A multifunctional plasmid-encoded replication initiation protein both recruits and positions an active helicase at the replication origin (original) (raw)
Abstract
The DnaA replication initiation protein has been shown to be essential for DNA strand opening at the AT-rich region of the replication origin of the_Escherichia coli_ chromosome as well as serving to recruit and position the DnaB replicative helicase at this open region. Homologues of the_dnaA_ gene of E. coli have been found in most bacterial species, and the DnaA protein has been shown to be required for the initiation of replication of both chromosomal and plasmid DNA. For several plasmid elements it has been found that a plasmid-encoded initiation protein is required along with the DnaA protein to bring about opening of the AT-rich region at the replication origin. The broad host range plasmid RK2 encodes two forms of its replication initiation protein (TrfA-33 and TrfA-44) that differ by an additional 98 aa at the N terminus of the larger (TrfA-44) form. Both forms initiate replication of RK2 in E. coli in vitro by a DnaA-dependent mechanism. However, as shown in this study, TrfA-44 specifically interacts with the DnaB replicative helicase of Pseudomonas putida and Pseudomonas aeruginosa and initiates the formation of a prepriming open complex in the absence of DnaA protein. Thus, the TrfA-44 initiation protein has the multifunctional properties of recruiting and positioning an active form of the DnaB helicase at the RK2 replication origin by a DnaA-independent process. This unique property for a replication initiation protein undoubtedly plays an important role in extending the host range of the RK2 antibiotic resistance plasmid.
The specific mechanism for replication initiation of a given replicon depends on both the structure of the replication origin and the nature of the replication initiation protein (Rep). For a number of replicons the initiation process begins with the binding of the Rep to specific iterated nucleotide sequences in the origin region, causing localized unwinding of an AT-rich region that is an integral part of most origin structures. This event has been studied in great detail with the Escherichia coli chromosomal origin of replication (oriC) (1,2) and studied extensively for the replication origin of the bacteriophage lambda (3), origins of simian virus 40 (4), herpes simplex virus 1 (5), Epstein–Barr virus (6), and bovine papillomavirus (7), and the replication origin of plasmids P1 (8,9), F (10), pSC101 (11), R6K (12,13), and RK2 (14). The unwound DNA bubble produced by Rep binding, described as an open complex, provides the entry site for the replicative helicase. Further unwinding of the double-stranded DNA requires proper positioning of the helicase on the separated DNA strand(s). The molecular mechanism(s) of this positioning event is not fully understood.
In E. coli, the key initiation protein, DnaA, binds to specific sequences (DnaA boxes) at the chromosomal origin of replication and melts three AT-rich, 13-mer direct repeats (1,15,16). Additional binding of the DnaA protein to single-stranded 6-mer boxes in the open region is thought to stabilize the open complex (17). The DnaA protein also specifically binds the DnaB hexameric helicase, and this binding is critical to the recruitment of the helicase to the origin (2,18). Specific association of the E. coli DnaB protein with the origin requires that this protein is complexed with the helicase loader protein DnaC, an accessory ATPase, which also serves, possibly in concert with the DnaA protein, to both deliver and position the helicase onto the single-stranded DNA in the open region (19–22). After loading, dissociation of DnaC from DnaB, coupled with ATP hydrolysis, releases the activity of the replicative helicase, resulting in further unwinding of the DNA and the formation of a prepriming complex (20). DnaA boxes and AT-rich regions are commonly found at the origin of a variety of prokaryotic replicons, and the DnaA protein has been shown to play a key role in the initiation of replication at these origins (2). Thus, the initiation process in E. coli is a paradigm for replication initiation events at the chromosomal origin of other bacteria that have been studied in addition to the replication origin of a number of plasmid elements. Although the DnaA protein has been shown to be both sufficient and indispensable in opening the AT-rich region at the origin of the bacterial chromosome, it alone is not sufficient for the formation of an open complex at the origin of plasmids RK2, P1, F, pSC101, or R6K (8–14). In these cases, efficient formation of an open complex requires binding of the plasmid Rep, although often in concert with the host DnaA protein and HU or IHF (integrated host factor) protein. The requirement for a plasmid-specified initiation protein for open complex formation is key to the plasmid's ability to exercise control over the frequency of initiation events at its replication origin and, therefore, its copy number.
Homologous sets of the DnaA and DnaB proteins of E. coli, Pseudomonas putida, and Pseudomonas aeruginosa have been shown to form a tight complex with each other (18,23). In addition, the E. coli DnaA protein has been found to interact with the Reps of plasmids R6K (24), pSC101 (25), and RK2 (26). The Reps of plasmids R6K and pSC101 have in turn been shown to interact with the DnaB helicase protein (11,12,25).
