The Coronavirus Spike Protein Is a Class I Virus Fusion Protein: Structural and Functional Characterization of the Fusion Core Complex (original) (raw)
Abstract
Coronavirus entry is mediated by the viral spike (S) glycoprotein. The 180-kDa oligomeric S protein of the murine coronavirus mouse hepatitis virus strain A59 is posttranslationally cleaved into an S1 receptor binding unit and an S2 membrane fusion unit. The latter is thought to contain an internal fusion peptide and has two 4,3 hydrophobic (heptad) repeat regions designated HR1 and HR2. HR2 is located close to the membrane anchor, and HR1 is some 170 amino acids (aa) upstream of it. Heptad repeat (HR) regions are found in fusion proteins of many different viruses and form an important characteristic of class I viral fusion proteins. We investigated the role of these regions in coronavirus membrane fusion. Peptides HR1 (96 aa) and HR2 (39 aa), corresponding to the HR1 and HR2 regions, were produced in Escherichia coli. When mixed together, the two peptides were found to assemble into an extremely stable oligomeric complex. Both on their own and within the complex, the peptides were highly alpha helical. Electron microscopic analysis of the complex revealed a rod-like structure ∼14.5 nm in length. Limited proteolysis in combination with mass spectrometry indicated that HR1 and HR2 occur in the complex in an antiparallel fashion. In the native protein, such a conformation would bring the proposed fusion peptide, located in the N-terminal domain of HR1, and the transmembrane anchor into close proximity. Using biological assays, the HR2 peptide was shown to be a potent inhibitor of virus entry into the cell, as well as of cell-cell fusion. Both biochemical and functional data show that the coronavirus spike protein is a class I viral fusion protein.
To successfully initiate an infection, viruses need to overcome the cell membrane barrier. Enveloped viruses achieve this by membrane fusion, a process mediated by specialized viral fusion proteins. Most viral fusion proteins are expressed as precursor proteins, which are endoproteolytically cleaved by cellular proteases, giving rise to a metastable complex of a receptor binding subunit and a membrane fusion subunit. Upon receptor binding at the cell membrane or as a result of protonation after endocytosis, the fusion proteins undergo a dramatic conformational transition. A hydrophobic fusion peptide becomes exposed and inserts into the target membrane. The free energy released upon subsequent refolding of the fusion protein to its most stable conformation is believed not only to facilitate the close apposition of viral and cellular membranes but also to effect the actual membrane merger (1, 47, 57). Knowledge about the molecular and biophysical events of this process is required for a thorough understanding of this essential step in the virus life cycle, as well as for the rational design of methods for intervention.
With a positive-stranded RNA genome of 28 to 32 kb, the Coronaviridae are the largest enveloped RNA viruses. Coronaviruses exhibit a broad host range, infecting mammalian and avian species. They are responsible for a variety of acute and chronic diseases of the respiratory, hepatic, gastrointestinal, and neurological systems (59). The spike (S) protein is the sole viral membrane protein responsible for cell entry. It binds to the receptor on the target cell and mediates subsequent virus-cell fusion (6). Spikes can be seen under the electron microscope as clear, 20-nm-long, bulbous surface projections on the virion membrane (14). The spike protein of mouse hepatitis virus strain A59 (MHV-A59) is a 180-kDa heavily N-glycosylated type I membrane protein which occurs in a homodimeric (38, 69) or homotrimeric (16) complex. In most MHV strains, the S protein is cleaved intracellularly into an N-terminal subunit (S1) and a membrane-anchored subunit (S2) of similar sizes which are noncovalently linked and have distinct functions. Binding to the MHV receptor (77) has been mapped to the N-terminal 330 amino acids (aa) of the S1 subunit (65), whereas the membrane fusion function resides in the S2 subunit (81). It has been suggested that the S1 subunit forms the globular head while the S2 subunit constitutes the stalk-like region of the spike (15). Binding of S1 to soluble MHV receptor, or exposure to 37°C and an elevated pH (pH 8.0), induces a conformational change which is accompanied by the separation of S1 and S2 and which might be involved in triggering membrane fusion (22, 28, 63). Cleavage of the S protein into S1 and S2 has been shown to enhance fusogenicity (26, 64), but cleavage is not absolutely required for fusion (2, 27, 62, 64).
The ectodomain of the S2 subunit contains two regions with a 4,3 hydrophobic (heptad) repeat (15), a sequence motif characteristic of coiled coils. These two heptad repeat (HR) regions, designated here HR1 and HR2, are conserved in position and sequence among the members of the three coronavirus antigenic clusters (Fig. 1). A number of studies have shown that the HR1 and HR2 regions are involved in viral fusion. First, a putative internal fusion peptide has been proposed to occur close to (7) or within (41) the HR1 region. Second, viruses with mutations in the membrane-proximal HR2 region exhibited defects in spike oligomerization and in fusion ability (40). Third, it has been suggested that the MHV-4 (JHM) strain can utilize both endosomal and nonendosomal pathways for cell entry but does not require acidification of endosomes for fusion activation (49). However, mutations found in MHVs which do require a low pH for fusion appeared to map to the HR1 region (24).
FIG. 1.
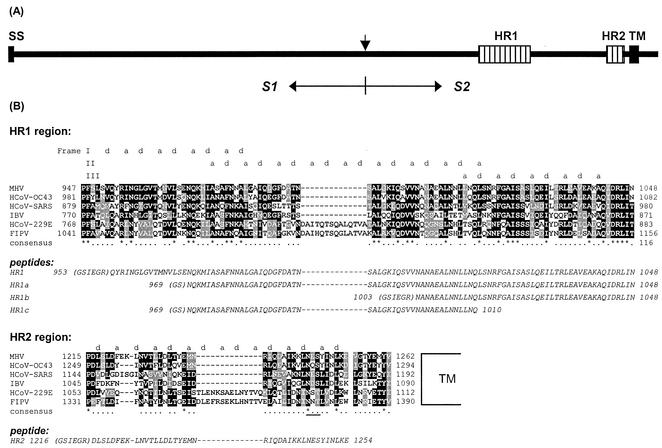
(A) Schematic representation of the coronavirus MHV-A59 spike protein structure. The glycoprotein has an N-terminal signal sequence (SS) and a transmembrane domain (TM) close to the C terminus. The protein is proteolytically cleaved (vertical arrow) into an S1 and an S2 subunit, which are noncovalently linked. S2 contains two HR regions (hatched bars), HR1 and HR2, as indicated. (B) Sequence alignment of HR1 and HR2 domains of MHV-A59 with those of HCoV-OC43, HCoV-229E, FIPV strain 79-1146, infectious bronchitis virus strain Beaudette (IBV), and the newly identified HCoV-SARS (strain TOR2). HCoV-229E and FIPV, MHV-A59 and HCoV-OC43, and IBV are representatives of groups 1, 2, and 3, respectively—the three coronavirus subgroups (59). Dark shading marks sequence identity, while lighter shading represents sequence similarity. The alignment shows a remarkable insertion of exactly two HRs (14 aa) in both HR1 and HR2 of HCV-229E and FIPV, a characteristic of all group 1 viruses. The predicted hydrophobic HR a and d residues are indicated above the sequence. The frame shifts in the predicted HRs in HR1 are caused by a stutter (51). The asterisks indicate conserved residues, and the dots represent similar residues. The amino acid sequences of the peptides HR1, HR1a, HR1b, HR1c, and HR2 used in this study are presented in italics below the alignments. N-terminal residues derived from the proteolytic cleavage site of the GST fusion protein are in parentheses. A conserved N-glycosylation sequence in the HR2 region is underlined.
