Phosphorylation of the α Subunit of Eukaryotic Initiation Factor 2 Is Required for Activation of NF-κB in Response to Diverse Cellular Stresses (original) (raw)
Abstract
Nuclear factor κB (NF-κB) serves to coordinate the transcription of genes in response to diverse environmental stresses. In this report we show that phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2) is fundamental to the process by which many stress signals activate NF-κB. Phosphorylation of this translation factor is carried out by a family of protein kinases that each respond to distinct stress conditions. During impaired protein folding and assembly in the endoplasmic reticulum (ER), phosphorylation of eIF2α by PEK (Perk or EIF2AK3) is essential for induction of NF-κB transcriptional activity. The mechanism by which NF-κB is activated during ER stress entails the release, but not the degradation, of the inhibitory protein IκB. During amino acid deprivation, phosphorylation of eIF2α by GCN2 (EIF2AK4) signals the activation of NF-κB. Furthermore, inhibition of general translation or transcription by cycloheximide and actinomycin D, respectively, elicits the eIF2α phosphorylation required for induction of NF-κB. Together, these studies suggest that eIF2α kinases monitor and are activated by a range of stress conditions that affect transcription and protein synthesis and assembly, and the resulting eIFα phosphorylation is central to activation of the NF-κB. The absence of NF-κB-mediated transcription and its antiapoptotic function provides an explanation for why eIF2α kinase deficiency in diseases such as Wolcott-Rallison syndrome leads to cellular apoptosis and disease.
Nuclear factor κB (NF-κB) is a dimer of the Rel family of proteins that regulates the transcription of genes involved in immune and inflammatory responses, stress remediation, cell growth, and apoptosis (2, 3, 9, 15, 33, 39, 47, 54). In its inert state, NF-κB is present in the cytoplasm in association with proteins known as inhibitors of NF-κB (IκB). In response to certain inducing conditions, IκB becomes phosphorylated, leading to its ubiquitination and subsequent degradation by the proteasome. Release and proteolysis of IκB facilitates NF-κB passage into the nucleus, where this protein binds to the κB consensus DNA sequence and regulates transcription.
Activation of NF-κB in cells occurs in response to a large variety of stress conditions, including exposure to proinflammatory cytokines, UV or γ irradiation, bacterial or viral infection, or impaired protein folding in the endoplasmic reticulum (ER) (47). The mechanisms by which these diverse stress signals are recognized and signal NF-κB induction have been the focus of much research. In the example of activation of NF-κB by tumor necrosis factor alpha (TNF-α), a cytokine that functions as an activator of the innate immune response, association of TNF-α with its receptor directs the activation of the IκB kinase (3, 15, 39). The IκB kinase (IKK)—containing two catalytic subunits, IKK α and β, as well as a regulatory subunit, IKKγ (NEMO)—phosphorylates Ser residues in IκB, thus contributing to its release from NF-κB (32). Elevated expression of many different proteins slated for the secretory pathway or exposure to ER stress agents was found to activate NF-κB (47-49). This signal transduction pathway, designated the ER overload response (EOR), is proposed to be distinct from another ER stress pathway—the unfolded protein response (UPR), which induces the transcription of a large number of genes involved in protein secretion and processing, such as the ER chaperones GRP78/BiP and GRP94 (22, 35, 47). The mechanism by which the EOR activates NF-κB, including the involvement of IκB, is currently unclear.
We have been interested in understanding the role of a family of protein kinases that phosphorylate the α subunit of eukaryotic initiation factor 2 (eIF2) in the early events of stress response pathways (12, 65). The eIF2 coupled with GTP and initiator Met-tRNAiMet participates in the ribosomal recognition of the start codon (27). During this translation initiation process, GTP associated with eIF2 is hydrolyzed to GDP and eIF2 is released from the ribosome. Recycling of eIF2-GDP to eIF2-GTP requires a guanine nucleotide exchange factor, designated eIF2B. Phosphorylation of eIF2α by PEK (also designated Perk and EIF2AK3) in response to impaired ER function converts this initiation factor from a substrate to an inhibitor of the eIF2B (24, 25, 42, 53, 55). The resulting reduction in eIF2-GTP levels can reduce general translation, allowing the cell sufficient time to correct the impaired protein folding resulting from ER stress prior to synthesizing additional proteins. Accompanying this reduction in global protein synthesis, phosphorylation of eIF2α by PEK can induce gene-specific translation, which is important for the expression of stress remedy genes (14, 22, 23, 35). Loss of PEK (Perk) in mouse embryonic stem (ES) cells exposed to ER stress leads to inappropriately elevated protein synthesis that further exacerbates protein misfolding in this organelle, thus leading to apoptosis (24). Loss of PEK (EIF2AK3) in humans leads to Wolcott-Rallison syndrome (WRS), a disorder involving neonatal insulin-dependent diabetes resulting from a characteristic destruction of pancreatic islet beta cells (11). WRS patients do not display autoantibodies characteristic of type 1 diabetes, and they also suffer from epiphyseal dysplasia, osteoporosis, growth retardation, recurrent hepatitis, and isolated central hypothyroidism (6, 8, 58). PEK−/− mice display many of these phenotypes and succumb to complications related to hyperglycemia within several weeks of birth (21, 72).
In addition to PEK, three other mammalian eIF2α kinases have been described, and each directly senses distinct stress signals and activates downstream response pathways by regulating translation. These eIF2α kinases include GCN2, which is activated by nutritional stress, including amino acid deprivation (23, 28, 66, 69, 73); HRI, which links protein synthesis to heme availability in erythroid cells and is also activated by oxidative and heat stresses and exposure to certain diffusible gases (10, 20, 40, 59, 71); and PKR, which controls an antiviral defense pathway that is induced by interferon (34, 67). To date, only PKR has been linked with activation of NF-κB by a mechanism independent of its eIF2α kinase activity (1, 17, 67). Initially, PKR was proposed to directly phosphorylate IκB (36). More recent studies suggest that PKR physically associates with the IKK complex and stimulates NF-κB transcriptional function through the action of IKK and NF-κB-inducing kinase (NIK) (7, 16, 18, 30, 70). The details of the PKR-dependent induction of NF-κB are controversial, as activation of NF-κB has been reported to be both dependent and independent of PKR kinase catalytic activity. Furthermore, the double-stranded RNA (dsRNA) binding properties of the PKR regulatory domain have also been suggested to be dispensable for the induction of NF-κB (30).
Given the diverse stress conditions activating both NF-κB and eIF2α kinases and their important medical implications, we were interested in establishing possible regulatory overlap between these two stress pathways. In this study, we used mouse embryo fibroblasts (MEFs) from which PEK or GCN2 had been deleted or that contained a homozygous mutation at the eIF2α phosphorylation site (Ser51Ala), and we demonstrate that phosphorylation of eIF2α is required for activation of NF-κB in response to either ER stress or amino acid starvation. The mechanism of NF-κB induction by the eIF2α kinases entails the release, but not the degradation, of IκB. These results support the idea that impaired NF-κB activation in PEK-deficient cells is an important underlying reason for their susceptibility to apoptosis upon exposure to stress.
MATERIALS AND METHODS
Cell culture and stress conditions.
MEF cells prepared from embryos generated from crosses between heterozygous PEK−/− (Perk−/−) mice (72), GCN2−/− animals (73), heterozygous eIF2α A/A (51), or their wild-type counterparts were immortalized by infection with a recombinant retrovirus expressing simian virus 40 (SV40) large T antigen as previously described (68). MEF p65/RelA+/+ and MEF p65/RelA−/− cells were obtained from Harikrishna Nakshatri (Indiana University School of Medicine) (4, 45). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; BioWhittaker) supplemented with 2 mM glutamine, 1 mM nonessential amino acids, 10% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Cultures of MEF A/A cells require additional amino acid supplements for growth viability (51), and therefore the A/A and the wild-type counterpart S/S cells were maintained under these enriched medium conditions. MEF cells were grown to 50 to 70% confluency and subjected to ER stress that involved the addition of thapsigargin at a concentration of 2 μM unless otherwise indicated or of 2 μg of tunicamycin per ml to DMEM for the specified incubation times. Confluent growth is itself a stress that can induce eIF2α phosphorylation. To ensure a confluency of less than 70%, MEF cells were shifted to DMEM with 0.5% fetal bovine serum 12 h prior to ER or nutritional stress, with results similar to those found in stress experiments performed using cells dividing continuously in DMEM supplemented with 10% fetal bovine serum. To address the role of transcription or protein synthesis in combination with ER stress, 50 μg of cycloheximide per ml or 10 μg of actinomycin D per ml was added to the MEF cells along with 2 μM thapsigargin, and the cells were incubated for 3 or 6 h prior to collection and analysis. Amino acid starvation was brought about by culturing MEF cells in DMEM without leucine (BioWhittaker). As a control for IκB phosphorylation and subsequent degradation, 10 ng of TNF-α per ml was added to MEF cells in DMEM for 30 min. The eIF2α phosphorylation induced during ER or nutrient or stress conditions was similar between the primary MEF cells and immortalized MEF cell lines.
EMSA.