In this study we examine initiation events in vitro at the replication origin of the plasmid RK2. Plasmid RK2 encodes resistance to the antibiotics ampicillin, kanamycin, and tetracycline, and it has the unusual property of being able to replicate and be stably maintained in a wide range of Gram-negative bacteria (27). Two forms of the Rep, 44 kDa (TrfA-44) and 33 kDa (TrfA-33), generated by two in-frame translational start sites are encoded by RK2 (28,29). Either form of the TrfA protein binds to the five 17-bp iterons at the replication origin (oriV) of this plasmid (30). This binding results in the formation of an open complex that is enhanced by either DnaA protein or HU (14,23). It has been shown that in_E. coli_ the group of four DnaA boxes present in the RK2 origin (14) serves as the site for the initial formation of the DnaA/DnaBC complex (26). Recruitment of the_E. coli_ DnaB helicase to RK2 oriV requires a specific DnaA–DnaB interaction (31). Subsequent positioning and activation of the helicase to form a prepriming complex, however, requires the TrfA protein bound to the essential iterons at oriV (31). Helicase activity at_oriV in vitro_ with E. coli and to some extent with P. putida replication proteins was shown to occur with either the 33- or 44-kDa form of the TrfA protein. P. aeruginosa DnaB helicase activity at oriV, however, was found to specifically require the TrfA-44 protein (23). In this same study it was shown that unlike the E. coli DnaB helicase, both_Pseudomonas_ helicases could form an active complex at oriV in the absence of a DnaC accessory protein. Thus, whereas the overall events of replication initiation at the origin of plasmid RK2 are similar to those found to occur at the replication origin of the E. coli chromosome and of several plasmids that are naturally found in this bacterium, studies with DnaB helicases purified from P. aeruginosa and P. putida have revealed an important difference with regard to a role for the DnaC accessory protein.
Most of the studies to date with plasmid RK2 have used the smaller (TrfA-33) form of the Rep. In the present study the requirement for the larger form of the protein (TrfA-44) for the initiation of replication of the plasmid_in vitro_ using DnaB helicases from P. putida and P. aeruginosa was examined further. It was found that this form of the protein, unlike the smaller TrfA-33 form, specifically binds to the DnaB helicases of the Pseudomonas species but not to the E. coli helicase. Furthermore, TrfA-44 is not only capable of recruiting the_Pseudomonas_ DnaB helicases to the replication origin independent of an accessory helicase loader protein but, surprisingly, it also recruits and properly positions an active form of the helicase at the open region by a process that does not require the DnaA protein. The multifunctional properties of the TrfA-44 Rep are not only novel but also have implications with regard to the broad host range replication properties of plasmid RK2.
Materials and Methods
Proteins, Plasmids, and Reagents. C-terminal 6×His-tagged DnaA and DnaB proteins from E. coli, P. putida, and P. aeruginosa were overexpressed in E. coli and purified as described (23,32). Purification of E. coli DnaC was carried out following an established protocol as described (31). E. coli DNA gyrase was purified by using an overexpression vector and a purification protocol kindly provided by N. E. Dixon (Australian National University, Canberra). The previously characterized monomeric mutant form of the 33-kDa TrfA protein, 6×His-TrfA33(G254D/S267L) (33,34), and the 44-kDa version of this protein, 6×His-TrfA44(M98L/G254D/S267L) (23), were purified as described (23,34). These proteins are designated as TrfA-33 and TrfA-44 and were used throughout this study. Commercially available proteins were HU and single-stranded DNA-binding protein from Enzyco (Denver), creatine kinase and BSA (fraction V) from Sigma, and goat anti-rabbit IgG from BioRad. Plasmid pKD19L1 (35) carrying the minimal RK2 origin region and plasmid pKD19L1Δ1–4 (36) containing an origin in which all four DnaA-box sequences were deleted were used throughout this study.
Helicase Unwinding Assay (FI*). Assays (FI* formation) were performed essentially as described (31). Plasmid pKD19L1 (300 ng) or pKD19L1Δ1-4 (300 ng) was used as the DNA template. Unless noted otherwise, proteins were added in the following amounts: DnaA (10 ng), DnaB (600 ng), DnaC (120 ng), TrfA-33 (125 ng), TrfA-44 (125 ng), HU (5 ng), gyrase (120 ng), and single-stranded DNA-binding protein (230 ng).
Gel Exclusion Chromatography. Gel filtration chromatography using Sepharose CL-4B resin was used to isolate helicase complexes on the pKD19L1Δ1-4 plasmid template as described (31). Fractions eluting from the column were analyzed by SDS/ PAGE, followed by Western transfer and immunoblotting with rabbit antiserum specific against the E. coli DnaB protein. Bound rabbit antibody was detected by a colorimetric reaction with a horseradish peroxidase-conjugated goat anti-rabbit IgG.
Surface Plasmon Resonance (SPR). Standard SPR analyses were performed by using a BiaCore 1000 as described (23,26,32). A double-stranded 64-bp linear fragment containing the four DnaA boxes from oriV (32) or a 131-bp double-stranded linear fragment containing the five iterons from oriV (5′-GCCCTGCGGTATTGACACTTGAGGGGCGCGACTACTGACAGATGAGGGGCGCGATCCTTGACACTTGAGGGGCAGAGTGATGACAGATGAGGGGCGCACCTATTGACATTTGAGGGGCTGTCCACAGGCAG-3′) was used as indicated. DNA fragments were immobilized on a streptavidin matrix-coated Sensor Chip SA by biotin covalent linkage as described by the manufacturer (Pharmacia Biosensor AB). The results are presented as sensograms obtained after subtraction of the background response signal obtained in control experiments. As a control for nonspecific protein–DNA interactions, complementary oligonucleotides with scrambled sequences were synthesized, annealed, and used for analysis. No significant response signal was detected in the control experiments.
