Transcription and Analysis of Polymorphism in a Cluster of Genes Encoding Surface-Associated Proteins of Clostridium difficile (original) (raw)
Abstract
Recent investigations of the Clostridium difficile genome have revealed the presence of a cluster of 17 genes, 11 of which encode proteins with similar two-domain structures, likely to be surface-anchored proteins. Two of these genes have been proven to encode proteins involved in cell adherence: slpA encodes the precursor of the two proteins of the S-layer, P36 and P47, whereas cwp66 encodes the Cwp66 adhesin. To gain further insight into the function of this cluster, we further focused on slpA, cwp66, and cwp84, the latter of which encodes a putative surface-associated protein with homology to numerous cysteine proteases. It displayed nonspecific proteolytic activity when expressed as a recombinant protein in Escherichia coli. Polymorphism of cwp66 and cwp84 genes was analyzed in 28 strains, and transcriptional organization of the three genes was explored by Northern blots. The slpA gene is strongly transcribed during the entire growth phase as a bicistronic transcript; cwp66 is transcribed only in the early exponential growth phase as a polycistronic transcript encompassing the two contiguous genes upstream. The putative proteins encoded by the cotranscribed genes have no significant homology with known proteins but may have a role in adherence. No correlation could be established between sequence patterns of Cwp66 and Cwp84 and virulence of the strains. The cwp84 gene is strongly transcribed as a monocistronic message. This feature, together with the highly conserved sequence pattern of cwp84, suggests a significant role in the physiopathology of C. difficile for the Cwp84 protease, potentially in the maturation of surface-associated adhesins encoded by the gene cluster.
Clostridium difficile, a gram-positive spore-forming anaerobic bacterium, is a significant nosocomial enteric pathogen, causing pseudomembranous colitis (PMC) and many cases of antibiotic-associated diarrhea (AAD) (29). The two main virulence factors are exotoxins, toxins A and B, both of which damage the human colonic mucosa and are potent tissue-damaging enzymes (3, 23). C. difficile takes advantage of the disturbance of the normal colonic flora, following antibiotic treatment, to colonize the gastrointestinal tract.
C. difficile has been shown to adhere in vitro to a variety of cultured cell lines, including Vero (37) and Caco-2 cells (9, 14). Interest for adherence determinants has recently risen, and numerous studies have characterized factors involved in the adherence and colonization processes, such as S-layer proteins (5, 6, 7), the adhesin Cwp66 (37), flagella (34), the heat-shock protein GroEL (17), and hydrolytic enzymes (28, 31).
C. difficile expresses on its surface an S-layer, which forms a regular two-dimensional array visible by electron microscopy (7). Each strain carries an S-layer, which is composed of two distinct proteins, one of high molecular weight called P47 and another of low molecular weight called P36. Both of these subunits are encoded by the slpA gene and are produced from the posttranslational cleavage of a precursor (6, 19). P36, encoded by the variable 5′-terminal part of the slpA gene, is immunodominant, and its variability could play a role in the antigenic variation of the bacteria (7, 8, 19). P47 is encoded by the conserved 3′-terminal part of slpA, displays significant homology to the cell wall-anchoring domain of the autolysin CwlB of Bacillus subtilis (6, 7, 19), and shows strong and specific binding to gastrointestinal tissues and some extracellular matrix proteins (collagen I, thrombospondin, and vitronectin) (5). Recently the sequences of the variable region of the slpA gene were found to be strictly identical within a given serogroup (except for serogroup A) but divergent between serogroups (20).
Cwp66 (clostridial wall protein of 66 kDa) is a surface-associated protein with a two-domain structure. The N-terminal part of the protein presents homology to the cell wall-anchoring domain of the autolysin CwlB of B. subtilis; the C-terminal domain is cell surface exposed. Cwp66 has been shown to mediate adherence of C. difficile to Vero cells (37).
Since antibodies directed against P47 (5), P36 (19), and Cwp66 but also against the flagellar cap FliD (34) and GroEL (17) partially inhibit adherence of C. difficile, it seems very likely that the binding of C. difficile to host cells involves several proteins.
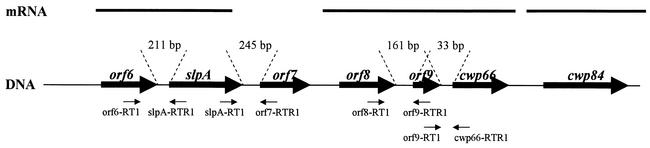
The genes encoding Cwp66 and the S-layer precursor are located close to each other in a 37-kb DNA fragment (Fig. 3 in reference 19) in the C. difficile genome (http://www.sanger.ac.uk/Projects.C_difficile/). This genetic locus carries 17 open reading frames (ORFs) in the same orientation, 11 of which encode proteins which present a two-domain structure, as described above: a domain homologous to the cell wall-anchoring domain of CwlB, present in either the N-terminal or the C-terminal part, and a second domain (named the functional domain) displaying remote homologies with different enzymes or structural proteins from gram-positive bacteria (4, 6, 19).
FIG. 3.
Genetic organization of the DNA cluster carrying the slpA, cwp66, and cwp84 genes from the genome of C. difficile 630. The thick horizontal arrows represent the open reading frames with their names indicated above. Thick lanes above represent transcripts that are monocistronic (for cwp84) or polycistronic (for _orf6_-slpA and _orf8_-_orf9_-cwp66). The sizes of intergenic regions, determined from analysis of the available genome sequence of strain 630, are indicated between the dotted lines. Thin arrows below DNA indicate the localization of primers used in intergenic RT-PCR.
