Accurate and reliable high-throughput detection of copy number variation in the human genome (original) (raw)
Abstract
This study describes a new tool for accurate and reliable high-throughput detection of copy number variation in the human genome. We have constructed a large-insert clone DNA microarray covering the entire human genome in tiling path resolution that we have used to identify copy number variation in human populations. Crucial to this study has been the development of a robust array platform and analytic process for the automated identification of copy number variants (CNVs). The array consists of 26,574 clones covering 93.7% of euchromatic regions. Clones were selected primarily from the published “Golden Path,” and mapping was confirmed by fingerprinting and BAC-end sequencing. Array performance was extensively tested by a series of validation assays. These included determining the hybridization characteristics of each individual clone on the array by chromosome-specific add-in experiments. Estimation of data reproducibility and false-positive/negative rates was carried out using self–self hybridizations, replicate experiments, and independent validations of CNVs. Based on these studies, we developed a variance-based automatic copy number detection analysis process (CNVfinder) and have demonstrated its robustness by comparison with the SW-ARRAY method.
Until recently, the importance of large-scale copy number changes in the genomes of humans and other vertebrates has been under-appreciated. Two reports in 2004, using comparative genomic hybridization with DNA microarrays (array-CGH), highlighted the widespread nature of this normal copy number variation (Iafrate et al. 2004; Sebat et al. 2004). Other studies have now confirmed and further detailed the extent of copy number variation (CNV) in human and primate genomes (Newman et al. 2005; Sharp et al. 2005; Tuzun et al. 2005; Conrad et al. 2006; Perry et al. 2006). The key to the identification of the extent of CNV was the use of array-CGH. In the initial studies, the microarrays used were of limited resolution. Iafrate et al. (2004) used a commercial BAC array with one clone approximately every 1 Mb across the genome, whereas Sebat et al. (2004) used long-oligonucleotide arrays with an effective resolution of >90 kb. Recent advances in array technology are continuing to improve the resolution of microarrays for array-CGH. For example, a large-insert clone set has been developed using DNA fingerprinting overlaps, which has allowed the production of arrays with a resolution of ∼60 kb (Ishkanian et al. 2004). Furthermore, long-oligonucleotide arrays are now available with as many as 385,000 elements (e.g., Agilent, Inc., Nimblegen, Inc.), but array-CGH using this type of platform is generally noisy and multiple probes must be averaged in order to call CNVs (Ylstra et al. 2006). Although the superior signal-to-noise ratio of large-insert clone arrays allows CNVs to be called from a single clone, to date there has not been a detailed analysis of the false-positive and false-negative calling rates using this type of array.
In this paper, we describe the construction of a whole-genome tiling path resolution array that has been used to survey CNV in the human genome (Redon et al. 2006). The clones have been largely selected from the “Golden Path” used to generate the reference human sequence (Lander et al. 2001) and have been subjected to high levels of validation. Furthermore, we have developed an algorithm (CNVfinder) for calling significant copy number changes based on estimates of variation in each hybridization and tested the performance of this algorithm against the Smith-Waterman approach (Price et al. 2005). To enable accurate testing of the algorithms, we have sampled CNV calls using different statistically defined thresholds and validated the calls from individual comparison of two publicly available normal DNA samples using independent methods. These data allow estimates of the false-positive and false-negative rates of CNV calling for not only the algorithms tested in this study but also for other array-CGH platforms.
Results
Clone selection, validation, and array construction
Our initial set of 26,678 large insert clones was selected predominantly from the “Golden Path” used to sequence the human genome (Lander et al. 2001). This set has the advantage that the majority of the clones have been completely sequenced, in contrast to the previously published 32K clone set that was identified from fingerprinting overlaps (Ishkanian et al. 2004). The clones were amplified using three different DOP-PCR primers before arraying onto glass slides. This approach has been shown to improve the reliability and reproducibility of array-CGH data (Fiegler et al. 2003). All clones were validated initially by fingerprinting and subsequently by end sequencing. For the majority of clones, the mapping position was confirmed. However, for 15.6% of clones, end sequencing failed or end reads could not be placed on the reference sequence. As these clones were verified only by fingerprinting, they were mapped by the original sequence. Discrepant mapping locations were found for 7.3% of clones by end sequencing, and the positions of these clones were reassigned. The clone set and mapping information can be accessed and downloaded using the Ensembl genome browser (http://www.ensembl.org/Homo_sapiens/index.html) (Kent et al. 2002) by activating the “30K TPA clones” decoration within the graphical overview.
To validate the microarray further, the hybridization characteristics of all clones were assessed using chromosome-specific add-in experiments. This approach uses self–self hybridizations where the “test” probe is spiked with extra copies of DNA from a specific chromosome. Clones mapping to the chromosome spiked into a particular hybridization will respond in a linear fashion to the number of extra chromosome copies. The majority of clones responded as expected to increased copy number. After consideration of the chromosome add-in results and selected fluorescence in situ hybridization experiments, 104 clones (0.34%) failing to respond or responding inappropriately were excluded from analysis.
Final clone selection and array performance
The final validated set of 26,574 clones covers 93.7% of the euchromatin of the human genome, leaving 2237 gaps. Difficulties in obtaining sequenced clones from inaccessible libraries reduced the coverage for chromosomes 19 and 21 (68.4% coverage with 112 gaps and 83.6% coverage with 50 gaps, respectively).
The array was subjected to a series of initial validation experiments including self–self and male–female hybridizations using DNA derived from normal individuals. Estimated standard deviations of all ratios were between 0.019 and 0.028 for self–self hybridizations (n = 4) and 0.033 and 0.053 for male–female hybridizations (n = 5).
For each clone, we calculated the standard deviation of the log2 ratios from the four self–self experiments. The distribution of these standard deviations was unimodal, with a median value of 0.020 (Supplemental Fig. 1A). Furthermore, the distribution of the standardized log2 ratio variances (clone variance/global clone variance × degrees of freedom) followed a χ2 distribution, as would be expected from an underlying normal distribution of ratios (Supplemental Fig. 1B). This analysis demonstrates the absence of a subpopulation of inherently noisier clones.