Results
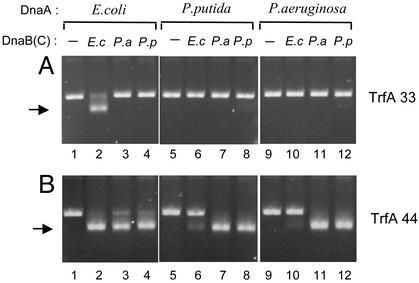
Activity of Homologous and Heterologous Helicase Complexes at oriV. We have recently shown that helicase activation at oriV of plasmid RK2 has differing protein requirements depending on the source of the DnaA and DnaB proteins (23). To determine the molecular basis of these differences we performed in vitro helicase assays with DnaA and DnaB proteins from the same bacterium (homologous) or different bacteria (heterologous) in the presence of the naturally occurring shorter (TrfA-33) or longer (TrfA-44) versions of the TrfA initiation protein. Helicase activity on the mini-RK2 plasmid template was identified electrophoretically by the formation of a substantially unwound form of the supercoiled plasmid DNA, designated FI* (37). The formation of the FI* form of supercoiled DNA requires the combined activities of plasmid and bacterial replication proteins, including helicase, and is a result of origin opening, helicase recruitment and translocation to the open region, and helicase-dependent extensive unwinding of the DNA template. When TrfA-33 and_E. coli_ DnaA protein were present in the reaction mixture, FI* formation was observed with E. coli DnaB and DnaC (Fig. 1_A_, lane 2). In contrast, heterologous combinations of E. coli DnaA with either_Pseudomonas_ sp. helicase were not active (Fig. 1 A, lanes 3 and 4). Helicase activity also was not detected when P. putida or P. aeruginosa DnaA proteins were tested together with the E. coli helicase (Fig. 1 A, lanes 6 and 10). Only very faint FI* bands were visible when homologous and heterologous combinations of Pseudomonas helicase with P. putida or P. aeruginosa DnaA protein were tested for activity in the presence of the TrfA-33 protein (Fig. 1_A_, lanes 7, 8, 11, and 12). The low level of FI* formation observed with TrfA-33 and P. putida DnaA and DnaB compared with previously reported results (23) is caused by the lower concentrations of TrfA and DnaA used (125 ng as compared with 500 ng TrfA and 10 ng compared with 320 ng DnaA) to avoid nonspecific activation on_oriV_.
Fig. 1.
Helicase activity at oriV with homologous and heterologous DnaA proteins. The FI* assay was used to test the activity of E. coli,P. putida, and P. aeruginosa DnaB helicase with the various DnaA proteins. The position of the FI* DNA band, the extensively unwound covalently closed circular DNA generated by helicase activity on the template DNA, is indicated by an arrow. All reactions included the supercoiled form of plasmid pKD19L1 containing the RK2 minimal replication origin, E. coli DNA gyrase, E. coli single-stranded DNA-binding protein,E. coli HU, creatine kinase, and other components of the standard reaction mixture as described in Materials and Methods. The 33-kDa TrfA protein (A) or the 44-kDa TrfA protein (B) and E. coli, P. putida, or P. aeruginosa DnaB helicases were used with homologous or heterologous combinations of the E. coli,P. putida, or P. aeruginosa DnaA proteins as indicated.E. coli DnaC (120 ng) was added to the reaction mixtures run in lanes 2, 6, and 10.
The activity of heterologous DnaA/DnaB combinations in the presence of TrfA-44, however, showed a different pattern (Fig. 1_B_). In contrast to experiments with TrfA-33, heterologous combinations of E. coli DnaA with Pseudomonas helicases were active when TrfA-44 was present in the reaction mixture (compare Fig. 1_A_, lanes 3 and 4 withFig. 1_B_, lanes 3 and 4). Very efficient conversion of the supercoiled plasmid DNA to the FI* form was also observed with homologous or heterologous combinations of the_Pseudomonas_ DnaA proteins with the Pseudomonas helicases (Fig. 1_B_, lanes 7, 8, 11, and 12); however, only weak FI* conversion was observed when the_Pseudomonas_ DnaA was tested with the E. coli DnaBC complex (Fig. 1_B_, lanes 6 and 10).