The aim of this study was to characterize this putative virulence cluster by focusing on three genes: slpA, cwp66, and cwp84. The cwp84 gene, located immediately downstream from cwp66, encodes a putative 84-kDa protein with a characteristic signal peptide, whose functional domain displays homologies with several cysteine proteases, and shows the conserved Pept_C1 domain of the papain family. Since extracellular proteases have been described to be virulence factors in many bacteria (22), we cloned and expressed the corresponding gene to permit functional studies. We studied the transcription of slpA, cwp66, and cwp84 genes by Northern blotting to determine whether these genes are expressed as part of an operon. We also investigated the polymorphism of cwp66 and cwp84 genes in different strains of C. difficile in an attempt to see if a correlation between pattern of sequences and virulence exists.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Twenty-eight C. difficile isolates belonging to nine different serogroups were studied (Table 1). The strains include the nine reference strains for specific serogroups (11), clinical isolates from PMC, AAD, or asymptomatic carriers, and one strain of animal origin. Clostridial strains were grown under anaerobic conditions in TGY (tryptone glucose yeast infusion broth; Difco Laboratories) and on Columbia agar plates supplemented with 4% horse blood (Biomerieux).
TABLE 1.
Strains of C. difficile used in this study
The BL21 Escherichia coli strain, used as a host for cwp84 cloning, was grown on Luria broth or brain heart infusion (BHI) agar or in broth (Difco Laboratories), supplemented with 100 μg of ampicillin/ml to maintain the pGEX plasmid.
Serratia sp. and Streptococcus pyogenes strains, used as positive controls in proteolytic assays, were grown on BHI agar or in broth, at 37°C, under aerobic conditions.
Typing of strains.
Typing of strains belonging to serogroup C was done by PCR ribotyping and randomly amplified polymorphic DNA (RAPD) analysis with primers AP3, AP4 (2), and APRB11 (10), as described previously.
Cloning of cwp84 into the vector pGEX-6P-1 and protein expression.
To clone the cwp84 gene into the pGEX-6P-1 expression vector (Amersham Biosciences), two oligonucleotide primers, pGEXcwp84-_Eco_RI (5′GGGTAGAATTCAGAAAGTATAAATCA3′) and pGEXcwp84-_Xho_I (5′TCTCTCGAGTCACTATTTTCCTAAAAG3′), incorporating an _Eco_RI and _Xho_I site, respectively (underlined), were used to amplify by PCR the full-length coding region of the cwp84 gene of the 79685 strain. The resulting 2.4-kb DNA fragment was digested with the two enzymes and ligated (1 U of T4 ligase; Invitrogen) between the _Eco_RI and _Xho_I sites of pGEX-6P-1. Sequencing of the insert was done, with primers pGEX-3′ and pGEX-5′ (Amersham Biosciences) and internal primers, to ensure that no sequence mismatch occurred during the cloning. The plasmid carrying an in-frame fusion between gst and cwp84 was transformed into E. coli BL21 (Amersham Biosciences). Subsequent protein expression and purification steps were performed by induction of the tac promoter with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), following by a single-step affinity chromatography employing glutathione-Sepharose-4B, as described in protocols from Amersham Biosciences. Purification attempts were done with or without cysteine protease inhibitors, such as leupeptin and E64 (Sigma). Before cleavage from the glutathione _S_-transferase (GST), purified fractions were tested for the presence of recombinant protein by immunoblotting, as previously described (37), with antibodies directed against GST (Amersham Biosciences) and against the N-terminal part of Cwp66, which is 56% homologous to the C-terminal part of Cwp84, as shown by amino acid sequence alignment.
Proteolytic assays.
Proteolytic assays were done with the clone E. coli BL21(pGEXΩ_cwp84_) and the strain without insert BL21(pGEX) as a negative control. Serratia sp. and S. pyogenes were used as positive controls for gelatinase and cysteine protease activity, respectively. Assays were performed on gelatin, skimmed milk, and azocoll. To test for gelatinase activity, clones were grown aerobically for 24 h in BHI broth, and then 0.1 mM IPTG and a photographic film containing gelatin and charcoal were added, and cultures were continued for 24 h under anaerobic conditions. Gelatinase activity was assessed by release of charcoal in the culture medium after lysis of gelatin. Caseinase activity was determined on skimmed milk supplemented or not with 0.1 mM IPTG by the measure of the clear halo around the streak culture. Azocoll degradation was determined by measuring optical density at 520 nm (OD520) after 48 h of culture following by 6 h of contact with azocoll, as previously described (28).
RNA manipulations.
Total RNA extraction was performed at the beginning (OD600, ∼0.3) and the middle (OD600, ∼0.7) of the exponential growth phase and during the stationary phase from C. difficile strain 630 (18-h culture) according to the Trizol (Invitrogen) procedure previously described (13), with some modifications. Briefly, after bacterial lysis in prelysis buffer containing 50 mM Tris (pH 8.0), 25% sucrose, and 10 mg of lysozyme/ml and RNA extraction with trizol, RNA was precipitated in absolute ethanol and stored until utilization at −20°C. Before every manipulation, an aliquot was removed, washed with 70% ethanol, and dissolved in RNase-free water at 60°C. The RNA concentration was measured optically at 260 nm. Fifteen to thirty micrograms of RNA was electrophoresed in 20 mM guanidine thiocyanate (15; I. Podglajen, personal communication)-1.8% agarose gel in 1× Tris-borate-EDTA at 65 V for 4 to 5 h in a horizontal gel electrophoresis apparatus. RNA was then transferred directly to Nylon membrane Hybond-N+ (Amersham Biosciences) by the downflow capillarity method.
(i) Generation of probes.
PCR products obtained with primers described in Table 2 were purified and labeled using the ECL direct labeling and detection system (Amersham Biosciences), according to the manufacturer's instructions.
TABLE 2.