Nuclear extracts were prepared from MEF cells as described previously (31). This preparation involved resuspending cultured MEF cells subjected to either stress or nonstress conditions in 1 ml of cold hypotonic RSB buffer (10 mM Tris [pH 7.4], 10 mM NaCl, and 3 mM MgCl2) supplemented with 0.5% NP-40 and protease inhibitors (100 μM phenylmethylsulfonyl fluoride, 0.15 μM aprotinin, 1 μM leupeptin, and 1 μM pepstatin). Cells were lysed with a Dounce homogenizer, and after centrifugation at 14,000 × g, the nuclei pellets were resuspended in two packed nuclear volumes of extraction buffer C (420 mM KCl, 20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.2 mM EDTA, and 20% glycerol) supplemented with protease inhibitors. Protein concentrations were determined by using the Bio-Rad protein assay. The sequence of the double-stranded DNA fragment containing the NF-κB binding element derived from c-myc (URE) was 5′-GATCCAAGTCCGGGTTTTCCCCAACC-3′ and for Octomer-1 (OCT-1) binding was 5′-GATCTGTCGAATGCAAATCACTAGAA-3′ (31). In the binding reactions, 32P-labeled DNA fragments (20,000 to 25,000 cpm), 5 μg of nuclear extract, and 2.5 μg of poly (dI-dC) as nonspecific competitor were added to a solution of 10 mM HEPES (pH 7.9), 4 mM dithiothreitol, 0.5% Triton X-100, 100 mM KCl, and 2.5% glycerol in a final assay volume of 25 μl. Binding assays were performed at room temperature for 30 min, and the DNA-protein complexes were separated by gel electrophoresis and visualized by autoradiography as previously described (57). To address NF-κB binding specificity, unlabeled competitor DNA fragments were added at the indicated stoichiometry to the binding mixture. CREB competitor DNA included a published ATF consensus binding sequence, i.e., TGACGTCA (61). Supershift studies were carried out by including p65 (Upstate Biotechnology)- and/or p50-specific polyclonal antibody (Santa Cruz) in the binding mixture, followed by electrophoretic mobility shift assay (EMSA) and autoradiography.
Preparation of protein lysates and immunoblot analyses.
MEF cells subjected to the indicated stress conditions (or to no stress) were washed twice with ice-cold phosphate-buffered saline solution and lysed using a solution of 50 mM Tris-HCl (pH 7.9), 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate (SDS), 100 mM NaF, 17.5 mM β-glycerolphosphate, and 10% glycerol supplemented with protease inhibitors and subjected to sonication for 30 s. Lysates were clarified by centrifugation and measured for protein content by using the Bio-Rad protein quantitation kit for detergent lysis. Equal amounts of each protein sample were separated by electrophoresis in a SDS-polyacrylamide gel and transferred to nitrocellulose filters. Filters were incubated in TBS-T solution (20 mM Tris-HCl [pH 7.9], 150 mM NaCl, and 0.2% Tween 20 supplemented with 4% nonfat milk) and antibodies that recognize the specified protein. ATF4, p65, IκBα, IκBβ, and Chop antibodies was purchased from Santa Cruz Biotechnology. ATF4 studies were confirmed by using ATF4 polyclonal antibody that was provided by David Ron (New York University School of Medicine). Phosphorylation of eIF2α and IκBα was measured with polyclonal antibody that recognizes eIF2α phosphorylated at Ser-51 (Research Genetics) or IκBα phosphorylated at Ser-32 (Cell Signaling). Total eIF2α levels were measured by using monoclonal antibody provided by Scot Kimball (Pennsylvania State University College of Medicine). Filters were washed three times in TBS-T to remove unbound antibody and incubated with TBS-T containing secondary antibody conjugated to horseradish peroxidase (Bio-Rad). Protein-antibody complexes were visualized by using horseradish peroxidase-labeled secondary antibody and chemiluminescent substrate. Low- and high-range polypeptide markers (Bio-Rad) were used to measure the sizes of proteins detected in the immunoblots. Linearity in the immunoblot assays was established by serially diluting proteins in the SDS-polyacrylamide gel electrophoresis (PAGE) and by carrying out multiple autoradiographic exposures for increasing lengths of time. Quantitation of visualized bands was carried out by densitometry.
Immunofluorescence and confocal microscopy.
PEK+/+ MEF cells were grown overnight on glass slides and then treated with 2 μM thapsigargin for 6 h or were not subjected to stress. Alternatively, PEK−/− MEF cells were cultured in 150-mm petri dishes to 50 to 70% confluence, and 5 μg of PEK expression plasmid pKM10 (41) was cotransfected with a GFP-expressing plasmid by using FUGENE (Roche). After 24 h, the transfected cultures were divided into eight-well chamber slides and grown overnight followed by treatment with 2 μM thapsigargin or with no stress. Cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.5% Triton X-100 for 10 min. p65 was visualized by using polyclonal antibodies directed specifically against this transcription factor, followed by goat anti-rabbit immunoglobulin G (IgG) conjugated with Rhodamine Red (Molecular Probes, Inc.) as previously described (31). Immunofluorescence was carried out by using a laser confocal microscope LSM510 (Carl Zeiss). Secondary antibodies were visualized by using an argon-krypton laser producing excitation bands at 568 nm for Rhodamine Red and by using monochromatic light for differential interference contrast images. Fluorescent images were collected with emission filters for 585 to 610 nm for Rhodamine Red and for 488 to 520 nm for fluorescein isothiocyanate. Images were stored digitally by using Adobe Photoshop. For visualization of nuclei, cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) mountain medium (Jason Lab).
Dual luciferase assays.
To measure NF-κB transcriptional activity, PEK+/+ and PEK−/− MEF cells were transfected in triplicate with an NF-κB element, i.e., a firefly luciferase reporter plasmid that contains three κB elements derived from upstream of the MHC class I promoter (50). Luciferase assays were carried out with the Dual-Luciferase Reporter Assay System (Promega) per the manufacturer's instructions. MEF cells were transfected by using Lipofectamine (Invitrogen Life Technologies). One day prior to transfection, MEF cells were plated at 105 cells per well on 24-well plates. The NF-κB-directed luciferase construct and Renilla luciferase expressed from the SV40-derived promoter (Promega) (as an internal control) were transfected into the MEF cells. To determine whether transient expression of PEK rescued NF-κB transcriptional activity in the PEK−/− cells, the PEK expression plasmid pKM10 was cotransfected with the luciferase reporter genes. Cells were incubated for 48 h and treated with 0 to 2.0 μM thapsigargin as indicated for 6 h. Cells were then washed twice with PBS and harvested with 900 μl of 1× passive lysis buffer (Promega). The cell lysate in a volume of 200 μl was mixed with 100 μl of Luciferase Assay Reagent II for measuring the firefly luciferase activity, and 100 μl of Stop & Glo Reagent (Promega) was added to measure the Renilla luciferase activity. Light units of both luciferase activities were assayed for 10 s. Luciferase activity was measured as relative light units (RLU) (Monolight Luminometer, model 2010). The luciferase activity for the NF-κB promoter constructs was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity for each transfection. All data are presented as the mean relative luciferase activity as calculated by RLU for the NF-κB-directed luciferase divided by the RLU for the reference construct. Luciferase activity was then normalized to that activity determined for PEK+/+ MEF cells not subjected to thapsigargin treatment. Three independent experiments were carried out.
RESULTS
Phosphorylation of eIF2α by PEK is required for activation of NF-κB in response to ER stress.
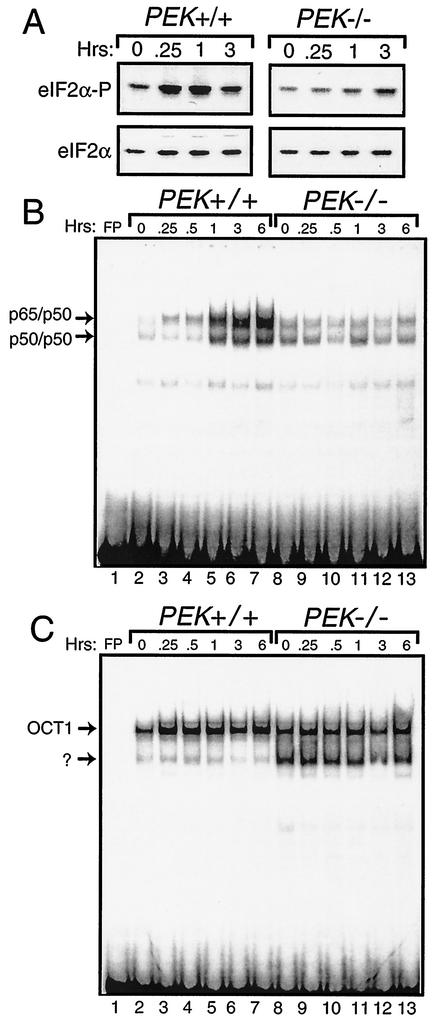
Phosphorylation of eIF2α is an early signal in the coordinate response to many different environmental stress conditions. In the case of ER stress, PEK phosphorylates eIF2α in MEF cells within 15 min of treatment with thapsigargin, which is a standard ER stress agent that triggers the release of calcium from this organelle (Fig. 1A). Phosphorylation of eIF2α was measured by using immunoblot analysis and polyclonal antibody specific to eIF2α phosphorylated at Ser-51. By comparison, minimal induction of eIF2α phosphorylation was found in PEK−/− MEF cells. Similar levels of total eIF2α, as judged by immunoblotting using antibody that recognizes both phosphorylated and nonphosphorylated forms of eIF2α, were found between the different lysate preparations, thus demonstrating that changes in eIF2α phosphorylation were not due to changes in protein levels (Fig. 1A). To address the contribution of eIF2α phosphorylation in ER stress-mediated induction of NF-κB, we measured the activity of this transcription factor by using the EMSA and a radiolabeled DNA fragment containing a NF-κB binding site as previously described (31) (Fig. 1B). Nuclear lysates were prepared from PEK+/+ and PEK−/− MEF cells treated with thapsigargin or with no stress. Significant DNA binding attributed to NF-κB dimers p65/p50 and p50/p50 was detected in PEK+/+ cells after 1 h of thapsigargin treatment, with further enhanced binding observed following 3 and 6 h of exposure to this ER stress (Fig. 1B). No induction of NF-κB binding during ER stress was detected in the PEK−/− cells. As a control, we also measured DNA binding of OCT1 by using the PEK+/+ and PEK−/− nuclear lysates, and we found minimal differences in the levels of binding of this transcription factor (Fig. 1C). It is noteworthy that in the PEK−/− sample, there was an additional faster-migrating band using the radiolabeled DNA containing the OCT1 binding site that was present only at a much reduced intensity in the PEK+/+ lysates.
