Role of Mammalian Rad54 in Telomere Length Maintenance (original) (raw)
Abstract
The homologous recombination (HR) DNA repair pathway participates in telomere length maintenance in yeast but its putative role at mammalian telomeres is unknown. Mammalian Rad54 is part of the HR machinery, and Rad54-deficient mice show a reduced HR capability. Here, we show that Rad54-deficient mice also show significantly shorter telomeres than wild-type controls, indicating that Rad54 activity plays an essential role in telomere length maintenance in mammals. Rad54 deficiency also resulted in an increased frequency of end-to-end chromosome fusions involving telomeres compared to the controls, suggesting a putative role of Rad54 in telomere capping. Finally, the study of mice doubly deficient for Rad54 and DNA-PKcs showed that telomere fusions due to DNA-PKcs deficiency were not rescued in the absence of Rad54, suggesting that they are not mediated by Rad54 activity.
Unrepaired double-strand breaks (DSBs) are a threat to the genome, because they disrupt the integrity of the DNA molecule and lead to genomic instability, a hallmark of cancer, or cause lethality, underlining the importance of these processes in the organism (21). Cells have evolved two different pathways for repairing DSBs; homologous recombination (HR), which requires large regions of homology, and DNA nonhomologous end joining (NHEJ), which does not require extensive DNA homology regions (20). In yeast, HR is the major pathway involved in DSB repair, whereas NHEJ is the main pathway for DSB repair in mammalian cells (20). In yeast cells, HR is mediated by the Rad52 group of genes, which includes the Rad51, Rad54, and Rad57 genes, as well as by the Rad50 complex, which includes Xrs2 and Mre11 (36). These proteins have homologues in mammalian cells; however, the contribution of each gene to HR may differ between yeasts and mammals (29). The mammalian NHEJ pathway includes six essential components, three of which (Ku70, Ku86, and DNA-PKcs) compose the so-called DNA-dependent protein kinase (DNA-PK) (33). Functional homologues of both subunits of Ku have been identified in yeast cells, but the catalytic subunit, DNA-PKcs, is absent (33). In addition to their role in DSB repair, components of the HR and NHEJ pathways have been shown to be present at telomeres both in yeast and in mammals (8, 15, 24).
Telomeres are unique structures at the ends of eukaryotic chromosomes that serve to limit the loss of genetic material that occurs during DNA replication and keep the chromosome ends from being detected as DSBs by the cell DNA repair machinery (4). Vertebrate telomeres are composed of tandem repeats of the TTAGGG sequence and an array of associated proteins (4). In addition, telomeres end in a 3′ overhang, known as the G-strand overhang (8). The 3′-G-strand overhang is able to fold back and invade the double-stranded region of the telomere, forming a loop, the so-called T-loop, which is stabilized by a set of specialized proteins. T-loops facilitate the formation of a higher-order structure that has been proposed to mediate telomere capping by contributing to mask DNA ends from being recognized by the DNA repair system, thus preventing end-to-end fusions and loss of cell viability (8). Telomeres may also regulate the access of telomerase to the telomere (8, 15).
Telomerase is a nucleoprotein complex with reverse transcriptase activity known as Tert and a RNA molecule or Terc (telomerase RNA component), which serves as a template for the synthesis of new telomere repeats (7). Telomere dysfunction, either due to exhaustion of TTAGGG repeats in the absence of telomerase activity or due to disruption of the end-capping structure, leads to chromosomal instability and impacts on both cancer and aging, as demonstrated initially by using mouse models for telomere dysfunction, such as the telomerase-deficient mouse (6, 15).
The role of NHEJ at telomeres has been extensively studied in yeast, plants, and mammals. Whereas in yeast defects in either Ku subunit result in loss of telomeric repeats, loss of telomere clustering, loss of telomeric silencing, and deregulation of the G-strand overhang (24), plants and rodents show extended telomeres (28, 30). This telomere elongation is mediated by telomerase, indicating that Ku is a negative regulator of telomerase-dependent telomere elongation (10, 28). In addition, DNA-PKcs and Ku86 play in mice important roles in telomere capping, as well as in the response to dysfunctional telomeres. On the one hand, deficiency in either Ku86 or DNA-PKcs lead to end-to-end chromosome fusions that involve long tracks of TTAGGG sequences, suggesting that in the absence of these proteins telomeres are long but dysfunctional (2, 3, 13, 14, 19, 30). In this regard, a role for DNA-PKcs in the processing of the telomeres produced by leading-strand synthesis has been proposed (3). In addition, DNA-PKcs has been shown to functionally interact with telomerase in maintaining telomere length; in particular, mice doubly deficient for telomerase and DNA-PKcs undergo a faster rate of telomere attrition than corresponding single-mutant controls (11). On the other hand, when Ku86 and DNA-PKcs deficiencies have been studied in the context of telomerase deficiency, a role for these factors in mediating end-to-end fusions and apoptosis due to short telomeres was also clearly established (10, 11). More recently, the study of functional interactions between DNA-ligase IV, which is central to the NHEJ pathway, and a telomere-binding protein, TRF2, has also established an essential role of NHEJ in mediating end-to-end fusions of unprotected chromosome ends (34, 37).
HR has been also shown to regulate telomere length, as well as to mediate telomerase-independent telomere elongation in yeast (24). However, the role of HR in telomere length maintenance in mammals, remains unknown. Similarly, the putative role of HR in mediating chromosome end-to-end fusions due to unprotected telomeres has not been addressed to date.
Rad54 is a Rad51-interacting DNA-dependent ATPase involved in homologous DNA pairing (27, 32). Its role in HR in yeast is also conserved in rodents (12), as demonstrated by the reduced efficiency of DSB repair and reduced sister chromatid gene conversion (GC) observed in Rad54-deficient mice (9, 12). By using Rad54-defective animals, generated by disrupting the gene by HR, we provide evidence for the role of this gene product in telomere length maintenance and telomere capping. Interestingly, generation of mice simultaneously deficient in Rad54 and DNA-PKcs activities also served as a means to demonstrate the inability of Rad54 deficiency in rescuing chromosome-type end-to-end fusions produced by the lack of DNA-PKcs. This in turn suggests that Rad54, in contrast to NHEJ activities, does not play a key role in the generation of fusions due to unprotected chromosome ends.
MATERIALS AND METHODS
Mice and cells.