HR regions appear to be a common motif in many viral fusion proteins (60). There are usually two of them; one N-terminal HR region (HR1) adjacent to the fusion peptide and a C-terminal HR region (HR2) close to the transmembrane anchor. Structural studies of viral fusion proteins reveal that the HR regions form a six-helix bundle structure implicated in viral entry (reviewed in reference 19). The structure consists of a homotrimeric coiled coil of HR1 domains, in the exposed hydrophobic grooves of which the HR2 regions are packed in an antiparallel manner. This conformation brings the N-terminal fusion peptide into close proximity to the transmembrane anchor. Because the fusion peptide inserts into the cell membrane during the fusion event, such a conformation facilitates a close apposition of the cellular and viral membrane (reviewed in reference 19). Recent evidence suggests that the actual six-helix bundle formation is directly coupled to the merging of the membranes (47, 57). The similarities in the structures of the six-helix bundle complexes elucidated for influenza virus hemagglutinin (HA) (4, 11), human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) gp41 (5, 8, 42, 66, 72, 79), Moloney murine leukemia virus type1 gp21 (20), Ebola virus GP2 (43, 71), human T-cell leukemia virus type I gp21 (32), Visna virus TM (44), simian parainfluenza virus 5 (SV5) F1 (1), and human respiratory syncytial virus (HRSV) F1 (83) all point to a common fusion mechanism for these viruses.
Based on structural similarities, two classes of viral fusion proteins have been distinguished (37). Proteins containing HR regions and an N-terminal or N-proximal fusion peptide are classified as class I viral fusion proteins. Class II viral fusion proteins (e.g., the alphavirus E1 and the flavivirus E fusion proteins) lack HR regions and have an internal fusion peptide. Their fusion protein is folded in tight association with a second protein as a heterodimer. Here, fusion activation takes place upon cleavage of the second protein.
The coronavirus fusion protein (S) shares several features with class I virus fusion proteins. It is a type I membrane protein, synthesized in the endoplasmic reticulum, and is transported to the plasma membrane. It contains two HR sequences, one located downstream of the fusion peptide and one in close proximity to the transmembrane region. Despite its similarity to class I fusion proteins, there are several characteristics that make the coronavirus S protein exceptional. One is the absence of an N-terminal or even N-proximal fusion peptide in the membrane-anchored subunit. Another peculiarity is the relatively large size of the HR regions (∼100 and ∼40 aa). Third, cleavage of the S protein is not required for membrane fusion; in fact, it does not occur at all in the group 1 coronaviruses.
In the present study, we have investigated the biochemical and functional characteristics of the HR regions of the MHV-A59 spike protein. We show that peptides corresponding to the HR regions assembled into a thermostable, oligomeric, alpha-helical rod-like complex, with the HR1 and HR2 helices oriented in an antiparallel manner. HR2 was found to be a strong inhibitor of both virus entry into the cell and cell-cell fusion. Our findings show that the coronavirus MHV spike fusion protein belongs to the class I viral fusion proteins.
MATERIALS AND METHODS
Plasmid constructions.
For the production of peptides corresponding to amino acid residues 953 to 1048 (HR1), 969 to 1048 (HR1a), 1003 to 1048 (HR1b), 969 to 1010 (HR1c), and 1216 to 1254 (HR2) of the MHV-A59 spike protein, PCR fragments were prepared using as a template the plasmid pTUMS, which contains the MHV-A59 spike gene (67). Primers were designed (Table 1) to introduce into the amplified fragment an upstream _Bam_HI site and a downstream _Eco_RI site, as well as a stop codon preceding the _Eco_RI site. The fragments corresponding to aa 953 to 1048 and 1216 to 1254 were additionally provided with sequences specifying a factor Xa cleavage site immediately downstream of the _Bam_HI site. Fragments were cloned into the _Bam_HI/_Eco_RI site of the pGEX-2T bacterial expression vector (Amersham Bioscience) in frame with the glutathione _S_-transferase (GST) gene just downstream of the thrombin cleavage site.
TABLE 1.
Primers used for PCR of HR regions
| Primer | Polarity | Sequence (5′-3′) | HR product |
|---|---|---|---|
| 973 | + | GTGGATCCATCGAAGGTCGTCAATATAGAATTAATGGTTTAG | HR1 |
| 974 | + | GTGGATCCATCGAAGGTCGTAATGCAAATGCTGAAGC | HR1b |
| 975 | − | GGAATTCAATTAATAAGACGATCTATCTG | HR1, HR1a, HR1b |
| 976 | − | CGAATTCATTCCTTGAGGTTGATGTAG | HR2 |
| 990 | + | GCGGATCCATCGAAGGTCGTGATTTATCTCTCGATTTC | HR2 |
| 1151 | + | GTGGATCCAACCAAAAGATGATTGC | HR1a, HR1c |
| 1152 | − | GGAATTCAATTGAGTGCTTCAGCATTTG | HR1c |
To establish a cell-cell fusion inhibition assay, the firefly luciferase gene was cloned under a T7 promoter and an encephalomyocarditis virus internal ribosome entry site. The luciferase gene-containing fragment was excised from the pSP-_luc_+ vector (Promega) by digestion with _Nco_I and _Eco_RV, treated with Klenow, and ligated into the _Bam_HI-linearized, Klenow-blunted pTN3 vector (68), yielding the pTN3-_luc_+ reporter plasmid.
Bacterial protein expression and purification.
Freshly transformed BL21 cells (Novagen) were grown in 2× yeast-tryptone medium to log phase (optical density at 600 nm, ∼1.0) and subsequently induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) (GIBCO BRL) to a final concentration of 0.4 mM. Two hours later, the cells were pelleted, resuspended in 1/25 volume of 10 mM Tris (pH 8.0)-10 mM EDTA-1 mM phenylmethylsulfonyl fluoride, and sonicated on ice (five times for 2 min each time). The cell homogenates were centrifuged at 20,000 × g for 60 min at 4°C. To each 50 ml of supernatant, 2 ml of glutathione-Sepharose 4B (Amersham Bioscience; 50% [vol/vol] in phosphate-buffered saline [PBS]) was added, and the mixtures were incubated overnight at 4°C under rotation. The beads were washed three times with 50 ml of PBS and resuspended in a final volume of 1 ml of PBS. Peptides were cleaved from the GST moiety on the beads using 20 U of thrombin (Amersham Bioscience) by incubation for 4 h at room temperature (RT). Peptides in the supernatant were purified by reversed-phase high-pressure liquid chromatography (RP HPLC) using a Phenyl-5PW RP column (Tosoh) with a linear gradient of acetonitrile containing 0.1% trifluoroacetic acid. The peptide-containing fractions were vacuum dried overnight and dissolved in water. The peptide concentration was determined by measuring the absorbance at 280 nm (25) and by bicinchoninic acid protein analysis (Micro BCA assay kit; Pierce).
Temperature stability of HR1-HR2 complex.
