Pseudogene.org: a comprehensive database and comparison platform for pseudogene annotation (original) (raw)
Abstract
The Pseudogene.org knowledgebase serves as a comprehensive repository for pseudogene annotation. The definition of a pseudogene varies within the literature, resulting in significantly different approaches to the problem of identification. Consequently, it is difficult to maintain a consistent collection of pseudogenes in detail necessary for their effective use. Our database is designed to address this issue. It integrates a variety of heterogeneous resources and supports a subset structure that highlights specific groups of pseudogenes that are of interest to the research community. Tools are provided for the comparison of sets and the creation of layered set unions, enabling researchers to derive a current ‘consensus’ set of pseudogenes. Additional features include versatile search, the capacity for robust interaction with other databases, the ability to reconstruct older versions of the database (accounting for changing genome builds) and an underlying object-oriented interface designed for researchers with a minimal knowledge of programming. At the present time, the database contains more than 100 000 pseudogenes spanning 64 prokaryote and 11 eukaryote genomes, including a collection of human annotations compiled from 16 sources.
INTRODUCTION
Pseudogenes, defined as non-functional copies of gene fragments incorporated into the genome by either retro-transposition of mRNA or duplication of genomic DNA, are found throughout the genomes of most eukaryotic organisms. Pseudogenes both help and hinder studies of genomic structure: they serve as a historical record, providing insight into the evolutionary history and past structure of individual genes and the genome as a whole. They also confuse and disrupt computational gene finding tools and can contribute to cross-hybridization artifacts in microarray experiments. Whether a researcher wishes to analyze or filter pseudogenes, there is a clear need for tools that allow quick identification of these sequences. Hence, it is important that pseudogene information be available and easily accessible.
There are a variety of online annotation databases available to the research community, each with its own focus. NCBI GenBank contains general information for numerous species (1), whereas UniProt has a tighter focus on protein annotations (2). Similarly, Ensembl details annotations for genes and their corresponding protein features, along with a limited amount of pseudogene annotation (3). The UCSC Genome Browser focuses on a wide range of nucleotide-level genomic information and is useful for comparing diverse sets of annotations from different sources (4). All of these databases contain pseudogene information, but lack any comprehensive collection of pseudogene annotation data. The Hoppsigen database provides more detailed annotations of processed pseudogenes, serving as a repository for the results of their specific pseudogene identification method as applied to the human and mouse genomes (5), and the University of Iowa presents their own set in a local online database (see http://genome.uiowa.edu/pseudogenes/).
Pseudogene.org is a searchable repository for all protein-coding derived pseudogenes identified in the literature, merging results originating from a variety of identification tools and other studies. However, in attempting to collect pseudogenes from such a wide range of sources we face challenges beyond those of tracking disparate genomic information. These difficulties arise because there is no consensus definition of a pseudogene. If we were instead investigating coding sequences, any segment predicted as such could be subjected to experimental verification. In the investigation of pseudogenes this is impossible; a computational tool might annotate a given segment as a pseudogene, but the prediction cannot be experimentally verified. Hence, there is no way to systematically validate the results of a given pseudogene identification tool or to resolve all the differences between two such tools. Different algorithms will produce different results, and to make use of these predictions researchers must have some means of tracking, merging and saving them.
The various pseudogene identification tools discussed in the literature are based on different computational approaches. Some methods rely only on homology searches and identification of sequence irregularities (e.g. finding a frame-shift or nonsense mutations) (5–12), while others use information such as the relative quantities of synonymous and non-synonymous coding mutations (dN/dS or Ka/Ks) (11,13). As any of these is of potential use to the research community, a database focusing on pseudogene information should integrate results from all these sources. Further complicating matters is the heterogeneity of the results; each identification method is associated with specific parameters and annotations that are unique to the method, and must be retained if researchers are to make effective use of the information. Thus, any pseudogene database must have the flexibility to store a variety of information in an efficient and accessible manner.
In the Pseudogene.org knowledgebase, we provide a publicly accessible online pseudogene repository that efficiently and transparently deals with the problems we have discussed. More specifically, the database has the following features:
- Integration of different identification methodologies: The database is designed to consolidate information from a variety of sources into a single database while retaining all necessary method-specific details.