The Rad54-deficient and DNA-PKcs mice used to generate doubly heterozygous Rad54+/− DNA-PKcs+/− mice were described elsewhere (12, 35). Mouse embryonic fibroblasts (MEFs) were derived from heterozygous crosses and, for all studies, littermates were compared. Rad54+/− mice were used to generate single Rad54−/− and Rad54+/+ MEFs when indicated. MEFs were prepared from day 13.5 embryos derived from heterozygous crosses as described previously (6). First-passage (passage 1) MEFs used in the different experiments corresponded to approximately two population doublings (PDL = 2).
Q-FISH.
First-passage MEFs were prepared for quantitative fluorescence in situ hybridrization (Q-FISH) as described previously (18). Q-FISH hybridization was carried out as described previously (18).
To correct for lamp intensity and alignment, images from fluorescent beads (Molecular Probes) were analyzed by using the TFL-Telo program. Telomere fluorescence values were extrapolated from the telomere fluorescence of LY-R and LY-S lymphoma cell lines (1) of known lengths of 80 and 10 kb (25). There was a linear correlation (_r_2 = 0.999) between the fluorescence intensities of the R and S telomeres, with a slope of 38.6. The calibration-corrected telomere fluorescence intensity was calculated as described previously (18).
Images were recorded by using a COHU charge-coupled device camera on a Leica Leitz DMRB fluorescence microscope. A Philips CS 100W-2 mercury vapor lamp was used as a source. Images were captured by using Leica Q-FISH software at a 400-ms integration time in a linear acquisition mode to prevent oversaturation of the fluorescence intensity.
Quantitative image analysis.
TFL-Telo software (a gift from P. Lansdorp) was used to quantify the fluorescence intensity of telomeres from at least 10 metaphases of each individual MEF culture. The integrated fluorescence intensity for each telomere was calculated after correction for image acquisition exposure time. Finally, the integrated fluorescence intensity of individual telomeres is expressed in a table for each chromosome, which can be subjected to editing. Each metaphase of 40 chromosomes (in the mouse) yields 160 telomere spots and a typical analysis of 10 metaphases produces several thousand telomere fluorescence values (Table 1). Because of the large number of datum points in Q-FISH analysis, the standard error of mean telomere fluorescence estimates is typically small (for example, less than a few percent of the average) despite considerable variation in individual telomere fluorescence values. Importantly, the analysis of thousands of individual telomere length values allows for very reliable statistical significance calculations.
TABLE 1.
Determination of telomere length by using Q-FISH in primary MEFsa
| Genotype and MEF group | No. of metaphases | Mean length (kb) ± SE | No. of telomeres | No. of signal-free ends (%)b | ||
|---|---|---|---|---|---|---|
| p-arm | q-arm | Avg | ||||
| Rad54+/− DNA-PKcs+/− | ||||||
| E | 10 | 45.4 ± 0.51 | 54.3 ± 0.58 | 50.0 ± 0.40 | 1,600 | 7 (0.44) |
| F | 10 | 42.0 ± 0.45 | 49.4 ± 0.56 | 45.7 ± 0.37 | 1,652 | 3 (0.18) |
| A | 10 | 43.4 ± 0.47 | 54.1 ± 0.55 | 48.8 ± 0.40 | 1,588 | 4 (0.25) |
| A+E+F | 30 | 43.6 ± 0.28 | 52.5 ± 0.33 | 48.1 ± 0.23 | 4,840 | 14 (0.29 ± 0.08) |
| Rad54−/− DNA-PKcs+/− | ||||||
| B2 | 10 | 31.6 ± 0.23 | 38.6 ± 0.55 | 35.2 ± 0.33 | 1,588 | 0 (0.0) |
| C2 | 10 | 31.3 ± 0.32 | 36.3 ± 0.47 | 33.8 ± 0.29 | 1,688 | 8 (0.47) |
| B | 10 | 30.0 ± 0.36 | 41.3 ± 0.49 | 35.7 ± 0.34 | 1,580 | 3 (0.19) |
| E2 | 10 | 25.3 ± 0.34 | 37.0 ± 0.44 | 31.2 ± 0.31 | 1,644 | 15 (0.9) |
| B2+C2+B+E2 | 40 | 29.6 ± 0.18 | 38.3 ± 0.25 | 33.9 ± 0.16 | 6,500 | 33 (0.39 ± 0.19) |
| Rad54+/− DNA-PKcs−/− | ||||||
| C | 10 | 39.3 ± 0.47 | 47.3 ± 0.57 | 48.8 ± 0.39 | 1,544 | 4 (0.26) |
| D | 10 | 36.0 ± 0.41 | 41.4 ± 0.50 | 38.7 ± 0.33 | 1,676 | 9 (0.54) |
| C+D | 20 | 37.6 ± 0.31 | 43.8 ± 0.36 | 40.7 ± 0.25 | 3,220 | 13 (0.40 ± 0.14) |
| Rad54−/− DNA-PKcs−/− | ||||||
| A2 | 10 | 29.4 ± 0.38 | 35.0 ± 0.50 | 32.2 ± 0.32 | 1,536 | 8 (0.52) |
| D2 | 10 | 37.0 ± 0.38 | 44.4 ± 0.45 | 40.7 ± 0.31 | 1,732 | 7 (0.40) |
| A2+D2 | 20 | 33.4 ± 0.29 | 40.0 ± 0.36 | 36.7 ± 0.24 | 3,268 | 15 (0.46 ± 0.06) |
The images from littermate metaphases were captured on the same day, in parallel, and blindly. All of the images from the MEFs were captured in a 3-day period after the hybridization.
Telomerase assay.
S-100 extracts were prepared from primary MEF cultures, and a modified version of the TRAP assay was used to measure telomerase activity (5). An internal control for PCR efficiency was included (TRAPeze kit Oncor).
Scoring of chromosomal abnormalities by Q-FISH.
The indicated numbers of metaphases (≥100) from each MEF culture were scored for chromosomal aberrations by superimposing the telomere image on the DAPI (4′,6′-diamidino-2-phenylindole) chromosome image in the TFL-Telo software. End-to-end fusions can be two chromosomes fused by their p-arms (Robertsonian-like fusions) or two chromosomes fused by their q-arms (dicentrics). Robertsonian-like fusions were classified as those that showed TTAGGG signal at the fusion point (+TTAGGG) or that lacked TTAGGG signal at the fusion point (−TTAGGG).
CO-FISH.