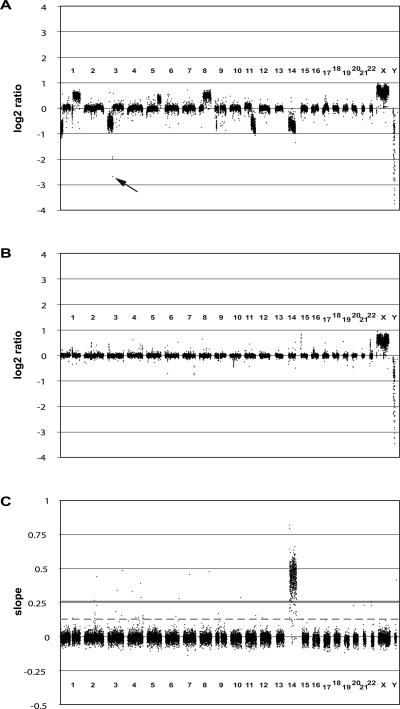
Figure 1.
Whole-genome tiling path array-CGH profiles. (A) Array-CGH profile for the renal cell carcinoma cell line 769P. The whole-genome tiling path array identified a previously undetected homozygous deletion on chromosome 3 (60.84–60.91 Mb, black arrow). (B) Array-CGH profile for NA15510 versus NA10851 (replicate A). (C) Chromosome 14 add-in profile. The slopes of response for each clone are plotted against the chromosomal position. (Gray continuous line) threshold of 10 times the standard deviation, (dashed gray line) threshold of five times the standard deviation (see Methods for details).
We also hybridized DNA from an extensively studied renal cell carcinoma cell line (769P) that displays multiple single-copy gains and losses across its genome. Compared with previous analysis of this cell line using an array with a resolution of one clone every 1 Mb (Fiegler et al. 2003), the tiling path array detected all previously identified copy number changes with refined resolution of copy number change breakpoints. Moreover, previously undetected changes were found, such as a small homozygous deletion on chromosome 3 (60.72–60.92 Mb; Fig. 1A).
Calling copy number variation
To allow robust automatic classification of CNVs, we have developed an algorithm (CNVfinder) based on the ratio variance of each array experiment. The algorithm is based on two working hypotheses. First, in whole-genome profiles from apparently normal individuals, the majority of observations are normally distributed around a log2 ratio of zero (representing normal diploid copy number in both test and reference genomes). This central distribution can be used to provide a good estimate of experimental variability (termed SDe). Second, the consequence of variation in DNA copy number will be ratio values that fall outside the central distribution.
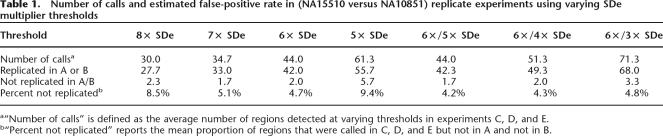
Multiples of the SDe can be used to define positive and negative thresholds beyond which ratios are unlikely to occur by chance in the absence of copy number variation. We have used this approach to develop an algorithm for calling CNVs, which is described in detail in the Methods. The starting point for the algorithm is the measurement of SDe by calculating the 68.2 percentile value of absolute dye-swap combined ratios on a chromosome-by-chromosome basis. In a normal distribution, 68.2% of values are contained within ±1 standard deviation from the mean. Thus, the 68.2 percentile value (SDe) provides an estimation of the standard deviation that is relatively insensitive to outlying values. As a description of the overall hybridization quality, we define the global SDe as the median of these values. To set significance thresholds, we determined the value of the SDe multiplier empirically using technical replicates and validated these values using replicate self–self hybridizations and a set of independently validated CNVs.
Five replicate experiments (A–E) of cell line NA15510 versus the reference cell line NA10851 were carried out on three different days and using three different batches of arrays. An example hybridization is shown in Figure 1B. Ordered by global SDe, experiments A and B displayed the best hybridization quality (SDe = 0.033), followed by experiments C (SDe = 0.036), D (SDe = 0.039), and E (SDe = 0.053).
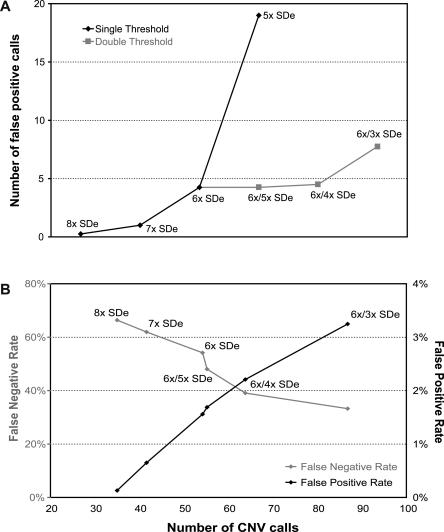
Our aim for calling significant copy number changes was to achieve a low false-positive call rate (<5%) while maximizing the number of calls beyond the thresholds. To estimate the false-positive rate, we calculated the number of calls made in the more variable experiments (C, D, and E) that were not called in the less variable experiments (A and B) for different thresholds. As greater confidence can be ascribed to small ratio changes when these are reported by multiple neighboring clones, we also investigated the use of dual thresholds, one for isolated clone calls and a second for consecutive clone calls. For single thresholds, the optimal setting of the SDe multiplier was achieved at a value of 6 (Table 1). However, the inclusion of a second, lower threshold for consecutive clone calls increased the number of calls without additional false positives. A combination of a single clone SDe multiplier of 6 and a consecutive clone multiplier of 3 was optimal.
Table 1.
Number of calls and estimated false-positive rate in (NA15510 versus NA10851) replicate experiments using varying SDe multiplier thresholds
To validate the optimal thresholds, we applied the same set of thresholds to an independent set of four self–self experiments. As by definition CNV cannot exist in self–self hybridizations, we would not expect to make calls using thresholds optimized for CNV detection. However, in practice, a small number of calls will be made due to differences in labeling bias and hybridization artifacts. We found that using the optimal dual threshold of 6×/3× SDe, a mean of 7.75 calls was made (Fig. 2A). Increasing the consecutive clone threshold to 4 SDe reduced the number of calls to 4.5, equivalent to a single threshold of 6 SDe. To avoid overcalling small consecutive changes, we decided to use conservative settings of a dual threshold of 6× SDe for isolated clones and 4× SDe for consecutive clones.