An equimolar mix of peptides HR1 and HR2 (80 μM each) in H2O was incubated at RT for 1 h. After the addition of an equal volume of 2× Tricine sample buffer (0.125 M Tris [pH 6.8], 4% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol, 10% glycerol, 0.004 g of bromophenol blue) (58), the mixtures were either left at RT or heated for 5 min at different temperatures and subsequently analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) in a 15% Tricine gel (58).
CD spectroscopy.
Circular dichroism (CD) spectra of peptides (25 μM in H2O) were recorded at RT on a Jasco J-810 spectropolarimeter, using a 0.1-mm path length, 1-nm bandwidth, 1-nm resolution, 0.5-s response time, and a scan speed of 50 nm/min. The alpha-helix content was calculated using the program CDNN (http://bioinformatik.biochemtech.uni-halle.de/cd_spec/).
Electron microscopy.
A preincubated equimolar mix of the peptides HR1 and HR2 was subjected to size exclusion chromatography (Superdex 75 HR 10/30; Amersham Pharmacia Biotech). A sample from the HR1-HR2 peptide complex-containing fraction was adsorbed onto a discharged carbon film, negatively stained with a 2% uranyl acetate solution, and examined with a Philips CM200 microscope at 100 kV.
Proteinase K treatment.
Stock solutions (1 mM) of the peptides HR1, HR1a, HR1b, HR1c, and HR2 in water were diluted to 80 μM in PBS. The peptides alone (80 μM) or after preincubation for 1 h at 37°C with HR2 (80 μM each) were subsequently subjected to proteinase K digestion (1% [wt/wt] proteinase K/peptide) for 2 h at 4°C. Samples were immediately subjected to Tricine SDS-PAGE analysis. Protease-resistant fragments were also separated and purified by RP HPLC and characterized by mass spectrometry.
Virus cell entry assay.
The potencies of HR peptides in inhibiting viral infection were determined using a recombinant MHV-A59, MHV-EFLM, which expresses the firefly luciferase gene (C. A. M. de Haan and P. J. M. Rottier, unpublished data). LR7 cells (35) were maintained as monolayer cultures in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS; GIBCO BRL). LR7 cells grown in 96-well plates were inoculated with MHV-EFLM in DMEM at a multiplicity of infection (MOI) of 5 in the presence of various concentrations of peptide ranging from 0.4 to 50 μM. After 1 h, the cells were washed with DMEM and the medium was replaced with DMEM containing 10% FCS. At 5 h postinfection (p.i.), the cells were harvested in 50 μl of 1× Passive Lysis buffer (Luciferase Assay System; Promega) according to the manufacturer's protocol. Upon mixing 10 μl of cell lysate with 40 μl of substrate, luciferase activity was measured using a Wallac Betalumino meter.
Cell-cell fusion assay.
LR7 cells (2 × 106), used as target cells, were washed with DMEM and overlaid with transfection medium consisting of 0.2 ml of DMEM containing 10 μl of Lipofectin (Life Technologies) and 4 μg of the plasmid pTN3-luc+. After 10 min at RT, 0.8 ml of DMEM was added and incubation was continued at 37°C. BSR T7/5 cells—BHK cells constitutively expressing T7 RNA polymerase (3) (a gift from K. K. Conzelmann)—were grown in BHK-21 medium supplemented with 10% FCS, 100 IU of penicillin/ml, and 1 mg of Geneticin (GIBCO BRL)/ml. BSR T7/5 cells (104), designated effector cells, were infected in 96-well plates with wild-type vaccinia virus at an MOI of 1 in DMEM at 37°C. After 1 h, the cells were washed with DMEM and incubated for 3 h at 37°C with transfection medium consisting of 50 μl of DMEM containing 1 μl of Lipofectin and 0.2 μg of the plasmid pTUMS (68), which carries the MHV-A59 spike gene under the control of a T7 promoter. Then, 3 × 104 target cells in 100 μl of DMEM were added, and the cells were incubated for another 4 h in the presence or absence of HR peptide. The cells were lysed, and luciferase activity was measured as described above.
RESULTS
HR1 and HR2 regions in coronavirus spike proteins.
The S2 subunit ectodomain of coronaviruses contains two HR domains, HR1 and HR2, which are conserved in sequence and position (15) (diagrammed in Fig. 1A). HR2 is located adjacent to the transmembrane domain, while HR1 occurs ∼170 aa upstream of HR2. Figure 1B shows a protein sequence alignment of the HR1 and HR2 regions of five coronaviruses from the three antigenic groups and the recently identified human coronavirus associated with severe acute respiratory syndrome (HCoV-SARS) (17, 34, 52, 53). The sequence alignment reveals a remarkable insertion of exactly two HRs (14 aa) in both the HR1 and HR2 domains of the spike protein of the group 1 coronaviruses HCoV strain 229E (HCoV-229E) and feline infectious peritonitis virus (FIPV) strain 79-1146. Alignment of all known coronavirus spike protein sequences shows these insertions in all group 1 coronaviruses. Another characteristic feature is that the length of the linker region between the HR2 region and the transmembrane region is strictly conserved in all coronavirus spike proteins.
HR1 and HR2 can form a hetero-oligomeric complex.
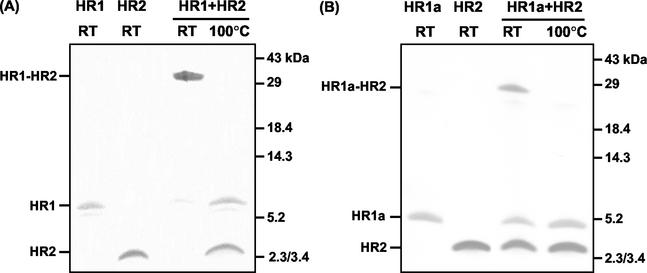
To study the HR regions in the S2 subunit of the MHV-A59 spike protein, peptides corresponding to the HR residues 953 to 1048 (HR1), 969 to 1048 (HR1a), 969 to 1048 (HR1b), 969 to 1003 (HR1c), and 1216 to 1254 (HR2) (Fig. 1B) were produced in bacteria as GST fusion proteins. The peptides were affinity purified using glutathione-Sepharose beads, proteolytically cleaved from the resin, and purified to homogeneity by RP HPLC. The masses of the peptides, as determined by mass spectrometry, matched their predicted _Mw_s (HR1, 10,873 Da; HR1a, 8,653 Da; HR1b, 5,631 Da; HR1c, 4,447 Da; and HR2, 5,254 Da). To study an interaction between the two HR regions, the purified peptides HR1 and HR2 were incubated alone (80 μM) or in an equimolar (80 μM each) mixture for 1 h at 37°C, and the samples were subjected to SDS-PAGE either directly or after being heated for 5 min at 95°C (Fig. 2A). While the peptides migrated according to their molecular masses after separate incubation, most of the protein of the preincubated mixture of HR1 and HR2 migrated as a higher-molecular-mass complex with a slightly lower mobility than the 29-kDa marker. Upon being heated, the complex dissociated, giving rise to the individual subunits HR1 and HR2. We also tested the other HR1 peptides for interaction with HR2. While we did not observe complexes upon mixing of HR2 with HR1b or HR1c (data not shown), a higher-molecular-mass species comigrating with the 29-kDa marker was found when HR1a was incubated with HR2 (Fig. 2B), though the extent of complex formation appeared to be lower than with peptide HR1. Higher-molecular-mass species were not seen. The results indicated that the HR1 region contains the information to associate with the HR2 region into a hetero-oligomeric complex and that this complex is stable in the presence of 2% SDS.