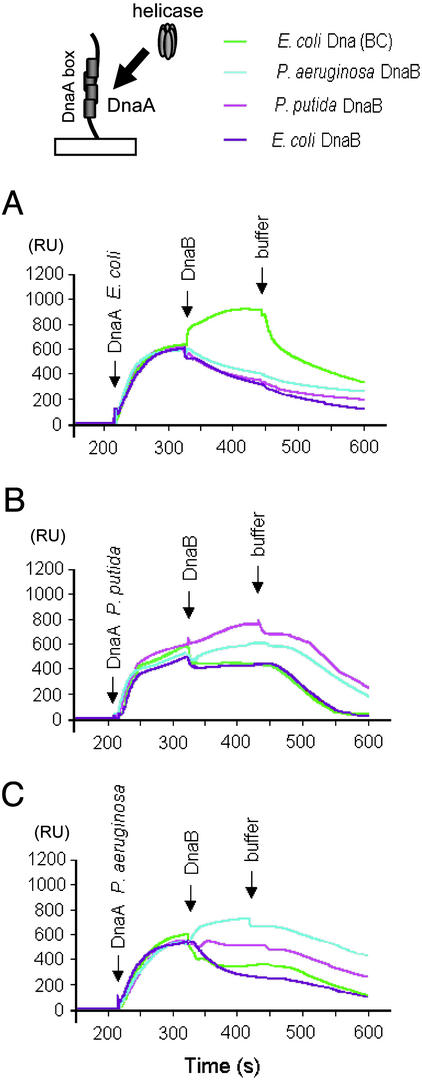
Analysis of Homologous and Heterologous Helicase Complex Formation at the DnaA-Box Region of oriV. The binding of E. coli DnaA protein to the DnaA-box sequences at oriV results in the formation of a specific nucleoprotein structure for the recruitment of the E. coli DnaBC helicase complex (23) as has been observed for oriC (19,37,38). Similarly, complexes were observed with the Pseudomonas DnaA and DnaB proteins at RK2_oriV_; however, in contrast to the E. coli proteins, the presence of DnaC, an accessory ATPase protein, was not required (23). We used the SPR technique to analyze the formation of heterologous helicase complexes with DnaA protein bound at the DnaA boxes of oriV. A 64-bp linear double-stranded DNA fragment, containing the sequences of the four DnaA boxes of oriV, was linked to the sensor chip. All possible combinations of purified DnaA and DnaB proteins from the three bacterial species, along with_E. coli_ DnaC as indicated, were injected sequentially over the sensor matrix and tested for binding. Sensograms, which indicate a change of mass at the attached DNA fragment, clearly showed signals from E. coli (Fig. 2_A_), P. putida (Fig. 2_B_), and P. aeruginosa (Fig. 2_C_) DnaA proteins bound to the DNA. Subsequent additions of the E. coli DnaBC complex or the Pseudomonas DnaB proteins resulted in the detection of a helicase complex when the linear DnaA-box DNA fragment was bound by the homologous DnaA protein (Fig. 2). When heterologous protein combinations were tested, E. coli helicase with or without added DnaC protein did not form a complex with either Pseudomonas DnaA protein bound to the DnaA-box fragment (Fig. 2 B and_C_), nor did either of the Pseudomonas helicases form a complex with E. coli DnaA (Fig. 2 A). This result indicated that the helicase activity observed as FI* formation with the_Pseudomonas_ helicases in the presence of E. coli DnaA and TrfA-44 proteins (Fig. 1_B_, lanes 3 and 4) was not caused by the formation of a heterologous helicase complex with E. coli DnaA protein bound to the DnaA boxes at oriV. This finding suggested the possibility that the TrfA-44 protein itself was capable of interacting with the_Pseudomonas_ DnaB proteins.
Fig. 2.
SPR analysis of helicase interaction with DnaA bound to a linear fragment containing the four DnaA boxes of RK2 oriV. SPR analysis was performed as described in Materials and Methods by injecting solutions of DnaA (65 ng) from E. coli (A), P. putida (B), or P. aeruginosa (C) followed by injection of the DnaB protein (120 ng) as noted. E. coli DnaC (22 ng) protein was added to preform an E. coli DnaBC complex as indicated. RU, response units.
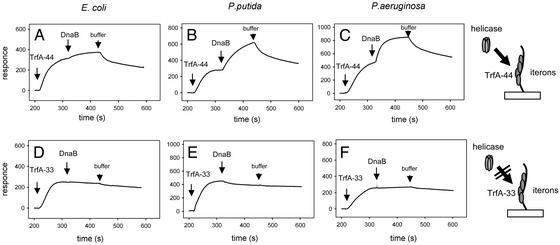
P. putida and P. aeruginosa Helicase Complex Formation at Iteron Sequences of oriV. Using SPR we analyzed the interaction of the three DnaB helicases with TrfA protein bound to a 131-bp linear fragment of double-stranded DNA containing the sequences of the five iterons present at_oriV_ immobilized on the sensor chip. Either form of the TrfA protein followed by each of the three bacterial helicases was sequentially injected over the sensor matrix and tested for binding. A strong signal, indicating TrfA protein binding to the iterons, was obtained after injection of TrfA-44 (Fig. 3_A_–C) or TrfA-33 (Fig. 3_D_–F). Subsequent injections of the helicases revealed significant differences between the two forms of the TrfA protein. A strong signal indicating complex formation was observed with both_Pseudomonas_ helicases when TrfA-44 was first bound to the iterons (Fig. 3 B and_C_). In contrast, under the same experimental conditions, only very weak binding by the E. coli DnaB helicase was observed (Fig. 3_A_). The addition of DnaC or varying the concentration of the E. coli DnaB protein did not significantly enhance the response signal (data not presented). Furthermore, when TrfA-33 was bound to the iteron sequences neither of the Pseudomonas helicases nor E. coli DnaB were found complexed with the DNA fragment (Fig. 3 D_–_F).