Chromosome orientation FISH (CO-FISH) was performed as described previously with some modifications (3). Briefly, confluent primary MEFs were subcultured in the presence of bromodeoxyuridine (Sigma) at a final concentration of 10−5 M and then allowed to replicate their DNA once at 37°C for 24 h. Colcemid was added at a concentration of 0.2 g/ml during the last 4 h. Cells were then recovered, and metaphases were prepared as described previously (30). Prior to hybridization of the single-stranded (TTAGGG)7 telomere probe, slides were treated with 0.5 mg of RNase A/ml for 10 min at 37°C and then stained with 0.5 μg of Hoechst 33258 (Sigma)/ml in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min at room temperature. Slides were then exposed to 365-nm UV light (Stratalinker 1800 UV irradiator) for 25 min. Enzymatic digestion of the bromodeoxyuridine-substituted DNA strands with 3 U of exonuclease III (Promega)/μl in buffer supplied by the manufacturer (50 mM Tris-HCl, 5 mM MgCl2, 5 mM dithiothreitol; pH 8.0) was allowed to proceed for 10 min at room temperature. An additional denaturation in 70% formamide-2× SSC at 70°C for 1 min was performed, followed by dehydration in a cold ethanol series (70, 85, and 100%). Probe hybridization and analysis was identical to that described for FISH experiments. This strategy facilitates identification of the telomere produced by leading-strand DNA synthesis and is described in detail elsewhere (3). After being washed five times (15 min each time) with 2× SSC at 42°C, slides were counterstained with DAPI (0.2 g/ml) in Vectashield (Vector Laboratories). Metaphase spreads were photographed on a Leitz Leica DMRB fluorescence microscope.
Scoring of chromosomal abnormalities by SKY.
Painting probes for each chromosome were generated from flow-sorted mouse chromosomes by using sequence-independent DNA amplification. Labeling was performed by incorporating four different dyes in a combination sequence that allows unique and differential identification of each chromosome. Slides were prepared from fixative-stored material and were hybridized and washed by the spectral karyotyping (SKY) method according to the manufacturer's protocol (Applied Spectral Imaging, Migdal Ha-Emck, Israel). Chromosomes were counterstained with DAPI. Images were captured and processed as described previously (10). The indicated number of metaphases of each culture were captured and analyzed by SKY, and chromosomal abnormalities were scored as described above.
Statistical analysis.
Statistical calculations to determine average telomere length and standard errors were determined by using Microsoft Excel. Because of the large number of datum points in Q-FISH analysis, the standard error of mean telomere fluorescence estimates is typically small despite considerable variation in individual telomere fluorescence values (Table 1). A Student t test with two tails, i.e., “two-samples of unequal variance” (or Welch's correction), was used to calculate the statistical significance of the observed differences in telomere length. GraphPad Prism v.3.0a and Microsoft Excel v.2001 were used for the calculations. Importantly, the analysis of thousands of individual telomere length values allows for very reliable statistical significance calculations.
A χ2 test was used to calculate the statistical significance of differences in chromosomal aberrations between different genotypes. The two-sided P values were obtained from a 2-by-2 contingency table analyzed by χ2 test (including Yates' continuity correction). GraphPad Instat v.2.03 was used for the calculations. With both the Student t test and the χ2 test, the differences are considered significant if P is <0.05, very significant is P is <0.01, highly significant if P is <0.001, and extremely significant if P is <0.0001.
RESULTS AND DISCUSSION
Rad54 deficiency in mice results in a significant shortening of telomeres.
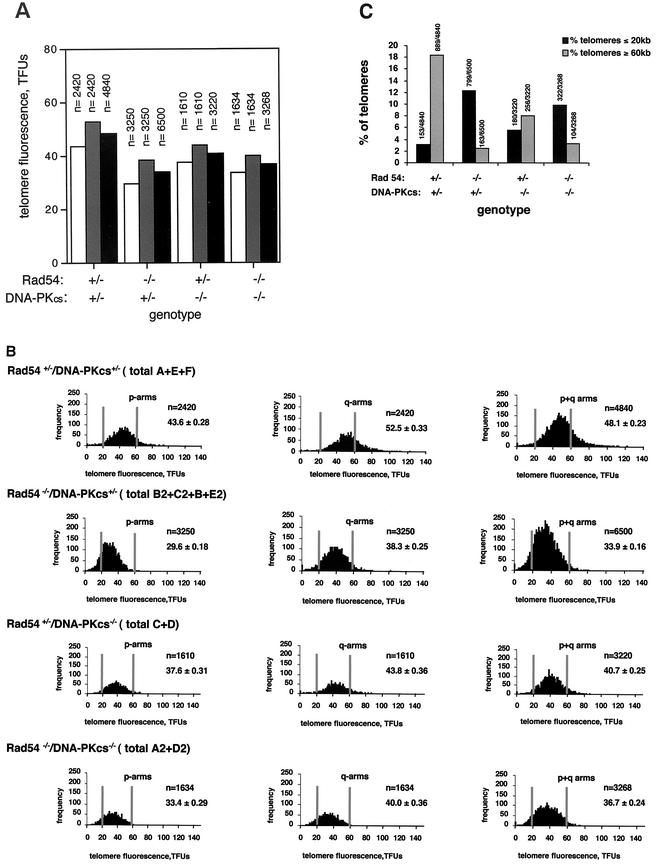
To study the putative role of HR in telomere length maintenance in mammalian cells, we determined telomere length in passage 1 MEFs derived from animals genetically deficient in either Rad54 (Rad54−/− DNA-PKcs+/−), DNA-PKcs (Rad54+/− DNA-PKcs−/−), or both Rad54 and DNA-PKcs (Rad54−/− DNA-PKcs−/−) and compared them with the corresponding controls. The MEFs used were littermates derived from Rad54+/− DNA-PKcs+/− intercrosses (see Materials and Methods). To measure telomere length, we used Q-FISH on metaphase chromosomes, which allows for the determination of thousands of individual telomere length values per individual MEF culture (see Materials and Methods). Two to four primary MEF cultures from each genotype were used for the analysis, and a total of 3,220 to 6,500 individual telomere length values per genotype were obtained and used for determination of the average telomere length and standard error (Table 1 and Fig. 1A) (see Materials and Methods). This high number of individual telomere length measurements allows calculation of the statistical significance of telomere length differences between genotypes. Q-FISH analysis revealed that MEFs lacking Rad54 (average of B2, C2, B, and E2 Rad54−/− DNA-PKcs+/− MEFs) showed a very marked decreases in average telomere length compared to the Rad54+/− DNA-PKcs+/− controls (average of E, F, and A MEFs): 33.9 ± 0.16 and 48.1 ± 0.23 kb, respectively (Table 1 and Fig. 1A). The decrease in average telomere length was seen both in the q- and p-chromosome arms (Table 1 and Fig. 1A). To calculate the statistical significance of this difference, we compared 6,500 telomere length values obtained from Rad54−/− DNA-PKcs+/− MEFs (n = 6500) with 4,840 telomere values obtained from the Rad54+/− DNA-PKcs+/− controls (n = 4840), a Student t test value P of <0.0001 indicated that the difference in telomere length between genotypes was extremely significant (see Materials and Methods). Figure 1B shows histograms of telomere length frequencies for p-arm, q-arm, and the sum of p+q-arm telomeres of primary MEFs grouped by genotype (Table 1). These histograms show a shift toward shorter telomere lengths in the Rad54−/− DNA-PKcs+/− MEFs compared to the Rad54+/− DNA-PKcs+/− controls (Fig. 1B). The gray vertical lines help to visualize the increased frequency of telomeres of ≤20 kb, along with the decreased frequency telomeres of ≥60 kb, in the Rad54−/− DNA-PKcs+/− MEFs compared to the Rad54+/− DNA-PKcs+/− controls (Fig. 1B). To allow direct comparison of telomere length in different genotypes, we calculated the percentage of telomeres that were shorter than 20 kb or longer than 60 kb out of the total number of telomeres analyzed per genotype (Fig. 1C). Rad54 deficiency in Rad54−/− DNA-PKcs+/− MEFs resulted in 12.3% (799 of 6,500) of telomeres of ≤20 kb and 2.5% (163 of 6,500) of telomeres of ≥60 kb compared to 3.2% of telomeres of ≤20 kb (153 out 4,840) and 18.4% of telomeres of ≥60 kb (889 of 4,840) in the Rad54+/− DNA-PKcs+/− controls (Fig. 1C). Altogether, these different analyses indicate a clear decrease in telomere length in Rad54−/− DNA-PKcs+/− MEFs compared to the Rad54+/− DNA-PKcs+/− controls.