Figure 2.
CNVfinder performance using varying SDe multiplier thresholds in WGTP array validation experiments. (A) Number of false-positive calls in self–self experiments in relation to SDe multiplier thresholds. Various single/dual SDe multiplier thresholds were applied to four replicate self–self experiments and the mean number of calls calculated. (Black diamonds) mean number of regions called by single SDe multiplier thresholds, (gray squares) mean number of regions called by dual SDe multiplier thresholds. (B) False-positive and false-negative rates against the number of CNVs called for varying single/dual SDe multiplier thresholds in NA15510 versus NA10851 experiments. False-negative and false-positive rates are based on the quantitative PCR results from 154 sampled clones.
Our final testing of the dual threshold of 6×/4× SDe involved independent validation of a set of regions by Quantitative Multiplex PCR of Short Fluorescent Fragments (QMPSF) or SYBR Green real-time PCR. The regions were selected by sampling randomly a subset of clones from two replicate experiments (NA15510 versus NA10851), one with a high SDe and one with a low SDe. We chose clones from five intervals based on multiples of SDe from 4× to greater than 6×, with 20 clones per interval per replicate. This selection ensured that the clone subset would include a high proportion of CNVs but also clones not containing CNVs, to allow estimates of false-negative as well as false-positive rates. A total of 154 clones was assayed using quantitative PCR for the presence of CNV (46 clones were selected in both replicates). CNVs were found in 123 of the selected regions (see Supplemental Table 1 for full details).
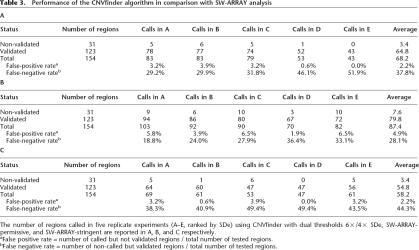
Calls against this set of 154 independently validated clones were then made with varying single or dual thresholds in the five replicate experiments (NA15510 versus NA10851). The number of CNV calls was calculated, and the false-positive and false-negative rates were determined (Fig. 2B). As expected, more permissive thresholds led to more CNV calls, a lower false-negative rate, and a higher false-positive rate. For a false-positive rate <1% (threshold of 7× SDe), only 42 CNVs could be called, resulting in a false-negative rate of 62%. In contrast, using a dual threshold of 6×/3× SDe, 86 CNVs were detected, with a false-negative rate of 33% and a false-positive rate of 3.2%. Our final dual threshold, 6×/4× SDe, enables the detection of 64 CNVs on average, a false-positive rate of 2.2%, and a false-negative rate of 38% (Table 3A, see below). The low false-positive rate demonstrates that the vast majority of calls made with these thresholds will represent real CNVs. The higher false-negative rate can be explained to some extent because quantitative PCR has a resolution defined by the amplicon size, while the array has a lower detection limit defined by the clone insert. Thus, quantitative PCR will detect CNVs that will be too small to produce a detectable ratio change in a large insert clone, particularly for more variable hybridizations. As the absolute value of the thresholds is proportional to SDe, the ability to call small ratio changes (smaller CNVs) decreases with increasing experimental variability.
Table 3.
Performance of the CNVfinder algorithm in comparison with SW-ARRAY analysis
Final merging of calls to CNVs
Application of the dual 6×/4× SDe thresholds to replicate experiments highlighted an additional problem with defining CNVs. We found that CNVs called in experiments with low SDe often became fragmented in higher SDe replicates. Due to varying repeat content, sequence homologies, and experimental variation, some clones underrespond to a specific copy number change and may fail to be called in higher SDe experiments, thus fragmenting the CNV. The final calling algorithm (CNVfinder) allows restricted extension of called regions with ratios >3× SDe and permits the incorporation of single, non-consecutive uncalled clones within the region (Fig. 3; for details, see Methods).
Figure 3.
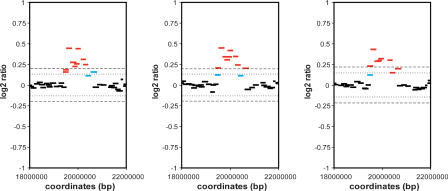
Definition of CNV boundaries by CNVfinder. A CNV on chromosome 3 detected in three different hybridizations, demonstrating the consistent detection of the boundaries of CNV. First, clones above the dual 6×/4× SDe thresholds are called (red bars). Secondly, CNVfinder allows restricted extension of called regions using lower thresholds of 3×/1× SDe (blue bars, see Fig. 5 for details). The 6×/4× SDe thresholds are shown as dashed (6× SDe) and dotted (4× SDe) lines.
Estimate of false-negative rate
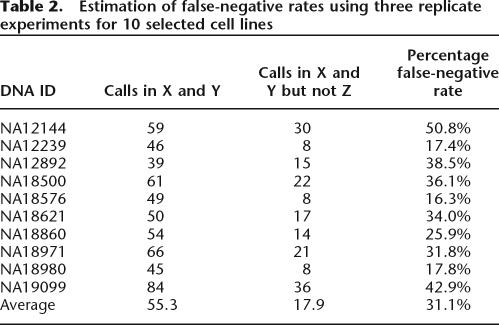
Using the final version of CNVfinder, we estimated the false-negative rate by using three independent replicate dye-swap experiments for 10 different cell line DNAs. Experiments were made at different dates and using different batches of arrays. The three replicate experiments for each cell line were ranked by the total number of called regions. Three data sets were generated, with set X containing the 10 replicates with the highest number of calls, set Z the 10 replicates with the lowest number of calls, and set Y the 10 remaining replicates.
As would be expected, the lowest global SDe was found in set X (0.035 on average) and the highest SDe in set Z (0.045). Moreover, the proportion of regions called in only one out of three replicate experiments is higher in set X (29.9%) than in sets Y (17.3%) and Z (13.1%). For each cell line, we then calculated the proportion of regions called in replicates X and Y, but not in replicate Z (Table 2). This gives an estimated false-negative rate of 31% on average in the worst experiment (range: 16%–51%).
Table 2.