- Flexible search capacity: The database interface provides efficient, flexible search capabilities that hide the heterogeneous nature of the data.
- Pre-computed search sets: The database allows the user to perform restricted searches on pre-defined sets of interest in the literature.
- Robust interaction with other databases: The database easily integrates supporting information from other databases.
- Temporal reconstructability: The database can be reconstructed as it existed at any point in time.
- Simplified accessibility: The database can be accessed through a simple Perl interface library (with supporting code available on the website).
This paper describes the database and some related challenges. In Section 2, we present an overview of the database contents and discuss the tools that are available to database users. In Section 3, we briefly describe the technical issues addressed in creating the database; details are provided in Supplementary Data.
Database contents and analysis tools
In Table 1 we present a breakdown of the pseudogene contents by organism; more organisms will be added as adequately sequenced genomes become available [i.e. 4x shotgun; for details see Zhang _et al_. (10)]. Results for human and mouse are complied from the works of Torrents et al. (13), Khelifi et al. (14), Zhang and Gerstein (8), Collins et al. (15), the UCSC browser (4) and other compilations (11,16–22), as well as new sequences arising from the application of PseudoPipe (6) and from manual annotations. For chimp, rat, dog, chicken, tetraodon, zebrafish, fly, mosquito and Plasmodium falciparum, the content results from the application the PseudoPipe tool to those organisms—the first detailed analysis of pseudogenes for any of those organisms.
Table 1.
Contents of the pseudogene database at time of submission (June 2006)
| Genome | Number of pseudogenes |
|---|---|
| Eukaryotes | |
| Homo sapiens (human) | 31 768 |
| Pan troglodytes (chimp) | 8355 |
| Mus musculus (mouse) | 15 320 |
| Rattus norvegicus (rat) | 10 750 |
| Canis familiaris (dog) | 2802 |
| Gallus gallus (chicken) | 4179 |
| Danio rerio (zebrafish) | 15 779 |
| Anopheles gambiae (mosquito) | 1713 |
| Drosophila melanogaster (fly) | 484 |
| Plasmodium falciparum | 5179 |
| Tetraodon nigroviridis | 3250 |
| Eukaryote total | 99 579 |
| Prokaryotes (sample) | |
| Thermotoga maritima | 37 |
| Borrelia burgdorferi | 10 |
| Pseudomonas aeruginosa | 187 |
| Escherichia coli K12 | 134 |
| Buchnera sp. APS | 18 |
| Bacillus subtilis | 203 |
| Chlamydia trachomatis | 11 |
| Thermoplasma acidophilum | 39 |
| Methanothermobacter thermautotrophicus | 35 |
| Sulfolobus solfataricus | 172 |
| Prokaryote total (including 54 genomes not shown) | 6890 |
| Database total | 106 469 |
Pseudogene classification
Each pseudogene in the database is classified into one of four categories:
- Processed: Segments clearly retro-transposed into the genome from mRNA. Pseudogenes are identified as processed if they reflect specific characteristics (e.g. lack of introns), as discussed in Harrison et al. (11). Note that such signals will degrade over time, preventing the identification of older pseudogenes in this category.
- Non-processed: Non-processed pseudogenes can be subdivided into two categories:
- Duplicated: Pseudogenes clearly created by the duplication of a genome segment containing a given gene, followed by the inactivation of one copy. They are often identified by the presence of an intron/exon structure, as well as features such as proximity to the parent gene.
- Other: Pseudogenes that are clearly not retro-genes (hence not processed), but were also not the result of a duplication event. Unitary pseudogenes, pseudogenes resulting from the decay of a previously functional gene, are a prime example. The caspase 12 pseudogene is one example (accession no. 76 507 in our database); other examples of unitary pseudogenes can be found in Wang et al. (23).
As before, many of these signals will degrade over time, preventing the assignment of older pseudogenes to this class.
- Unclassified: Pseudogenes that cannot be classified, either because of signal degradation (as would be the case with many ancient pseudogenes) or because of an inherent ambiguity in the structure (e.g. a pseudogene spawning from a single exon gene).
