Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly (original) (raw)
Abstract
The general transcription factor TFIID is composed of the TATA box binding protein (TBP) and a set of conserved TBP-associated factors (TAFs). Here we report the completion of genome-wide expression profiling analyses of yeast strains bearing temperature-sensitive mutations in each of the 13 essential TAFs. The percentage of the yeast genome dependent on each TAF ranges from 3% (TAF2) to 59–61% (TAF9). Approximately 84% of yeast genes are dependent upon one or more TAFs and 16% of yeast genes are TAF independent. In addition, this complete analysis defines three distinct classes of yeast promoters whose transcriptional requirements for TAFs differ substantially. Using this collection of temperature-sensitive mutants, we show that in all cases the transcriptional dependence for a TAF can be explained by a requirement for TBP recruitment and assembly of the preinitiation complex (PIC). Unexpectedly, these assembly experiments reveal that TAF11 and TAF13 appear to provide the critical functional contacts with TBP during PIC assembly. Collectively, our results confirm and extend the proposal that individual TAFs have selective transcriptional roles and distinct functions.
Keywords: chromatin immunoprecipitation/genome-wide transcription/preinitiation complex assembly/TAF/TBP
Introduction
Transcription initiation by RNA polymerase II involves the assembly of general transcription factors (GTFs) on the core promoter to form a preinitiation complex (PIC). Transcriptional activator proteins (activators) bind to specific _cis_-acting promoter elements within upstream activating sequences (UASs) and stimulate PIC assembly via a mechanism thought to involve direct interactions with one or more components of the transcription machinery (Orphanides et al., 1996; Roeder, 1996; Ptashne and Gann, 1997; Lee and Young, 2000). The first step in PIC assembly is binding of the GTF TFIID to the TATA box. TFIID is a multi-subunit complex consisting of the TATA box binding protein (TBP) and a set of TBP-associated factors (TAFs) (Burley and Roeder, 1996; Albright and Tjian, 2000; Green, 2000). Like other components of the transcription machinery, TAFs are highly conserved from yeast to humans. In yeast, 14 TAFs have been identified, 13 of which are required for viability (reviewed in Green, 2000).
Several TAFs are associated with large nuclear protein complexes in addition to TFIID. For example, yeast TAF5, TAF6, TAF9, TAF10 and TAF12 (formerly known as TAF90, TAF60, TAF17, TAF25 and TAF61/68, respectively; Tora, 2002) are also integral components of the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex (Grant et al., 1998), which is involved in transcription of a subset of RNA polymerase II-dependent genes. In yeast, promoters have been grouped into two extreme classes based on their requirement for TAFs (Kuras et al., 2000; Li et al., 2000). TAF-dependent (TAF-dep) promoters require TAFs for transcription and on these promoters TBP and TAFs are present at comparable levels. TAF-independent (TAF-ind) promoters do not require TAFs for activity, and on these promoters TAFs are either absent or present at levels far below that of TBP.
We have been carrying out a systematic analysis of TAF function through the isolation of temperature-sensitive mutants followed by genome-wide expression analysis. We previously reported the results for seven yeast TAFs (Apone et al., 1998; Holstege et al., 1998; Lee et al., 2000). The percentage of the genome affected by temperature-sensitive alleles of each of these seven TAFs was calculated by comparison with data obtained from a strain defective in the largest subunit of RNA polymerase II, rpb1-1 (Holstege et al., 1998). By this criterion, we found that none of the seven TAFs analyzed are globally required for transcription, and each has a distinct role in transcription of the yeast genome (Lee et al., 2000). In order to generalize these conclusions, it is essential to examine the complete set of TAFs. Here we present the analysis of the six remaining essential yeast TAFs, and then use the complete collection of taf temperature-sensitive mutants to study the mechanisms of transcription activation.
Results
Isolation of new temperature-sensitive taf mutants
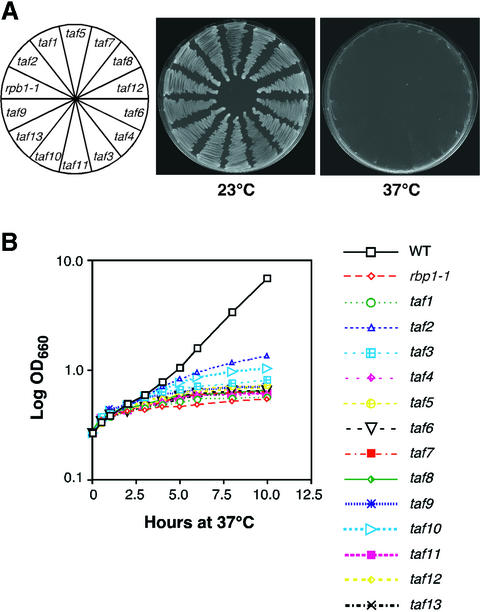
To complete the genome-wide transcription analysis for all essential yeast TAFs, we generated temperature-sensitive mutants of TAF3, TAF4, TAF7, TAF8, TAF11 and TAF13 (see Materials and methods for details). Strains bearing a taf temperature-sensitive allele were assessed for growth at 23 and 37°C, and those mutants that displayed a strong temperature-sensitive growth phenotype were selected for genome-wide transcription analysis. Growth of each of the six temperature-sensitive taf mutant strains used in this study is shown in Figure 1. As a comparison, growth of previously characterized taf mutant strains, as well as the RNA polymerase II mutant strain rbp1-1, was monitored in parallel. Figure 1A shows that all mutant strains grew at 23°C but not 37°C. To examine the mutant strains in greater detail, we analyzed the growth of each taf mutant strain in liquid culture. Figure 1B shows that each of the 13 taf mutant strains displayed a rapid growth arrest upon transfer from the permissive to the non-permissive temperature, comparable to that observed with the rpb1-1 strain.