Estimation of false-negative rates using three replicate experiments for 10 selected cell lines
This false-negative rate represents the proportion of CNVs that are not called in an experiment, but which potentially are detectable by the WGTP platform. To investigate the number and nature of CNVs that are not detectable by the WGTP array, we compared our results on 270 HapMap samples with those obtained using the Affymetrix GeneChip Human Mapping 500K Early Access (500K EA) Arrays (Redon et al. 2006). We found that the WGTP platform detected only 18% of the CNVs of <80 kb in size identified on the 500K EA platform, but detected 51% of those between 80 and 150 kb, and 90% of those >150 kb in size. Furthermore, the WGTP array identified larger CNVs more efficiently in complex regions of the genome, such as regions of segmental duplication (see Redon et al. 2006 for more detailed comparisons).
Comparison of CNVfinder with SW-ARRAY
To gauge the effectiveness of CNVfinder against other algorithms, we tested initially SW-ARRAY (Price et al. 2005) and DNAcopy (Olshen et al. 2004). We found that both methods gave very similar results, but DNAcopy did so at a greater computational price. Therefore, we compared SW-ARRAY with CNVfinder using the same set of experiments.
The SW-ARRAY method applies the Smith-Waterman algorithm (Smith and Waterman 1981) to identify segments within a set of ordered array log ratio values. Two parameters can be tuned in this method to adjust its sensitivity. First, a data transformation is applied to adjust the input data by subtracting (median + (const × median absolute deviation)) from each data point (data are processed by chromosome arm). This isolates positive ratio CNVs. The process is repeated on the data set after inverting the sign of the ratio to isolate negative-ratio CNVs. Different values of the constant, const, allow tuning of the calling sensitivity. Second, a score is associated with each CNV called, which is the sum of ratios within the CNV. Filtering by this score value provides a simple means to rank and select the most obvious CNVs from each set of input data. We modified this score to be a multiple of the median absolute deviation for each chromosome, hence allowing a single threshold to be used between chromosomes of differing variance.
Relative sensitivities of different parameter pairs were determined by generating receiver operator (ROC) curves against the quantitative PCR-validated clone data. ROC curves were generated with const values ranging from 0 to 5 and score filter values ranging from 0 to 100. Using the ROC curves, the optimal parameter set was chosen as being the point that maximized the number of CNVs called, while keeping the false-positive rate <5%. Two final parameter sets were chosen: a permissive set (const = 3, with a score filter ≥3) and a stringent set (const = 3, with a score filter ≥5). Overall, the method was robust to small changes in these parameters (data not shown), although it is worth noting that the optimal const value is almost an order of magnitude larger than that originally used by Price et al. (2005). The summary of CNV calls using the two SW-ARRAY settings (SW-ARRAY-permissive and SW-ARRAY-stringent) and CNVfinder for the clones included in the independent validation experiments (NA15510 versus NA10851) is shown in Table 3.
The greatest number of clones called was found using the SW-ARRAY-permissive algorithm (87.4/154), although with the highest false-positive rate of 4.9% but a low false-negative rate of 28.0%. The worst performing algorithm was SW-ARRAY-stringent (58.2/154 calls, a false-positive rate of 2.2% and a false-negative rate of 44.3%).
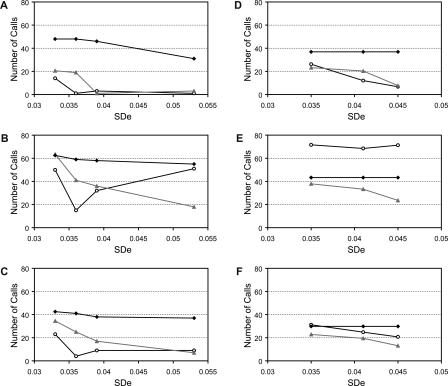
We then tested the effect of varying SDe on CNV calling by applying the three algorithms to the five replicate experiments of cell line NA15510 versus the reference cell line NA10851. For each replicate, we calculated the number of calls made in three or four other replicates, in one or two replicates, or not called in any other experiments and plotted these values against the SDe of the replicate (Fig. 4A,B,C).
Figure 4.
Comparison of CNVfinder with SW-ARRAY. CNV calling using CNVfinder (A), SW-ARRAY-permissive (B), and SW-ARRAY-stringent (C) in five replicate experiments. The number of regions called in three or four replicates (black diamonds), one or two replicates (gray triangles), or in none (white circles) of the other replicates is plotted against global SDe values. (D,E,F) CNV calling using CNVfinder (D), SW-ARRAY-permissive (E), and SW-ARRAY-stringent (F) in triplicate experiments of 10 different cell line DNAs. The replicate experiments were pooled into three different sets X, Y, and Z (see Table 3). The number of regions called in the two other replicates (black diamonds), one other replicate (gray triangles), or in none (white circles) is then plotted against the average of the global SDe values for each set.
For the lowest SDe hybridizations, SW-ARRAY-permissive called the greatest number of CNVs, but the fewest of these were found in at least four replicates (176 CNVs and 35% called in four out of five replicates). Corresponding values were 100 CNVs and 42.5% called in four out of five replicates for SW-ARRAY-stringent, and 82.5 CNVs and 58.2% called in four out of five replicates for CNVfinder. As expected, the number of calls decreased as the SDe increased. At higher SDe, CNVfinder continued to call fewer CNVs than SW-ARRAY, but these were consistently called in four out of five replicates (88.6%) compared with 69.8% for SW-ARRAY-stringent and 44.4% for SW-ARRAY-permissive. We concluded that, while CNVfinder calls fewer CNVs, its performance is more consistent across experiments with varying SDe.
A similar analysis was applied to the three independent replicate dye-swap experiments of 10 different cell line DNAs from the HapMap collection (Fig. 4D,E,F; see Supplemental Table 2 for full details). In this analysis, the replicates were ranked by the number of calls and combined to generate the three sets X, Y, and Z previously described. Similar results were obtained, although it is worth noting that the highest number of calls made by the SW-ARRAY-permissive algorithm was found in only one experiment. Again, CNVfinder gave consistent results across experiments with different SDe, and in particular tended to call only highly replicated CNVs when the SDe was high.