FIG. 2.
Hetero-oligomeric complex formation of HR1 and HR1a with HR2. (A) HR1 and HR2 on their own or as a preincubated equimolar (80 μM) mix were subjected to Tricine SDS-15% PAGE. Before gel loading, samples were either heated at 100°C or left at RT. The positions of HR1, HR2, and the HR1-HR2 complex are indicated on the left, while the positions of molecular mass markers are indicated on the right. (B) Same as panel A but with peptide HR1a instead of HR1.
HR1-HR2 complex is highly temperature resistant.
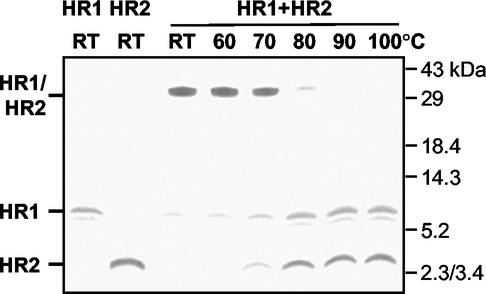
Next, we determined the stability of the HR1-HR2 complex at increasing temperatures. An equimolar (80 μM each) mix of the two peptides was again incubated for 1 h at 37°C and subsequently heated for 5 min at different temperatures in 1× Tricine sample buffer or left at RT. The complexes were analyzed by SDS-PAGE in a 15% gel. As Fig. 3 demonstrates, the high-molecular-mass complexes remained intact up to 70°C, partly dissociated at 80°C, and fully dissociated at 90°C. The stability of the complex at high temperatures indicates that the peptides are held together by strong interaction forces in an energetically favorable conformation.
FIG. 3.
Temperature stability of HR1-HR2 complex. An equimolar mix of HR1 and HR2 (80 μM) was incubated at RT for 1 h. Samples were subsequently heated for 5 min at the indicated temperatures in 1× Tricine sample buffer and analyzed by SDS-PAGE in a 15% Tricine gel, together with HR1 and HR2 alone. The positions of HR1, HR2, and the HR1-HR2 complex are indicated on the left, while the molecular mass markers are indicated on the right.
HR1, HR2, and the HR1-HR2 complex are highly alpha helical.
The secondary structure of the HR peptides was examined by CD, and the CD spectra of HR1 and HR2 and of an equimolar mixture of HR1 and HR2 were recorded (Fig. 4). The spectra showed clear minima at 208 and 222 nm, which is characteristic of alpha-helical structure. Calculations revealed that the alpha-helical contents of the individual HR1 and HR2 peptides and of the mixture of the two peptides were 89.2, 89.3, and 81.9%, respectively.
FIG. 4.
CD spectra (mean residue eliplicity [φ]) of the HR1 peptide (25 μM; open squares), the HR2 peptide (25 μM; solid triangles), and the HR1-HR2 complex (25 μM; solid squares) in water at RT. Note that the HR1 and HR2 spectra virtually coincide.
The HR1-HR2 complex has a rod-like structure.
The overall shape of the HR1-HR2 complex was examined by electron microscopy. Complexes were purified and viewed after negative staining. Electron micrographs revealed rod-like structures (Fig. 5). Based on measurements of 40 particles, an average length of 14.5 nm (±2 nm) was calculated. This length is consistent with an alpha helix ∼90 aa in length, which corresponds approximately to the predicted length of the HR1 coiled-coil region. Similar rod-shaped complexes have been reported for the influenza virus HA protein (12, 56), for portions of the HIV-1 gp41 protein (73), and for the Ebola virus GP2 protein (70).
FIG. 5.
Electron micrographs of HR1-HR2 complex.
HR1 and HR2 helices associate in an antiparallel manner.
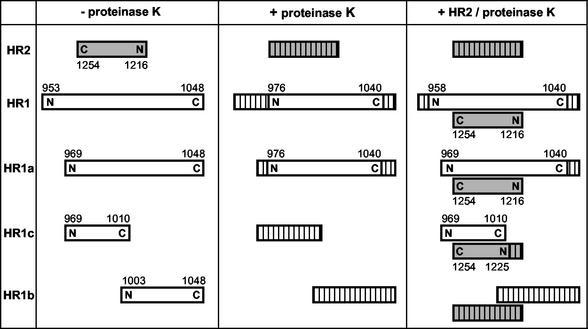
The relative orientation and position of HR2 with respect to HR1 in the complex were examined by limited proteolysis using proteinase K in combination with mass spectrometry. Complexes were generated by incubation of the HR2 peptide with each of the peptides HR1, HR1a, HR1b, and HR1c. The reaction mixtures, as well as the individual peptides, were then treated with proteinase K. Samples from each reaction were analyzed by Tricine SDS-PAGE (data not shown). Using RP HPLC, the protease-resistant fragments were purified, and their molecular masses were determined by mass spectrometry, which allowed us to identify the protease-resistant cores of the peptides. For each protease-resistant core, a unique amino acid composition could be deduced that allowed the unequivocal identification of the peptides in the different samples. Figure 6 gives a schematic overview of the proteinase K-resistant fragments. Digestion of HR1 alone left a protease-resistant fragment with a molecular mass of 6,801 Da, corresponding to residues 976 to 1040. Although CD spectra had indicated a folded structure, HR2 was completely degraded by proteinase K. However, in the presence of HR1, HR2 was fully protected from proteolytic degradation. HR2 was able to rescue 18 additional residues at the N terminus of HR1, leaving a fragment of 8,675 Da, corresponding to residues 958 to 1040.
FIG. 6.
Proteinase K treatment of HR peptides. The peptides HR2, HR1, HR1a, HR1b, and HR1c were subjected to proteinase K either individually in solution or after mixing of the different HR1 peptides with HR2 at equimolar concentration followed by a 1-h incubation at 37°C. Proteolytic fragments were separated and purified by HPLC and characterized by mass spectrometry. The peptides are represented by bars. The hatched bars indicate the protease-sensitive part(s) of the peptide. Shaded bars represent the HR2 peptide. The N- and C-terminal positions of the peptide and the amino acid numbering are indicated.
Proteolysis of the HR1a peptide alone generated the same fragment (residues 976 to 1040) obtained with HR1. In the HR1a-HR2 mixture, the HR2 peptide was completely protected by HR1a from degradation, while HR2 fully shielded the N terminus of HR1a from proteolysis, including the glycine and serine residues originating from the thrombin cleavage site.
Although a higher-molecular-mass species could not be detected by Tricine SDS-PAGE (data not shown), the protease treatment of the HR1c-HR2 complex left a protease-resistant core. HR1c was fully sensitive to proteinase K but was completely protected in the presence of HR2. HR2 itself was partly protected against proteolysis by HR1c, yielding a fragment of 3,583 Da that represents residues 1225 to 1254. Importantly, this HR2 fragment has an intact C terminus but is degraded at its N terminus. HR1c has the same N terminus as HR1a but is truncated at its C terminus. Thus, its inability to protect the HR2 N terminus combined with the full protection provided by HR1a implies an antiparallel association of the HR1 and HR2 helices in the hetero-oligomeric complex. The peptide HR1b was fully sensitive to proteinase K both by itself and when mixed with HR2. HR1b also could not prevent proteolysis of HR2. Altogether, the proteolysis results suggest that the antiparallel association of HR2 and HR1 occurs in the middle part of HR1.