Fig. 3.
Formation of helicase complexes at iterons bound by the TrfA proteins. SPR analysis was carried out on a linear oriV DNA fragment containing five iterons. The TrfA-44 protein (50 ng) (A_–_C) or the TrfA-33 protein (50 ng) (D_–_F) was injected first followed by injections of the E. coli (A and D),P. putida (B and E), or P. aeruginosa (C and F) DnaB helicases (120 ng).
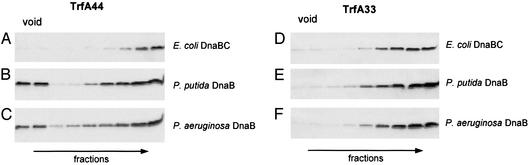
DnaA Boxes and DnaA Protein Are Not Required for Pseudomonas Helicase Complex Formation at oriV. The SPR analysis revealed Pseudomonas helicase complex formation at iterons bound by the TrfA-44 version of the initiation protein. Using size exclusion column chromatography we further analyzed helicase complex formation on a supercoiled DNA template containing an oriV with a deletion of all four DnaA-box sequences (plasmid pKD19L1Δ1-4). Experiments were performed in the absence of DnaA and the presence of either TrfA-33 or TrfA-44. The analyses of fractions eluted from the columns clearly demonstrated that both the P. putida and P. aeruginosa helicases formed a nucleoprotein complex with supercoiled pKD19L1Δ1-4 DNA, which appeared in the void volume when TrfA-44 was present in the incubation mixture (Fig. 4_B_ and C). By contrast, regardless of the presence or absence of DnaC, E. coli DnaB was not detected in the void volume of the column (Fig. 4_A_ and data not shown). When the same incubations were carried out with TrfA-33 protein neither E. coli nor_Pseudomonas_ helicase was detected in the void fractions (Fig. 4_D_–F). Thus, both SPR analysis with linear_oriV_ and gel exclusion chromatography with supercoiled oriV DNA showed that the TrfA-44 protein, but not the TrfA-33 protein, is capable of forming a complex with the DnaB helicases of P. putida and P. aeruginosa. These observations, therefore, raised the possibility that the DnaA protein, known to recruit the DnaB protein to a replication origin in a number of different systems, is not required for recruitment and activation of the Pseudomonas DnaB proteins in the presence of the TrfA-44 protein.
Fig. 4.
Formation of Pseudomonas sp. helicase complexes at oriV is not DnaA dependent when TrfA-44 is present. The formation of E. coli and Pseudomonas sp. helicase complexes at oriV was analyzed by gel exclusion chromatography on Sepharose CL4B. Experiments were performed in the presence of supercoiled pKD19L1Δ1-4 DNA containing the RK2 replication origin with a deletion of all four DnaA boxes (1,200 ng) and TrfA-44 (500 ng) (A_–_C) or TrfA-33 (500 ng) (D_–_F). E. coli DnaB (2,400 ng) plus DnaC (450 ng) (A and D), P. putida DnaB (2,400 ng) (B and E), or P. aeruginosa DnaB (2,400 ng) (C and F) were added to the reaction mixtures. Consecutive fractions from the column were analyzed by SDS/PAGE followed by Western blotting and detection of DnaB protein by using specific antiserum.
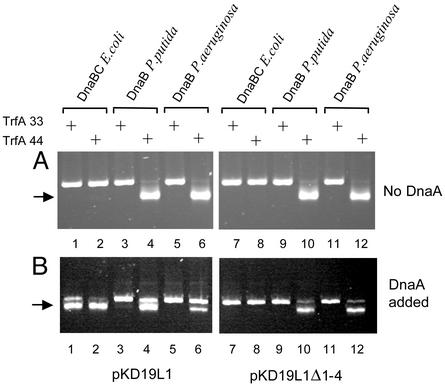
DnaA Protein Is Dispensable for the Recruitment and Positioning of an Active Pseudomonas Helicase at oriV. To explore the possibility that the DnaA protein is not essential for activity of the Pseudomonas helicases at oriV, we analyzed FI* formation with supercoiled DNA containing either the WT RK2 origin (pKD19L1) or an RK2 origin in which all four DnaA boxes were deleted (pKD19L1Δ1-4). Experiments were carried out with both versions of the TrfA protein and in the absence or presence of the homologous DnaA protein. The fact that the DnaB proteins of the_Pseudomonas_ species were purified after overexpression in E. coli precludes the possibility of their contamination with the_Pseudomonas_ DnaA proteins; their contamination with E. coli DnaA is extremely unlikely, as the Pseudomonas DnaB proteins do not stably associate with E. coli DnaA (Fig. 2 A).