We believe that it is very important to avoid false calling of CNVs, as validation of these will be time-consuming and expensive. It is therefore better to generate highly reproducible calls with a low false-positive rate than to call higher numbers of CNVs with accompanying increased absolute numbers of false positives. Overall, we consider that CNVfinder performs better in this respect than SW-ARRAY.
Conclusions
The identification of CNVs from large-insert clone arrays requires a well-validated clone set and an objective and accurate method for detecting significant copy number changes. Our whole-genome tile path clone set has not only been validated by fingerprinting and end sequencing, but also by the use of control “add-in” hybridizations that allow direct estimation of clone hybridization characteristics. The quality of hybridizations to these arrays varies, so it is important that the method used to detect CNVs is robust to differences in measurement variation, particularly with regard to false-positive calls. Our CNVfinder algorithm was trained using a series of replicate hybridizations of varying quality and using independently verified CNVs to maximize the number of calls while keeping false positives to <5%. Importantly, CNVfinder made more consistent calls across arrays with different ratio variance than SWarray. We found that using CNVfinder, experiments with higher SDe tended to produce an increased number of false negatives, but without an increase in false positives.
In conclusion, CNVfinder is a new tool for accurate and reliable high-throughput detection of copy number variation in the human genome. We have used CNVfinder to detect CNV in human populations using DNAs from the 270 cell lines extensively genotyped in the HapMap project. CNVfinder should find equal utility in studies of constitutional chromosomal imbalances associated with human syndromes.
Methods
Clone selection and verification
A total of 26,678 large insert clones was selected primarily from the published “Golden Path” to cover the human genome in tiling path resolution. As only the first two segments of the RPCI-11 BAC library were available for clone selection, “Golden Path” clones that were not available were replaced with equivalent clones identified by corresponding DNA fingerprints or end-sequence matches. These clones (5344 clones in total) were then picked from RPCI-1, RPCI-3, RPCI-4, RPCI-5, RPCI-6, RPCI-13 (http://bacpac.chori.org/) (Kent et al. 2002) libraries, the CalTech BAC libraries (http://informa.bio.caltech.edu/Bac_info.htlm), and the Lawrence Livermore National Laboratory libraries (http://www.llnl.gov/library/) held at the Wellcome Trust Sanger Institute. For chromosomes 6 and 22, BAC, fosmid, and cosmid clones were privately supplied by various institutes to fill in gaps. Clones were screened for T1 phage and Pseudomonas contamination and verified by fingerprinting (Marra et al. 1997; Soderlund et al. 2000) and end sequencing (Adams et al. 2005). Clone information can be obtained from http://www.ensembl.org/Homo_sapiens/cytoview.
Clone positions were mapped onto the reference sequence (Build NCBI35) using known accessions and end sequences. End sequencing verified the mapping position of 20,967 clones. 5611 clones failed to provide end sequence that could be mapped onto the reference sequence, but were taken forward for array construction and further validation. The final validated set consists of 26,574 clones. The clone set can be obtained from BACPAC resources (http://bacpac.chori.org/).
Preparation of clones for spotting
Large insert clone DNA was isolated as described previously (Marra et al. 1997; Humphray et al. 2001) and diluted to a final concentration of 1 ng/μL. For array construction, clone DNA was amplified in three separate DOP-PCR reactions using primers DOP1, DOP2, and DOP3 as described (Fiegler et al. 2003). After combining the appropriate DOP-PCR-amplified products, a secondary PCR reaction using a 5′-amine modified primer designed to match the 10 bases at the 5′-end of each DOP-PCR primer was performed. Twenty-nine microliters of 4× microarray spotting buffer (1 M sodium phosphate buffer, pH 8.5, 0.001% sarkosyl) were then added to 88 μL of PCR products and filtered by centrifugation at 2000 rpm for 10 min through 0.22 μm Millipore Multiscreen-GV filter plates (Millipore).
Array spotting
Arrays were printed in a HEPA-filtered and humidity-controlled environment (40%–45% RH) onto CodeLink activated slides (GE Healthcare UK Limited) using MicroGrid 610 robots equipped with tungsten 10K split pins (Genomic Solutions) and were stored desiccated at room temperature until use.
Genomic DNAs for hybridization
Genomic DNA derived from monochromosomal hybrid cell lines (hybrid mapping panel #2; http://locus.umdnj.edu/nigms/maps/map02.html) and DNA of the parental Chinese hamster (RJK88; NA10658) and mouse strains (3T6; NA05862) and genomic DNA of cell lines NA 15,510; NA 10,851; NA 12,144; NA 12,239; NA 12,892; NA 18,500; NA 18,576; NA 18,621; NA 18,860; NA 18,971; NA 18,980; NA 19,099 was obtained from the DNA Polymorphism Discovery Resource Collection (Coriell Cell Repositories; http://ccr.coriell.org).
The primary renal cell adenocarcinoma line (769P, ATCC No. CRL-1933) was obtained from the American Tissue Culture Collection (Manassas, VA).
Array hybridization
Test and reference DNA samples were differentially labeled using the Bioprime labeling kit (Invitrogen) with modifications of the nucleotide mix. Briefly, a 260-μL reaction was set up containing 300 ng of DNA and 120 μL of 2.5× random primer solution. After denaturing the DNA for 10 min at 100°C, 30 μL of 10× dNTP mix (1 mM dCTP, 2 mM dATP, 2 mM dGTP, and 2 mM dTTP in TE buffer), 3 μL of 1 mM Cy5-dCTP or Cy3-dCTP (NEN Life Science Products), and 6 μL of Klenow fragment were added on ice to a final reaction volume of 300 μL. The reaction was incubated overnight at 37°C and stopped by adding 30 μL of stop buffer supplied in the kit. Unincorporated nucleotides were removed by use of Microcon YM-30 Filter Devices (Millipore), according to the suppliers’ instructions.