HR2 strongly inhibits viral entry and syncytium formation.
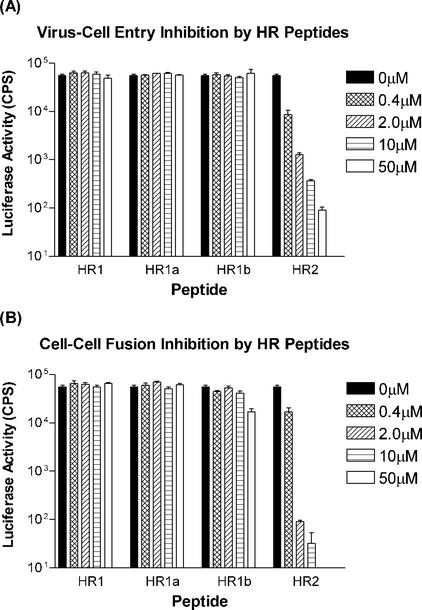
The formation of stable HR complexes is supposedly an essential step in the process of membrane fusion during viral cell entry. Thus, we evaluated the potencies of our HR peptides for inhibiting MHV entry, making use of a recombinant MHV-A59, MHV-EFLM, which expresses the firefly luciferase reporter gene. Cells were inoculated with MHV-EFLM in the presence of different concentrations of the peptides HR1, HR1a, HR1b, HR1c, and HR2. After 1 h, the cells were washed, and culture medium without peptide was added. At 4 h p.i., i.e., before syncytium formation takes place, the cells were lysed and tested for luciferase activity (Fig. 7A). HR1, HR1a, and HR1b were not able to inhibit viral entry up to concentrations of 50 μM. In contrast, HR2 blocked viral entry in a concentration-dependent manner, and inhibition was almost complete at a concentration of 50 μM.
FIG. 7.
Inhibition of virus cell entry and cell-cell fusion by HR peptides. (A) Virus cell entry inhibition by HR peptides using a luciferase gene-expressing MHV. LR7 cells were inoculated with virus at an MOI of 5 in the presence of various concentrations of peptide ranging from 0.4 to 50 μM. At 5 h p.i., the cells were lysed, and luciferase activity was measured. CPS, counts per second. The error bars indicate standard deviations. (B) Inhibition of spike-mediated cell-cell fusion by HR peptides. BSR T7/5 effector cells—BHK cells constitutively expressing T7 RNA polymerase (3)—were infected with vaccinia virus for 1 h and subsequently transfected with a plasmid containing the S gene under a T7 promoter. Three hours posttransfection, LR7 target cells transfected with a plasmid carrying the luciferase gene behind a T7 promoter were added to the effector cells. The cells were incubated for another 4 h in the presence or absence of HR peptide. The cells were lysed, and luciferase activity was measured.
We also studied the abilities of the HR peptides to block cell-cell fusion. To this end, we established a sensitive cell-cell fusion assay based on the coculturing of BHK cells expressing the bacteriophage T7 polymerase, as well as the MHV-A59 spike protein, with murine L cells transfected with a plasmid carrying a luciferase gene cloned behind a T7 promoter. Fusion of the cells was determined by measuring luciferase activity. The effects of adding the HR peptides during the coculturing of the cells are compiled in Fig. 7B. The HR2 peptide again appeared to be a potent inhibitor able to efficiently block cell-cell fusion. A 1,000-fold reduction in luciferase activity was measured at a concentration of 10 μM, whereas essentially no activity was observed at a concentration of 50 μM. Of the HR1 peptides, only the HR1b peptide had a minor effect at the highest concentration of 50 μM.
DISCUSSION
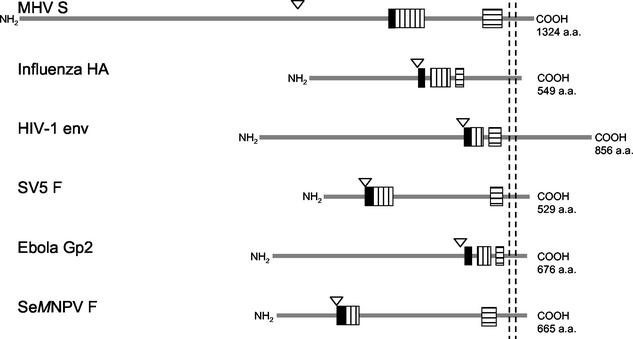
HR regions play a critical role in viral membrane fusion. Fusion proteins from widely disparate virus families have been shown to contain two such regions, one located close to the fusion peptide, the other generally in the vicinity of the viral membrane (7) (summarized in Fig. 8). Distances between the HR regions vary greatly, from some 50 aa, as in HIV-1, to ∼300 residues in Spodoptera exigua multicapsid nucleopolyhedrosis virus (74). The crystal structures resolved for influenza virus HA (4, 10, 78), HIV-1 and SIV gp41 (5, 8, 42, 66, 72, 79), Moloney murine leukemia virus gp21 (20), Ebola virus GP2 (43, 71), human T-cell leukemia virus type I gp21 (32), Visna virus TM (44), SV5 F1 (1), HRSV F1 (83), and Newcastle disease virus F (13) all show a central trimeric coiled coil constituted of three HR1 regions. In some of these structures (e.g., HIV-1 and SIV gp41, SV5 F1, Ebola virus gp2, Visna virus TM, and HRSV F1), a second layer of helices or elongated peptide chains was observed, contributed by HR2 domains which were packed in an antiparallel manner into the hydrophobic grooves of the HR1 coiled coil, forming a six-helix bundle. In the full-length protein, such a conformation brings the fusion peptide present at the N terminus of HR1 close to the transmembrane region that occurs C terminally of HR2. With the fusion peptide inserted in the cellular membrane and the transmembrane region anchored in the viral membrane, such a hairpin-like structure facilitates the close apposition of cellular and viral membranes and enables subsequent membrane fusion (reviewed in reference 19). Combined with the findings that peptides derived from these HR domains can act as potent inhibitors of fusion (reviewed in reference 19), the biological relevance of the HR regions in the viral life cycle is obvious. Our studies of the HR motifs in the MHV-A59 spike protein presented here indicate that coronaviruses use membrane fusion and cell entry mechanisms similar to those of the other viruses, allowing coronavirus spike proteins to be classified as class I viral fusion proteins (37).
FIG. 8.
Schematic representation (approximately to scale) of the viral fusion proteins of six different virus families: MHV-A59 S (Coronaviridae), influenza virus HA (Orthomyxoviridae), HIV-1 gp160 (Retroviridae), SV5 F (Paramyxoviridae), Ebola Gp2 (Filoviridae), and S. exigua multicapsid nucleopolyhedrosis virus (Se_M_NPV) F (Baculoviridae). Cleavage sites are indicated by triangles; the solid bars represent the (putative) fusion peptides, the vertically hatched bars represent the HR1 domains, and the horizontally hatched bars represent the HR2 domains. Transmembrane domains are indicated by the vertical dashed lines. For each polypeptide, the total length is given at the right.