QUERY CAPABILITIES
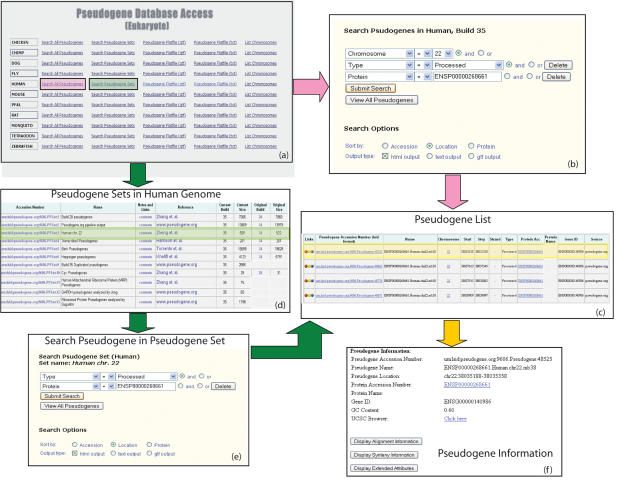
Researchers can interact with the Pseudogene.org database in several ways. They can download the entire content of the database in a variety of formats, but many users will be interested in only a small subset of the existing pseudogenes. To this end, we have provided web-based search capabilities and pre-computed annotated sets. Through this users may perform Boolean searches over a number of characteristics (e.g. location, associated protein or identifying source). In Figure 1, we illustrate a potential search, in which the user wishes to find all processed pseudogenes on chromosome 22 that correspond to the protein with Ensembl accession no. ENSP00000268661. By choosing the ‘search all pseudogenes’ link in the human row of the page displayed in Figure 1a, the user will reach the search page displayed in Figure 1b. In that picture, we see the specification of the three terms defining the search; then clicking the submit search button leads to the result list display in Figure 1c. Individual pseudogenes may be clicked to examine details, as shown in Figure 1f.
Figure 1.
A diagram of the Pseudogene.org search page (Eukaryote section), illustrating two ways a user might search for all processed pseudogenes on chromosome 22 that were created by the protein with Ensembl accession number ENSP00000268661. In (a) the user could choose to search all human pseudogenes, resulting in the search page shown (b), which can then be configured as shown. Or the user could look at all pre-computed sets as shown in (d), choose the set corresponding to the Zheng et al. analysis of chromosome 22 and resulting in the search page shown in (e). In this case both methods will result in the same list, as shown in (c), and by choosing an individual pseudogene the user will see the specific details as shown in (f).
PRE-COMPUTED SETS
It is often the case that a user may want to restrict a search to a specially annotated set of pseudogenes—one that cannot be characterized by any set of recorded attributes. Examples of such sets include the set of putatively transcribed pseudogenes (24), the set of known cytochrome c pseudogenes (20) and the set of mitochondrial ribosomal protein pseudogenes (22). Researchers investigating such collections frequently want to limit their search by excluding pseudogenes in the database outside of the target set. By the nature of a manual analysis this cannot be done within the framework of a general database search.
To this end the database provides a way of defining, annotating and managing a number of closed sets corresponding to annotations of interest to the research community. The collection of these sets can be searched by set name or recorded comments, and the user can perform searches over these sets as well as within the database as a whole. In Figure 1d, the user is conducting previously described search by considering only the set of pseudogenes list in the Zheng et al. analysis of chromosome 22 (25). By choosing that set the user researches the search page displayed in Figure 1e, and can then specify the search criteria to reach the result in the list shown in Figure 1c as before.
LAYERED SETS
When dealing with several disparate sets of pseudogenes, a research will frequently find it useful to construct the union of those sets. For example, a researcher who needs to consider all pseudogenes identified by any of several different identification algorithms would want to merge these results by computing the union of the result sets. This problem is complicated by the nature of pseudogene data: given the variability of the definition of pseudogenes, it is common to find that the different identification tools have identified the ‘same’ pseudogene in different ways. In such cases, there is a core region shared by the putative pseudogenes that differ in characteristics such as endpoints or exon structure. When computing the union of sets it is unclear how to resolve such conflicts; including all versions of the pseudogene is redundant, but there is no clear way to pick only one of the variants.