Hybridizations were carried out on a Tecan HS Hybridization Station (Tecan Group Ltd.) using 63 × 20-mm chambers. Cy3- and Cy5-labeled DNAs were combined, precipitated with 270 μg of human Cot1 DNA (Roche Diagnostics Ltd.), and resuspended in 165 μL of hybridization buffer (50% formamide, 5% dextran sulfate, 0.1% Tween 20, 2× SSCl, 10 mM Tris/HCl, pH 7.4, 10 mM Cysteamine). Pre-hybridization solution was prepared simultaneously by precipitating 100 μL of herring sperm DNA (10 mg/mL, Sigma Aldrich) and resuspending in 165 μL of hybridization buffer.
The prehybridization and hybridization solutions were then denatured for 10 min at 72°C. The prehybridization solution was injected into the Tecan chamber following instructions displayed on the station. During prehybridization (45 min at 37°C), the hybridization solution was incubated at 37°C. Hybridization was carried out for 21 h at 37°C with medium agitation frequency. Slides were washed with PBS/Tween 20/2mM cysteamine (wash time 0.5 min, soak time 0.5 min, 15 cycles at 37°C), 0.1× SSC (wash time 1.0 min, soak time 2.0 min, 5 cycles at 54°C), PBS/Tween 20/2mM cysteamine (wash time 0.5 min, soak time 0.5 min, 10 cycles at 23°C), and HPLC water (wash time 0.5, soak time 0.0, 1 cycle at 23°C) before drying for 2.5 min using nitrogen gas. All experiments were performed in duplicate with DNA labeling color reversal (dye swap).
Chromosome-specific add-in experiments
Chromosome-specific add-in experiments were performed for every chromosome as described previously with slight modifications (Fiegler et al. 2003; Rickman et al. 2006). Briefly, DNA derived from either the monochromosomal hybrid cell lines or the parental rodent strains was spiked prior to labeling into anonymous male blood DNA (test) for hybridization against the same DNA (reference). Dye-swap hybridizations were performed with the equivalent of one and two extra copies for each chromosome. Following calculation of combined dye-swap ratios, species-specific background from the parental cell line DNA was reduced by subtracting the dye-swap ratios of parental control hybridizations. The slope of the response curve to the additional copies of each chromosome was then calculated for all clones (see Fig. 1C). Thresholds to define the type of clone response were determined using multiples of the standard deviation of all slopes, excluding clones corresponding to the spiked chromosome. Clones assigned to the spiked chromosome with slopes below a threshold of five times the standard deviation were defined as non-responders. Clones not assigned to the spiked chromosome with slopes greater than 10 times the standard deviation were defined as responders. Clones not assigned to the spiked chromosome with slopes between the thresholds of five and 10 times the standard deviation were defined as cross-responding clones. These results were combined with end-sequence and fingerprint information to establish the final mapping information for each clone. For a more detailed description see Supplemental File 1.
Raw data analysis: Genome profiling
Array images were acquired using an Agilent laser scanner (Agilent Technologies). Fluorescence intensities and log2 ratio values were extracted using Bluefuse software (Bluegnome Ltd). Spots with low signal intensities (“amplitude” < 100 in both channels) or inconsistent fluorescence patterns (“confidence” < 0.5 or “quality” = 0) were excluded before normalizing all log2 ratio values by blocks (sub-arrays).
Fusion of dye-swap results and subsequent analyses were performed using custom Perl scripts. For each individual hybridization, the median of all ratio values was calculated chromosome by chromosome. Each ratio was then normalized by the corresponding chromosomal median. The ratios of each clone in the two dye-swap hybridizations were then averaged if replicate ratios differed by <50% (i.e., less than a difference of 0.585 on the log2 scale).
The 68.2th percentile of the absolute values for all combined ratios was then calculated chromosome by chromosome as an estimation of the standard deviation (SDe). Clones reporting replicates different by more than eight times the SDe were excluded from further analysis.
Dye-swap experiments were accepted for CNV calling only if the following criteria were fulfilled: (1) Global SDe < 0.06; (2) Global clone exclusion rate < 10%; (3) Clone exclusion rate per individual chromosome < 20%. Clones contained in chromosomes with a corresponding chromosomal median of >0.1 or < −0.1 were flagged and excluded from CNV calling. Thus, chromosomes X and Y in female versus male results, as well as chromosomal artifacts (aneusomies or very large imbalances) were automatically excluded from calling.
Array images, raw intensities, and normalized log2 ratios can be downloaded from http://www.sanger.ac.uk/humgen/cnv/data/. All validation results are also available through ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) by the accession numbers E-TABM-123 (self–self and validation replicate experiments) and E-TABM-124 (add-in experiments).
CNV-calling algorithm
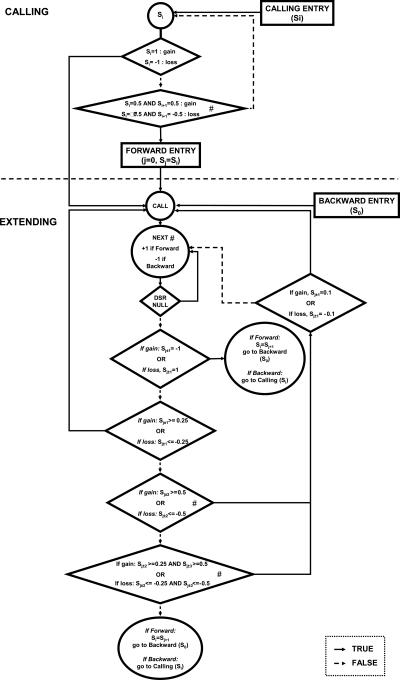
Initially, one score (Si) was assigned to each individual clone, reflecting the deviation of its combined ratio (cR) from the central distribution:
- If cR ≥ 6× SDe or cR ≤ −6× SDe, then Si = 1 or Si = −1, respectively.
- If cR ≥ 4× SDe or cR ≤ −4× SDe, then Si = 0.5 or Si = −0.5, respectively.
- If cR ≥ 3× SDe or cR ≤ −3× SDe, then Si = 0.25 or Si = −0.25, respectively.
- If cR ≥ 1× SDe or cR ≤ −1× SDe, then Si = 0.1 or Si = −0.1, respectively.
All subsequent analysis steps were carried out for each chromosome independently, as described in Figure 5.
Figure 5.
The CNVfinder algorithm. (#) In case of a gap in genome coverage, two clones are considered as consecutive only if their closest extremities are separated by <1.3 Mb of sequence.