FIG. 1.
eIF2α kinase PEK is required for activation of NF-κB in response to ER stress. (A) PEK+/+ and PEK−/− MEF cells were exposed to thapsigargin for 0.25 to 3 h (as indicated) or in the absence of this ER stress agent (0 h). Phosphorylation of eIF2α was measured by immunoblot analysis by using polyclonal antibody specific to eIF2α phosphorylated at Ser-51 (eIF2α∼P). Levels of total eIF2α were assayed by using antibody that recognizes both phosphorylated and nonphosphorylated versions of the translation initiation factor. (i.e., eIF2α). Nuclear lysates were prepared from PEK+/+ (lanes 2 to 7) and PEK−/− (lanes 8 to 13) MEF cells treated with thapsigargin for the indicated times and incubated with radiolabeled DNA containing a NF-κB (B) or OCT1 (C) binding sites. Binding mixtures were separated by electrophoresis, and bound DNAs were visualized by autoradiography. Arrows indicate DNA complexed with p65/p50 or p50/p50 as defined in experiments shown in Fig. 2. OCT1 bound to DNA and an unknown protein complex are also indicated by arrows. Radiolabeled DNA at the bottom of panels B and C are unbound probe. Free probe (FP) indicates the radiolabeled NF-κB or OCT1 DNA fragments without nuclear lysate.
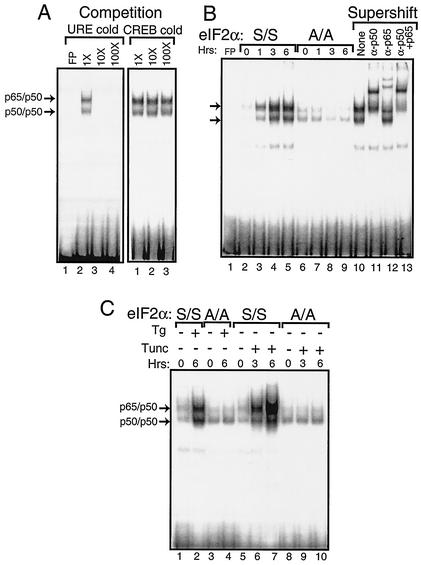
The specificity of the NF-κB DNA binding was confirmed by first adding nonradiolabeled competitor DNA to the EMSA reaction mixture. We found that excess nonradiolabeled probe containing the NF-κB binding site (i.e., URE) reduced binding, while no competition was observed with nonradiolabeled DNA containing the CREB binding element (Fig. 2A). Furthermore, we assessed the mobility shift following the addition of polyclonal antibody specific for p50 or p65. The addition of p50 antibody completely retarded the migration of the radiolabeled DNA designated p50/p50 and reduced the migration of a portion of the p65/p50 band (Fig. 2B, lanes 10 to 13). By comparison, addition of antibody specific to p65 to the EMSA mixture elicited a supershift of a portion of the designated p65/p50 radiolabeled DNA and had no impact on the p50/p50 band. Together, the p50 and p65 antibodies elicited a migration shift of both NF-κB bands. These results indicate that PEK activity is required for induction of these NF-κB dimers during ER stress conditions.
FIG. 2.
Phosphorylation of eIF2α at Ser-51 facilitates activation of NF-κB during ER stress. Nuclear lysates were prepared from MEF cells containing eIF2α with wild-type Ser-51 (S/S) or with alanine substituted for the eIF2α phosphorylation site (A/A) subjected to ER stress or to no stress. Equal amounts of nuclear lysate were used in each EMSA mixture containing radiolabeled DNA with an NF-κB binding site. Instances of DNA complexed with NF-κB dimers p65/p50 and p50/p50 (indicated by arrows) were visualized following autoradiography. (A) To determine the specificity for the NF-κB binding site, nonradiolabeled DNA containing the NF-κB site, URE (lanes 2 to 4), or the unrelated CREB DNA binding site (lanes 1 to 3) was added to EMSA binding mixtures containing nuclear lyates prepared from S/S MEF cells treated with thapsigargin for 6 h. Competition indicates that nonradiolabeled competitor DNA was added at a 1×, 10×, or 100× molar excess. Free probe (FP) indicates only radiolabeled NF-κB DNA fragments without nuclear lysate. Radiolabeled DNA at the bottom of panel is unbound probe. (B) Nuclear lysates were prepared from S/S (lanes 2 to 5) and A/A (lanes 6 to 9) MEF cells subjected to thapsigargin for between 0 and 6 h and assayed for NF-κB binding in the EMSA. In lanes 10 to 13, supershift indicates that polyclonal antibodies that specifically recognize p50 and/or p65 were added to the EMSA binding mixture. “None” indicates that no antibody was used in the assay (lane 10). (C) MEF cells were exposed to either thapsigargin (Tg) or tunicamycin (Tunc) as indicated by the “+” or “−” for the indicated number of hours. Nuclear lysates prepared from S/S and A/A MEF cells (as indicated) were analyzed for binding to the NF-κB probe in the EMSA.
The role of phosphorylation of the eIF2α at Ser-51 in the activation of NF-κB was directly addressed by using MEF cells containing wild-type eIF2α (S/S) or a mutant version containing Ala substituted for the phosphorylated Ser-51 residue (A/A) (Fig. 2B, lanes 2 to 9). While activation of NF-κB occurred in the S/S cells exposed to thapsigargin following a similar time course described above, no increase in NF-κB binding was detected in the A/A cells. Finally, we addressed the importance of eIF2α phosphorylation in the activation of NF-κB in response to a different ER stress agent, tunicamycin, which inhibits protein glycosylation in this organelle and activates PEK (25). Enhanced NF-κB binding was detected within 3 h of tunicamycin exposure of S/S cells, with a further increase following 6 h of ER stress treatment (Fig. 2C). No activation of NF-κB was observed in A/A MEF cells. We conclude that phosphorylation of eIF2α by PEK is required for activation of NF-κB in response to different stress conditions in the ER.
eIF2α kinase GCN2 facilitates activation of NF-κB during amino acid deprivation.
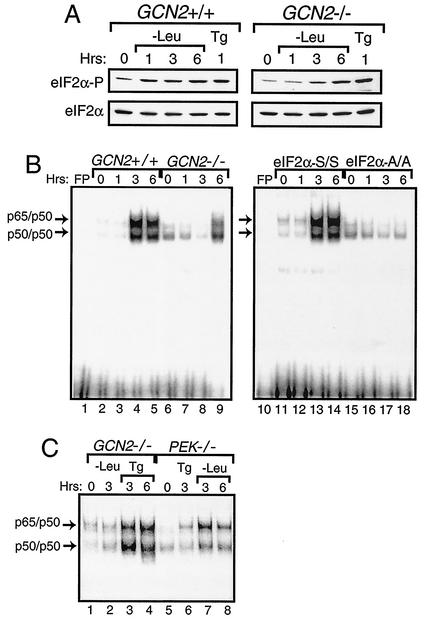
We next addressed whether other members of the eIF2α kinase family mediate activation of NF-κB. GCN2 is the predominant eIF2α kinase activated during amino acid deprivation, with elevated phosphorylation of eIF2α in GCN2+/+ MEF cells occurring within 1 h of this stress (Fig. 3A) (13, 23, 56, 73). By contrast, significant eIF2α phosphorylation was detected in GCN2−/− MEF cells only following 6 h of leucine limitation, thus indicating that one or more alternative eIF2α kinases can be activated during extended nutrient stress (Fig. 3A). Induction of eIF2α phosphorylation by ER stress was observed in both the GCN2+/+ and GCN2−/− MEF cells. Using nuclear lysates prepared from GCN2+/+ MEF cells, we found enhanced NF-κB binding in the EMSA following 3 h of leucine deprivation, and this binding continued to be elevated after 6 h of this stress condition (Fig. 3B, lanes 2 to 5). By contrast, only following 6 h of leucine starvation was there a modest increase in NF-κB binding in the GCN2−/− cells (Fig. 3B, lanes 6 to 9). Further emphasizing the regulatory specificity for GCN2 and PEK for amino acid starvation or ER stress, respectively, we found activation of NF-κB in GCN2−/− cells in response to ER stress, and NF-κB was induced in PEK−/− cells in response to leucine depletion but not ER stress (Fig. 3C). In the MEF cells containing eIF2α with Ala substituted for Ser-51 (A/A), there was no activation of NF-κB during the amino acid limitation—even after 6 h of leucine starvation (Fig. 3B, lanes 15 to 18). These results indicate that eIF2α phosphorylation is required for induction of NF-κB in response to amino acid limitation as well as ER stress. While GCN2 is the predominant eIF2α kinase in response to amino acid starvation, one or more alternative eIF2α kinases can be induced during prolonged leucine starvation, and this elevated eIF2α phosphorylation leads to the activation of NF-κB.