Pseudogene.org address this problem by allowing the computation of layered sets. A layered set is computed by considering a user-specified prioritizing of the sets. They are constructed using the set union operator, but conflicts are resolved by choosing the pseudogene from the set of highest priority. The primary motivation for this tool is in the construction of a customized ‘canonical’ set of pseudogenes. That is, it allows the user to create a ‘full set’ of pseudogenes based on their own estimation of the quality of different outputs, thus ensuring that pseudogenes from a particular method will be prioritized over other methods the user believes to be less reliable.
SET COMPARISONS
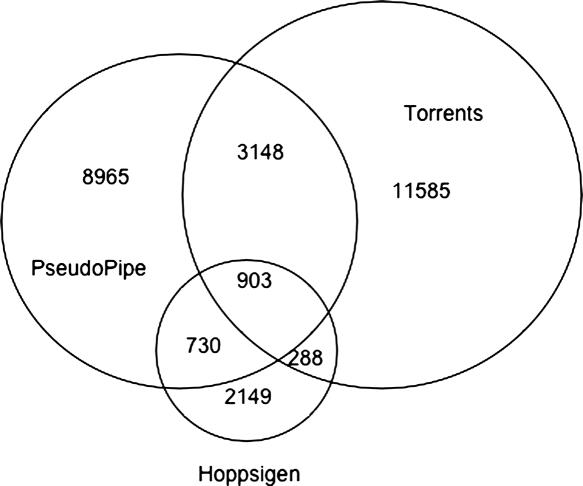
As we claim that the Pseudogene database is necessary due to a significant disparity between different pseudogene sets, we include Figure 2 to illustrate the extent of this disparity. In the figure we have selected three large sets of pseudogenes: those identified by the PseudoPipe tool (6), those identified by the method of Torrents et al. (13) and those identified by the method associated with the Hoppsigen database (14). We consider pseudogenes from two different sets as equivalent if one sequence covers at least 90% of the other; reducing the required overlap makes no appreciable difference in the diagram. A significant fraction of pseudogenes predicted by any one of the search methods are not found by the other methods, reflecting the lack of a uniform definition of a pseudogene.
Figure 2.
Venn diagrams representing the intersections between the sets corresponding to PseudoPipe pipeline, the Torrents identification method and the Hoppsigen method. (Not drawn to scale.) We define two pseudogenes as equivalent if there exists more than a 90% overlap between them.
If a consensus definition of pseudogenes existed, we would expect automated search methodologies to identify the same core set of elements; smaller differences would occur due to varying computational techniques and parameters. From Figure 2 we can see that this does not happen. For each set, be it automated or manually curated, we find the majority of identified elements to be unique to that set. Nor is there any reason to accept the results of one set over the others. The loose definition of the problem does not allow for any definitive quantitative ranking, and the nature of pseudogenes forestalls the possibility of experimental verification. These results highlight the problems arising from the lack of a definitive pseudogene definition and underscore both the need for a composite database and the need for such a database to provide the searchable sets structure.
DATABASE STRUCTURE, INTERFACE AND MAINTENANCE
The database was designed using an object-oriented approach, with information stored in an MySQL database. We developed an interface for the Perl code to make the structure accessible to users unfamiliar with the SQL language and to provide a mapping of conceptual objects onto the relational database. A detailed discussion of the database structure and implementation is beyond the scope of this paper, though more details are presented in Supplementary Data. However, certain aspects are worth reviewing. Specifically, we review the pseudogene class (the central focus of the database) and discuss the problems of synchronization and versioning.
PSEUDOGENE CLASS
A pseudogene is a collection of (genome) fragments; processed pseudogenes are composed of a single fragment, while duplicate pseudogenes are composed of one or more fragments. A description of a pseudogene is a list of its fragments and the values of certain ‘data attributes’. The latter includes important aspects of a pseudogene that cannot be efficiently calculated on the fly, such as the parent protein and the relevant fragment/protein alignments. Other core data attributes include chromosomal location information, associated gene information, GC-content, pseudogene type, identifying source and information on the protein alignment.
Given the heterogeneous nature of pseudogene information, it is frequently necessary to record data specific to the identification method used to find a given pseudogene. In order to improve storage efficiency, such information is recorded using the entity attribute value (EAV) database technique (26). Such elements include the Ka/Ks ratio, CpG content, distance from query protein, proximity to CpG islands and relevant PCR tiling microarray results.