Quantitative PCR
Quantitative PCR was performed using Quantitative Multiplex PCR of Short Fluorescent Fragments (QMPSF), as previously described (Charbonnier et al. 2000), and the SYBR Green method. For QMPSF, PCR products were designed to be 120–210 bp in length. One primer in each primer pair was labeled with a FAM moiety in the 5′-end, while a stabilizing GTTTCTT tail was added to the 5′-end of the other primer. A region in the CFTR gene was used as an internal control, with forward primer 5′-GGGCC TGTGCAAGGAAGTGTTA-3′ and reverse primer 5′-gtttcttAGTC ACCAAAGCAGTACAGC-3′. The amplified samples were run on an ABI3730×L genetic analyzer (Applied Biosystems) using the POP7 polymer. Results were analyzed using the software GeneMapper v3.5. One to three regions were amplified for each candidate CNV from DNAs of both NA15510 and the reference cell line NA10851. All assays were run in triplicate. The ratio between the target sequence and the internal control was compared between the samples, using Student’s _t_-test to determine significance. Results with P < 0.05 were considered to validate a copy number difference between the two samples. In cases where more than one assay was designed within a putative CNV region, one significant result was considered sufficient to confirm the presence of a CNV.
For the SYBR Green method, PCR products were designed to be 100–150 bp in length. A fragment from the TP53 gene was used as an internal control, with forward primer 5′-CCCTTCC CAGAAAACCTACC-3′ and reverse primer 5′-CAGGCATTGA AGTCTCATGG-3′. Samples were run in 25-μL reactions using iQ SYBR Green Supermix (Bio-Rad) in a 96-well plate on a Bio-Rad iCycler Thermal Cycler with initial denaturation for 2 min at 93°C, followed by 40 cycles of 15 sec at 93°C and 30 sec at 60°C. For each test sample (NA15510) replicate, 2 ng of genomic DNA was used. Standard curves were created by using dilutions of genomic DNA from the reference individual (NA10851). Test samples were run in triplicate for both the test fragment and the TP53 internal control fragment, and standards were run in duplicate. The final standard deviation was calculated from the standard deviations for the test fragment and the TP53 internal control fragment according to the manufacturer’s instructions, and results were considered sufficient to confirm the presence of a CNV when the NA15510 relative copy number (to NA10851) was significantly different than 1 and agreed on the direction of change with the array-CGH results, based on the 95% confidence interval (±2 standard deviations).
Primer sequences and quantitative PCR results are detailed in Redon et al. 2006.
Acknowledgments
We thank A.V. Cox, J.W. Stalker, and J. Smith for making the whole-genome tile-path clone set available via Ensembl, Shotgun Sequencing Team 42 of the Wellcome Trust Sanger Institute for end sequencing, Fengtang Yang and the FISH core group for additional clone verification, and Chris Barnes for his help in statistical analysis. This work was funded by the Wellcome Trust. R.R. was supported by a Sanger Institute Postdoctoral Fellowship.
Footnotes
References
- Adams D.J., Quail M.A., Cox T., van der Weyden L., Gorick B.D., Su Q., Chan W.I., Davies R., Bonfield J.K., Law F., Quail M.A., Cox T., van der Weyden L., Gorick B.D., Su Q., Chan W.I., Davies R., Bonfield J.K., Law F., Cox T., van der Weyden L., Gorick B.D., Su Q., Chan W.I., Davies R., Bonfield J.K., Law F., van der Weyden L., Gorick B.D., Su Q., Chan W.I., Davies R., Bonfield J.K., Law F., Gorick B.D., Su Q., Chan W.I., Davies R., Bonfield J.K., Law F., Su Q., Chan W.I., Davies R., Bonfield J.K., Law F., Chan W.I., Davies R., Bonfield J.K., Law F., Davies R., Bonfield J.K., Law F., Bonfield J.K., Law F., Law F., et al. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics. 2005;86:753–758. doi: 10.1016/j.ygeno.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Charbonnier F., Raux G., Wang Q., Drouot N., Cordier F., Limacher J.M., Saurin J.C., Puisieux A., Olschwang S., Frebourg T., Raux G., Wang Q., Drouot N., Cordier F., Limacher J.M., Saurin J.C., Puisieux A., Olschwang S., Frebourg T., Wang Q., Drouot N., Cordier F., Limacher J.M., Saurin J.C., Puisieux A., Olschwang S., Frebourg T., Drouot N., Cordier F., Limacher J.M., Saurin J.C., Puisieux A., Olschwang S., Frebourg T., Cordier F., Limacher J.M., Saurin J.C., Puisieux A., Olschwang S., Frebourg T., Limacher J.M., Saurin J.C., Puisieux A., Olschwang S., Frebourg T., Saurin J.C., Puisieux A., Olschwang S., Frebourg T., Puisieux A., Olschwang S., Frebourg T., Olschwang S., Frebourg T., Frebourg T. Detection of exon deletions and duplications of the mismatch repair genes in hereditary nonpolyposis colorectal cancer families using multiplex polymerase chain reaction of short fluorescent fragments. Cancer Res. 2000;60:2760–2763. [PubMed] [Google Scholar]
- Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K., Carter N.P., Hurles M.E., Pritchard J.K., Hurles M.E., Pritchard J.K., Pritchard J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Fiegler H., Carr P., Douglas E.J., Burford D.C., Hunt S., Scott C.E., Smith J., Vetrie D., Gorman P., Tomlinson I.P., Carr P., Douglas E.J., Burford D.C., Hunt S., Scott C.E., Smith J., Vetrie D., Gorman P., Tomlinson I.P., Douglas E.J., Burford D.C., Hunt S., Scott C.E., Smith J., Vetrie D., Gorman P., Tomlinson I.P., Burford D.C., Hunt S., Scott C.E., Smith J., Vetrie D., Gorman P., Tomlinson I.P., Hunt S., Scott C.E., Smith J., Vetrie D., Gorman P., Tomlinson I.P., Scott C.E., Smith J., Vetrie D., Gorman P., Tomlinson I.P., Smith J., Vetrie D., Gorman P., Tomlinson I.P., Vetrie D., Gorman P., Tomlinson I.P., Gorman P., Tomlinson I.P., Tomlinson I.P., et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- Humphray S.J., Knaggs S.J., Ragoussis I., Knaggs S.J., Ragoussis I., Ragoussis I. Contiguation of bacterial clones. Methods Mol. Biol. 2001;175:69–108. doi: 10.1385/1-59259-235-X:069. [DOI] [PubMed] [Google Scholar]