FIG. 1.
Telomere length analysis by Q-FISH. (A) Average telomere fluorescence and standard error of q-telomeres, p-telomeres, or all (p+q) telomeres in primary MEFs grouped by genotype. Fluorescence is expressed in telomere fluorescence units (TFUs), where 1 TFU corresponds to 1 kb of TTAGGG repeats (30). Each value represents the mean of 10 metaphases and of the indicated number (n) of individual telomere values. n refers to the total number of telomere values used for calculation of the average telomere fluorescence for each chromosome arm, as well as for the sum of both arms. Bars: ▪, average of q- and p-telomeres; Created by potrace 1.16, written by Peter Selinger 2001-2019 , q-telomeres; □, p-telomeres. Despite the wide heterogeneity in individual telomere fluorescence intensity values (see, for example, Fig. 1B), the standard errors of the mean are very small due to the large number of datum points (see “_n_” values). As a result, the error bars are not visible in the graphs (see Table 1 for standard error values). (B) Histograms showing the telomere length frequencies for p-arms, q-arms, or the sum of p+q-arms of primary MEFs grouped by genotype. The letters indicate the individual MEFs of each genotype used for the analysis. n is the total number of p-telomeres, q-telomeres, or the sum of p+q-telomeres per genotype that are represented in each histogram. One TFU corresponds to 1 kb of TTAGGG repeats (30). To facilitate visualization of the telomere length values, two vertical lines indicate the position of the 20- and 60-kb telomeres for each histogram. The telomere length frequency distribution in each histogram is an indication of the standard deviation of telomere length values and not of the standard error. (C) Percentage of telomeres of the total number of telomeres analyzed for each genotype that are ≤20 kb or ≥60 kb, as indicated. The absolute numbers of telomeres used for the analysis are also shown on top of the bars.
The absence of DNA-PKcs in Rad54+/− DNA-PKcs−/− MEFs resulted in a modest decrease of the average telomere length (average of C and D MEFs) compared to the Rad54+/− DNA-PKcs+/− controls (average of E, F and A MEFs), i.e., 40.7 ± 0.25 and 48.1 ± 0.23 kb, respectively (Table 1 and Fig. 1A). This difference was statistically significant as indicated by a Student t test value P < 0.0001, which was calculated comparing 3,220 telomere values obtained from Rad54+/− DNA-PKcs−/− MEFs with 4,840 individual telomere length values obtained from the Rad54+/− DNA-PKcs+/− controls. The slight decrease in average telomere length due to DNA-PKcs deficiency in Rad54+/− DNA-PKcs−/− MEFs is also illustrated in the histograms of telomere length frequencies (Fig. 1B), as well as by the percentages of telomeres of ≤20 and ≥60 kb shown in Fig. 1C, i.e., 5.6% of telomeres of ≤20 kb (180 of 3,220) and 8% of telomeres of ≥60 kb (256 of 3,223) for Rad54+/− DNA-PKcs−/− MEFs compared to 3.2% of telomeres of ≤20 kb (153 of 4,840) and 18.4% of telomeres of ≥60 kb (889 of 4,840) for the Rad54+/− DNA-PKcs+/− controls (Fig. 1C). It has been previously shown that deficiency in DNA-PKcs alone does not result in a significant telomere shortening compared to wild-type controls (13, 14). The results obtained here, however, indicate that in a Rad54+/− DNA-PKcs−/− background, with only one Rad54 allele, DNA-PKcs deficiency results in shorter telomeres than those of the Rad54+/− DNA-PKcs+/− controls. These results, together with recent findings from our group that showed that simultaneous deletion of telomerase and DNA-PKcs in mice results in a dramatic shortening of telomeres compared to the single mutant controls (11), suggest that DNA-PKcs has an important role in telomere length maintenance that is unveiled by deficiencies in activities important for telomere length maintenance. It is important to point out, however, that Rad54 deficiency rather than DNA-PKcs haploinsuficiency is responsible for the shorter telomeres present in Rad54−/− DNA-PKcs+/− MEFs, as suggested by the fact that single Rad54−/− MEFs (wild-type for DNA-PKcs) showed significantly shorter telomeres than the corresponding single Rad54+/+ controls, 40.0 ± 0.4 and 49.3 ± 0.5 kb, respectively (Student _t_ test, _P_ > 0.0001).