Fig. 1. Isolation of new temperature-sensitive taf mutants. (A) Growth of taf temperature-sensitive mutant strains on solid media. Plates were incubated at 23 and 37°C for 48 h. As a comparison, growth of the RNA polymerase II mutant strain rbp1-1 was monitored in parallel. (B) Growth of taf temperature-sensitive mutant strains in liquid culture. Growth of the strains was monitored by measuring OD660 at various time points following the temperature shift to 37°C.
TAFs are selectively required for genome-wide transcription
To examine the role of each TAF in genome-wide transcription, RNA prepared from mutant strains grown at the restrictive temperature was used to interrogate high-density DNA oligonucleotide arrays as described previously (Holstege et al., 1998). The genome-wide expression profile of each taf mutant strain was compared to an isogenic wild-type strain 45 min after the shift to 37°C. The results for the six new taf temperature-sensitive mutants are shown in Table I, along with our results for the seven previously characterized TAFs. The percentage of yeast genes requiring an individual TAF ranged from 3% (TAF2) to 59–61% (TAF9), indicating that none of the 13 essential yeast TAFs is required for global transcription. Collectively, 84% of yeast genes required at least one of the 13 TAFs for transcription and the remaining 16% of genes were independent of all 13 TAFs. This analysis also revealed that 65% of yeast genes require at least one of the eight TFIID-specific TAFs. We have previously reported that 70% of yeast genes require at least one of the five TAFs shared between TFIID and SAGA (Lee et al., 2000).
Table I. Summary of genome-wide analyses of all 13 essential TAFs.
| TAF protein | Percent genome dependent |
|---|---|
| TAF1 (145) | |
| ts1* | 9 |
| ts2 | 14 |
| ts3* | 9 |
| TAF2 (150) | 3 |
| TAF3 (47) | 10 |
| TAF4 (48) | 11 |
| TAF5 (90) | 8 |
| TAF6 (60) | 18 |
| TAF7 (67) | 24 |
| TAF8 (65) | 51 |
| TAF9 (17) | |
| ts1 | 59 |
| ts2* | 61 |
| TAF10 (25) | |
| ts1 | 16 |
| ts2* | 19 |
| TAF11 (40) | 18 |
| TAF12 (61/68) | 9 |
| TAF13 (19) | 16 |
Several studies have reported that different temperature-sensitive mutants of a single TAF can behave variably (see, for example, Durso et al., 2001; Kirschner et al., 2002). We therefore analyzed three temperature-sensitive alleles of TAF1, two temperature-sensitive alleles of TAF9 and two temperature-sensitive alleles of TAF10 (Table I). We found that the percentage of the genome affected by inactivation ranged from 9 to 14% for TAF1, from 59 to 61% for TAF9 and from 16 to 19% for TAF10. In all instances, there was a >50% overlap in the genes affected by different mutants of the same TAF.
Three classes of yeast promoters defined by their TAF requirements
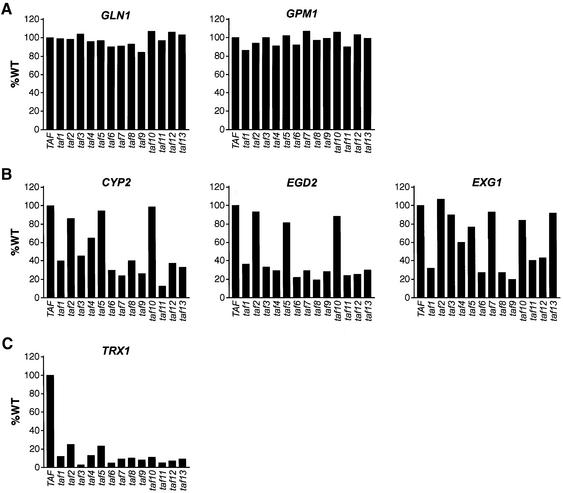
Previous classification of yeast promoters as TAF-dep or TAF-ind was based on the analysis of six TAFs (Kuras et al., 2000; Li et al., 2000). We therefore used the complete collection of taf temperature-sensitive mutants to confirm and extend the classification of yeast promoters. We selected six genes that, based upon genome-wide transcription data, appeared to represent three classes of promoters: those whose transcription was dependent upon all or almost all TAFs (TAF-dep; TRX1), those whose transcription did not require any TAFs (TAF-ind; GLN1 and GPM1) and those whose transcription was dependent upon a subset of TAFs (CYP2, EGD2 and EXG1).