The MHV-A59-derived HR peptides exhibited a number of typical class I characteristics. First, the purified HR1 and HR2 peptides assembled spontaneously into unique, homogeneous multimeric complexes. These complexes were highly stable, surviving, for instance, high concentrations (2%) of SDS and high temperatures (70 to 80°C). The peptides apparently associate with great specificity into an energetically very favorable structure. Another typical feature was the observed secondary structures in the peptides. As for HR peptides of other class I viruses, the CD spectra of both the individual and the complexed HR1 and HR2 peptides showed patterns characteristic of alpha-helical structure. The alpha-helix contents of the separate peptides were calculated to be ∼89%, and that of their equimolar mixture was calculated to be ∼82%. Consistent with these observations, the HR complex revealed a rod-like structure when examined by electron microscopy. The length of this structure (∼14.5 nm) correlates well with the length predicted for an alpha helix the size of HR1 (96 aa). Similar rod-like structures have been observed for other class I virus fusion proteins, such as the influenza virus HA protein (12, 56), portions of the HIV-1 gp41 protein (73), and the Ebola virus GP2 protein (70), but the lengths of the MHV-A59-derived structures are substantially larger. This is presumably even more the case for group I coronaviruses, which have an insertion of two HRs (14 aa) (Fig. 1) in both HR regions. These insertions into otherwise conserved areas suggest that these additional sequences associate with each other in the HR1-HR2 complex, thereby extending the alpha-helical complex by exactly four turns. We can only speculate about the significance of the exceptional lengths of coronavirus HR complexes. It is conceivable that the supposedly higher energy gain of their formation corresponds with higher energy requirements for membrane fusion by these viruses.
Another important characteristic of class I viral fusion proteins is the formation of a heterotrimeric six-helix bundle during the membrane fusion process, resulting in a close colocation of the fusion peptide and the transmembrane domain. Consistently, protein dissection studies using proteinase K demonstrated an antiparallel organization of the HR1 and HR2 alpha-helical peptides in the MHV-A59 HR complex. So far, no fusion peptides have been identified in any coronavirus spike protein, but predictions for MHV S have located such fusion sequences at (7) or in (41) the N terminus of HR1. In both cases, an antiparallel orientation of the HR1 and HR2 alpha helices ensures that the fusion peptide is brought into close proximity to the transmembrane region. Sequence analysis reveals that the e and g positions in the HR1 regions of all coronaviruses are primarily occupied by hydrophobic residues, unlike the e and g positions in the HR2 regions, which are mostly polar (Fig. 1). The HR2 region also contains a strictly conserved N-linked glycosylation sequence, indicating its surface accessibility. Preliminary X-ray data on the HR1-HR2 complex show a six-helix bundle structure in the electron-dense region (B. J. Bosch, P. J. M. Rottier, and F.A. Rey, unpublished results). The combined observations suggest a packing analogous to those of the fusion proteins of other class I viruses (e.g., HIV and SV5), where the HR1 and HR2 peptides can form a six-helix bundle with the long HR1 peptide in the middle as a three-stranded coiled-coil with the hydrophobic a and d residues in its inner core. The shorter HR2 peptide packs with its apolar interface in the hydrophobic grooves of the HR1 coiled coil, which exposes the mostly hydrophobic residues on e and g positions.
Peptides derived from the HR regions of retrovirus (30, 39, 48, 50, 61, 75, 76; S. Jiang, K. Lin, N. Strick, and A. R. Neurath, Letter, Nature **365:**113, 1993) and paramyxovirus (29, 36, 54, 80, 82) fusion proteins have been shown to strongly interfere with the fusion activities of these proteins. We observed the same effect when we tested the HR2 peptide of the MHV-A59 spike protein. With a recombinant luciferase-expressing MHV-A59, the peptide acted as an effective inhibitor of virus entry at micromolar concentrations. Cell-cell fusion inhibition was even more efficiently blocked by the peptide, as tested in a cell fusion luciferase assay system. However, peptides derived from the HR1 region had no or only a minor effect on virus entry and syncytium formation. HIV-1 gp41-derived HR peptides that inhibit membrane fusion have been shown not to bind to the native protein or to the six-helix bundle. They can bind only to an intermediate stage of gp41 occurring during the fusion process (9, 21, 31). Repeated passage of HIV in the presence of the inhibitory peptide DP178, which is derived from the C-terminal gp41 HR region, resulted in resistant viruses containing mutations in the N-terminal HR region (55). By analogy to HIV-1 and other class I viruses, inhibition of membrane fusion by the MHV HR2 peptide most likely takes place during an intermediate stage of the fusion process by binding of the peptide to the HR1 region in the spike protein. This binding, which may occur before, during, or after the association of the HR1 regions into the inner trimeric coiled coil, presumably inhibits the subsequent interaction with native HR2 and, consequently, membrane fusion. For the HIV-1 gp41 and SV5 F protein also, peptides corresponding to the HR1 region show membrane fusion inhibition, supposedly by binding to the native HR2 region (29, 75). It has been reported for HIV-1 that the HR1 peptide aggregates in solution (39) and that its inhibitory activity could be enhanced by fusing it to a designed soluble trimeric coiled coil, making the HR1 peptide more soluble (18). The MHV-A59 HR1 peptide is soluble in water but appeared to precipitate in salt solutions (data not shown). We cannot exclude the possibility that this solubility feature obscured the inhibitory potencies of our HR1-derived peptides and that it accounts for the negative results with these peptides in our fusion assays. The HR2 peptide (as well as, possibly, soluble forms of HR1) may well provide powerful antivirals for the therapy of coronavirus-induced diseases both in animals and humans.
Membrane fusion mediated by class I fusion proteins is accompanied by dramatic structural rearrangements within the viral polypeptide complexes (19). Though little is known of the coronavirus membrane fusion process (for a review, see reference 23), the occurrence of conformational changes induced by various conditions has been described for MHV spikes (46). While MHV-A59 is quite stable at mildly acidic pH, it is rapidly and irreversibly inactivated at pH 8.0 and 37°C (63). Under these conditions, the S1 subunit dissociates from the virions and the S2 subunit aggregates concomitantly, resulting in the aggregation of the particles. Due to the structural rearrangements in the spike, virions can bind to liposomes and the S2 protein becomes sensitive to protease degradation (28). Similar conformational changes can apparently also be induced at pH 6.5 by the binding of spikes to the (soluble) MHV receptor (22, 28) as this interaction enhances liposome binding and protease sensitivity as well (28). Virion binding to liposomes is presumably caused by the exposure of hydrophobic protein surfaces or of the fusion peptide as a result of the conformational change. It appears that the structural rearrangements in the spikes, whether elicited by elevated pH or soluble receptor interaction, reflect the process that naturally gives rise to the fusion of viral and cellular membranes. Accordingly, cell-cell fusion induced by MHV-A59 was maximal at slightly basic pH (63).