Primers used in this study
| Purpose, relevant gene, or primer name | Oligonucleotide sequence (5′ → 3′) | Position (bp) |
|---|---|---|
| Probe generation | ||
| slpA | ||
| slpA-NB1 | GCT GCT CCT GTT TTT GCT | +54/+71 |
| slpA-NBR1 | CTT CTT TTG CAT TTA TAA C | +828/+810 |
| cwp66 | ||
| cwp66-R1 | AAT CCA TCA TCT GTA GCG | +1818/+1801 |
| cwp66-NB1 | CTC AAA TTG GTG GCT TAG G | +915/+933 |
| cwp66-NBR1 | ATG GCT CTT CAT CTG TTG G | +1712/+1694 |
| cwp84 | ||
| cwp84-NB1 | CTC TAG ATG GAG TAG AAA CT | +104/+123 |
| cwp84-NBR1 | GAC CAG CAT ATT CAA GTT G | +1030/+1012 |
| gdh | ||
| gdh-NB1 | GAT GTA AAT GTC TTC GAG ATG | +13/+33 |
| gdh-NBR1 | GGT CCA TTA GCA GCC TCA C | +974/+956 |
| RT-PCR | ||
| slpA | ||
| orf6-RT1 | AGA TGG ACA AGT GTA TGC | +1545/+1562 |
| slpA-RTR1 | CAG TCG TTT TTA ACT ACA G | +115/+97 |
| slpA-RT1 | ATG GTG GAA CTA ACT TAG | +2077/+2094 |
| orf7-RTR1 | TTG CTC ATC TGC TTT GTC | +41/+24 |
| cwp66 | ||
| orf8-RT1 | AGT TGA ATT GAC AGT AAT AC | +1824/+1843 |
| orf9-RTR1 | CTG TGC ATA ATA TGA CAT GT | +60/+41 |
| orf9-RT1 | GCT ATA GGA TAT CAT TCA G | +576/+594 |
| slpA-RTR1 | GAT AAA GCA TCT GCT ATG G | +195/+177 |
| Amplification of functional gene domains | ||
| cwp84 | ||
| orfE-ATG | GGG GTA AAC ATG AGA AAG | −9/+9 |
| cwp84-TAA | GGA ACT CCA TTT ACT ACT G | +1178/+1150 |
| cwp66 | ||
| cwp66-1 | AGC AGT GGG TGT ATT AGC | +805/+822 |
| cwp66-R1 | AAT CCA TCA TCT GTA GCG | +1818/+1801 |
| cwp66-NB1 | CTC AAA TTG GTG GCT TAG G | +915/+933 |
| cwp66-NBR1 | ATG GCT CTT CAT CTG TGG G | +1712/+1694 |
| cwp66-ATG | CGA AAG AAT TAG GAG GTA AGA | −35/−15 |
| cwp66-TAA | TAT GTA TGT AAT GAT TGA TTT GC | +1992/+1970 |
(ii) Northern blot.
Membranes were prehybridized for 1 h at 42°C in hybridization buffer (Amersham Biosciences) prior to the addition of probe. Hybridizations were performed overnight at 42°C and followed by two washes (20 min each) in 6 M urea-0.4% sodium dodecyl sulfate (SDS)-0.5×, 0.2×, or 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), depending on the probe used, and two washes in 2× SSC (10 min each). Detection was carried out with the ECL direct labeling and detection system (Amersham Biosciences). A probe for the glutamate-deshydrogenase gdh gene of C. difficile was generated for use as a positive control for the hybridization experiments, as previously described (16).
RT-PCR.
RNA were treated by RNase free-DNase (Amersham Biosciences). Reverse transcription (RT)-PCR was performed using the SuperscriptOne-Step RT-PCR with the Platinum Taq kit (Invitrogen), as recommended by the manufacturer, with 100 ng of RNA. Primers used were those designed to generate the probes for Northern blots and some others chosen to check the polycistronic nature of mRNA (Table 2). Simultaneously, PCR was performed on samples with the same oligonucleotides to exclude false-positive amplification from residual DNA.
Analysis of cotranscribed ORFs.
Homologies of cotranscribed ORFs with known proteins were searched with BLAST (http://www.ncbi.nlm.nih.gov/BLAST). Particular motifs and secondary structures were analyzed with the PROSITE and SOPMA programs (http://www.pbil.univ-lyon1.fr/).
PCR amplifications.
DNA from the 28 isolates was extracted according to the protocol provided in the DNeasy tissue kit (Qiagen). The primers used for amplification of the functional regions of the cwp66 and cwp84 genes (corresponding to their C-terminal and N-terminal domains, respectively [Fig. 3 in reference 19]), are listed in Table 2. PCR amplification was performed in a Perkin-Elmer GeneAmp PCR System 2400 thermocycler, with one bead of Ready-To-Go PCR kit (Amersham Biosciences), each primer at a final concentration of 0.4 μM, and 100 ng of genomic DNA (in a 25-μl reaction mixture). Initial denaturation was carried out at 95°C for 5 min, following by 35 cycles of amplification: denaturation at 95°C (1 min), annealing at 50 or 55°C (depending on primers used) for 1 min, and extension at 72°C for 1 min, 1 min 30 s, or 2 min. An additional step of extension for 10 min at 72°C was performed at the end of the amplification. Samples (3 μl) of amplified products were analyzed by electrophoresis in a 1.0% (wt/vol) agarose gel.
cwp66 and cwp84 sequencing.
PCR products with the expected size were purified with the High Pure PCR purification kit (Roche). Automatic DNA sequencing was performed on the two strands with the Big Dye Terminator cycle-sequencing kit (Applied Biosystems) and analyzed with an ABI PRISM 310 genetic analyzer (Perkin-Elmer). Initial sequencing was carried out with the same primers as used for PCR, and more sequence was acquired by the DNA walking strategy.
Nucleotide and protein sequence alignments were performed with the DNA CLUSTAL W program (http://www.ebi.ac.uk/clustalw/).
Nucleotide sequence accession numbers.
The GenBank nucleotide sequence accession numbers of the functional domain of the genes cwp66 and cwp84 from the C. difficile strains studied are given in Table 1.
RESULTS
Expression of Cwp84 and proteolytic assays.