FIG. 3.
Activation of NF-κB during amino acid conditions requires phosphorylation of eIF2α by GCN2 protein kinase. (A) GCN2+/+ and GCN2−/− MEF cells were deprived of leucine for between 1 and 6 h and subjected to thapsigargin for 1 h or to no stress (0 h) as indicated. Phosphorylation of eIF2α was assayed for by immunoblot analysis using antibody specific to eIF2α phosphorylated at Ser-51 (eIF2α∼P), and total eIF2α levels (eIF2α) were measured using antibody that recognizes both phosphorylated and nonphosphorylated versions of eIF2α. (B) Nuclear lysates were prepared from GCN2+/+ (lanes 2 to 5), GCN2−/− (lanes 6 to 9), S/S (lanes 11 to 14), and A/A (lanes 15 to 18) MEF cells deprived of leucine for the indicated number of hours and were assayed for binding with radiolabeled DNA containing a NF-κB binding site by the EMSA. (C) NF-κB binding was measured by the EMSA using nuclear lysates prepared from GCN2−/− and PEK−/− MEF cells that were starved for leucine or exposed to thapsigargin for the indicated number of hours. Arrows indicate DNA complexed with p65/p50 or p50/p50. Free probe (FP) indicates that only the radiolabeled NF-κB DNA was used in the assay, and the radiolabeled DNA at the bottom of panel B is unbound probe.
ER stress induces NF-κB localization to the nucleus and transcriptional activation.
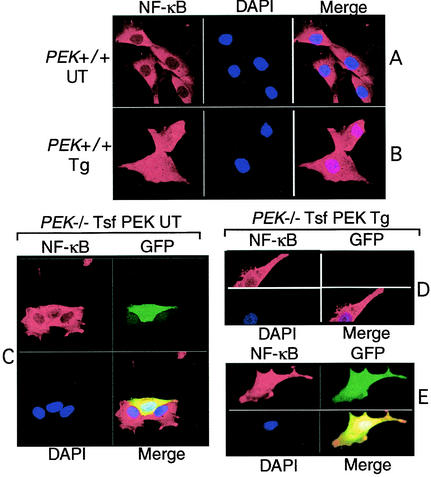
Nuclear targeting of NF-κB is an important step in the mechanism of activation of this transcription factor in response to a wide range of stress conditions. To characterize the cellular localization of NF-κB in response to ER stress, MEF cells with functional PEK were grown on glass slides and treated with thapsigargin for 6 h or were not subjected to ER stress. NF-κB was visualized by indirect immunofluorescence using primary antibody specific to p65 and secondary antibody conjugated to Rhodamine Red. NF-κB was readily visible in PEK+/+ cells in the absence of thapsigargin treatment, with a predominant cytoplasmic localization (Fig. 4A). During ER stress, NF-κB was uniformly present in both the nucleus and cytoplasm (Fig. 4B). Similar characterization of ATF4 and Chop/GADD153 in PEK+/+ cells revealed that these transcription factors were also induced by ER stress and were predominantly present in the nucleus (data not shown) (23). To address the importance of PEK in this nuclear targeting, we looked at NF-κB localization in PEK−/− MEF cells and in those cells transiently expressing this eIF2α kinase. The PEK-expressing cells were delineated by cotransfecting a plasmid encoding green fluorescent protein (GFP). In MEF cells devoid of PEK function, there was no nuclear localization of NF-κB in response to ER stress (Fig. 4D). By contrast, in cells containing GFP—the presence of which is indicative of PEK expression—there was nuclear localization of NF-κB (Fig. 4E). In the absence of ER stress, NF-κB was restricted to the cytoplasm in either PEK−/− cells or in those MEF cells transiently expressing PEK (Fig. 4C).
FIG. 4.
eIF2α kinase PEK is required for translocation of NF-κB into the nucleus during ER stress. (A and B) PEK+/+ MEF cells grown on glass slides were treated with thapsigargin for 6 h (Tg; panel A) or no stress (UT; panel B). Cells were prepared and the p65 subunit of NF-κB was visualized using rabbit polyclonal antibodies specific for this transcription factor, followed by goat anti-rabbit IgG conjugated with Rhodamine Red. NF-κB linked with Rhodamine Red was visualized by laser confocal microscopy. Cell nuclei were stained with DAPI mountain medium, and electronic images from the Rhodamine Red and DAPI were merged in the right figures. (C through F) PEK−/− cells transfected with a plasmid expressing PEK (PEK) in combination with plasmid encoding GFP were grown on glass slides. Transfected cells were exposed to thapsigargin (D and E) or were untreated (C). NF-κB was visualized by using the secondary antibody conjugated with Rhodamine Red, nuclear DNA by DAPI stain, and GFP by fluorescein isothiocyanate. All three panels were merged as indicated. PEK−/− cells transiently expressing PEK were identified by coexpression with GFP. The PEK−/− MEF cell in panel D was expressing no PEK or GFP and served as a control for eIF2α kinase dependence for NF-κB translocation to the nucleus during ER stress.
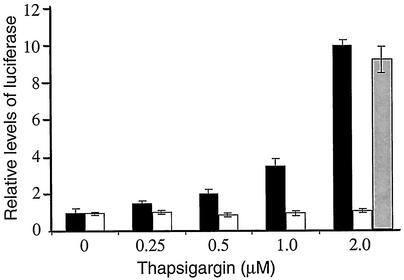
Nuclear localization contributes to transcriptional regulation of genes containing a NF-κB binding element(s) in their promoters. We introduced a NF-κB-directed luciferase reporter gene containing multiple upstream NF-κB binding sites into PEK+/+ or PEK−/− MEF cells. These transiently transfected cells were exposed to thapsigargin at a concentration range of 0 to 2 μM for 6 h. With a thapsigargin concentration of 0.5 μM, there was a twofold increase in luciferase activity in the PEK+/+ cells compared to their nonstressed counterparts (Fig. 5). With exposure to higher concentrations—1 μM to 2 μM of thapsigargin—there was a 3.5- to 10-fold increase, respectively, in luciferase expression in the PEK+/+ cells. By comparison, luciferase activity remained unchanged in PEK−/− cells that were exposed to these different concentrations of thapsigargin. As a control, we also transfected a cDNA encoding wild-type PEK using a CMV promoter in plasmid pcDNA3 into the PEK−/− cells exposed to 2 μM thapsigargin, and we found a nearly 10-fold increase in luciferase activity in response to ER stress (Fig. 5). Together, these results are consistent with the idea that eIF2α phosphorylation by PEK facilitates nuclear localization of NF-κB during ER stress, thus leading to transcriptional activation of its targeted genes.
FIG. 5.
eIF2α kinase PEK is required for NF-κB transcriptional activity in response to ER stress. An NF-κB firefly luciferase reporter gene was cotransfected with a Renilla luciferase control plasmid into PEK+/+ (black bar) or PEK−/− (white bar) MEF cells. MEF cells were treated with from 0 to 2 μM thapsigargin for 6 h, and the dual luciferase assay was carried out as described in Materials and Methods. To address whether transient expression of PEK in the PEK−/− MEF cells restored NF-κB transcriptional activity, we cotransfected a PEK expression plasmid with the luciferase genes and subjected these cells to 2 μM thapsigargin (gray bar). Relative NF-κB-luciferase activity is presented in the histogram, which is normalized for PEK+/+ cells not subjected to thapsigargin treatment.
ER stress activates NF-κB by a mechanism that involves release, but not degradation, of IκB.
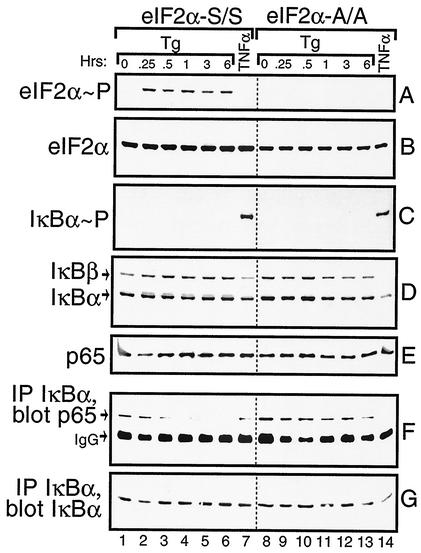
One mechanism facilitating nuclear localization of NF-κB involves phosphorylation and proteolysis of IκB. We wished to measure IκB phosphorylation and protein levels in the S/S and A/A MEF cells in response to ER stress. As a control, we also analyzed these MEF cells in response to treatment with TNF-α, a cytokine shown to induce phosphorylation of IκB at Ser residues 32 and 36, thus leading to IκB ubiquitin-mediated proteolysis. Within 15 min of thapsigargin exposure, there was an induction of eIF2α phosphorylation that was sustained over the 6-h time course (Fig. 6A). No phosphorylation of eIF2α was detected after 30 min of exposure to TNF-α (Fig. 6A, lanes 7 and 14). As expected, no eIF2α phosphorylation was detected in the A/A cells. To determine whether ER stress facilitates phosphorylation of IκBα, we used a polyclonal antibody specific to this inhibitory protein phosphorylated at Ser-32 in an immunoblot analysis. TNF-α induced phosphorylation of IκBα in both the S/S and A/A cells (Fig. 6C); however, no phosphorylation was detected over the 6-h time course of ER stress in either MEF cell line.