With replication proteins from E. coli, helicase activity was not detected unless the WT oriV template and DnaA protein were present in the reaction mixture (compare Fig. 5_B_, lanes 1 and 2 withFig. 5_A_, lanes 1, 2, 7, and 8 or Fig. 5_B_, lanes 7 and 8). E. coli helicase activity was observed with both versions of the TrfA protein (Fig. 5_B_, lanes 1 and 2). In contrast to these results, both_Pseudomonas_ helicases were active at WT oriV (pKD19L1) and at the oriV DnaA-box deletion mutant (pKD19L1Δ1-4), regardless of the presence or absence of DnaA proteins, when TrfA-44 protein was present (Fig. 5 A and_B_, lanes 4, 6, 10, and 12). In the presence of TrfA-33, without DnaA proteins, Pseudomonas helicases were inactive when tested on WT template (pKD19L1) or the oriV DnaA-box deletion mutant (pKD19L1Δ1-4) (Fig. 5_A_, lanes 3 and 5 andFig. 5_A_, lanes 9 and 11). A very faint FI* band was observed when P. putida helicase was tested with the WT oriV plasmid in the presence of TrfA-33 and P. putida DnaA (Fig. 5_B_, lane 3).
Fig. 5.
The activity of Pseudomonas sp. helicase is not DnaA dependent in the presence of the TrfA-44 protein. Activities of E. coli, P. putida, and P. aeruginosa helicases were tested by analysis of DNA unwinding of supercoiled pKD19L1, containing the RK2 minimal replication origin, or pKD19L1Δ1-4, containing the RK2 replication origin with a deletion of the DnaA-box region, in the absence (A) or presence (B) of the homologous DnaA protein. TrfA-33 or TrfA-44 was added as indicated. The position of FI* is marked by the arrow.
Discussion
An essential role for the DnaA protein in replication has now been established for the chromosome of several bacterial species and for several plasmid elements (2). In addition to the DnaA protein, DnaB and DnaC have been shown to be required for the formation of a prepriming complex at the E. coli chromosomal origin of replication (19–22). On the basis of amino acid sequences derived from genomic sequences, homologues of two of these proteins, DnaA and DnaB, have been found in all eubacteria where the chromosomal DNA sequence is known (2,39). However, homologues to the E. coli DnaC protein are not readily found in other bacteria (23). The formation of a stable complex between the DnaA and DnaB proteins has been shown not only for the E. coli proteins but also for the homologous sets of the DnaA and DnaB proteins of P. aeruginosa and P. putida (23). Thus, it is clear that the genetic determinants of the DnaA and DnaB proteins are likely to be widespread among bacteria and that the two proteins exhibit similar property of association with each other regardless of their bacterial origin. Furthermore, DnaA boxes with varying degrees of homology to the nucleotide sequence of the 9-mer E. coli DnaA boxes are also present at the chromosomal replication origin of a number of bacterial species and a variety of plasmids that replicate by the theta mode, suggesting a role for DnaA replication initiation in a wide range of replicons (2).
Largely as a result of the detailed biochemical analyses of the initiation of replication of the E. coli chromosome, the sequence of events resulting in the formation of a prepriming complex has for the most part been defined. These biochemical events appear to account for the initiation of chromosomal DNA replication for several bacterial species that have been studied in addition to E. coli and for several plasmid elements (2). However, as shown in the present in vitro study involving plasmid RK2, the TrfA-44 Rep can bring about the key replication initiation events at the plasmid origin in the absence of the DnaA protein. These events include specific unwinding of the 13-mers within the AT-rich region, recruitment of the Pseudomonas DnaB helicases to the origin region, and the positioning of an active form of the helicase within the open complex. Although the RK2 TrfA-33 protein, lacking the N-terminal 98 aa present in TrfA-44, is also capable of bringing about specific strand opening at the AT-rich region, it fails to stably bind to the DnaB helicase, regardless of source, and requires the host DnaA protein for the formation of a prepriming complex with the E. coli DnaB and DnaC proteins or, albeit weakly, with the P. putida DnaB helicase. It is interesting that the multifunctional TrfA-44 protein lacks the ability to form a stable association with the DnaB or DnaB/DnaC complex of E. coli. It, therefore, depends on the E. coli DnaA protein to recruit the E. coli helicase to the origin region and, probably in concert with the DnaA protein, to load the DnaB/DnaC complex at the open origin region of plasmid RK2. It is of further interest that in the case of the Pseudomonas helicases, not only does the TrfA-44 protein form a stable association with the Pseudomonas DnaB proteins, but it is capable of carrying out the translocation and positioning of the helicase_in vitro_ in the absence of the DnaC protein (23).