Transcription of these six genes in each of the 13 taf mutant backgrounds was analyzed by northern blotting and the results are quantified and summarized in Figure 2. Consistent with the microarray data, transcription of GLN1 and GPM1 did not require any of the 13 TAFs analyzed (Figure 2A), confirming that these are truly TAF-ind promoters. In striking contrast, transcription of TRX1 was dependent upon all 13 TAFs (Figure 2C), confirming that TRX1 is a bona fide TAF-dep promoter. In fact, transcription of TRX1 was dependent upon more TAFs than other genes, such as RPS5, that have been previously used as representative TAF-dep promoters (see below). Finally, transcription of CYP2, EGD2 and EXG1 displayed a differential requirement for the 13 essential yeast TAFs (Figure 2B), again consistent with the genome-wide expression analysis. Thus, these results define three distinct classes of yeast promoters whose transcriptional requirements for TAFs differ substantially.
Fig. 2. Three classes of yeast promoters defined by their TAF requirements. (A) Transcription of the TAF-ind genes GLN1 and GPM1 in each of the 13 taf mutant backgrounds was analyzed by northern blotting and the results were quantified. (B) Transcription of CYP2, EGD2 and EXG1, whose transcription was dependent upon a subset of TAFs, was analyzed as described in (A). (C) Transcription of the TAF-dep gene TRX1 was analyzed as described in (A). The results are presented as the level of transcription observed in a taf mutant strain relative to an isogenic wild-type strain.
TAFs are generally required for TBP recruitment and PIC assembly
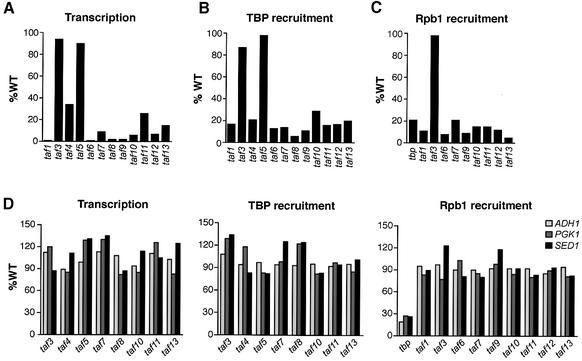
We have shown previously that TAF1 and TAF6 are required for recruitment of TBP to the RPS5 promoter (Li et al., 2002). To study the basis by which the other TAFs are required for RPS5 transcription, we used a chromatin immunoprecipitation (ChIP) assay to analyze TBP recruitment in strains bearing various temperature-sensitive taf mutants. Figure 3 shows that inactivation of TAF1, TAF4, TAF6, TAF7, TAF8, TAF9, TAF10, TAF11, TAF12 and TAF13 significantly diminished RPS5 transcription (Figure 3A) and recruitment of TBP to the RPS5 promoter (Figure 3B), whereas inactivation of TAF3 or TAF5 did not affect RPS5 transcription or TBP recruitment. Figure 3C shows that in all cases analyzed, inactivation of TAFs that were required to support transcription and TBP recruitment also resulted in a failure to recruit RNA polymerase II and thus to assemble a functional PIC. We conclude that the general basis by which TAFs are required for transcription is, at least in part, to facilitate TBP recruitment and assemble a functional PIC. In contrast, Figure 3D shows that on the ADH1, PGK1 and SED1 promoters, which are TAF independent (Li et al., 2000), none of the TAFs analyzed were required for transcription or recruitment of TBP and RNA polymerase II.
Fig. 3. TAFs are generally required for TBP recruitment and PIC assembly. (A) Transcription of the TAF-dep RPS5 gene in strains bearing various temperature-sensitive taf alleles was monitored by primer extension analysis. The results are presented as the level of transcription observed in a taf mutant strain relative to an isogenic wild-type strain. (B) ChIP assay to analyze TBP recruitment to the RPS5 promoter in taf mutant strains. The results are presented as the percentage of DNA immunoprecipitated relative to wild type. (C) ChIP assay to monitor recruitment of the RNA polymerase II subunit Rbp1 to the RPS5 promoter in taf mutant strains. (D) Analysis of transcription (left panel), TBP recruitment (middle panel) and Rpb1 recruitment (right panel) to the TAF-ind ADH1, PGK1 and SED1 promoters in various taf mutant strain backgrounds.