A number of studies of the MHV spike protein have shown the importance of the HR regions in membrane fusion. Three codon mutations (Q1067H, Q1094H, and L1114R) in or close to the HR1 region of the spike protein were found to be responsible for the low pH requirement for fusion of some MHV-JHM variants isolated from persistently infected cells (24). Analysis of soluble receptor-resistant variants of this virus also pointed to an important role for the HR1 region in fusion activity and suggested that it interacts somehow with the N-terminal domain (S1N330-III; aa 278 to 288) of the spike protein (45). In yet another MHV-JHM variant, a great reduction in cell-cell fusion was attributed to the occurrence of two mutations in the spike protein, one of which was again located in the HR1 region (A1046V), while the other (V870A) was in a small nonconserved HR region (N helix) close to the S cleavage site (33). Acidification resulted in a clear enhancement of fusion by this double mutant. It was speculated that the three predicted helical regions (N helix, HR1, and HR2) all collapse into a low-energy coiled coil during the process of membrane fusion (33). This paper provides evidence that the HR1 and HR2 regions indeed can form such a low-energy coiled coil. However, the role of the small N helix, although not conserved in group I and III coronaviruses, remains to be determined. Studies with the MHV-A59 S protein showed that mutations introduced at a and d positions in an N-terminal part of the HR1 region, a fusion peptide candidate, severely affected cell-cell fusion ability (41). This effect was not due to defects in spike maturation or cell surface expression. Finally, codon mutations in the HR2 region were also found to significantly reduce cell-cell fusion (40). Though these mutant spike proteins were apparently impaired in oligomerization, their surface expression was hardly affected.
In conclusion, our structural and functional studies indicate that the coronavirus spike protein can be classified as a class I viral fusion protein. The protein has, however, several unusual features that set it apart. An important characteristic of all class I virus fusion proteins known so far is the cleavage of the precursor by host cell proteases into a membrane-distal subunit and a membrane-anchored subunit, an event essential for membrane fusion. Consequently, the hydrophobic fusion peptide is then located at or close to the newly generated N terminus of the membrane-anchored subunit, just preceding the HR1 region. In contrast, the MHV-A59 spike does not have a hydrophobic stretch of residues at the distal end of S2 but carries a fusion peptide internally at a location that has yet to be determined (7, 41). Unlike other class I fusion proteins, cleavage of the S protein into S1 and S2 has been shown to enhance fusogenicity (26, 64) but not to be absolutely required (2, 27, 62, 64). In fact, spikes belonging to group 1 coronaviruses are not cleaved at all.
Acknowledgments
We are grateful to Mayken Grosveld and Alida Noordzij for their technical assistance with the HPLC and to Maurits de Planque, Bianca van Duyl, and Antoinette Killian for their assistance with the CD spectophotometer. We thank Raoul de Groot and Bert Jan Haijema for helpful discussions and Jean Lepault for his assistance with the electron microscope.
These investigations were supported by financial aid from The Netherlands Foundation for Chemical Research (CW) and The Netherlands Organization for Scientific Research (NWO) to B.J.B. and P.J.M.R.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3**:**309-319. [DOI] [PubMed] [Google Scholar]
- 2.Bos, E. C., L. Heijnen, W. Luytjes, and W. J. Spaan. 1995. Mutational analysis of the murine coronavirus spike protein: effect on cell-to-cell fusion. Virology 214**:**453-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73**:**251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371**:**37-43. [DOI] [PubMed] [Google Scholar]
- 5.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17**:**4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanagh, D. 1995. The coronavirus surface glycoprotein. Plenum Press, New York, N.Y.
- 7.Chambers, P., C. R. Pringle, and A. J. Easton. 1990. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 71**:**3075-3080. [DOI] [PubMed] [Google Scholar]
- 8.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89**:**263-273. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. H., T. J. Matthews, C. B. McDanal, D. P. Bolognesi, and M. L. Greenberg. 1995. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J. Virol. 69**:**3771-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95**:**409-417. [DOI] [PubMed] [Google Scholar]
- 11.Chen, J., J. J. Skehel, and D. C. Wiley. 1999. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 96**:**8967-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J., S. A. Wharton, W. Weissenhorn, L. J. Calder, F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1995. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc. Natl. Acad. Sci. USA 92**:**12205-12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9**:**255-266. [DOI] [PubMed] [Google Scholar]
- 14.Davies, H. A., and M. R. MacNaughton. 1979. Comparison of the morphology of three coronaviruses. Arch. Virol. 59**:**25-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Groot, R. J., W. Luytjes, M. C. Horzinek, B. A. van der Zeijst, W. J. Spaan, and J. A. Lenstra. 1987. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 196**:**963-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delmas, B., and H. Laude. 1990. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 64**:**5367-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drosten, C., S. Gunther, W. Preiser, S. Van Der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348**:**1967-1976. [DOI] [PubMed] [Google Scholar]
- 18.Eckert, D. M., and P. S. Kim. 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. USA 98**:**11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70**:**777-810. [DOI] [PubMed] [Google Scholar]
- 20.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 3**:**465-469. [DOI] [PubMed] [Google Scholar]
- 21.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5**:**276-279. [DOI] [PubMed] [Google Scholar]
- 22.Gallagher, T. M. 1997. A role for naturally occurring variation of the murine coronavirus spike protein in stabilizing association with the cellular receptor. J. Virol. 71**:**3129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279**:**371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallagher, T. M., C. Escarmis, and M. J. Buchmeier. 1991. Alteration of the pH dependence of coronavirus-induced cell fusion: effect of mutations in the spike glycoprotein. J. Virol. 65**:**1916-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182**:**319-326. [DOI] [PubMed] [Google Scholar]
- 26.Gombold, J. L., S. T. Hingley, and S. R. Weiss. 1993. Fusion-defective mutants of mouse hepatitis virus A59 contain a mutation in the spike protein cleavage signal. J. Virol. 67**:**4504-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hingley, S. T., I. Leparc-Goffart, and S. R. Weiss. 1998. The spike protein of murine coronavirus mouse hepatitis virus strain A59 is not cleaved in primary glial cells and primary hepatocytes. J. Virol. 72**:**1606-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmes, K. V., B. D. Zelus, J. H. Schickli, and S. R. Weiss. 2001. Receptor specificity and receptor-induced conformational changes in mouse hepatitis virus spike glycoprotein. Adv. Exp. Med. Biol. 494**:**173-181. [DOI] [PubMed] [Google Scholar]
- 29.Joshi, S. B., R. E. Dutch, and R. A. Lamb. 1998. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 248**:**20-34. [DOI] [PubMed] [Google Scholar]
- 30.Judice, J. K., J. Y. Tom, W. Huang, T. Wrin, J. Vennari, C. J. Petropoulos, and R. S. McDowell. 1997. Inhibition of HIV type 1 infectivity by constrained alpha-helical peptides: implications for the viral fusion mechanism. Proc. Natl. Acad. Sci. USA 94**:**13426-13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kliger, Y., and Y. Shai. 2000. Inhibition of HIV-1 entry before gp41 folds into its fusion-active conformation. J. Mol. Biol. 295**:**163-168. [DOI] [PubMed] [Google Scholar]