The present experiments showing that the Pseudomonas helicases, in contrast to E. coli DnaB, are recruited and loaded via a direct interaction with TrfA-44 protein bound to the iterons are consistent with an earlier in vivo study showing that the RK2 derivative pKD19L1Δ1-4, which contains a deletion of all four DnaA boxes, is functional in P. aeruginosa but not in E. coli (36). Furthermore, it has been shown that stable replication of RK2 in P. aeruginosa is supported only by the 44-kDa TrfA protein and not by the 33-kDa TrfA protein (29,40,41). RK2 replication in vivo in P. putida requires the DnaA boxes (36) and can use either version of TrfA (40,41). It is possible that in_P. putida_ RK2 replication requires the DnaA-box region and binding of the DnaA protein for a more extended plasmid origin opening, which may be critical for P. putida helicase loading. It is also possible that the DnaA–DnaB interaction additionally stabilizes the P. putida DnaB helicase complex at the RK2 origin in vivo.
In addition to the finding of unique multiple functions for a plasmid Rep and the unusual independence from the host DnaA protein for the recruitment of a replicative helicase, this work advances our understanding of the different strategies used by a broad host range replicon for the initiation of replication in different bacterial species. It is now evident that both the structure of its replication origin and the presence of two versions of the essential plasmid-encoded Rep provides plasmid RK2 with alternative pathways for host helicase recruitment, positioning, and activation. One of these pathways involving the initiation of replication of plasmid RK2 with the TrfA-33 protein in E. coli is similar to that described for the initiation of replication of the E. coli chromosomal origin with the important exception that the plasmid Rep is required for opening at the AT-rich region. In the case of replication initiation with the P. putida DnaB protein, the TrfA-33 protein is only weakly effective in the formation of FI* in the presence and ineffective in the absence of the host DnaA protein. A distinctly different pathway involves the TrfA-44 protein, which, in association with the DnaB protein of either P. putida or_P. aeruginosa_, is active in the formation of a prepriming complex independent of the DnaA protein.
Although the interaction of a plasmid Rep with a host DnaB protein has been shown earlier in the case of the Reps of two narrow host range plasmids, pSC101 and R6K, in both of these cases this interaction with the E. coli helicase did not obviate the need for the DnaA protein (11,12,25). Also, unlike the pSC101 and R6K Reps, the TrfA-44 protein does not form a stable association with the_E. coli_ DnaB protein. Recent experiments introducing specific mutations in the N-terminal region of TrfA-44 have established a correlation between a loss of DnaB binding properties and an inability to replicate in_P. aeruginosa_ (Z. Zhong, D.R.H., and A.T., unpublished work). Thus, it is clear that the additional 98 aa at the N terminus of TrfA-44 not only provides this protein with added biochemical functions but also extends the host range of the RK2 antibiotic resistance plasmid. It will be of interest to examine the mechanism of formation of a prepriming complex at the chromosomal replication origin of P. aeruginosa or P. putida in view of the failure to identify an E. coli dnaC homologue in the genome of these Pseudomonas species and the properties of the_Pseudomonas_ replicative helicases described in this study.
Acknowledgments
This work was supported by National Institutes of Health Research Grant AI-07194 (to D.R.H.), Polish State Committee for Scientific Research Grant 3P04A01422 (to I.K.), and the European Molecular Biology Organization Young Investigator Program (I.K.). M.P. is a recipient of the Annual Stipend for Young Scientists provided by the Foundation for Polish Science.
Abbreviations: Rep, replication initiation protein; SPR, surface plasmon resonance.
References
- 1.Bramhill, D. & Kornberg, A. (1988) Cell 52**,** 743–755. [DOI] [PubMed] [Google Scholar]
- 2.Messer, W. (2002) FEMS Microbiol. Rev. 26**,** 355–374. [DOI] [PubMed] [Google Scholar]
- 3.Schnos, M., Zahn, K., Inman, R. B. & Blattner, F. R. (1988) Cell 52**,** 385–395. [DOI] [PubMed] [Google Scholar]
- 4.Borowiec, J. A., Dean, F. B., Bullock, P. A. & Hurwitz, J. (1990) Cell 60**,** 181–184. [DOI] [PubMed] [Google Scholar]