FIG. 6.
Activation of NF-κB in response to ER stress involves release but not proteolysis of IκB. S/S (lanes 1 to 7) and A/A (lanes 8 to 14) MEF cells were treated with thapsigargin for up to 6 h, with TNF-α for 30 min, or with no stress (0 h). Equal amounts of whole-cell lysates were separated by SDS-PAGE; phosphorylated eIF2α (A), total eIF2α (B), IκBα phosphorylated at Ser-32 (C), total levels of IκBα and IκBβ (D), and p65 (E) were measured by immunoblotting using specific antibodies. IκBα was immunoprecipitated and the levels of p65 (F) or IκBα (G) in the immunocomplexes were measured by immunoblot analysis. The IgG present in the immunocomplex migrated near p65 in the SDS-PAGE as indicated in panel F. Each panel is derived from one immunoblotting experiment, and the dotted line between lanes 7 and 8 is shown only for alignment purposes.
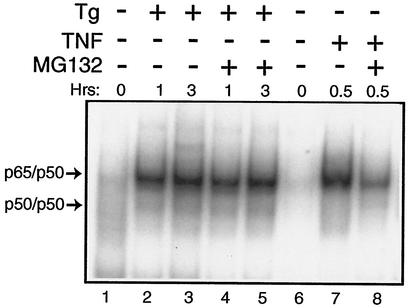
Levels of the α and β isoforms of IκB were measured by using polyclonal antibodies specific to these proteins. Consistent with the TNF-α-mediated phosphorylation of IκB, there was a reduction of both IκB α and β in the S/S and A/A MEF cells following treatment with this cytokine (Fig. 6D, lanes 7 and 14). At the time point assayed, TNF-α induced a greater reduction in IκB α and β levels in the A/A MEF cells compared with that measured in the S/S cells. By comparison, levels of the IκB isoforms were not appreciably reduced in these MEF cells, even after 6 h of thapsigargin treatment (Fig. 6D). To further address whether the increase in NF-kB activity in response to ER stress involved degradation of IκB by proteasomes, MG132—a proteasome inhibitor—was added to MEF cells along with TNF-α or thapsigargin. While inhibition of the proteasome significantly lowered TNF-α activation of NF-κB, there was no deleterious effect on induced NF-κB activity in response to ER stress (Fig. 7). We conclude that PEK-mediated activation of NF-κB in response to ER stress occurs by a mechanism that does not involve degradation of IκB α and β. Further emphasizing the separation between eIF2α kinase and TNF-α pathways is our observation that deletion of an eIF2α kinase, such as PEK, has no adverse impact on the activation of NF-κB in response to treatment of MEF cells with this cytokine (data not shown).
FIG. 7.
Reduced protein degradation impairs TNF-α-directed induction of NF-κB but does not reduce NF-κB activity during ER stress. PEK+/+ and PEK−/− MEF cells were treated with thapsigargin (Tg), TNF-α, and MG132 as indicated by the “+” or “−” symbols for the indicated number of hours or were subjected to no stress (0 h). Equal amounts of nuclear lysates prepared from MEF cells were analyzed for binding to the NF-κB probe by the EMSA. DNA complexed with p65/p50 or p50/p50 is indicated by arrows.
Dissociation from IκB is important for localization of NF-κB to the nucleus and subsequent transcriptional regulation of its target genes. To address whether IκB release is critical for activation of NF-κB in response to ER stress, we immunoprecipitated IκBα from lysates prepared from S/S and A/A MEF cells treated with thapsigargin for up to 6 h. Proteins from the immunocomplexes were subjected to SDS-PAGE and analyzed by immunoblotting. Similar IκBα levels were measured in each immunocomplex derived from the thapsigargin-treated lysates (Fig. 6G). By contrast, the levels of coprecipitated NF-κB as measured by p65 polyclonal antibody were reduced in S/S cells in response to ER stress (Fig. 6F). After 30 min of thapsigargin treatment, there was an appreciable reduction in p65 associated with IκBα, which was further lowered following 1 to 3 h of ER stress. In contrast, in A/A cells p65 was uniformly associated with IκBα independent of thapsigargin treatment. After TNF-α exposure, there was a reduction in coprecipitation of p65 with IκBα in both the S/S and A/A cells. Levels of p65 in the total cell lysate were uniform in each sample, demonstrating that this reduction in NF-κB association with IκB cells upon the induction of ER stress was not due to decreased amounts of p65 in the in PEK+/+ MEF cells (Fig. 6E).
Phosphorylation of eIF2α is required to activate NF-κB in response to inhibition of general transcription or translation.
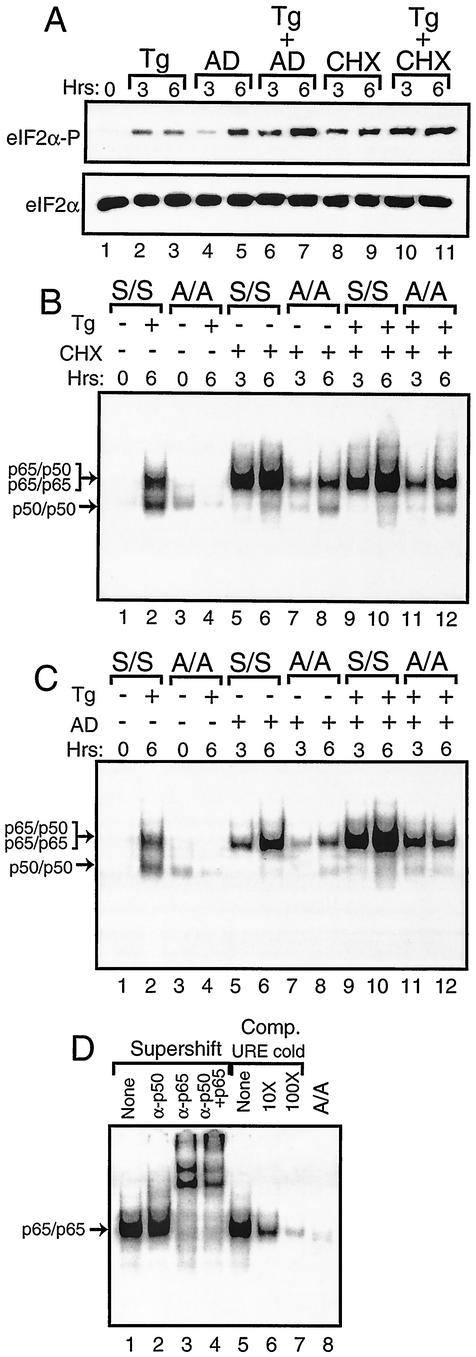
During the course of experiments designed to address the role of protein and mRNA synthesis in stress responses, we found that treatment with cycloheximide or actinomycin D induces eIF2α phosphorylation (Fig. 8A). Cycloheximide induced a high level of eIF2α phosphorylation in MEF cells following 3 h of exposure. By comparison, treatment of MEF cells with actinomycin D showed a modest level eIF2α phosphorylation after 3 h of exposure, a level which was significantly elevated after 6 h. The levels of eIF2α phosphorylation were further increased by combining either chemical inhibitor with thapsigargin (Fig. 8A). Similar amounts of total eIF2α were found in each lysate.
FIG. 8.
Activation of NF-κB in response to cycloheximide or actinomycin D exposure requires phosphorylation of eIF2α. (A) S/S MEF cells were exposed for the indicated number of hours to thapsigargin (Tg), actinomycin D (AD), or cycloheximide (CHX) as indicated or to no stress agent (0 h). Phosphorylation of eIF2α was measured by immunoblot analysis using polyclonal antibody specific to eIF2α phosphorylated at Ser-51 (eIF2α∼P). Levels of total eIF2α were assayed by using antibody that recognizes both phosphorylated and nonphosphorylated versions of the translation initiation factor (eIF2α). S/S and A/A MEF cells were exposed to thapsigargin, cycloheximide (B), or actinomycin D (C) as indicated by the “+” or “−” for the indicated number of hours. Nuclear lysates prepared from S/S and A/A MEF cells, as indicated, were analyzed for binding to the NF-κB probe in the EMSA. Arrows indicate DNA complexed with p65/p50 or p50/p50 in thapsigargin-stressed preparations or with p65/p65 in cycloheximide or actinomycin D-stressed cells as defined in experiments shown in panel D. In panel D, S/S MEF cells were treated with actinomycin D for 6 h and analyzed for binding with the NF-κB DNA. In lanes 1 to 4, supershift designates that antibodies that specifically recognize p50 and/or p65 were added to the EMSA binding mixture. “None” indicates that no antibody was added to the assay. In lanes 5 to 7, competition (Comp) indicates that nonradiolabeled NF-κB URE competitor DNA was added at a 10× or 100× molar excess or was not added (None). In lane 8, A/A MEF cells were similarly analyzed using actinomycin D.
Given that inhibition of protein synthesis by cycloheximide treatment has been reported to also induce NF-κB (52), we asked whether eIF2α phosphorylation in response to this chemical inhibitor was linked to NF-κB activation as observed for ER and nutritional stress. Following exposure of S/S MEF cells to cycloheximide for 3 h, there was a significant activation of NF-κB as measured by enhanced binding in the EMSA that was further increased after 6 h of treatment (Fig. 8B, lanes 5 and 6). Unlike the ER stress condition, the induction of NF-κB by cycloheximide involved only detection of the higher-molecular-weight radiolabeled band. A combined addition of thapsigargin and cycloheximide to the S/S MEF cells also induced only the larger band at levels similar to that observed for cycloheximide alone (Fig. 8B, lanes 9 and 10). In similar experiments involving A/A, there was a marked reduction in NF-κB binding, which demonstrated that eIF2α phosphorylation is required for full activation of NF-κB in response to cycloheximide (Fig. 8B).