TBP is dispensable for activator-mediated recruitment of TAFs
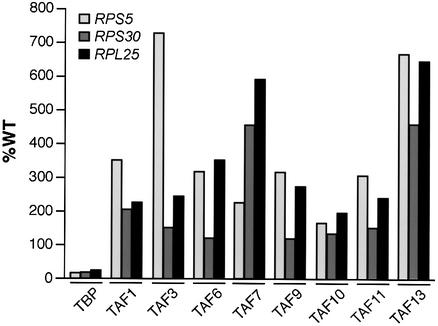
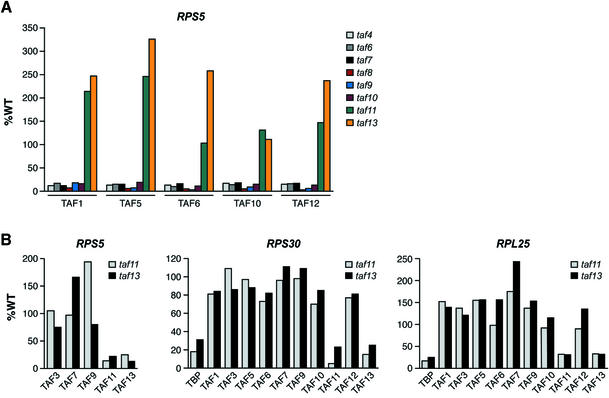
We have shown previously that following temperature-sensitive inactivation of TBP, TAF1, TAF6, TAF10 and TAF12 are still recruited to the core promoter of the RPS5 and ACT1 promoters in an activator-dependent fashion (Li et al., 2000). To determine the generality of this result, we analyzed recruitment of eight TAFs to three promoters following TBP inactivation. Figure 4 shows, as expected, that following temperature-sensitive inactivation of TBP, recruitment of TBP to the core promoters of RPS5, RPS30 and RPL25 substantially decreased, as did transcription of these genes (data not shown). In contrast, recruitment of TAF1, TAF3, TAF6, TAF7, TAF9, TAF10, TAF11 and TAF13 was not compromised in the absence of functional TBP, and in most cases actually increased several fold. Thus, although TAFs are generally required for recruitment of TBP to TAF-dep promoters (Figure 3), TBP is dispensable for recruitment of TAFs.
Fig. 4. TBP is dispensable for activator-mediated recruitment of TAFs. Recruitment of a set of TAFs to the core promoters of RPS5, RPS30 and RPL25 was monitored by ChIP analysis following temperature- sensitive inactivation of TBP.
TAF11 and TAF13 have a specialized role in TBP recruitment
To understand in greater detail the role of TAFs in TFIID complex assembly, we examined the consequences of inactivating an individual TAF on the recruitment of other TAFs. Figure 5A shows that inactivation of TAF4, TAF6, TAF7, TAF8, TAF9 or TAF10 substantially compromised recruitment of a representative set of TAFs (TAF1, TAF5, TAF6, TAF10, TAF12) to the RPS5 promoter. Unexpectedly, inactivation of TAF11 and TAF13 did not appear to affect recruitment of other TAFs, although as shown in Figure 3B, TBP recruitment was significantly reduced. Further analysis of TAF11 and TAF13 at RPS5 and two other promoters, RPS30 and RPL25, revealed that inactivation of TAF11 resulted in a loss of TAF13 recruitment and, conversely, inactivation of TAF13 resulted in a loss of TAF11 recruitment, whereas recruitment of other TAFs was unaffected (Figure 5B). Thus, TAF11 and TAF13 are required for the recruitment of TBP and each other but not other TAFs. In conjunction with previous studies (see Discussion), these results suggest that TAF11 and TAF13 may provide the major functional contacts with TBP during activator-mediated recruitment.
Fig. 5. TAF11 and TAF13 have a specialized role in TBP recruitment. (A) ChIP analysis monitoring TAF recruitment to the RPS5 promoter following temperature-sensitive inactivation of various TAFs. (B) ChIP analysis monitoring TAF recruitment to the promoters of RPS5, RPS30 and RPL25 in taf11 and taf13 mutant strains.
Discussion
Here we report the completion of genome-wide expression profiling analyses of yeast strains bearing temperature-sensitive mutations in the 13 essential TAFs. We find that no TAF is universally required for transcription, in contrast to GTFs such as the large subunit of RNA polymerase II, Srb4 and Kin28 (Holstege et al., 1998). Rather, each TAF is required for the expression of a subset of genes, ranging from 3% to 59–61% of the yeast genome. Collectively, 84% of yeast genes require at least one of the 13 essential TAFs; 70% of yeast genes require at least one of the five TAFs shared between TFIID and SAGA (Lee et al., 2000) and 65% of yeast genes require at least one of the eight TFIID-specific TAFs.
Although most studies are in agreement that yeast TAFs are required for selective, not global, transcription, there have been differences in the specific conclusions drawn (Green, 2000). These discrepancies are most likely a result of the different experimental approaches and methods of analysis used in each study (reviewed in Green, 2000). For example, yeast TAFs have been inactivated by several different approaches including temperature-sensitive mutations (Walker et al., 1996), conditional depletion (Apone et al., 1996; Li and Reese, 2000) and targeted protein degradation (Moqtaderi et al., 1996). The effects on transcription following TAF inactivation have also been determined by multiple approaches including analysis of either specific genes, total poly(A)+ mRNA (Thompson and Young, 1995; Kuldell and Buratowski, 1997; Michel et al., 1998), or the whole-genome using high-density oligonucleotide arrays (Holstege et al., 1998; Lee et al., 2000).
Several studies have reported that different temperature-sensitive mutants of a single TAF can differentially affect transcription (see, for example, Durso et al., 2001; Kirschner et al., 2002). Significantly, however, in these reports none of the various taf temperature-sensitive mutants analyzed compromised global transcription, consistent with our main conclusion. We have analyzed multiple temperature-sensitive mutants of three TAFs and found they affected genome-wide transcription to a comparable extent (Table I). The consistency of our results may be due to several factors, including selection of the temperature-sensitive mutants based on very tight growth phenotypes, analysis of transcription by expression profiling and normalization of the results to the expression profile of the RNA polymerase II rpb1-1 mutant.