SYNCHRONIZATION AND IDENTIFICATION
The database is intended to record and present pseudogene information. In order to fully present the details of each pseudogene we rely on other established databases for supporting information: the UCSC Genome Browser (4) for genome sequence information, Ensembl (27) for gene annotations and UniProt (2) for protein information. However, these databases are undergoing constant changes, and modifications must be incorporated carefully if we are to achieve our goal of temporal reconstructability. Updated information must be regularly downloaded and inserted into our database, but existing objects in our database cannot be modified if we are to retain the ability to reconstruct older versions.
The problem is solved with a versioning system that allows us to add a new version of a pseudogene that reflects updated data (as opposed to modifying the existing version), and maintaining a relation between the unmodified object and its replacement. The scheme works on the basis of a identifier system composed of an accession number/version id pair; accession numbers specify a set of versions, are distinguished by version numbers and provide the necessary association between versions of the same pseudogene. Accession identifiers are based on the LSID naming convention (28), a system designed with the intent of creating a unified naming convention usable by any database.
Build remapping
Integrating new genome builds is particularly difficult. Updating the database to conform to the new build requires the modification of significant portions of the data; tasks such as updating coordinates, recomputing alignments and determining the effect on set content must be performed. The UCSC liftOver tool is used for automatically recomputing coordinates (4), and the rest of the tasks can be automated as well. The result is an automated system for updating the contents to conform to the new build, allowing researchers still working with previous builds to easily map the new data back to the older versions as needed.
INTERFACE SOFTWARE
This database is intended to be accessible to users with no knowledge of MySQL and a limited knowledge of programming; it was designed with the idea that a user could maintain their own version of such a database through simple command-line Perl scripts or other tools of their own creation. Although the database structure is complicated, we have developed a comprehensive interface tool that hides the complex structure and renders the database accessible to automatic queries or maintenance routines written by such users.
DISCUSSION
This paper is an overview of Pseudogene.org, a repository for detailed pseudogenic information compiled from a variety of sources. Currently (as of June 2006), Pseudogene.org contains a compilation of pseudogenes that includes the following:
- 31 768 pseudogene records on the Human genome, including those identified by several sources in the literature (5,8,11,13) and by the PseudoPipe identification tool (6).
- 15 063 pseudogenes on the Mouse genome, compiled from the literature (17) and PseudoPipe results.
- 51 491 pseudogenes on the chimp, rat, dog, chicken, mosquito, tetradon, zebrafish, falciparum and fly genomes, all newly identified by PseudoPipe.
- 6890 pseudogenes from 64 prokaryote genomes, as compiled by Liu et al. (29).
- Thirty pre-computed sets corresponding to manual analysis of human and mouse pseudogenes discussed in the literature and other work.
New pseudogenes and organisms are added as they become available, existing results are updated to reflect updated annotations and the annotations of new identification methods can be easily integrated. The pre-computed sets can accommodate manual annotations of interest, allowing users to either search the entire database or to limit their search to a combination of these sets.
In addition to serving as a useful resource, we believe that the underlying implementation is of use to the community. We have developed and made public a database infrastructure that is easily adaptable by someone with a basic understanding of database techniques, while hiding the MySQL details so as to make it usable by researchers with no knowledge of database programming and only a basic knowledge of Perl. We believe this implementation could be easily adapted for a number of other uses, such as the creation of a database of transcriptionally active regions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank Duncan Milburn, Mihali Felipe and Richard Burhans for their technical support, and Yuen-Jong Liu, Paul Bertone, Joel Rozowsky, Prachi Shah, Ross Hardison and Webb Miller for their advice and support. Mark Gerstein and John Karro acknowledge support from the NIH (P50 HG02357-01 and 5K01HG003315). Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khelifi A., Duret L., Mouchiroud D. HOPPSIGEN: a database of human and mouse processed pseudogenes. Nucleic Acids Res. 2005;33:D59–D66. doi: 10.1093/nar/gki084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z., Carriero N., Zheng D., Karro J., Harrison P.M., Gerstein M. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics. 2006;12:1437–1439. doi: 10.1093/bioinformatics/btl116. [DOI] [PubMed] [Google Scholar]