In an attempt to produce a recombinant protein corresponding to the cwp84 ORF in the cwp gene cluster of C. difficile (4, 6, 19), encoding a potential protease, the cwp84 gene of the strain 79685 was expressed as an in-frame fusion with GST in E. coli. The E. coli clone pGEXΩ_cwp84_ presented nonspecific proteolytic activity on gelatin and skimmed milk compared to the negative control (data not shown). A light degradation of azocoll could be observed for this clone, as measured by the mean of OD520 of three distinct experiments: OD520 for E. coli pGEXΩ_cwp84_ = 0.074; OD520 for E. coli pGEX = 0.022; OD520 for Serratia sp. = 0.379; OD520 for S. pyogenes = 0.109.
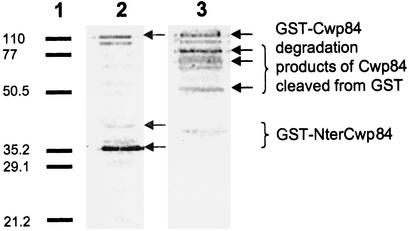
Purification of the recombinant protein was carried out with 0.1 mM IPTG induction, since use of 0.5 and 1.0 mM concentrations led to formation of inclusion bodies. Immunoblot analysis of purified fractions from various assays showed that the two antibodies used (anti-GST and anti-Cwp66 N-ter) revealed one very faint band of 110 kDa, the size of which corresponds to that of the fusion protein GST-Cwp84 (Fig. 1). Numerous other bands were recognized alternatively with the two antibodies. These bands, especially those specifically recognized by anti-Cwp66 N-ter**,** probably correspond to degradation products of Cwp84. A very tight band of approximately 35 kDa was detected only by anti-GST, which may correspond to the GST (29 kDa) bound to a small N-terminal part of Cwp84. Cysteine protease inhibitors used did not prevent degradation of the protein of interest. Purified fractions did not have proteolytic activity on nonspecific substrates, and various modifications in purification conditions were not more successful in inducing this activity.
FIG. 1.
Immunoblot analysis of purified fraction GST-Cwp84. Equivalent amounts of the purified fraction were separated by SDS-12% polyacrylamide gel electrophoresis, transferred to a Hybond-P membrane (Amersham Biosciences), and incubated either with anti-GST antibody (lane 2) or anti-Cwp66Nter (lane 3). Revelation was done with phosphatase alkaline-conjugate antigoat antibodies or phosphatase alkaline-conjugate antirabbit antibodies, respectively, with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (Invitrogen). Lane 1, low-range SDS-polyacrylamide gel electrophoresis standard (Bio-Rad). Size markers are given in kilodaltons on the left.
Transcriptional analysis of the cwp cluster.
The organization and structure of the genes in the cwp cluster suggest that some of the genes could be cotranscribed and form an operon. Therefore, the transcription of several genes of the cluster was investigated by Northern blotting and RT-PCR.
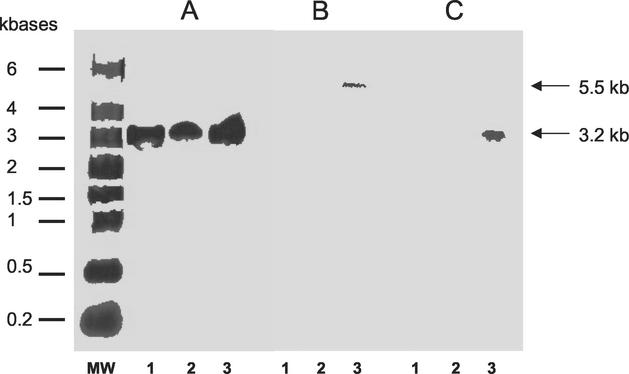
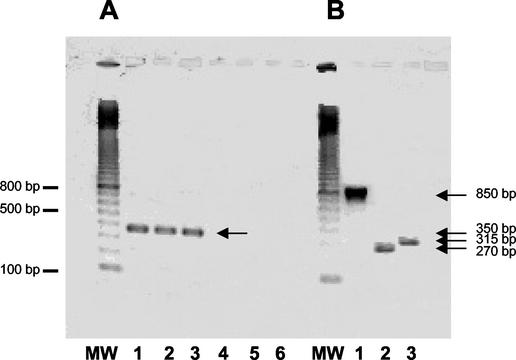
Hybridizations with _slpA_-specific probe are shown in Fig. 2A. Transcripts of slpA were detected during all phases of growth and were estimated to be 3.2 kb, whereas size of the slpA gene is only 2,160 bp. RT-PCR with primers encompassing intergenic regions between orf6 and slpA and between slpA and orf7 was performed (Fig. 3). Amplifications were positive with intergenic primers encompassing orf6 and slpA, indicating the bicistronic feature of the slpA transcript (Fig. 4A).
FIG. 2.
Transcriptional analysis of cwp cluster. Hybridization DNA-membrane-immobilized RNA on strain 630; 15, 30, and 25 μg of total RNA were used for hybridization with probes specific to slpA (A), cwp66 (B), and cwp84 (C), respectively. MW, RNA molecular weight marker (Sigma); lanes 1A, 1B, and 1C, RNA from stationary growth phase (18-h culture); lanes 2A, 2B, and 2C, RNA from the middle of the exponential growth phase (OD600 of ∼0.7); lanes 3A, 3B, and 3C: RNA from the beginning of the exponential growth phase (OD600 of ∼0.3). Sizes of transcripts are indicated with arrows. The probe specific for the transcript of the gdh gene hybridized with an estimated 1.3-kb transcript in the sample corresponding to the different growth phase, as expected (16) (data not shown).
FIG. 4.