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Qi Y., Scherer S.W., Lee C., Scherer S.W., Lee C., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Ishkanian A.S., Malloff C.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Malloff C.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Albertson D.G., Pinkel D., Marra M.A., Pinkel D., Marra M.A., Marra M.A., et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Marra M.A., Kucaba T.A., Dietrich N.L., Green E.D., Brownstein B., Wilson R.K., McDonald K.M., Hillier L.W., McPherson J.D., Waterston R.H., Kucaba T.A., Dietrich N.L., Green E.D., Brownstein B., Wilson R.K., McDonald K.M., Hillier L.W., McPherson J.D., Waterston R.H., Dietrich N.L., Green E.D., Brownstein B., Wilson R.K., McDonald K.M., Hillier L.W., McPherson J.D., Waterston R.H., Green E.D., Brownstein B., Wilson R.K., McDonald K.M., Hillier L.W., McPherson J.D., Waterston R.H., Brownstein B., Wilson R.K., McDonald K.M., Hillier L.W., McPherson J.D., Waterston R.H., Wilson R.K., McDonald K.M., Hillier L.W., McPherson J.D., Waterston R.H., McDonald K.M., Hillier L.W., McPherson J.D., Waterston R.H., Hillier L.W., McPherson J.D., Waterston R.H., McPherson J.D., Waterston R.H., Waterston R.H. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman T.L., Tuzun E., Morrison V.A., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Tuzun E., Morrison V.A., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Morrison V.A., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., McGrath S.D., Rocchi M., Eichler E.E., Rocchi M., Eichler E.E., Eichler E.E. A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 2005;15:1344–1356. doi: 10.1101/gr.4338005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen A.B., Venkatraman E.S., Lucito R., Wigler M., Venkatraman E.S., Lucito R., Wigler M., Lucito R., Wigler M., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Perry G.H., Tchinda J., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Tchinda J., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Tyler-Smith C., Scherer S.W., Eichler E.E., Scherer S.W., Eichler E.E., Eichler E.E., et al. Hotspots for copy number variation in chimpanzees and humans. Proc. Natl. Acad. Sci. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price T.S., Regan R., Mott R., Hedman A., Honey B., Daniels R.J., Smith L., Greenfield A., Tiganescu A., Buckle V., Regan R., Mott R., Hedman A., Honey B., Daniels R.J., Smith L., Greenfield A., Tiganescu A., Buckle V., Mott R., Hedman A., Honey B., Daniels R.J., Smith L., Greenfield A., Tiganescu A., Buckle V., Hedman A., Honey B., Daniels R.J., Smith L., Greenfield A., Tiganescu A., Buckle V., Honey B., Daniels R.J., Smith L., Greenfield A., Tiganescu A., Buckle V., Daniels R.J., Smith L., Greenfield A., Tiganescu A., Buckle V., Smith L., Greenfield A., Tiganescu A., Buckle V., Greenfield A., Tiganescu A., Buckle V., Tiganescu A., Buckle V., Buckle V., et al. SW-ARRAY: A dynamic programming solution for the identification of copy-number changes in genomic DNA using array comparative genome hybridization data. Nucleic Acids Res. 2005;33:3455–3464. doi: 10.1093/nar/gki643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Chen W., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Chen W., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Chen W., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Chen W., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Chen W., Andrews T.D., Fiegler H., Shapero M.H., Chen W., Fiegler H., Shapero M.H., Chen W., Shapero M.H., Chen W., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006 doi: 10.1038/nature05329. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman L., Fiegler H., Shaw-Smith C., Nash R., Cirigliano V., Voglino G., Ng B.L., Scott C., Whittaker J., Adinolfi M., Fiegler H., Shaw-Smith C., Nash R., Cirigliano V., Voglino G., Ng B.L., Scott C., Whittaker J., Adinolfi M., Shaw-Smith C., Nash R., Cirigliano V., Voglino G., Ng B.L., Scott C., Whittaker J., Adinolfi M., Nash R., Cirigliano V., Voglino G., Ng B.L., Scott C., Whittaker J., Adinolfi M., Cirigliano V., Voglino G., Ng B.L., Scott C., Whittaker J., Adinolfi M., Voglino G., Ng B.L., Scott C., Whittaker J., Adinolfi M., Ng B.L., Scott C., Whittaker J., Adinolfi M., Scott C., Whittaker J., Adinolfi M., Whittaker J., Adinolfi M., Adinolfi M., et al. Prenatal detection of unbalanced chromosomal rearrangements by array CGH. J. Med. Genet. 2006;43:353–361. doi: 10.1136/jmg.2005.037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lundin P., Maner S., Massa H., Walker M., Chi M., Maner S., Massa H., Walker M., Chi M., Massa H., Walker M., Chi M., Walker M., Chi M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Clark R.A., Schwartz S., Segraves R., Schwartz S., Segraves R., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Waterman M.S., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Soderlund C., Humphray S., Dunham A., French L., Humphray S., Dunham A., French L., Dunham A., French L., French L. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 2000;10:1772–1787. doi: 10.1101/gr.gr-1375r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Haugen E., Hayden H., Albertson D., Pinkel D., Hayden H., Albertson D., Pinkel D., Albertson D., Pinkel D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Ylstra B., van den Ijssel P., Carvalho B., Brakenhoff R.H., Meijer G.A., van den Ijssel P., Carvalho B., Brakenhoff R.H., Meijer G.A., Carvalho B., Brakenhoff R.H., Meijer G.A., Brakenhoff R.H., Meijer G.A., Meijer G.A. BAC to the future! Or oligonucleotides: A perspective for micro array comparative genomic hybridization (array CGH) Nucleic Acids Res. 2006;34:445–450. doi: 10.1093/nar/gkj456. [DOI] [PMC free article] [PubMed] [Google Scholar]