There was also enhanced NF-κB binding in S/S MEF cells exposed to actinomycin D for 3 h, with a further increase following 6 h of treatment (Fig. 8C, lanes 6 and 7). As described for cycloheximide, this induction of NF-κB by actinomycin D involved only the higher-molecular-weight version. Addition of actinomycin D and thapsigargin together led to an even greater induction of NF-κB (Fig. 8C, lanes 9 and 10). By comparison, this activation of NF-κB was significantly diminished in A/A cells treated with actinomycin D or with the combination of actinomycin D and thapsigargin (Fig. 8C, lanes 7, 8, 11, and 12).
To address the composition of the NF-κB subunits, we added polyclonal antibody specific to p50 to the EMSA mixture and found no change in migration of this band (Fig. 8D). By contrast, addition of antibody that recognizes p65 resulted in a complete supershift of the radioactive band, thus suggesting that this version of NF-κB represents a probable p65 homodimer. Addition of nonradiolabeled URE DNA effectively competed for binding of this NF-κB. Together, these results indicate that inhibition of general translation or transcription can induce NF-κB by a mechanism requiring phosphorylation of eIF2α. Induction of eIF2α phosphorylation by actinomycin D or cycloheximide occurs not only in MEF cells but also in other cell types, including NIH 3T3 and human embryonic kidney 293 cells (data not shown). This increased phosphorylation of eIF2α is not dependent on either PEK or GCN2 alone, as there is elevated eIF2α phosphorylation in PEK−/− and GCN2−/− MEF cells in response to cycloheximide treatment (data not shown). Furthermore, activation of PEK, as judged by the migration shift accompanying autophosphorylation of this eIF2 kinase (25, 41), does not occur in response to this protein synthesis inhibitor. Therefore, alternative eIF2α kinases may be activated in response to cycloheximide exposure, or there may be a reduction in eIF2α phosphatase activity.
p65 is dispensable for eIF2α kinase-dependent expression of ATF4 and Chop.
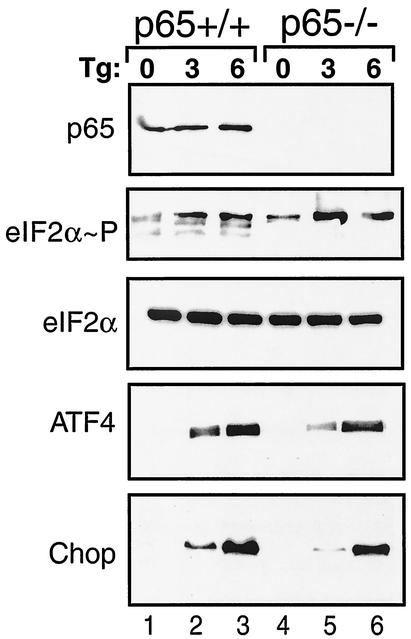
The eIF2α kinases induce a gene expression cascade that includes bZIP transcription factors ATF4 and Chop in response to diverse stress conditions (22, 23, 35). Given that eIF2α phosphorylation also facilitates activation of NF-κB, we asked whether p65 function is required for induction of the eIF2α kinase stress pathway. Induced phosphorylation of eIF2α occurs in response to thapsigargin treatment of MEF cells, thus contributing to elevated expression of ATF4 and Chop (Fig. 9). Deletion of p65 function did not affect the induction of eIF2α phosphorylation, although eIF2α phosphorylation in nonstressed p65−/− cells was slightly higher compared to its wild-type counterpart. ER stress induction of ATF4 and Chop expression occurred in both p65+/+ and p65−/− cells. After 3 h of thapsigargin exposure, induction of ATF4 and Chop expression was slightly lower in the p65−/− cells compared to p65+/+ cells, while 6 h of ER stress elicited comparable levels of these transcriptional factors (Fig. 9). These results suggest that the eIF2α kinase stress response can occur independent of p65 function.
FIG. 9.
p65 function is dispensable for increased expression of eIF2α kinase pathway genes in response to ER stress. p65 (RelA)+/+ (lanes 1 to 3) and p65−/− (lanes 4 to 6) MEF cells were exposed to thapsigargin (Tg) for 3 or 6 h or were subjected to no stress (0 h). Equal amounts of whole-cell lysates were separated by SDS-PAGE, and levels of p65, phosphorylated eIF2α, total eIF2α, ATF4, and Chop were measured by immunoblotting using antibodies specific to the indicated protein.
DISCUSSION
eIF2α phosphorylation mediates induction of NF-κB in response to diverse stress conditions.
NF-κB is a central regulator of stress response pathways, serving to coordinate the transcription of genes whose products serve to alleviate the underlying stress condition. In this study, we showed that phosphorylation of eIF2α is fundamental to the process by which diverse physiological stresses are monitored and relayed to the activation of NF-κB. For example, in response to impaired protein folding and assembly in the ER, phosphorylation of eIF2α by PEK is essential for the induction of NF-κB activity (Fig. 1, 2, 4, and 5). The mechanism by which NF-κB is activated during ER stress involves release of IκB but not degradation of this inhibitory protein (Fig. 6 and 7). During amino acid starvation, phosphorylation of eIF2α by GCN2 protein kinase signals activation of NF-κB (Fig. 3). Furthermore, inhibition of general translation or transcription by cycloheximide and actinomycin D, respectively, elicits the eIF2α phosphorylation required for the induction of NF-κB (Fig. 8). Together, these studies suggest that eIF2α kinases recognize and are activated by a range of stress conditions that have an impact on transcription and protein synthesis and assembly, and the resulting eIF2α phosphorylation is central to the activation of the NF-κB. This linkage between eIF2α phosphorylation and activation of NF-κB also provides an important explanation for why eIF2α kinase deficiency in diseases such as WRS lead to apoptotic episodes. In most cell types, NF-κB activates genes with antiapoptotic function, including the IAP family, TRAF1 and TRAF2, and the Bcl-2 homologues Bfl-1/A1 and Bcl-XL (2, 4, 19, 26, 38, 60, 62-64, 75). The absence of eIF2α phosphorylation and the directed NF-κB-mediated transcription during stressful conditions, such as those leading to elevated protein misfolding in the ER, would significantly enhance programmed cell death.
Role of eIF2α phosphorylation in the regulation of NF-κB function.
Transcriptional activity of NF-κB is regulated via DNA binding and transactivation. The ankyrin repeat domains of IκB family members associate with the Rel homology domain of NF-kB, thereby masking the NF-κB nuclear localization signal and DNA binding domain. Certain stresses can activate the IKK complex, contributing to phosphorylation of two residues in IκBα, Ser-32 and -36, and in IκBβ, Ser-19 and Ser-23, which triggers ubiquitination and IκB degradation by the 26S proteasome (32). In the example of activation of NF-κB by ER stress, we found no phosphorylation of Ser-32 of IκBα, nor did we observe any appreciable reduction in the levels of IκBα or IκBβ (Fig. 6). However, ER stress did signal the release of IκB from NF-κB, thus leading to the translocation of NF-κB into the nucleus and enhanced transcriptional activation (Fig. 4 to 6). We do not yet understand how eIF2α phosphorylation contributes to release of IκB. Previous studies have observed that tyrosine phosphorylation of IκBα, which can occur in reoxygenated hypoxic cells or in cells exposed to the tyrosine phosphatase inhibitor pervanadate, induces dissociation from NF-κB without proteolytic degradation of IκBα (29). The mechanism by which tyrosine phosphorylation aids IκB clearance from NF-κB is not clear, but it may involve sequestration through interaction of the modified IκB with the regulatory subunit (p85α) of phosphoinositide 3-kinase (5). Another mechanism for enhancing NF-κB activity involves direct phosphorylation of p65 at multiple residues in its carboxy terminus (15, 32, 39). Such phosphorylation of p65 enhances the transactivation potential of NF-κB by modulating its association with transcriptional regulators such as the CREB-binding protein (BCP)/p300 or the histone deacetylase HDAC-1.
Phosphorylation of eIF2α in response to inhibition of general mRNA and protein synthesis mediates NF-kB activation.
We found that eIF2α phosphorylation in response to general inhibition of transcription or translation by actinomycin D and cycloheximide, respectively, is required for activation of NF-κB (Fig. 8). Given the broad spectrum of stress conditions that are observed to induce NF-κB activity, it is inviting to speculate that stress agents that directly or indirectly reduce gene expression regulate the function of NF-κB by inducing eIF2α kinases or blocking eIF2α phosphatases. It is interesting that while phosphorylation of eIF2α during ER stress enhanced DNA binding by p65/p50 and p50/p50 dimers (Fig. 1 and 2), elevated eIF2α phosphorylation during cycloheximide or actinomycin D treatment enhanced the activity of only p65 homodimers (Fig. 8). This difference in NF-κB dimer induction suggests that eIF2α phosphorylation can work in conjunction with additional stress factors to program NF-κB activities. Such additional stress factors may be controlled by alternative stress pathways that are independent of eIF2α phosphorylation. Combinatorial interactions would provide for different patterns of gene expression, as p65 homodimers are thought to have different binding properties for κB-related sites than those of the p65/p50 dimer (37, 47).