RT-PCR analysis of the genes located upstream from slpA (A) and cwp66 (B) in strain 630. (A) MW, molecular weight marker (100-bp ladder; Amersham Biosciences). Lanes 1 to 3, amplification between orf6 and slpA with orf6-RT1/slpA-RTR1 primers, demonstrating the presence of an intergenic mRNA. Lanes 4 to 6, no amplification was obtained with the primers slpA-RT1/orf7-RTR1, showing absence of intergenic RNA messenger. RNA was extracted from the stationary phase (lanes 1 and 4), middle exponential growth phase (lanes 2 and 5), and beginning of the exponential growth phase (lanes 3 and 6). (B) MW, molecular weight marker (100 bp ladder). Lanes 1 to 3, RNA extracted from the beginning of the exponential growth phase. Lane 1, cwp66-NB1/cwp66NBR1 primers; lane 2, orf8-RT1/orf9-RTR1 primers; lane 3, orf9-RT1/cwp66-RTR1 primers.
Hybridizations with a probe specific for the cwp66 gene functional domain showed the presence of a transcript only at the beginning of the exponential growth phase (Fig. 2B). The size of the transcript was estimated to 5.5 kb, whereas the cwp66 gene size is only 1.8 kb. However, this size is compatible with a polycistronic transcript encompassing the two genes immediately upstream of cwp66 in the sequenced genome of strain 630 (Fig. 3). Primers encompassing regions between orf8 and orf9 and between orf9 and cwp66 gave positive results in RT-PCR, confirming the occurrence of a polycistronic transcript _orf8-orf9_-cwp66 (Fig. 4B).
Hybridizations with a probe specific for cwp84 showed the presence of a prominent transcript in the early exponential growth phase (Fig. 2C). The size was estimated to be 3.2 kb, corresponding to a monocistronic transcript. The presence of this transcript was confirmed by RT-PCR (data not shown).
Analysis of the putative Orf6, Orf8, and Orf9.
The deduced peptidic sequence of orf6 shows a 528-amino acid protein with a two-domain structure similar to that described for the other proteins encoded by the genes of the cluster. The cell wall-anchoring domain, located on the N-terminal part, has 53% homology with the cell wall-anchoring domain of CwlB. The predicted secondary structure of the C-terminal part is characterized by a mostly α-helical conformation without any particular motif and does not show any homology with known proteins. The deduced peptidic sequence of orf8 shows again a two-domain structure. The cell wall-anchoring domain, 52% homologous to the autolysin CwlB of B. subtilis, is located in the C-terminal part. The predicted secondary structure of the functional domain shows a strikingly α-helical conformation (71%) with a putative transmembrane segment but shares no significant homology with known proteins. Orf9 does not possess the two-domain structure. Its predicted secondary structure is also rich in α-helices (44%). The deduced protein has no significant homology with known proteins.
Polymorphism of cwp84 and cwp66 genes.
The functional region of the cwp84 gene was easily amplified and sequenced from the 28 strains studied. This domain shows a high degree of conservation, since there were only 51 polymorphic nucleotide sites over 1,043 nucleotides sequenced, including 14 that would result in amino acid replacements (Fig. 5). The nucleotide sequences were 100% identical within each serogroup of C, D, I, K, and X. In contrast, the four strains belonging to serogroup A each had a unique cwp84 functional region sequence, as previously observed for the S-layer precursor slpA (20).
FIG. 5.
Polymorphism of the functional domain of Cwp84, from amino acid 17 to 370. The seven sequences represents the different alleles recovered from the 28 strains studied. Only amino acid differences from serogroup C reference strain (ATCC 43596) are indicated. Identical amino acids are represented by dashes. RefC, same sequence than following strains (serogroup): 630(C), C253(C), 1075(C), Kohn(A), 95938(G), 53444(H), 57027, RefI, 56026(I), RefK, 94416(K), RefX, 36678(X). RefH, same sequence as the following: 93369(H), 90204(H), 68750(A), and 79685(S3). RefG, same sequence as strain CD268. Ex560(B), same sequence as CO109(B). RefA, same sequence as RefB. RefD, same sequence as 93136(D).
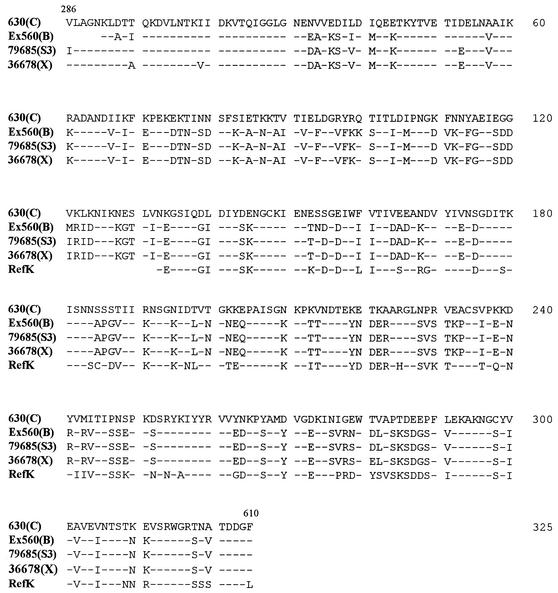
Amplification of the C-terminal functional domain of the cwp66 gene was successful only for 14 strains (RefC, 630, C253, 1075, CO109, Ex560, 68750, 56026, 95938, RefK, 94416, RefX, 36678, and 79685), and sequencing revealed numerous nucleotide changes, which resulted in nine distinct deduced peptidic sequences. The deduced peptidic sequences were 100% identical within serogroup C (four strains), serogroup X (two strains), and two strains of serogroup B (RefB could not be amplified). The genes sequenced from strains 56026 (serogroup I) and 95938 (serogroup G) were highly divergent from the strain 630 cwp66 gene. It is noteworthy that the Cwp66 sequence of serogroup C seems to diverge significantly from the other patterns of sequence: percentages of homology for serogroup C are of 61% with serogroup B strains, 62% with serogroup X strains, 59% with serogroup K strains, and 50% with strain 95938 and 56026. On the contrary, there is a high degree of homology (>90%) among serogroups B, X, and K (Fig. 6).
FIG. 6.
