 13, 4291–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peverali F.A., Isaksson,A., Papavassiliou,A.A., Plastina,P., Staszewski,L.M., Mlodzik,M. and Bohmann,D. (1996) Phosphorylation of Drosophila Jun by the MAP kinase rolled regulates photoreceptor differentiation. EMBO J., 15, 3943–3950. [PMC free article] [PubMed] [Google Scholar]
- Pfleger C.M. and Kirschner,M.W. (2000) The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev., 14, 655–665. [PMC free article] [PubMed] [Google Scholar]
- Pu R.T. and Osmani,S.A. (1995) Mitotic destruction of the cell cycle regulated NIMA protein kinase of Aspergillus nidulans is required for mitotic exit. EMBO J., 14, 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti S.E., Kim,S.Y. and Tansey,W.P. (1999) Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J., 18, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W.F., Rozenfeld,S., Lawrence,H.J. and Largman,C. (1997) The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J. Biol. Chem., 272, 8198–8206. [DOI] [PubMed] [Google Scholar]
- Shi X., Yang,X., Chen,D., Chang,Z. and Cao,X. (1999) Smad1 interacts with homeobox DNA-binding proteins in bone morphogenetic protein signaling. J. Biol. Chem., 274, 13711–13717. [DOI] [PubMed] [Google Scholar]
- Sigrist S., Jacobs,H., Stratmann,R. and Lehner,C.F. (1995) Exit from mitosis is regulated by Drosophila fizzy and the sequential destruction of cyclins A, B and B3. EMBO J., 14, 4827–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon H.G. and Tabin,C.J. (1993) Analysis of Hox-4.5 and Hox-3.6 expression during newt limb regeneration: differential regulation of paralogous Hox genes suggest different roles for members of different Hox clusters. Development, 117, 1397–1407. [DOI] [PubMed] [Google Scholar]
- Stroschein S.L., Bonni,S., Wrana,J.L. and Luo,K. (2001) Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev., 15, 2822–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suemori H., Takahashi,N. and Noguchi,S. (1995) Hoxc-9 mutant mice show anterior transformation of the vertebrae and malformation of the sternum and ribs. Mech. Dev., 51, 265–273. [DOI] [PubMed] [Google Scholar]
- Tada S., Li,A., Maiorano,D., Mechali,M. and Blow,J.J. (2001) Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol., 3, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley F.M., Aristarkhov,A., Beck,S., Hershko,A. and Ruderman,J.V. (1997) Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc. Natl Acad. Sci. USA, 94, 2362–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier M., Staszewski,L.M. and Bohmann,D. (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell, 78, 787–798. [DOI] [PubMed] [Google Scholar]
- van Oostveen J., Bijl,J., Raaphorst,F., Walboomers,J. and Meijer,C. (1999) The role of homeobox genes in normal hematopoiesis and hematological malignancies. Leukemia, 13, 1675–1690. [DOI] [PubMed] [Google Scholar]
- Vidwans S.J. and Su,T.T. (2001) Cycling through development in Drosophila and other metazoa. Nat. Cell Biol., 3, E35–E39. [DOI] [PubMed] [Google Scholar]
- Wakefield J.G., Huang,J.Y. and Raff,J.W. (2000) Centrosomes have a role in regulating the destruction of cyclin B in early Drosophila embryos. Curr. Biol., 10, 1367–1370. [DOI] [PubMed] [Google Scholar]
- Wan Y., Liu,X. and Kirschner,M.W. (2001) The anaphase-promoting complex mediates TGF-β signaling by targeting SnoN for destruction. Mol. Cell, 8, 1027–1039. [DOI] [PubMed] [Google Scholar]
- Yamanaka A., Hatakeyama,S., Kominami,K., Kitagawa,M., Matsumoto,M. and Nakayama,K. (2000) Cell cycle-dependent expression of mammalian E2-C regulated by the anaphase-promoting complex/cyclosome. Mol. Biol. Cell, 11, 2821–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur A. and Brandeis,M. (2001) Securin degradation is mediated by fzy and fzr and is required for complete chromatid separation but not for cytokinesis. EMBO J., 20, 792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]