- 5.Lee, S. S. & Lehman, I. R. (1997) Proc. Natl. Acad. Sci. USA 94**,** 2838–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frappier, L. & O'Donnell, M. (1992) J. Virol. 66**,** 1786–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillette, T. G., Lusky, M. & Borowiec, J. A. (1994) Proc. Natl. Acad. Sci. USA 91**,** 8846–8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukhopadhyay, G., Carr, K. M., Kaguni, J. M. & Chattoraj, D. K. (1993) EMBO J. 12**,** 4547–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park, K., Mukhopadhyay, S. & Chattoraj, D. K. (1998) J. Biol. Chem. 273**,** 24906–24911. [DOI] [PubMed] [Google Scholar]
- 10.Kawasaki, Y., Matsunaga, F., Kano, Y., Yura, T. & Wada, C. (1996) Mol. Gen. Genet. 253**,** 42–49. [DOI] [PubMed] [Google Scholar]
- 11.Sharma, R., Kachroo, A. & Bastia, D. (2001) EMBO J. 20**,** 4577–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu, Y. B., Datta, H. J. & Bastia, D. (1998) EMBO J. 17**,** 5192–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruger, R., Konieczny, I. & Filutowicz, M. (2001) J. Mol. Biol. 306**,** 945–955. [DOI] [PubMed] [Google Scholar]
- 14.Konieczny, I., Doran, K. S., Helinski, D. R. & Blasina, A. (1997) J. Biol. Chem. 272**,** 20173–20178. [DOI] [PubMed] [Google Scholar]
- 15.Fuller, R. S., Funnell, B. E. & Kornberg, A. (1984) Cell 38**,** 889–900. [DOI] [PubMed] [Google Scholar]
- 16.Gille, H. & Messer, W. (1991) EMBO J. 10**,** 1579–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Speck, C. & Messer, W. (2001) EMBO J. 20**,** 1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marszalek, J. & Kaguni, J. M. (1994) J. Biol. Chem. 269**,** 4883–4890. [PubMed] [Google Scholar]
- 19.Baker, T. A., Funnell, B. E. & Kornberg, A. (1987) J. Biol. Chem. 262**,** 6877–6885. [PubMed] [Google Scholar]
- 20.Funnell, B. E., Baker, T. A. & Kornberg, A. (1987) J. Biol. Chem. 262**,** 10327–10334. [PubMed] [Google Scholar]
- 21.Allen, G. C., Jr., & Kornberg, A. (1991) J. Biol. Chem. 266**,** 22096–22101. [PubMed] [Google Scholar]
- 22.Learn, B. A., Um, S. J., Huang, L. & McMacken, R. (1997) Proc. Natl. Acad. Sci. USA 94**,** 1154–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caspi, R., Pacek, M., Consiglieri, G., Helinski, D. R., Toukdarian, A. & Konieczny, I. (2001) EMBO J. 20**,** 3262–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratnakar, P. V., Mohanty, B. K., Lobert, M. & Bastia, D. (1996) Proc. Natl. Acad. Sci. USA 93**,** 5522–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta, H. J., Khatri, G. S. & Bastia, D. (1999) Proc. Natl. Acad. Sci. USA 96**,** 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pacek, M., Konopa, G. & Konieczny, I. (2001) J. Biol. Chem. 276**,** 23639–23644. [DOI] [PubMed] [Google Scholar]
- 27.Thomas, C. M. & Helinski, D. R. (1989) in Promiscuous Plasmids of Gram-Negative Bacteria, ed. Thomas, C. M. (Academic, San Diego), pp. 1–25.
- 28.Kornacki, J. A., West, A. H. & Firshein, W. (1984) Plasmid 11**,** 48–57. [DOI] [PubMed] [Google Scholar]
- 29.Shingler, V. & Thomas, C. M. (1984) J. Mol. Biol. 175**,** 229–249. [DOI] [PubMed] [Google Scholar]
- 30.Perri, S., Helinski, D. R. & Toukdarian, A. (1991) J. Biol. Chem. 266**,** 12536–12543. [PubMed] [Google Scholar]
- 31.Konieczny, I. & Helinski, D. R. (1997b) J. Biol. Chem. 272**,** 33312–33318. [DOI] [PubMed] [Google Scholar]
- 32.Caspi, R., Helinski, D. R., Pacek, M. & Konieczny, I. (2000) J. Biol. Chem. 275**,** 18454–18461. [DOI] [PubMed] [Google Scholar]
- 33.Toukdarian, A. E., Helinski, D. R. & Perri, S. (1996) J. Biol. Chem. 271**,** 7072–7078. [DOI] [PubMed] [Google Scholar]
- 34.Blasina, A., Kittell, B. L., Toukdarian, A. E. & Helinski, D. R. (1996) Proc. Natl. Acad. Sci. USA 93**,** 3559–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doran, K. S., Konieczny, I. & Helinski, D. R. (1998) J. Biol. Chem. 273**,** 8447–8453. [DOI] [PubMed] [Google Scholar]
- 36.Doran, K. S., Helinski, D. R. & Konieczny, I. (1999) Mol. Microbiol. 33**,** 490–498. [DOI] [PubMed] [Google Scholar]
- 37.Baker, T. A., Sekimizu, K., Funnell, B. E. & Kornberg, A. (1986) Cell 45**,** 53–64. [DOI] [PubMed] [Google Scholar]
- 38.Kobori, J. A. & Kornberg, A. (1982) J. Biol. Chem. 257**,** 13763–13769. [PubMed] [Google Scholar]
- 39.Leipe, D. D., Aravind, L., Grishin, N. V. & Koonin, E. V. (2000) Genome Res. 10**,** 5–16. [PubMed] [Google Scholar]
- 40.Durland, R. H. & Helinski, D. R. (1987) Plasmid 18**,** 164–169. [DOI] [PubMed] [Google Scholar]
- 41.Fang, F. C. & Helinski, D. R. (1991) J. Bacteriol. 173**,** 5861–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]