Polymorphism of the functional domain of Cwp66, from amino acids 286 to 610. The five sequences represents the various alleles recovered from 12 strains studied (the sequences of strains 56026 and 95938 are too divergent to appear in this alignment). Only differences from strain 630 (serogroup C) are indicated. Identical amino acids are represented by dashes.
Typing of strains.
Typing of strains belonging to serogroup C was done to check for the nonclonality of these strains, since the sequences of cwp84 and cwp66 were perfectly identical among the four strains of this serogroup. PCR ribotyping generated only two profiles, one for the three toxigenic strains and one corresponding to the nontoxigenic strain. In contrast, patterns of RAPD obtained with three different primers readily differentiated the three toxigenic strains (data not shown). A higher level of discrimination for RAPD compared to PCR ribotyping has already been described for toxigenic strains belonging to serogroup C (2, 36).
DISCUSSION
The cluster carrying genes studied in this study was previously characterized (6, 19). Two of these genes encode well-characterized surface-exposed proteins involved in adherence of C. difficile to host cells: adhesin Cwp66 (37) and S-layer proteins (5, 7). The transcription of some of these genes has been recently studied by RT-PCR (6), but no information on their transcriptional organization was available in the beginning of our study. So, the first aim of our study was to analyze transcription of the three genes, slpA, cwp66, and cwp84, to investigate if an organization in operon exists in this cluster as has been described for the toxigenic element of C. difficile (16).
Transcriptional analysis of slpA, encoding the precursor of the S-layer proteins, revealed that this gene is strongly transcribed during the whole growth phase, reflecting the fact that S-layer proteins are the major surface proteins (7, 8). More surprisingly, the putative 57.5-kDa protein encoded by orf6 may also be strongly expressed. This protein may be of importance for the bacterium, but no putative role could be assessed in view of its structure and homologies. DNA sequence analysis showed that a ribosome binding site exists just upstream from orf6 (AGGAGG) and also sequences which could be part of a promoter: TATAAA (−10) and TTTTAG (−35). Intriguingly, we also found putative promoter sequences immediately upstream of slpA. As has been demonstrated for toxins A and B (13, 16), a monocistronic transcript of slpA could also exist in conditions other than those used in this work, increasing production of the S-layer precursor, which could be important for the adaptation of C. difficile to its environment.
The cwp66 gene is transcribed only in the early exponential phase, at a low level, as a polycistronic transcript encompassing three contiguous orf genes in the following order: orf8-orf9-cwp66 (Fig. 3). Cotranscription of orf9 and cwp66 is not surprising because the intergenic region is short (<50 bp) and there is no putative promoter upstream from cwp66. orf8 and orf9 are each preceded by likely promoter consensus sequences at −10 and −35. We are unable to explain this simultaneous transcription, since the putative proteins encoded by orf8 and orf9 do not exhibit any homology to known proteins. Nevertheless, one hypothesis could be that the two or three proteins could associate in vivo to form a complexed adhesin, as has been already described for Porphyromonas gingivalis (27). In this bacterium, the associated proteins (cysteine protease, adhesin, and hemagglutinin) are processed from a large polyprotein encoded by a single gene (26). It is possible that Orf8, which displays a putative transmembrane segment, could anchor the adhesin in the cytoplasmic membrane. The fact that cwp66, considered as a putative colonization factor, is transcribed only in the beginning of the exponential phase, while no transcription of toxins could be detected (13, 16), is somewhat surprising. But this phenomenon has already been described for Staphyloccocus aureus, in which cell wall-associated adhesins, such as the fibronectin binding-proteins, are expressed during the exponential phase and repressed postexponentially when synthesis of exoproteins is induced (30, 39).
In a recent publication, Calabi and Fairweather (4) found no polycistronic RNA transcript for this cluster. This discrepancy could be explained first by the different strain studied and, second, by different culture conditions (especially for slpA, which could be transcribed from its own promoter), but lack of details about the experiments undertaken by Calabi and Fairweather prevents us from drawing definitive conclusions.
As the Cwp66 protein has already been demonstrated to be involved in adherence of C. difficile to cultured cells and could therefore be involved in pathogenesis of the bacterium, it seemed interesting to compare sequences of this gene from different clinical isolates in order to establish virulence profiles. Unfortunately, although six different couples of primers were used, we obtained a specific amplification product only for 14 strains. The absence of amplification of the 3′ part of cwp66 in some strains, regardless of serogroup or toxinotype, could be explained by the high variability of this domain, as previously described by dot blot experiments (37). However, we cannot exclude the possibility that this domain could be deleted in some strains. The sequence of cwp66 does not display any obvious correlation with serogroup or virulence. However, it is noteworthy that the four nonrelated serogroup C strains shared the same conserved cwp66 gene sequence, which is divergent from those of the other strains: this serogroup is known to contain the most outbreak-related strains (36), and conservation of this adhesin could be of importance for colonization and dissemination of these strains.
To further our understanding of the role of all the component genes of this cluster, we investigated cwp84, a gene located just downstream from cwp66. The anchoring domain of Cwp84 is located in the C-terminal part, and the N-terminal domain displays significant homologies to cysteine proteases of archaea (e.g., Methanosarcina mazei, GenBank number NC_003901), eukaryotes (e.g., papain, GenBank number M15203), and bacteria (e.g., PepC, Streptococcus thermophilus, GenBank number Q56115), especially around the active amino acids of the enzymes. Alignments allow us to determine the putative catalytic triad: cysteine in position 216, histidine in 262, and asparagine in 287. C. difficile is not currently considered a proteolytic bacterium, but some studies have shown that this bacterium displays some surface-associated proteolytic activity (28, 32), and most proteolytic strains have been shown to be the most virulent in the hamster model (31). Moreover, surface-associated proteolytic activity seems to be mainly due to a thiol-protease (32). Proteases are a well-known virulence factor for some important pathogenic bacteria (22), and especially the cysteine proteases produced by Porphyromonas gingivalis or Streptococcus pyogenes (21, 35).