To study the effect on telomere length of the simultaneous deletion of Rad54 and DNA-PKcs, we also determined telomere length in MEFs doubly deficient for Rad54 and DNA-PKcs (Rad54−/− DNA-PKcs−/−) and compared it to the corresponding controls. As shown in Table 1 and Fig. 1A, the average telomere length in Rad54−/− DNA-PKcs−/− MEFs (average of A2 and D2 MEFs) was significantly decreased compared to the Rad54+/− DNA-PKcs+/− controls (shown above), i.e., 36.7 ± 0.24 and 48.1 ± 0.23 kb, respectively. Again, the difference was highly significant, as indicated by a Student t test value (P < 0.0001) obtained after comparison of 3,268 and 4,840 individual telomere values for Rad54−/− DNA-PKcs−/− MEFs and Rad54+/− DNA-PKcs+/− MEFs, respectively. These results are in agreement with a role of Rad54 in telomere length maintenance. Importantly, the average telomere length in Rad54−/− DNA-PKcs−/− MEFs was comparable to that of Rad54−/− DNA-PKcs+/− MEFs, 36.7 ± 0.24 and 33.9 ± 0.16 kb, respectively. Likewise, the similar percentages of telomeres shorter than 20 kb or longer than 60 kb reflects the same findings (Fig. 1B and C). We conclude from these results that Rad54 deficiency leads to a severe telomere shortening and that this decrease in telomere length is not aggravated by DNA-PKcs deficiency. In addition, the fact that doubly deficient Rad54−/− DNA-PKcs−/− MEFs show a telomere length similar to that of Rad54−/− DNA-PKcs+/− MEFs also supports the notion that Rad54 deficiency rather than DNA-PKcs haploinsufficiency is responsible for the shorter telomeres present in Rad54−/− DNA-PKcs+/− MEFs (shown above). Finally, the observation that simultaneous absence of Rad54 and DNA-PKcs renders a similar degree of telomere loss to that produced by Rad54 deficiency alone could also support a model in which both proteins may participate in different aspects of a similar process at the telomere. Of notice, we have not detected differences in the length of the G-strand overhang between the different genotypes studied here (not shown), a finding in agreement with previous results that show that DNA-PKcs deficiency does not lead to changes in the average length of the G-strand overhang, even when it occurs in combination with telomerase deficiency (11, 13, 14).
Altogether, these data indicate that Rad54 deficiency in mice results in a significant loss of telomere sequences, suggesting that Rad54 is important for telomere length maintenance in mice. Since Rad54 is a central player in HR, these results suggest, although they do not demonstrate, that HR participates in telomere length maintenance in mammals. Alternatively, the fact that Rad54 is required for strand invasion during HR-mediated repair of DSBs could also suggest a role for Rad54 in the formation of a proper telomere capping structure (see below).
Rad54 deficiency does not lead to increased undetectable telomere signals.
Q-FISH on the metaphase chromosome allows determination of the percentage of telomeres that lack detectable TTAGGG signal out of the total number of telomeres analyzed per genotype. It has been demonstrated that the presence of undetectable telomeres correlates with increased end-to-end fusions and loss of cell viability in mice (16, 31). The percentage of undetectable telomeres (signal-free ends in Table 1) was similar for all of the different genotypes studied, indicating that either DNA-PKcs or Rad54 deficiency, or a combination of both, did not lead to a significant increase in the percentage of telomeres without detectable TTAGGG signal (Table 1). These results indicate that, although Rad54 deficiency leads to a significant shortening of average telomere length, chromosome ends are not TTAGGG exhausted, as shown by the low percentages of TTAGGG signal-free ends in Rad54-deficient cells. This finding can be explained by the fact that these cells have proficient telomerase activity (see below), which in turn has been shown to preferentially elongate short telomeres, thus preventing critical telomere loss (16, 31).
Normal levels of in vitro telomerase activity in Rad54-deficient cells.
To investigate whether the shortening of telomeres identified in MEFs isolated from Rad54 deficient animals may be the result of down regulation telomerase activity, we measured telomerase levels by using the TRAP assay in cells isolated from mutants and controls (see Materials and Methods). As shown in Fig. 2 telomerase activity was indistinguishable in the different genotypes analyzed, suggesting that Rad54 deficiency does not alter the activity of telomerase in the cell, as detected by an in vitro assay. This does exclude the possibility, however, that Rad54 could have complex regulatory effects on the action of telomerase at telomeres. In this regard, lack of Rad54 may interfere with the recruitment of telomerase to the telomere, thus leading to telomere shortening independently of changes in telomerase activity levels.
FIG. 2.
Telomerase activity in MEFs. S-100 extracts were prepared from MEFs of the indicated genotype and assayed for telomerase activity. For some genotypes more than 1 MEF was assayed for TRAP activity. Different letters refer to independent MEFs. Extracts were pretreated (+R) or not with RNase. The protein concentration used is indicated. The arrow indicates the internal control (IC) for PCR efficiency.
Importantly, the fact that Rad54-deficient cells show normal levels of telomerase activity confers on them the ability to prevent critical telomere loss. This, in turn, could explain the low frequency of signal-free ends determined by Q-FISH in Rad54-deficient despite the significant decrease in average telomere length shown by these cells (Table 1).
HR activities have been also demonstrated to play an essential role in telomere length maintenance in yeast (24). In addition, HR activities are required for telomere elongation in the absence of telomerase activity in yeast (22, 23). Telomerase-independent telomere elongation also occurs in mammalian cells and is generally referred as ALT (17, 26). Although we did not aim to directly address the role of Rad54 in ALT here, it is worth noting that the fact that Rad54 participates in telomere length maintenance in mice is consistent with the idea that Rad54 may also be part of ALT in mammalian cells. Future generation and study of mice simultaneously deficient in telomerase and Rad54 will help to address this possibility.
Role of Rad54 in telomere protection.
As discussed above, the fact that Rad54 deficiency results in a significant loss of telomeric sequences may also suggest a role for Rad54 in telomere protection. To investigate this possibility, we studied chromosomal aberrations spontaneously arising in primary MEFs (passage 1) by using three independent techniques: Q-FISH (Table 2), CO-FISH (Fig. 3), and SKY (Table 3) (see Materials and Methods).
TABLE 2.