In light of the multi-functional nature of TAFs, it would not be surprising if certain temperature-sensitive alleles had a larger effect on genome-wide transcription than others. For example, a temperature-sensitive taf mutant that is defective in maintaining TFIID complex integrity may compromise the function of other TAFs, resulting in an apparently broad effect on transcription. Consistent with this notion, some temperature-sensitive taf mutants result in the degradation of other TAFs and even TBP (Walker et al., 1996; Apone et al., 1998). Moreover, some TAFs are present in multiple complexes (Grant et al., 1998; Ogryzko et al., 1998; Martinez et al., 2001), and various mutant alleles could differentially affect the function of these complexes and thus genome-wide transcription.
The genetic intractability of higher eukaryotes has made it challenging to assess whether the results with yeast TAFs can be generalized to higher eukaryotes. However, several studies have suggested that, as in yeast, TAFs are also not required for general transcription in animal cells. For example, mammalian cell lines harboring a temperature-sensitive TAF1 allele do not exhibit a global defect in RNA polymerase II transcription under non-permissive conditions (Wang and Tjian, 1994; Suzuki-Yagawa et al., 1997). Similarly, murine cells in which both TAF10 genes have been deleted by targeted homologous recombination are not compromised for global transcription (Metzger et al., 1999; Mohan et al., 2003). Finally, inactivation of TAF9 in the DT40 chicken cell line results in a highly selective transcriptional defect (Chen and Manley, 2000).
Our analysis of the complete set of essential TAFs has confirmed previous reports that yeast promoters can be classified into distinct categories based on their transcriptional requirement for TAFs (Kuras et al., 2000; Li et al., 2000). TFIID assembly experiments indicated that on TAF-dep promoters TAFs are, in general, required for recruitment of TBP as well as other TAFs. In contrast, TBP is dispensable for TAF recruitment, consistent with the proposal that on TAF-dep promoters TAFs, and not TBP, are targeted by activators. Interestingly, we found that several genes encoding GTFs (e.g. TBP, SRB4, TOA1) are TAF independent. Our analysis also revealed a previously unrecognized third category of genes that are dependent on some, but not all, TAFs.
By combining the results of the transcription and ChIP analyses, we found that in some cases TAFs can be recruited to a promoter even though they are not required for transcription of that particular gene. For example, transcription of RPS5 is dependent on all TAFs except for TAF3 and TAF5. Using ChIP analysis we found that the entire TFIID complex, including TAF3 and TAF5, is recruited to the RPS5 promoter (Figure 4; S.R.Bhaumik and M.R.Green, unpublished observations). Analogous results have been found for the SAGA complex: for example, the SAGA subunit Gcn5 is recruited to the GAL1 UAS, even though it is not required for GAL1 transcription (Bhaumik and Green, 2001).
The results of our study have provided insight into the function of specific TAFs. Our results suggest that TAF11 and TAF13 make the primary functional contacts with TBP during activator-mediated recruitment. This role for TAF11 and TAF13 was unexpected, because a variety of biochemical studies have indicated that TAF1 makes the principle contact with TBP in the TFIID complex (Reese et al., 1994; Bai et al., 1997; Kokubo et al., 1998; Ogryzko et al., 1998). The fact that all other TAFs are still recruited following inactivation of TAF11, TAF13 and TBP further suggests that one or more of these other TAFs, and not TAF11, TAF13 or TBP, are targeted by activators. This model is consistent with several observations, including physical and genetic interactions between TAF11 and TAF13 (Birck et al., 1998; Komarnitsky et al., 1999), as well as physical interactions between TBP and TAF11 (Mengus et al., 1995; Lavigne et al., 1996, 1999; Kraemer et al., 2001) and between TBP and TAF13 (Mengus et al., 1995; Lavigne et al., 1996, 1999).
In summary, we have systematically analyzed the complete set of essential yeast TAFs and found that none of the 13 TAFs is required for global transcription. Transcription complex assembly experiments using the complete collection of taf temperature-sensitive mutants have revealed specific and unanticipated roles for individual TAFs in TBP recruitment. Collectively, our results confirm and extend the proposal that individual TAFs have selective and distinct functions.
Materials and methods
Yeast strains and media
Growth of yeast cells in rich (YPD) or synthetic media was performed according to standard procedures (Guthrie et al., 1991). The PCR-mediated mutagenesis procedure used to generate plasmid-borne temperature-sensitive alleles of taf3, taf4, taf7, taf8, taf11 and taf13 was performed essentially as described previously (Leung et al., 1989). Heterozygous tafx/TAFx diploid strains were created through targeted gene deletion using PCR-based modules (Longtine et al., 1998); the diploid strain was then transformed with a CEN URA3 plasmid carrying a wild-type copy of the TAFx gene. Haploid taf mutant strains were then generated by standard tetrad dissection followed by plasmid shuffle. The detailed genotypes of the 13 haploid yeast strains harboring a temperature-sensitive taf allele are listed in Table II. The rbp1-1 strain (Nonet et al., 1987), as well as tbp-ts1 and its corresponding isogenic wild-type strain (Cormack and Struhl, 1992), has been described previously.