Cloning of the cwp84 gene into an expression system in an E. coli strain with deletions of major proteases demonstrated nonspecific proteolytic activity of this protein on gelatin, skimmed milk, and azocoll. Purification of this protease failed, likely due to an autocatalytic process of the protease, leading to the cleavage between the GST and the catalytic domain of Cwp84. This phenomenon has been described for other extracellular cysteine proteases (38), such as SpeB, a well-characterized virulence factor of S. pyogenes (12). The fact that leupeptin and E64 do not inhibit the autocatalytic process of Cwp84 is somewhat surprising, but other cysteine proteases, like Lys-gingipain and clostripain, respectively, are also not inhibited by these molecules. Further purification attempts with different strategies are in process.
The transcription of the cwp84 gene could be detected only in the early exponential phase, but in a pronounced fashion. Sequence analysis of the functional domain of the cwp84 gene indicated that this gene is highly conserved, particularly the amino acids potentially involved in the active site. No correlation between Cwp84 sequence patterns and serogroups or virulence profiles was found. In fact, strains from different serogroups or with different toxinotypes shared the same peptidic sequence. It is noteworthy that SpeB and gingipains, cysteine proteases involved in virulence of P. gingivalis, have been shown to be highly conserved and expressed among all the strains (24, 25, 33). Taken together, the highly conserved pattern of this gene and its strong expression, at least in the strain studied and in our experimental conditions, indicate that Cwp84 might have an important function in the physiology of C. difficile. A tempting hypothesis would be that Cwp84 acts as a maturation protease for various cell surface-associated proteins, as it has been shown for SpeB in S. pyogenes (1) and the gingipains in P. gingivalis (18). In particular, Cwp84 could play a role in the processing of the S-layer precursor or of Cwp66, for which some cleavage products have been detected in surface extracts of C. difficile (37).
The study of the transcription of the genes located on this putative virulence cluster is an important step in the characterization of the colonization process by C. difficile and will be further confirmed by in vivo experiments, in which transcription of genes coding for adhesins and toxins will be studied in a C. difficile monoxenic mouse model.
Acknowledgments
We are grateful to Isabelle Podglajen (European Hospital Georges Pompidou, Paris, France) for her valuable advice on RNA extraction and Northern blot experiments.
REFERENCES
- 1.Berge, A., and L. Björck. 1995. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J. Biol. Chem. 270**:**9862-9867. [DOI] [PubMed] [Google Scholar]
- 2.Bidet, P., V. Lalande, B. Salauze, B. Burghoffer, V. Avesani, M. Delmée, A. Rossier, F. Barbut, and J.-C. Petit. 2000. Comparison of PCR-ribotyping, arbitrarily primed PCR and pulsed-field gel electrophoresis for typing Clostridium difficile. J. Clin. Microbiol. 38**:**2484-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bongaerts, G. P. A., and D. M. Lyerly. 1994. Role of toxins A and B in the pathogenesis of Clostridium difficile disease. Microb. Pathog. 17**:**1-12. [DOI] [PubMed] [Google Scholar]
- 4.Calabi, E., and N. Fairweather. 2002. Pattern of sequence conservation in the S-layer proteins and related sequences in Clostridium difficile. J. Bacteriol. 184**:**3886-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calabi, E., F. Calabi, A. D. Phillips, and N. F. Fairweather. 2002. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect. Immun. 70**:**5770-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calabi, E., S. Ward, B. Wren, T. Paxton, M. Panico, H. Morris, A. Dell, G. Dougan, and N. Fairweather. 2001. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40**:**1187-1199. [DOI] [PubMed] [Google Scholar]
- 7.Cerquetti, M., A. Molinari, A. Sebastianelli, M. Diociaiuti, R. Petruzzelli, C. Capo, and P. Mastrantonio. 2000. Characterization of surface layer proteins from different Clostridium difficile clinical isolates. Microb. Pathog. 28**:**363-372. [DOI] [PubMed] [Google Scholar]
- 8.Cerquetti, M., A. Panstoti, P. Stefanelli, and P. Mastrantonio. 1992. Purification and characterization of an immunodominant 36-kDa antigen present on the cell surface of Clostridium difficile. Microb. Pathog. 13**:**271-279. [DOI] [PubMed] [Google Scholar]
- 9.Cerquetti, M., A. Serafino, A. Sebastianelli, and P. Mastrantonio. 2002. Binding of Clostridium difficile to Caco-2 epithelial cell line and to extracellular matrix proteins. Immun. Med. Microbiol. 32**:**211-218. [DOI] [PubMed] [Google Scholar]
- 10.Collier, M. C., F. Stock, P. C. DeGirolami, M. H. Samore, and C. P. Cartwright. 1996. Comparison of PCR-based approaches to molecular epidemiologic analysis of Clostridium difficile. J. Clin. Microbiol. 34**:**1153-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmée, M., and V. Avesani. 1990. Virulence of ten serogroups of Clostridium difficile in hamsters. J. Med. Microbiol. 33**:**85-90. [DOI] [PubMed] [Google Scholar]
- 12.Doran, J. D., M. Nomizu, S. Takebe, R. Ménard, D. Griffith, and E. Ziomek. 1999. Autocatalytic processing of the streptococcal cysteine protease zymogen. Eur. J. Biochem. 263**:**145-151. [DOI] [PubMed] [Google Scholar]
- 13.Dupuy, B., and A. L. Sonenshein. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol. Microbiol. 27**:**107-120. [DOI] [PubMed] [Google Scholar]