Spontaneously arising chromosomal aberrations in primary MEFs of the indicated genotypes as determined by Q-FISH
| Parametera | Results obtained with MEF type: | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rad54+/− DNA-PKcs+/− | Rad54+/− DNA-PKcs−/− | Rad54−/− DNA-PKcs+/− | Rad54−/− DNA-PKcs−/− | ||||||||||||
| A | E | F | Total | C | D | Total | B | E2 | B2 | C2 | Total | A2 | D2 | Total | |
| No. of metaphases | 100 | 100 | 100 | 300 | 100 | 100 | 200 | 100 | 100 | 100 | 100 | 400 | 100 | 100 | 200 |
| No. DIC* | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0.02 | 0 | 0.0075 | 0.04 | 0.02 | 0.03 |
| No. RL (+TTAGGG)*† | 0 | 0 | 0 | 0 | 0.02 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0.02 | 0.02 | 0.02 |
| No. RL (−TTAGGG)*† | 0 | 0 | 0 | 0 | 0 | 0.02 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| No. of R* | 0 | 0 | 0 | 0 | 0 | 0.0 | 0 | 0 | 0 | 0 | 0.02 | 0.005 | 0.02 | 0.02 | 0.02 |
| Totalb fusions (DIC+RL+R)* | 0 | 0 | 0 | 0 | 0.02 | 0.02 | 0.02 | 0 | 0.01 | 0.02 | 0.02 | 0.0125 | 0.08 | 0.06 | 0.07 |
| No. of chromosome breaks* | 0.01 | 0.02 | 0 | 0.01 | 0 | 0.06 | 0.03 | 0.03 | 0.01 | 0.04 | 0 | 0.02 | 0.16 | 0.02 | 0.09 |
| No. of chromatid breaks* | 0.03 | 0.02 | 0.04 | 0.03 | 0.04 | 0.06 | 0.05 | 0.15 | 0.11 | 0.12 | 0.32 | 0.175 | 0.16 | 0.2 | 0.18 |
| No. of gaps* | 0.01 | 0.02 | 0.02 | 0.017 | 0 | 0.02 | 0.01 | 0.02 | 0.07 | 0.04 | 0.14 | 0.067 | 0.08 | 0.04 | 0.06 |
| No. of fragments* | 0.02 | 0.02 | 0 | 0.01 | 0 | 0 | 0 | 0.03 | 0.03 | 0.04 | 0.02 | 0.03 | 0.02 | 0.02 | 0.02 |
| Double minutes* | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.04 | 0.05 | 0 | 0.08 | 0.0425 | 0.06 | 0.06 | 0.06 |
FIG. 3.
CO-FISH primary MEFs. (Left panel) Frequency of chromosome-type or chromatid-type fusions involving telomeres produced by leading-strand synthesis in metaphases of the indicated genotypes. (+)TTAGGG, fusions showing TTAGGG signal at the fusion point; (−)TTAGGG, fusions lacking TTAGGG signal at the fusion point. (Right panels) Representative CO-FISH images of chromosome-type telomere fusions containing TTAGGG repeats at the fusion point (yellow arrows) in the indicated individual MEF. Blue, DAPI; red, TTAGGG signal.
TABLE 3.
SKY analysis of end-to-end fusions in primary MEFs
| Parameter | Resultsa for MEF genotype group: | ||
|---|---|---|---|
| Rad54−/− DNA-PKcs+/− E2 | Rad54+/− DNA-PKcs−/− C | Rad54−/− DNA-PKcs−/− D2 | |
| No. of metaphases | 101 | 78 | 89 |
| No. of end-to-end fusionsb | 1 (9;13) | 3 (X;3), (1;4), (3;15) | 3 (X;4), (X;12), (1;4) |
| No. of chromosome breaks | 1 (14) | 0 | 0 |
| No. of chromatid breaks | 3 (14), (2), (9) | 2 (5), (ND) | 3 (4), (3), (ND) |
| No. of fragments | 1 (9) | 0 | 1 (centromeric [ND]) |
| No. of gaps | 1 (1) | 0 | 0 |
First, we performed telomere Q-FISH on at least 100 metaphases from each individual MEF (Table 2) (Materials and Methods). Rad54 deficiency in MEFs (Rad54−/− DNA-PKcs+/−) resulted in a significant increase in chromatid-type breaks (χ2 test, P > 0.0001) and gaps (χ2 test, P = 0.0119), as well as double minutes (χ2 test, P = 0.003) compared to the controls (Rad54+/− DNA-PKcs+/−) (Table 2). The increased frequencies of chromatid breaks and gaps in Rad54-deficient cells are in agreement with the known role of Rad54 in HR. When we extended these studies to doubly deficient Rad54−/− DNA-PKcs−/− MEFs, the frequency of chromatid breaks and gaps was not further increased by a mutation in DNA-PKcs (Table 2), suggesting that these types of aberrations are reminiscent of the Rad54 deficiency.
Furthermore, Q-FISH on metaphase chromosomes showed that Rad54 deficiency in Rad54−/− DNA-PKcs+/− MEFs resulted in an average frequency of 0.0125 end-to-end fusions per metaphase (dicentrics plus Robertsonian-like fusions plus rings) compared to undetectable end-to-end fusions in the Rad54+/− DNA-PKcs+/− controls (Table 2) (found to be significant as determined by χ2 test [P = 0.0401]). A similar frequency of end-to-end fusions was found in DNA-PKcs deficient MEFs, Rad54+/− DNA-PKcs−/−, of 0.02 fusions per metaphase compared to no fusions in the Rad54+/− DNA-PKcs+/− controls (Table 2) (found to be significant as determined by χ2 test [P = 0.0026]). The increase in end-to-end fusions in Rad54-deficient MEFs is consistent with a role for this protein in telomere capping (see discussion below of CO-FISH and SKY data). Interestingly, MEFs doubly deficient in Rad54 and DNA-PKcs (Rad54−/− DNA-PKcs−/−) showed a further increase in the frequency of end-to-end fusions, i.e., 0.07 fusions per metaphase, compared to Rad54−/− DNA-PKcs+/− MEFs, with 0.0125 fusions per metaphase, and Rad54+/− DNA-PKcs−/− MEFs, with 0.02 fusions per metaphase (Table 2), which was statistically significant (χ2 test, P < 0.0001 and _P_ = 0.0118, respectively). This finding suggests a synergism between both deficiencies in the loss of telomere protection. It is important to note that many (>50%) of the Robertsonian-like fusions associated with either Rad54 or DNA-PKcs deficiencies or with a combination of both deficencies showed detectable telomeres at the fusion point (Table 2; see also Fig. 3 for CO-FISH data), suggesting that they are not the result of TTAGGG exhaustion, a finding in agreement with the low percentages of undetectable telomeres present in these cells (Table 1). Of note, Rad54+/− DNA-PKcs+/− MEFs did not show telomere fusions, suggesting that heterozygosity of either DNA-PKcs or Rad54 is sufficient to prevent the loss of telomere protection (Table 2).