Table II. List of Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| WCS132 | _MAT_α ade2-101a his3_Δ_200 leu2_-Δ_1 lys2-801a trp1_-Δ_63 ura3-52 taf2::TRP1 [pRS313 _(CEN HIS3 taf2-1)_] | Lee et al. (2000) |
| WCS169 | MATa ade2-101a his3_Δ_200 leu2_-Δ_1 lys2-801a trp1_-Δ_63 ura3-52 taf11::TRP1 [pRS413 _(CEN HIS3 taf11-ts2)_] | This study |
| WCS179 | MATa ade2-101a his3_Δ_200 leu2_-Δ_1 lys2-801a trp1_-Δ_63 ura3-52 taf13::HIS3 [pRS414 _(CEN TRP1 taf13-ts2)_] | This study |
| WCS189 | MATa ade2-101 his3_Δ_200 leu2-3,112 trp1_Δ_901 ura3-52 suc2_Δ_9::LYS2 taf10::TRP1 [pRS413 _(CEN HIS3 taf10-ts1)_] | Lee et al. (2000) |
| WCS203 | MATa ade2-101a his3_Δ_200 leu2_-Δ_1 lys2-801a trp1_-Δ_63 ura3-52 taf3::HIS3 [pRS414 _(CEN TRP1 taf3-ts3)_] | This study |
| WCS213 | MATa ade2-101a his3_Δ_200 leu2_-Δ_1 lys2-801a trp1_-Δ_63 ura3-52 taf7::TRP1 [pRS413 _(CEN HIS3 taf7-ts1)_] | This study |
| WCS299 | MATa ade2-101a his3_Δ_200 leu2_-Δ_1 lys2-801a trp1_-Δ_63 ura3-52 taf8::TRP1 [pRS413 _(CEN HIS3 taf8-ts7)_] | This study |
| WCS301 | MATa ade2-101a his3_Δ_200 leu2_-Δ_1 lys2-801a trp1_-Δ_63 ura3-52 taf4::TRP1 [pRS413 _(CEN HIS3 taf4-ts18)_] | This study |
| LY189 | MATa ade2-101 his3_Δ_200 leu2_Δ_1 lys2-801 ura3_Δ_99 taf5::HIS3 [Lp25 _(CEN LEU2 taf5-ts9-12)_] | Apone et al. (1996) |
| LY761 | MATa ade2-1 his3-11, 15 leu2-3,112 trp1-1 ura3-1 taf9::LEU2 [pRS413 _(CEN HIS3 taf9-ts2)_] | Apone et al. (1998) |
| YSB552 | MATa his3_Δ_200 leu2_Δ_1 or leu2-3,112 ura3-52 taf6_Δ::LEU2_ [pRS313 _(CEN HIS3 taf6-19)_] | Lee et al. (2002) |
| YSB547 | MATa ade2 his3_Δ_200 leu2::PET56 trp1_Δ_1 ura3-52 taf12_Δ_259::LEU2 [pRS313 _(CEN HIS3 taf12-23 ts)_] | Michel et al. (1998) |
| YSW93 | MATa ade2-101 his3_Δ_200 leu2_-Δ_1 lys2-801 ura3_Δ_99 GAL2 GAL3 taf1::LEU2 [pSW104 _(CEN HIS3 taf1-ts2)_] | Walker et al. (1996) |
Genome-wide expression profiling and data analysis
The oligonucleotide microarrays and GeneChip software used in this study were manufactured by Affymetrix. Expression data collection, normalization, correction and profiling were performed essentially as described previously (Holstege et al., 1998); detailed protocols are available at http://web.wi.mit.edu/young/expression/tech.html. A compiled database including datasets for the six TFIID-specific TAFs analyzed in this study and for the seven TAFs previously reported (Lee et al., 2000) is available at http://staffa.wi.mit.edu/cgi-bin/young/setlistWS.cgi.
Transcription assays
Wild-type and taf mutant strains were grown at 23°C; after shifting to the non-permissive temperature (37°C) for 1 h, cells were harvested and total cellular RNA was prepared. For the experiments shown in Figure 2, transcription was analyzed by northern blotting as described previously (Shen and Green, 1997). For the experiments shown in Figure 3, primer extension analysis was carried out as described previously (Li et al., 2000) using the following primers: RPS5, 5′-GACTGGGGTGAATTCTTCAACAACTTC-3′ and ADH1, 5′-TATCCTTGTGTTCCAATTTACCGTGG-3′; PGK1, 5′-AAATCTTGGACAGACAACT TTGAAG-3′; and SED1, 5′-AGTAGTCGAGGCTAAACCGG-3′. The results were quantitated and presented as the ratio of signal obtained in the taf mutant relative to that obtained in an isogenic wild-type strain.