- 14.Eveillard, M., V. Fourel, M.-C. Barc, S. Kernéis, M.-H. Coconnier, T. Karjalainen, P. Bourlioux, and A. L. Servin. 1993. Identification and characterization of adhesive factors of Clostridium difficile involved in adhesion to human colonic enterocyte-like Caco-2 and mucus-secreting HT29 cells in culture. Mol. Microbiol. 7**:**371-381. [DOI] [PubMed] [Google Scholar]
- 15.Goda, S. K., and N. P. Minton. 1995. A simple procedure for gel electrophoresis and Northern-blotting of RNA. Nucleic Acids Res. 23**:**3357-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond, G. A., D. M. Lyerly, and J. L. Johnson. 1997. Transcriptional analysis of the toxigenic element of Clostridium difficile. Microb. Pathog. 22**:**143-154. [DOI] [PubMed] [Google Scholar]
- 17.Hennequin, C., F. Porcheray, A.-J. Waligora-Dupriet, A. Collignon, M.-C. Barc, P. Bourlioux, and T. Karjalainen. 2001. GroEL (Hsp60) of Clostridium difficile is involved in cell adherence. Microbiology 147**:**87-96. [DOI] [PubMed] [Google Scholar]
- 18.Kadowaki, T., K. Nakayama, F. Yoshimura, K. Okamoto, N. Abe, and K. Yamamoto. 1998. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 273**:**29072-29076. [DOI] [PubMed] [Google Scholar]
- 19.Karjalainen, T., A.-J. Waligora-Dupriet, M. Cerquetti, P. Spigaglia, A. Maggioni, P. Mauri, and P. Mastrantonio. 2001. Molecular and genomic analysis of genes encoding surface-anchored proteins from Clostridium difficile. Infect. Immun. 69**:**3442-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karjalainen, T., N. Saumier, M.-C. Barc, M. Delmée, and A. Collignon. 2002. Clostridium difficile genotyping based on slpA variable region in S-layer gene sequence: an alternative to serotyping. J. Clin. Microbiol. 40**:**2452-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukomski, S., S. Sreevatsan, C. Amberg, W. Reichardt, M. Woischnik, A. Podbielski, and J. M. Musser. 1997. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J. Clin. Investig. 99**:**2574-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda, H. 1996. Role of microbial proteases in pathogenesis. Microbiol. Immunol. 40**:**685-699. [DOI] [PubMed] [Google Scholar]
- 23.Mastrantonio, P., A. Pantosti, M. Cerquetti, C. Fiorentini, and G. Donelli. 1996. Clostridium difficile: an update on virulence mechanisms. Anaerobe 2**:**337-343. [DOI] [PubMed] [Google Scholar]
- 24.Mikolajczyk-Pawlinska, J., T. Kordula, N. Pavloff, P. A. Pemberton, W.-C. A. Chen, J. Travis, and J. Potempa. 1998. Genetic variations of Porphyromonas gingivalis genes encoding gingipains, cysteine proteinases with arginine and lysine specificity. Biol. Chem. 379**:**205-211. [DOI] [PubMed] [Google Scholar]
- 25.Musser, J. M., A. R. Hauser, M. H. Kim, P. M. Schlievert, K. Nelson, and R. K. Selander. 1991. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. USA 88**:**2668-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavloff, N., J. Potempa, R. N. Pike, V. Prochazka, M. C. Kiefer, J. Travis, and P. J. Barr. 1995. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. J. Biol. Chem. 270**:**1007-1010. [DOI] [PubMed] [Google Scholar]
- 27.Pike, R., W. McGraw, J. Potempa, and J. Travis. 1994. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. J. Biol. Chem. 269**:**406-411. [PubMed] [Google Scholar]
- 28.Poilane, I., T. Karjalainen, M.-C. Barc, P. Bourlioux, and A. Collignon. 1998. Protease activity of Clostridium difficile strains. Can. J. Microbiol. 44**:**157-161. [PubMed] [Google Scholar]
- 29.Riley, T. V. 1998. Clostridium difficile: a pathogen of the nineties. Eur. J. Clin. Microbiol. Infect. Dis. 17**:**137-141. [DOI] [PubMed] [Google Scholar]
- 30.Saravia-Otten, P., H.-P. Müller, and S. Arvidson. 1997. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an _agr_-independent mechanism. J. Bacteriol. 179**:**5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seddon, S. V., I. Hemingway, and S. P. Borriello. 1990. Hydrolytic enzyme production by Clostridium difficile and its relationship to toxin production and virulence in the hamster model. J. Clin. Microbiol. 31**:**169-174. [DOI] [PubMed] [Google Scholar]
- 32.Seddon, S. V., and S. P. Borriello. 1992. Proteolytic activity of Clostridium difficile. J. Med. Microbiol. 36**:**307-311. [DOI] [PubMed] [Google Scholar]
- 33.Stockbauer, K. E., L. Magoun, M. Liu, E. H. Burns, Jr., S. Gubba, S. Renish, X. Pan, S. C. Bodary, E. Baker, J. Coburn, J. M. Leong, and J. M. Musser. 1999. A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins αVβ3 and αIIbβ3. Proc. Natl. Acad. Sci. USA 96**:**242-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tasteyre, A., M.-C. Barc, A. Collignon, H. Boureau, and T. Karjalainen. 2001. Role of FliC et FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69**:**7937-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tokuda, M., T. Karunakaran, M. Duncan, N. Hamada, and H. Kuramitsu. 1998. Role of Arg-gingipain A in virulence of Porphyromonas gingivalis. Infect. Immun. 66**:**1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Dijck, P., V. Avesani, and M. Delmée. 1996. Genotyping of outbreak-related and sporadic isolates of Clostridium difficile belonging to serogroup C. J. Clin. Microbiol. 34**:**3049-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waligora, A.-J., C. Hennequin, P. Mullany, P. Bourlioux, A. Collignon, and T. Karjalainen. 2001. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect. Immun. 69**:**2144-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wandersman, C. 1989. Secretion, processing and activation of bacterial extracellular proteases. Mol. Microbiol. 3**:**1825-1831. [DOI] [PubMed] [Google Scholar]
- 39.Wolz, C., P. Pöhlmann-Dietze, A. Steinhuber, Y.-T. Chien, A. Manna, W. Van Wamel, and A. Cheung. 2000. _Agr_-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36**:**230-243. [DOI] [PubMed] [Google Scholar]