Contributions of eIF2α kinases and NF-κB in the regulation of stress-induced apoptosis.
WRS patients have characteristic diseases related to many secretory tissues. The affected cell types include pancreatic islet beta cells, leading to early onset of diabetes, osteoblasts contributing to epiphyseal dysplasia, pancreatic acinar cells leading to digestive disorders, and thyroid cells promoting hypothyroidism (6, 8, 58). It is thought that normal developmental processes occurring after birth contribute to cellular ER stress that is magnified in tissues with extensive secretory organelles. Many of these pathologies have also been documented in PEK (Perk)−/− mice (21, 72). ER stress activates the processing of the ER-resident proapoptotic cysteine protease, caspase 12 (44). Treatment of PEK (Perk)−/− ES cells with ER stress agents, e.g., tunicamycin or thapsigargin, contributes to accumulation of higher levels of activated caspase 12 and increased programmed cell death compared to wild-type cells (24). How loss of PEK heightens activation of this caspase and apoptosis is not well understood. In fact, Chop, one of the best-documented transcription factors induced by eIF2α phosphorylation during ER stress, has a potent proapoptotic function (43, 46, 74).
One explanation for how the loss of PEK activity facilitates apoptosis centers on the idea that PEK−/− cells cannot phosphorylate eIF2α, thus leading to continued high levels of translation initiation compared to wild-type cells. Continued synthesis of secretory proteins during ER stress would exacerbate the protein assembly load in the ER and thwart the ability of the cell to remedy protein misfolding. Interestingly, treatment with cycloheximide in combination with ER stress was reported to elicit a partial suppression of apoptosis in PEK−/− ES cells (24). This observation led to the proposal that reduced protein synthesis was an obligatory response to the ER stress. Our results indicate that the absence of NF-κB activity, and its potent antiapoptotic function in many cell types, is also an important contributor to enhanced programmed cell death in many eIF2α kinase-deficient cells.
It is curious that the eIF2α kinase PKR is said to promote apoptosis as part of the antiviral defense pathway (1, 17, 67). A further difference is that PKR is thought to physically associate with IKK and to enhance IκB phosphorylation and ubiquitin-mediated degradation (7, 16, 18, 30, 70). The mechanism by which PKR activates NF-κB is not thought to involve eIF2α phosphorylation. The differences between the apparent pro- and antiapoptotic functions of PKR and PEK, respectively, may reflect their different physiological stress contexts and associated stress factors that work in concert with these two eIF2α kinases. These differences may also explain the distinct mechanisms by which PKR and PEK regulate NF-κB.
Acknowledgments
We thank Gail Sonenshein for generously sharing reagents, Harikrishna Nakshatri, Lawrence Quilliam, and members of our laboratory for helpful comments on the manuscript, and the Biochemistry Biotechnology Facility at Indiana University for technical support.
This study was supported in part by grants RO1GM49164 and R01GM643540 (R.C.W.) and AI42394 (R.J.K.) from the National Institutes of Health.
REFERENCES
- 1.Barber, G. N. 2001. Host defense, viruses and apoptosis. Cell Death Differ. 8**:**113-126. [DOI] [PubMed] [Google Scholar]
- 2.Barkett, M., and T. D. Gilmore. 1999. Control of apoptosis by Rel/NF-κB transcription factors. Oncogene 18**:**6910-6924. [DOI] [PubMed] [Google Scholar]
- 3.Baud, V., and M. Karin. 2001. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 11**:**372-377. [DOI] [PubMed] [Google Scholar]
- 4.Beg, A. A., and D. Baltimore. 1996. An essential role for NF-κB in preventing TNF-α-induced cell death. Science 274**:**782-784. [DOI] [PubMed] [Google Scholar]
- 5.Beraud, C., W. J. Henzel, and P. A. Baeuerle. 1999. Involvement of regulatory and catalytic subunits of phosphoinositide 3-kinase in NF-κB activation. Proc. Natl. Acad. Sci. USA 96**:**429-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bin-Abbas, B., A. Al-Mulhim, and A. Al-Ashwai. 2002. Wolcott-Rallison syndrome in two siblings with isolated central hypothyrroidism. Am. J. Med. Genet. 111**:**187-190. [DOI] [PubMed] [Google Scholar]
- 7.Bonnet, M. C., R. Weil, E. Dame, A. G. Hovanessian, and E. F. Meurs. 2000. PKR stimulates NF-κB irrespective of its kinase function by interacting with IκB kinase complex. Mol. Cell. Biol. 20**:**4532-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castelnau, P., M. Le Merrer, C. Diatlof-Zito, E. Marquis, M. J. Tete, and J. J. Robert. 2000. Wolcott-Rallison syndrome: a case with endocrine and exocrine pancreatic deficiency and pancreatic hypotrophy. Eur. J. Pediatr. 159**:**631-633. [DOI] [PubMed] [Google Scholar]
- 9.Chen, F. E., and G. Ghosh. 1999. Regulation of DNA binding by Rel/NF-κB transcription factors: structural views. Oncogene 18**:**6845-6852. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J.-J. 2000. Heme-regulated eIF2α kinase, p. 529-546. In N. Sonenberg, J. W. B. Hershey, and M. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Delepine, M., M. Nicolino, T. Barrett, M. Golamaully, G. M. Lathrop, and C. Julier. 2000. EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 25**:**406-409. [DOI] [PubMed] [Google Scholar]
- 12.Dever, T. E. 2002. Gene-specific regulation by general translation factors. Cell 108**:**545-556. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez, J., I. Yaman, W. C. Merrick, A. Koromilas, R. C. Wek, R. Sood, J. Hensold, and M. Hatzoglou. 2002. Regulation of internal ribosome entry site-mediated translation by eukaryotic initiation factor-2α phosphorylation and small upstream open reading frame. J. Biol. Chem. 277**:**2050-2058. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez, J., I. Yaman, P. Sarnow, M. D. Snider, and M. Hatzoglou. 2002. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2α. J. Biol. Chem. 277**:**19198-19205. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh, H., and M. Karin. 2002. Missing pieces in the NF-κB puzzle. Cell 109**:**S81-S96. [DOI] [PubMed] [Google Scholar]
- 16.Gil, J., J. Alcami, and M. Esteban. 2000. Activation of NF-κB by the dsRNA-dependent protein kinase PKR involves the IκB complex. Oncogene 19**:**1369-1378. [DOI] [PubMed] [Google Scholar]
- 17.Gil, J., and M. Esteban. 2000. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis 5**:**107-114. [DOI] [PubMed] [Google Scholar]
- 18.Gil, J., J. Rullas, M. A. Garcia, J. Alcami, and M. Esteban. 2001. The catalytic activity of dsRNA-dependent protein kinase, PKR, is required for NF-κB activation. Oncogene 20**:**385-394. [DOI] [PubMed] [Google Scholar]
- 19.Grumont, R. J., I. J. Rourke, and S. Gerondakis. 1999. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Genes Dev. 13**:**400-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, A., C. Yu, L. Lu, Y. Fujiwara, C. Browne, G. Chin, P. Fleming, P. Leboulch, S. H. Orkin, and J.-J. Chen. 2001. Heme-regulated eIF2α kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20**:**6909-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harding, H., H. Zeng, Y. Zhang, R. Jungreis, P. Chung, H. Plesken, D. D. Sabatini, and D. Ron. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in Perk −/− mice reveals a role for translational control in secretory cell survival. Mol. Cell 7**:**1153-1163. [DOI] [PubMed] [Google Scholar]
- 22.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18**:**575-599. [DOI] [PubMed] [Google Scholar]
- 23.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6**:**1099-1108. [DOI] [PubMed] [Google Scholar]
- 24.Harding, H. P., Y. Zhang, A. Bertolotti, H. Zeng, and D. Ron. 2000. Perk is essential for translation regulation and cell survival during the unfolded protein response. Mol. Cell 5**:**897-904. [DOI] [PubMed] [Google Scholar]
- 25.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397**:**271-274. [DOI] [PubMed] [Google Scholar]
- 26.Herr, I., and K.-M. Debatin. 2001. Cellular stress response and apoptosis in cancer therapy. Blood 98**:**2603-2614. [DOI] [PubMed] [Google Scholar]
- 27.Hershey, J. W. B., and W. C. Merrick. 2000. Pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Hinnebusch, A. G. 2000. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes, p. 185-244. In N. Sonenberg, J. W. B. Hershey, and M. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Imbert, V., R. A. Rupec, A. Livolsi, H. L. Pahl, E. B. Traenckner, C. Mueller-Dieckmann, D. Farahifar, B. Rossi, P. Auberger, P. A. Baeuerle, and J. F. Peyron. 1996. Tyrosine phosphorylation of IκB-α activates NF-κB without proteolytic degradation of IκB-α. Cell 86**:**787-798. [DOI] [PubMed] [Google Scholar]
- 30.Ishii, T., H. Kwon, J. Hiscott, G. Mosialos, and A. E. Koromilas. 2001. Activation of the IκBα kinase (IKK) complex by double-stranded RNA-binding defective and catalytic inactive mutants of the interferon-inducible protein kinase PKR. Oncogene 20**:**1900-1912. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, H. Y., C. Petrovas, and G. E. Sonenshein. 2002. RelB-p50 NFκB complexes are selectively induced by cytomegalovirus immediate-early protein 1: differential regulation of Bcl-xL promoter activity by NFκB family members. J. Virol. 76**:**5737-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18**:**621-663. [DOI] [PubMed] [Google Scholar]