A role for DNA-PKcs in the postreplicative processing of telomeres produced by leading-strand synthesis has been proposed as a possible mechanism by which DNA-PKcs deficiency leads to loss of telomere capping and increased end-to-end chromosome fusions (3). To study a putative role of Rad54 in leading-strand DNA processing and in the generation of these fusions, we performed CO-FISH on primary MEFs from the different genotypes, which allows identification of telomeres produced by leading-strand DNA synthesis (see Materials and Methods). Chromatid-type fusions are informative on the occurrence of end-to-end fusions involving telomeres produced by leading-strand synthesis (leading-to-leading chromatid fusions) (3). We were not able to detect any clear chromatid-type fusion of the leading-to-leading type in Rad54-deficient primary MEFs after analysis of 144 metaphases (Fig. 3); therefore, we could not demonstrate a role for Rad54 in the postreplicative processing of the leading-strand telomere. Rad54−/− DNA-PKcs+/− MEFs, however, showed an increased frequency of chromosome-type fusions containing detectable TTAGGG repeats at the fusion point, i.e., 0.014 fusions/metaphase, compared to no fusions in Rad54+/− DNA-PKcs+/− control MEFs (Fig. 3). The fact that these fusions contained TTAGGG repeats at the fusion point, as detected by CO-FISH (Fig. 3), suggests that they involve at least one leading-strand telomere and that they are produced by the loss of telomere capping rather than by TTAGGG exhaustion. Similarly, Rad54+/− DNA-PKcs−/− MEFs also showed a similar frequency of chromosome-type fusions with TTAGGG repeats at the fusion point after CO-FISH: 0.015 fusions/metaphase (Fig. 3). This frequency of end-to-end fusions is similar to that previously described for single DNA-PKcs deficiency (14). Importantly, double-mutant Rad54−/− DNA-PKcs−/− MEFs resulted in a further increase in the frequency of chromosome-type fusions with TTAGGG signals at the fusion point (0.05 fusions/metaphase), suggesting a cooperative effect on both activities in preventing this type of fusion (see Fig. 3). The fact that these fusions occurred in cells doubly deficient for DNA-PKcs and Rad54 indicates that the mechanism underlying this particular type of end-to-end chromosome fusion does not involve the activity of Rad54, although we cannot exclude that other HR components may be mediating these fusions.
To further characterize the nature of the fusions resulting from Rad54 and DNA-PKcs deficiencies, we performed SKY on metaphases derived from MEFs of the different genotypes; this allowed identification of the chromosomes involved in the fusion (Table 3). All end-to-end fusions present in DNA-PKcs deficient MEFs (Rad54+/− DNA-PKcs−/−) involved random pairs of chromosomes (Table 3). A similar outcome has been previously described for Ku86-deficient MEFs (10). Likewise, the only end-to-end fusion identified in Rad54-deficient MEFs (Rad54−/− DNA-PKcs+/−) was a chromatid-type fusion involving nonhomologous chromosomes (chromosomes 9 and 13) (Table 3). Similarly, when we extended the present study to Rad54−/− DNA-PKcs−/− MEFs, fusions also involved random pairs of chromosomes. The fact that end-to-end fusions due to either Rad54 or DNA-PKcs deficiency involve random pairs of chromosomes indicates that they are not the result of sister chromatid fusion and is consistent with previously published results (3, 10, 11).
The final frequencies of end-to-end fusions per metaphase considering all metaphases analyzed for each genotype by the different FISH techniques (Q-FISH plus CO-FISH plus SKY) were 0 fusions/metaphase after analysis of 408 metaphases from Rad54+/− DNA-PKcs+/− MEFs, 0.014 fusions/metaphase after analysis of 645 metaphases from Rad54−/− DNA-PKcs+/− MEFs, 0.02 fusions/metaphase after analysis of 415 metaphases from Rad54+/− DNA-PKcs−/− MEFs, and 0.06 fusions per metaphase after analysis of 464 metaphases from Rad54−/− DNA-PKcs−/− MEFs. The very high numbers of metaphases analyzed per genotype support a role for both Rad54 and DNA-PKcs in telomere capping.
In summary, the results presented here suggest that chromosome-type end-to-end fusions due to DNA-PKcs deficiency, which involve random pair of chromosomes, are not likely the result in HR events since they are not prevented in the absence of Rad54. In addition, Rad54 deficiency also results in chromosome fusions involving random pairs of chromosomes and containing telomere signals at the fusion point similar to those produced by DNA-PKcs deficiency, suggesting a role for Rad54 in telomere protection.
Final remarks and significance.
The results presented here demonstrate a role for Rad54, an essential component of the homologous recombination machinery, in telomere length maintenance and telomere capping in mammalian cells. In particular, Rad54 deficiency results in a significant loss of telomeric sequences in the presence of normal levels of telomerase activity, as well as in a significant increase in chromosome end-to-end fusions. Finally, we show that Rad54 does not play an important role in mediating end-to-end fusions due to unprotected telomeres, in contrast to the role shown for NHEJ activities.
Acknowledgments
We thank R. Serrano and E. Santos for mouse care and genotyping.
I.J. is a predoctoral fellow of Gulbenkian Foundation (Portugal), and P.M. is a Ramón y Cajal Senior Scientist. Research at the laboratory of M.A.B. is funded by grant PM97-0133 from the MCYT, grant 08.1/0030/98 from CAM, and grants EURATOM/991/0201, FIGH-CT-1999-00002 and FIS5-1999-00055 from the European Union and by the DIO. The DIO was founded and is supported by the Spanish Research Council (CSIC) and by Pharmacia. G.T. is a scholar of the Leukemia and Lymphoma Society. The G.T. laboratory is supported by National Institutes of Health (NIH) grant CA76409 and the Human Frontier Science Program. The S.B. laboratory is supported by grants from the U.S. Department of Energy (ER632339) and the NIH (CA43322).