Formaldehyde-based in vivo cross-linking and ChIP
Yeast strains harboring temperature-sensitive taf alleles were grown in YPD at 23°C to an OD600 of 0.8 and then transferred to 37°C for 1 h. Formaldehyde-based in vivo cross-linking and ChIP was performed essentially as described previously (Bhaumik and Green, 2002). Specific polyclonal rabbit antibodies recognizing TAF3, TAF7, TAF9, TAF11 and TAF13 were obtained from P.Anthony Weil (Sanders et al., 1999; Sanders and Weil, 2000), and polyclonal antibodies against TBP, TAF1, TAF5, TAF6, TAF10 and TAF12 were generated in our laboratory. Immunoprecipitated DNA was quantitated and presented as the ratio of immunoprecipitated to input relative to wild type. The following sets of primer pairs were used for PCR analysis: RPS5 (core), 5′-GGCCAACTTCTACGCTCACGTTAG-3′ and 5′-CGGTGTCAGACATCTTTGGAATGGTC-3′; RPS30 (core), 5′-TGTCGTGAGG TTGTCGGAAACCTC-3′ and 5′-ACCATATTTGCGTAGTTTTTGTATGG-3′; RPL25 (core), 5′-TTGGGGCTTCCCAATGCAAC-3′ and 5′-AGTTCACATACCAGATGGAGCC-3′; ADH1 (core), 5′-GGTATACGGCCTTCCTTCCAGTTAC-3′ and 5′-GAACGAGAACAATGA CGAGGAAACAAAAG-3′; PGK1 (core), 5′-CATCTAAGAACTTGAAAAACTACG-3′ and 5′-CAGCCTGTTCTCACACACTC-3′; and SED1 (core), 5′-TTTAGATTGGCCGTAGGGGC-3′ and 5′-CAAGAGAATAGAAAAAGAGAGGTGAG-3′.
Acknowledgments
Acknowledgements
We thank P.Anthony Weil for the anti-TAF antibodies; Lynne Apone for helpful discussions; Jin Wang and Lynn Jones for technical assistance; and Sara Evans for editorial assistance. This work was supported in part by grants from the NIH (M.R.G.) and the Leukemia and Lymphoma Society (W.-C.S.). M.R.G. is an investigator and S.R.B. is an associate of the Howard Hughes Medical Institute.
References
- Albright S.R. and Tjian,R. (2000) TAFs revisited: more data reveal new twists and confirm old ideas. Gene, 242, 1–13. [DOI] [PubMed] [Google Scholar]
- Apone L.M., Virbasius,C.M., Reese,J.C. and Green,M.R. (1996) Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev., 10, 2368–2380. [DOI] [PubMed] [Google Scholar]
- Apone L.M., Virbasius,C.A., Holstege,F.C., Wang,J., Young,R.A. and Green,M.R. (1998) Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell, 2, 653–661. [DOI] [PubMed] [Google Scholar]
- Bai Y., Perez,G.M., Beechem,J.M. and Weil,P.A. (1997) Structure–function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAF(II)130. Mol. Cell. Biol., 17, 3081–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S.R. and Green,M.R. (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev., 15, 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S.R. and Green,M.R. (2002) Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol., 22, 7365–7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birck C., Poch,O., Romier,C., Ruff,M., Mengus,G., Lavigne,A.C., Davidson,I. and Moras,D. (1998) Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell, 94, 239–249. [DOI] [PubMed] [Google Scholar]
- Burley S.K. and Roeder,R.G. (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem., 65, 769–799. [DOI] [PubMed] [Google Scholar]
- Chen Z. and Manley,J.L. (2000) Robust mRNA transcription in chicken DT40 cells depleted of TAF(II)31 suggests both functional degeneracy and evolutionary divergence. Mol. Cell. Biol., 20, 5064–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B.P. and Struhl,K. (1992) The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell, 69, 685–696. [DOI] [PubMed] [Google Scholar]
- Durso R.J., Fisher,A.K., Albright-Frey,T.J. and Reese,J.C. (2001) Analysis of TAF90 mutants displaying allele-specific and broad defects in transcription. Mol. Cell. Biol., 21, 7331–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P.A., Schieltz,D., Pray-Grant,M.G., Steger,D.J., Reese,J.C., Yates,J.R.,III and Workman,J.L. (1998) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell, 94, 45–53. [DOI] [PubMed] [Google Scholar]
- Green M.R. (2000) TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci., 25, 59–63. [DOI] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (eds) (1991) Methods in Enzymology: Guide to Yeast Genetics and Molecular and Cell Biology. Academic Press, New York, NY. [Google Scholar]
- Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Kirschner D.B., vom Baur,E., Thibault,C., Sanders,S.L., Gangloff,Y.G., Davidson,I., Weil,P.A. and Tora,L. (2002) Distinct mutations in yeast TAF(II)25 differentially affect the composition of TFIID and SAGA complexes as well as global gene expression patterns. Mol. Cell. Biol., 22, 3178–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo T., Swanson,M.J., Nishikawa,J.I., Hinnebusch,A.G. and Nakatani,Y. (1998) The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol., 18, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P.B., Michel,B. and Buratowski,S. (1999) TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev., 13, 2484–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer S.M., Ranallo,R.T., Ogg,R.C. and Stargell,L.A. (2001) TFIIA interacts with TFIID via association with TATA-binding protein and TAF40. Mol. Cell. Biol., 21, 1737–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuldell N.H. and Buratowski,S. (1997) Genetic analysis of the large subunit of yeast transcription factor IIE reveals two regions with distinct functions. Mol. Cell. Biol., 17, 5288–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L., Kosa,P., Mencia,M. and Struhl,K. (2000) TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science, 288, 1244–1248. [DOI] [PubMed] [Google Scholar]