- 7.Ohshima K., Hattori M., Yada T., Gojobori T., Sakaki Y., Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z., Gerstein M. Large-scale analysis of pseudogenes in the human genome. Curr. Opin. Genet. Dev. 2004;14:328–335. doi: 10.1016/j.gde.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z., Harrison P., Gerstein M. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res. 2002;12:1466–1482. doi: 10.1101/gr.331902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z., Harrison P.M., Liu Y., Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison P.M., Hegyi H., Balasubramanian S., Luscombe N.M., Bertone P., Echols N., Johnson T., Gerstein M. Molecular fossils in the human genome: identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res. 2002;12:272–280. doi: 10.1101/gr.207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng D., Gerstein M.B. A computational approach for identifying pseudogenes in the ENCODE regions. Genome Biol. 2006;7(Suppl. 1):S13.1–S13.10. doi: 10.1186/gb-2006-7-s1-s13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torrents D., Suyama M., Zdobnov E., Bork P. A genome-wide survey of human pseudogenes. Genome Res. 2003;13:2559–2567. doi: 10.1101/gr.1455503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khelifi A., Duret L., Mouchiroud D. HOPPSIGEN: a database of human and mouse processed pseudogenes. Nucleic Acids Res. 2005;33:D59–D66. doi: 10.1093/nar/gki084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins J.E., Goward M.E., Cole C.G., Smink L.J., Huckle E.J., Knowles S., Bye J.M., Beare D.M., Dunham I. Reevaluating human gene annotation: a second-generation analysis of chromosome 22. Genome Res. 2003;13:27–36. doi: 10.1101/gr.695703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison P.M., Milburn D., Zhang Z., Bertone P., Gerstein M. Identification of pseudogenes in the Drosophila melanogaster genome. Nucleic Acids Res. 2003;31:1033–1037. doi: 10.1093/nar/gkg169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z., Carriero N., Gerstein M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 2004;20:62–67. doi: 10.1016/j.tig.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z., Gerstein M. Reconstructing genetic networks in yeast. Nat. Biotechnol. 2003;21:1295–1297. doi: 10.1038/nbt1103-1295. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z., Gerstein M. Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res. 2003;31:5338–5348. doi: 10.1093/nar/gkg745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z., Gerstein M. The human genome has 49 cytochrome c pseudogenes, including a relic of a primordial gene that still functions in mouse. Gene. 2003;312:61–72. doi: 10.1016/s0378-1119(03)00579-1. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z., Gerstein M. Of mice and men: phylogenetic footprinting aids the discovery of regulatory elements. J. Biol. 2003;2:11. doi: 10.1186/1475-4924-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z., Gerstein M. Identification and characterization of over 100 mitochondrial ribosomal protein pseudogenes in the human genome. Genomics. 2003;81:468–480. doi: 10.1016/s0888-7543(03)00004-1. [DOI] [PubMed] [Google Scholar]
- 23.Wang X., Grus W.E., Zhang J. Gene losses during human origins. PLoS Biol. 2006;4:e52. doi: 10.1371/journal.pbio.0040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison P.M., Zheng D., Zhang Z., Carriero N., Gerstein M. Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 2005;33:2374–2383. doi: 10.1093/nar/gki531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng D., Zhang Z., Harrison P.M., Karro J., Carriero N., Gerstein M. Integrated pseudogene annotation for human chromosome 22: evidence for transcription. J. Mol. Biol. 2005;349:27–45. doi: 10.1016/j.jmb.2005.02.072. [DOI] [PubMed] [Google Scholar]
- 26.Nadkarni P.M., Marenco L., Chen R., Skoufos E., Shepherd G., Miller P. Organization of heterogeneous scientific data using the EAV/CR representation. J. Am. Med. Inform. Assoc. 1999;6:478–493. doi: 10.1136/jamia.1999.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birney E. Ensembl: a genome infrastructure. Cold Spring Harb. Symp. Quant. Biol. 2003;68:213–215. doi: 10.1101/sqb.2003.68.213. [DOI] [PubMed] [Google Scholar]
- 28.Dennis Q., Sean M., Grossman D. ISWC Bioinformatics. 2003 [Google Scholar]
- 29.Liu Y., Harrison P.M., Kunin V., Gerstein M. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 2004;5:R64. doi: 10.1186/gb-2004-5-9-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]