REFERENCES
- 1.**Alexander, P., and Z. B. Mikulski.1961. Mouse lymphoma cells with different radiosensitivities. Nature 192:**572-573. [DOI] [PubMed] [Google Scholar]
- 2.**Bailey, S. M., J. Meyne, D. J. Chen, A. Kurimasa, G. C. Li, B. E. Lehnert, and E. H. Goodwin.1999. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. USA 96:**14899-14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.**Bailey, S. M., M. N. Conforth, A. Kurimasa, D. J. Chen, and E. H. Goodwin.2001. Strand-specific postreplicative processing of mammalian telomeres. Science 293:**2462-2465. [DOI] [PubMed] [Google Scholar]
- 4.**Blackburn, E. H.2001. Switching and signaling at the telomere. Cell 106:**661-673. [DOI] [PubMed] [Google Scholar]
- 5.**Blasco, M. A., M. Rizen, C. W. Greider, and D. Hanahan.1996. Differential regulation of telomerase activity and telomerase RNA during multi-stage tumorigenesis. Nat. Genet. 12:**200-204. [DOI] [PubMed] [Google Scholar]
- 6.**Blasco, M. A., H.-W. Lee, P. Hande, E. Samper, P. Lansdorp, R. DePinho, and C. W. Greider.1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:**25-34. [DOI] [PubMed] [Google Scholar]
- 7.**Collins, K., and J. R. Mitchell.2002. Telomerase in the human organism. Oncogene 21:**564-579. [DOI] [PubMed] [Google Scholar]
- 8.**De Lange, T.2002. Protection of mammalian telomeres. Oncogene 21:**532-540. [DOI] [PubMed] [Google Scholar]
- 9.**Dronkert, M. L., H. B. Beverloo, R. D. Johnson, J. H. Hoeijmakers, M. Jasin, and R. Kanaar.2000. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 20:**3147-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.**Espejel, S., S. Franco, S. Rodríguez-Perales, S. D. Bouffler, J. C. Cigudosa, and M. A. Blasco.2002. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 21:**2207-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.**Espejel, S., S. Franco, A. Sgura, D. Gae, S. Bailey, G. Taccioli, and M. A. Blasco.2002. Functional interaction between DNA-PKcs and telomerase in telomere length maintenance. EMBO J. 21:**6275-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.**Essers, J., R. W. Hendriks, S. M. Swagemakers, C. Troelstra, J. de Wit, D. Bootsma, J. H. Hoeijmakers, and R. Kanaarm.1997. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell 89:**195-204. [DOI] [PubMed] [Google Scholar]
- 13.**Gilley, D., H. Tanaka, M. P. Hande, A. Kurimasa, G. C. Li, M. Oshimura, and D. J. Chen.2001. DNA-PKcs is critical for telomere capping. Proc. Natl. Acad. Sci. USA 98:**15084-15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.**Goytisolo, F., E. Samper, S. Edmonson, G. E. Taccioli, G. E. Taccioli, and M. A. Blasco.2001. Absence of DNA-PKcs in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol. 21:**3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.**Goytisolo, F. A., and M. A. Blasco.2002. Many ways to telomere dysfunction: in vivo studies using mouse models. Oncogene 21:**584-591. [DOI] [PubMed] [Google Scholar]
- 16.**Hemann, M. T., M. A. Strong, L. Y. Hao, and C. W. Greider.2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107:**67-77. [DOI] [PubMed] [Google Scholar]
- 17.**Henson, J. D., A. A. Neumann, T. R. Yeager, and R. R. Reddel.2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21:**598-610. [DOI] [PubMed] [Google Scholar]
- 18.**Herrera, E., E. Samper, J. Martín-Caballero, J. M. Flores, H.-W. Lee, and M. A. Blasco.1999. Disease states associated to telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 18:**2950-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.**Hsu, H.-L., D. Gilley, S. A. Galande, M. P. Hande, B. Allen, S.-H. Kim, G. C. Li, J. Campisi, T. Kowhi-Shigematsu, and D. J. Chen.2000. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 14:**2807-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.**Kanaar, R., J. H. Hoeijmakers, and D. C. van Gent.1998. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 8:**483-489. [DOI] [PubMed] [Google Scholar]
- 21.**Khanna, K. K., and S. P. Jackson.2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27:**247-254. [DOI] [PubMed] [Google Scholar]
- 22.**Le, S., J. K. Moore, J. E. Haber, and C. W. Greider.1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:**143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.**Lundblad, V., and E. H. Blackburn.1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:**347-360. [DOI] [PubMed] [Google Scholar]
- 24.**Lundblad, V.2002. Telomere maintenance without telomerase. Oncogene 21:**522-531. [DOI] [PubMed] [Google Scholar]
- 25.**McIlrath, J., S. Bouffler, E. Samper, A. Cuthbert, A. Wojcik, I. Szumiel, P. E. Bryant, A. C. Riches, A. Thompson, M. A. Blasco, R. F. Newbold, and P. Slijepcevic.2001. Telomere length anormalities in mammalian radiosensitive cells. Can. Res. 61:**912-915. [PubMed] [Google Scholar]
- 26.**Neumann, A. A., and R. R. Reddel.2002. Opinion—telomere maintenance and cancer: look, no telomerase. Nat. Rev. Cancer 2:**879-884. [DOI] [PubMed] [Google Scholar]
- 27.**Petukhova, G., S. Van Komen, S. Vergano, H. Klein, and P. Sung.1999. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double-helix conformation. J. Biol. Chem. 274:**29453-29462. [DOI] [PubMed] [Google Scholar]
- 28.**Riha, K., J. M. Watson, J. Parkey, and D. E. Shippen.2002. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21:**2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.**Rijkers, T., J. Van Den Ouweland, B. Morolli, A. G. Rolink, W. M. Baarends, P. P. Van Sloun, P. H. Lohman, and A. Pastink.1998. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18:**6423-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.**Samper, E., F. Goytisolo, P. Slijepcevic, P. van Buul, and M. A. Blasco.2000. Mammalian Ku86 prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1:**244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.**Samper, E., J. M. Flores, and M. A. Blasco.2001. Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc−/− mice with short telomeres. EMBO Rep. 2:**800-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.**Sigurdsson, S., S. Van Komen, G. Petukhova, and P. Sung.2002. Homologous DNA pairing by human recombination factors rad51 and rad54. J. Biol. Chem. 277:**42790-42794. [DOI] [PubMed] [Google Scholar]
- 33.**Smith, G. C. M., and S. P. Jackson.1999. The DNA-dependent protein kinase. Genes Dev. 13:**916-934. [DOI] [PubMed] [Google Scholar]
- 34.**Smogorzewska, A., J. Karlseder, H. Holtgreve-Grez, A. Jauch, and T. de Lange.2002. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 12:**1635-1644. [DOI] [PubMed] [Google Scholar]
- 35.**Taccioli, G. E., A. G. Amatucci, H. J. Beamish, D. Gell, X. H. Xiang, M. I. Torres-Arzayus, A. Priestley, S. P. Jackson, A. Marshak-Rothstein, P. A. Jeggo, and V. L. Herrera.1998. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity 3:**355-366. [DOI] [PubMed] [Google Scholar]
- 36.**van den Bosch, M., P. H. Lohman, and A. Pastink.2002. DNA double-strand break repair by homologous recombination. Biol. Chem. 383:**873-892. [DOI] [PubMed] [Google Scholar]
- 37.**van Steensel, B., A. Smogorzewska, and T. de Lange.**1998. TRF2 protects human telomeres from end-to-end fusions. Cell **92:**401-413. [DOI] [PubMed]