- 33.Karin, M., Y. Cao, F. R. Greten, and Z. W. Li. 2002. NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer 2**:**301-310. [DOI] [PubMed] [Google Scholar]
- 34.Kaufman, R. J. 2000. Double-stranded RNA-activated protein kinase, p. 503-528. In N. Sonenberg, J. W. B. Hershey, and M. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Kaufman, R. J., D. Scheuner, M. Schroder, X. Shen, K. Lee, C. Y. Lin, and S. M. Arnold. 2002. The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell Biol. 3**:**411-421. [DOI] [PubMed] [Google Scholar]
- 36.Kumar, A., J. Haque, J. Lacoste, J. Hiscott, and B. R. Williams. 1994. Double-stranded RNA-dependent protein kinase activates transcription factor NF-κB by phosphorylating IκB. Proc. Natl. Acad. Sci. USA 91**:**6288-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunsch, C., S. M. Ruben, and C. A. Rosen. 1992. Selection of optimal κB/Rel DNA-binding motifs interaction of both subunits of NF-κB with DNA is required for transcriptional activation. Mol. Cell. Biol. 12**:**4412-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, H. H., H. D. Dadgostar, Q. Cheng, J. Shu, and G. Cheng. 1999. NF-κB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc. Natl. Acad. Sci. USA 96**:**9136-9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, Q., and I. M. Verma. 2002. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2**:**725-734. [DOI] [PubMed] [Google Scholar]
- 40.Lu, L., A. P. Han., and J.-J. Chen. 2001. Translation initiation control by heme-regulated eukaryotic initiation factor 2α kinase in erythroid cells under cytoplasmic stresses. Mol. Cell. Biol. 21**:**7971-7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma, K., K. M. Vattem, and R. C. Wek. 2002. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J. Biol. Chem. 277**:**18728-18735. [DOI] [PubMed] [Google Scholar]
- 42.Ma, Y., J. W. Brewer, J. A. Diehl, and L. M. Hendershot. 2002. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 318**:**1351-1365. [DOI] [PubMed] [Google Scholar]
- 43.McCullough, K. D., J. L. Martindale, L. O. Klotz, T. Y. Aw, and N. J. Holbrook. 2001. Gadd153 sensitizes cells to endoplasmic reticulum stress by downregulating BcL2 and perturbing the cellular redox state. Mol. Cell. Biol. 21**:**1249-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakagawa, T., H. Zhu, N. Morishima, E. Li, J. Xu, B. A. Yankner, and J. Yuan. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature 403**:**98-103. [DOI] [PubMed] [Google Scholar]
- 45.Nozaki, S., G. W. Sledge, and H. Nakshatri. 2001. Repression of GADD153/CHOP by NF-κB: a possible cellular defense against endoplasmic reticulum stress-induced cell death. Oncogene 20**:**2178-2185. [DOI] [PubMed] [Google Scholar]
- 46.Oyadomari, S., A. Koizumi, K. Takeda, T. Gotoh, S. Akira, E. Araki, and M. Mori. 2002. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J. Clin. Investig. 109**:**525-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pahl, H. L. 1999. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18**:**6853-6866. [DOI] [PubMed] [Google Scholar]
- 48.Pahl, H. L., and P. A. Baeuerle. 1997. The ER-overload response: activation of NF-κB. Trends Biochem. Sci. 22**:**63-67. [DOI] [PubMed] [Google Scholar]
- 49.Pahl, H. L., and P. A. Baeuerle. 1995. A novel signal transduction pathway from the endoplasmic reticulum to the nucleus is mediated by transcription factor NF-κB. EMBO J. 14**:**2580-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romieu-Mourez, R., E. Landesman-Bollag, D. C. Sheldin, and G. E. Sonenshein. 2002. Protein kinase CK2 promotes aberrant activation of nuclear factor-κB, transformed phenotype, and survival of breast cancer cells. Cancer Res. 62**:**6770-6778. [PubMed] [Google Scholar]
- 51.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7**:**1165-1176. [DOI] [PubMed] [Google Scholar]
- 52.Sen, R., and D. Baltimore. 1986. Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell 47**:**921-928. [DOI] [PubMed] [Google Scholar]
- 53.Shi, Y., K. M. Vattem, R. Sood, J. An, J. Liang, L. Stramm, and R. C. Wek. 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translation control. Mol. Cell. Biol. 18**:**7499-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonenshein, G. E. 1997. Rel/NF-κB transcription factors and the control of apoptosis. Semin. Cancer Biol. 8**:**113-119. [DOI] [PubMed] [Google Scholar]
- 55.Sood, R., A. C. Porter, K. Ma, L. A. Quilliam, and R. C. Wek. 2000. Pancreatic eukaryotic initation factor 2α kinase (PEK) homologues in humans, Drosophila melanogaster and Caenorhabditis elegans that mediate translational control in response to ER stress. Biochem. J. 346**:**281-293. [PMC free article] [PubMed] [Google Scholar]
- 56.Sood, R., A. C. Porter, D. Olsen, D. R. Cavener, and R. C. Wek. 2000. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor 2α. Genetics 154**:**787-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sovak, M. A., R. E. Bellas, D. W. Kim, G. J. Zanieski, A. E. Rogers, A. M. Traish, and G. E. Sonensheim. 1997. Aberrant nuclear factor-κB/Rel expression and the pathogenesis of breast cancer. J. Clin. Investig. 100**:**2952-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thornton, C. M., D. J. Carson, and F. J. Stewart. 1997. Autopsy findings in the Wolcott-Rallison syndrome. Pediatric Pathol. Lab. Med. 17**:**487-496. [PubMed] [Google Scholar]
- 59.Uma, S., B. G. Yun, and R. L. Matts. 2001. The heme-regulated eukaryotic initiation factor 2α kinase—a potential regulatory target for control of protein synthesis by diffusible gases. J. Biol. Chem. 276**:**14675-14783. [DOI] [PubMed] [Google Scholar]
- 60.Van Antwerp, D. J., S. J. Martin, T. Kafri, D. R. Green, and I. M. Verma. 1996. Suppression of TNF-α-induced apoptosis by NF-κB. Science 274**:**787-789. [DOI] [PubMed] [Google Scholar]
- 61.Vinson, C. R., T. Hai, and S. M. Boyd. 1993. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev. 7**:**1047-1058. [DOI] [PubMed] [Google Scholar]
- 62.Wang, C.-Y., D. C. Guttridge, M. W. Mayo, and A. S. Baldwin. 1999. NF-κB induces expression of the Bcl-1 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol. Cell. Biol. 19**:**5923-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, C. Y., M. W. Mayo, and A. S. Baldwin. 1996. TNF- and cancer-therapy-induced apoptosis: potentiation by inhibition by NF-κB. Science 274**:**784-787. [DOI] [PubMed] [Google Scholar]
- 64.Wang, C. Y., M. W. Mayo, R. G. Korneluk, D. V. Goeddel, and A. S. Baldwin. 1998. NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-AP2 to suppress caspase-8 activation. Science 281**:**1680-1683. [DOI] [PubMed] [Google Scholar]
- 65.Wek, R. C. 1994. eIF-2 kinases: regulators of general and gene-specific translation initiation. Trends Biochem. Sci. 19**:**491-496. [DOI] [PubMed] [Google Scholar]
- 66.Wek, S. A., S. Zhu, and R. C. Wek. 1995. The histidyl-tRNA synthetase-related sequence in eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15**:**4497-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams, B. R. 1999. PKR: a sentinel kinase for cellular stress. Oncogene 18**:**6112-6120. [DOI] [PubMed] [Google Scholar]
- 68.Williams, D. A., M. F. Rosenblatt, D. R. Beier, and R. D. Cone. 1988. Generation of murine stromal cell lines supporting hematopoietic stem cell proliferation by use of recombinant retrovirus vectors encoding simian virus 40 large T antigen. Mol. Cell. Biol. 8**:**3864-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang, R., S. A. Wek, and R. C. Wek. 2000. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell. Biol. 20**:**2706-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zamanian-Daryoush, M., T. J. Mogensen, J. A. DiDonato, and B. R. G. Williams. 2000. NF-κB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-κB-inducing kinase and IκB kinase. Mol. Cell. Biol. 20**:**1278-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhan, K., K. M. Vattem, B. N. Bauer, T. E. Dever, J.-J. Chen, and R. C. Wek. 2002. Phosphorylation of eukaryotic initiation factor 2 by HRI-related protein kinases in Schizosaccharomyces pombe is important for resistance to environmental stresses. Mol. Cell. Biol. 22**:**7134-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, P., B. McGrath, S. Li, A. Frank, F. Zambito, J. Reinert, M. Gannon, K. Ma, K. McNaughton, and D. R. Cavener. 2002. The PERK eukaryotic initiation factor 2α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell. Biol. 22**:**3864-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, P., B. C. McGrath, J. Reinert, D. S. Olsen, L. Lei, S. Gill, S. A. Wek, K. M. Vattem, R. C. Wek, S. R. Kimball, L. S. Jefferson, and D. R. Cavener. 2002. The GCN2 eIF2α kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell. Biol. 22**:**6681-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zinszner, H., M. Kuroda, X. Z. Wang, N. Batchvarova, R. T. Lightfoot, H. Remotti, J. L. Stevens, and D. Ron. 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12**:**982-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zong, W.-X., L. C. Edelsterin, C. Chen, J. Bash, and C. Gelinas. 1999. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-κB that blocks TNF-α-induced apoptosis. Genes Dev. 13**:**382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]