- Lavigne A.C., Mengus,G., May,M., Dubrovskaya,V., Tora,L., Chambon,P. and Davidson,I. (1996) Multiple interactions between hTAFII55 and other TFIID subunits. Requirements for the formation of stable ternary complexes between hTAFII55 and the TATA-binding protein. J. Biol. Chem., 271, 19774–19780. [DOI] [PubMed] [Google Scholar]
- Lavigne A.C., Gangloff,Y.G., Carre,L., Mengus,G., Birck,C., Poch,O., Romier,C., Moras,D. and Davidson,I. (1999) Synergistic transcriptional activation by TATA-binding protein and hTAFII28 requires specific amino acids of the hTAFII28 histone fold. Mol. Cell. Biol., 19, 5050–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I. and Young,R.A. (2000) Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet., 34, 77–137. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Causton,H.C., Holstege,F.C., Shen,W.C., Hannett,N., Jennings,E.G., Winston,F., Green,M.R. and Young,R.A. (2000) Redundant roles for the TFIID and SAGA complexes in global transcription. Nature, 405, 701–704. [DOI] [PubMed] [Google Scholar]
- Leung D.W., Chen,E. and Goeddel,D.V. (1989) A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique, 1, 11–15. [Google Scholar]
- Li B. and Reese,J.C. (2000) Derepression of DNA damage-regulated genes requires yeast TAF(II)s. EMBO J., 19, 4091–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.Y., Bhaumik,S.R. and Green,M.R. (2000) Distinct classes of yeast promoters revealed by differential TAF recruitment. Science, 288, 1242–1244. [DOI] [PubMed] [Google Scholar]
- Li X., Bhaumik,S., Zhu,X., Li,L., Shen,W., Dixit,B. and Green,M. (2002) Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr. Biol., 12, 1240–1244. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A.,III, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Martinez E., Palhan,V.B., Tjernberg,A., Lymar,E.S., Gamper,A.M., Kundu,T.K., Chait,B.T. and Roeder,R.G. (2001) Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol., 21, 6782–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengus G., May,M., Jacq,X., Staub,A., Tora,L., Chambon,P. and Davidson,I. (1995) Cloning and characterization of hTAFII18, hTAFII20 and hTAFII28: three subunits of the human transcription factor TFIID. EMBO J., 14, 1520–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D., Scheer,E., Soldatov,A. and Tora,L. (1999) Mammalian TAFII30 is required for cell cycle progression and specific cellular differentiation programmes. EMBO J., 18, 4823–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Komarnitsky,P. and Buratowski,S. (1998) Histone-like TAFs are essential for transcription in vivo. Mol. Cell, 2, 663–673. [DOI] [PubMed] [Google Scholar]
- Mohan W.S. II, Scheer,E., Wendling,O., Metzger,D. and Tora,L. (2003) TAF10 [TAF(II)30] is necessary for TFIID stability and early embryogenesis in mice. Mol. Cell. Biol., 23, 4307–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z., Bai,Y., Poon,D., Weil,P.A. and Struhl,K. (1996) TBP-associated factors are not generally required for transcriptional activation in yeast. Nature, 383, 188–191. [DOI] [PubMed] [Google Scholar]
- Nonet M., Scafe,C., Sexton,J. and Young,R. (1987) Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol., 7, 1602–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko V.V., Kotani,T., Zhang,X., Schiltz,R.L., Howard,T., Yang,X.J., Howard,B.H., Qin,J. and Nakatani,Y. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Lagrange,T. and Reinberg,D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- Ptashne M. and Gann,A. (1997) Transcriptional activation by recruitment. Nature, 386, 569–577. [DOI] [PubMed] [Google Scholar]
- Reese J.C., Apone,L., Walker,S.S., Griffin,L.A. and Green,M.R. (1994) Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature, 371, 523–527. [DOI] [PubMed] [Google Scholar]
- Roeder R.G. (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci., 21, 327–335. [PubMed] [Google Scholar]
- Sanders S.L. and Weil,P.A. (2000) Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem., 275, 13895–13900. [DOI] [PubMed] [Google Scholar]
- Sanders S.L., Klebanow,E.R. and Weil,P.A. (1999) TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem., 274, 18847–18850. [DOI] [PubMed] [Google Scholar]
- Shen W.-C. and Green,M.R. (1997) Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell, 90, 615–624. [DOI] [PubMed] [Google Scholar]
- Suzuki-Yagawa Y., Guermah,M. and Roeder,R.G. (1997) The ts13 mutation in the TAF(II)250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol. Cell. Biol., 17, 3284–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C.M. and Young,R.A. (1995) General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl Acad. Sci. USA, 92, 4587–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tora L. (2002) A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev., 16, 673–675. [DOI] [PubMed] [Google Scholar]
- Walker S.S., Reese,J.C., Apone,L.M. and Green,M.R. (1996) Transcription activation in cells lacking TAFIIS. Nature, 383, 185–188. [DOI] [PubMed] [Google Scholar]
- Wang E.H. and Tjian,R. (1994) Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science, 263, 811–814. [DOI] [PubMed] [Google Scholar]