- 32.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. USA 96**:**4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krueger, D. K., S. M. Kelly, D. N. Lewicki, R. Ruffolo, and T. M. Gallagher. 2001. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion. J. Virol. 75**:**2792-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348**:**1953-1966. [DOI] [PubMed] [Google Scholar]
- 35.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74**:**1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka, and S. R. Petteway, Jr. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 93**:**2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105**:**137-148. [DOI] [PubMed] [Google Scholar]
- 38.Lewicki, D. N., and T. M. Gallagher. 2002. Quaternary structure of coronavirus spikes in complex with carcinoembryonic antigen-related cell adhesion molecule cellular receptors. J. Biol. Chem. 277**:**19727-19734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2**:**1075-1082. [DOI] [PubMed] [Google Scholar]
- 40.Luo, Z., A. M. Matthews, and S. R. Weiss. 1999. Amino acid substitutions within the leucine zipper domain of the murine coronavirus spike protein cause defects in oligomerization and the ability to induce cell-to-cell fusion. J. Virol. 73**:**8152-8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo, Z., and S. R. Weiss. 1998. Roles in cell-to-cell fusion of two conserved hydrophobic regions in the murine coronavirus spike protein. Virology 244**:**483-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malashkevich, V. N., D. C. Chan, C. T. Chutkowski, and P. S. Kim. 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. USA 95**:**9134-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malashkevich, V. N., B. J. Schneider, M. L. McNally, M. A. Milhollen, J. X. Pang, and P. S. Kim. 1999. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc. Natl. Acad. Sci. USA 96**:**2662-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malashkevich, V. N., M. Singh, and P. S. Kim. 2001. The trimer-of-hairpins motif in membrane fusion: Visna virus. Proc. Natl. Acad. Sci. USA 98**:**8502-8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuyama, S., and F. Taguchi. 2002. Communication between S1N330 and a region in S2 of murine coronavirus spike protein is important for virus entry into cells expressing CEACAM1b receptor. Virology 295**:**160-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuyama, S., and F. Taguchi. 2002. Receptor-induced conformational changes of murine coronavirus spike protein. J. Virol. 76**:**11819-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151**:**413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munoz-Barroso, I., S. Durell, K. Sakaguchi, E. Appella, and R. Blumenthal. 1998. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140**:**315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nash, T. C., and M. J. Buchmeier. 1997. Entry of mouse hepatitis virus into cells by endosomal and nonendosomal pathways. Virology 233**:**1-8. [DOI] [PubMed] [Google Scholar]
- 50.Nehete, P. N., R. B. Arlinghaus, and K. J. Sastry. 1993. Inhibition of human immunodeficiency virus type 1 infection and syncytium formation in human cells by V3 loop synthetic peptides from gp120. J. Virol. 67**:**6841-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parry, D. A. 1978. Fibrinogen: a preliminary analysis of the amino acid sequences of the portions of the alpha, beta, and gamma-chains postulated to form the interdomainal link between globular regions of the molecule. J. Mol. Biol. 248**:**180-189. [DOI] [PubMed] [Google Scholar]
- 52.Peiris, J., S. Lai, L. Poon, Y. Guan, L. Yam, W. Lim, J. Nicholls, W. Yee, W. Yan, M. Cheung, V. Cheng, K. Chan, D. Tsang, R. Yung, T. Ng, and K. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361**:**1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Brunham, and A. J. McGeer. 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348**:**1995-2005. [DOI] [PubMed] [Google Scholar]
- 54.Rapaport, D., M. Ovadia, and Y. Shai. 1995. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 14**:**5524-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72**:**986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruigrok, R. W., A. Aitken, L. J. Calder, S. R. Martin, J. J. Skehel, S. A. Wharton, W. Weis, and D. C. Wiley. 1988. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J. Gen. Virol. 69**:**2785-2795. [DOI] [PubMed] [Google Scholar]
- 57.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20**:**4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166**:**368-379. [DOI] [PubMed] [Google Scholar]
- 59.Siddell, S. G. 1995. The Coronaviridae; an introduction. Plenum Press, New York, N.Y.
- 60.Skehel, J. J., and D. C. Wiley. 1998. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95**:**871-874. [DOI] [PubMed] [Google Scholar]
- 61.Slepushkin, V. A., G. V. Kornilaeva, S. M. Andreev, M. V. Sidorova, A. O. Petrukhina, G. R. Matsevich, S. V. Raduk, V. B. Grigoriev, T. V. Makarova, V. V. Lukashov, and E. V. Karamov. 1993. Inhibition of human immunodeficiency virus type 1 (HIV-1) penetration into target cells by synthetic peptides mimicking the N-terminus of the HIV-1 transmembrane glycoprotein. Virology 194**:**294-301. [DOI] [PubMed] [Google Scholar]
- 62.Stauber, R., M. Pfleiderera, and S. Siddell. 1993. Proteolytic cleavage of the murine coronavirus surface glycoprotein is not required for fusion activity. J. Gen. Virol. 74**:**183-191. [DOI] [PubMed] [Google Scholar]
- 63.Sturman, L. S., C. S. Ricard, and K. V. Holmes. 1990. Conformational change of the coronavirus peplomer glycoprotein at pH 8.0 and 37°C correlates with virus aggregation and virus-induced cell fusion. J. Virol. 64**:**3042-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taguchi, F. 1993. Fusion formation by the uncleaved spike protein of murine coronavirus JHMV variant cl-2. J. Virol. 67**:**1195-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taguchi, F. 1995. The S2 subunit of the murine coronavirus spike protein is not involved in receptor binding. J. Virol. 69**:**7260-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94**:**12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15**:**2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vennema, H., R. Rijnbrand, L. Heijnen, M. C. Horzinek, and W. J. Spaan. 1991. Enhancement of the vaccinia virus/phage T7 RNA polymerase expression system using encephalomyocarditis virus 5′-untranslated region sequences. Gene 108**:**201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vennema, H., P. J. Rottier, L. Heijnen, G. J. Godeke, M. C. Horzinek, and W. J. Spaan. 1990. Biosynthesis and function of the coronavirus spike protein. Adv. Exp. Med. Biol. 276**:**9-19. [DOI] [PubMed] [Google Scholar]
- 70.Weissenhorn, W., L. J. Calder, S. A. Wharton, J. J. Skehel, and D. C. Wiley. 1998. The central structural feature of the membrane fusion protein subunit from the Ebola virus glycoprotein is a long triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 95**:**6032-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2**:**605-616. [DOI] [PubMed] [Google Scholar]
- 72.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387**:**426-430. [DOI] [PubMed] [Google Scholar]
- 73.Weissenhorn, W., S. A. Wharton, L. J. Calder, P. L. Earl, B. Moss, E. Aliprandis, J. J. Skehel, and D. C. Wiley. 1996. The ectodomain of HIV-1 env subunit gp41 forms a soluble, alpha-helical, rod-like oligomer in the absence of gp120 and the N-terminal fusion peptide. EMBO J. 15**:**1507-1514. [PMC free article] [PubMed] [Google Scholar]
- 74.Westenberg, M., H. Wang, W. F. IJkel, W. Goldbach, J. M. Vlak, and D. Zuidema. 2002. Furin is involved in baculovirus envelope fusion protein activation. J. Virol. 76**:**178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89**:**10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91**:**9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams, R. K., G. S. Jiang, and K. V. Holmes. 1991. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. USA 88**:**5533-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289**:**366-373. [DOI] [PubMed] [Google Scholar]
- 79.Yang, Z. N., T. C. Mueser, J. Kaufman, S. J. Stahl, P. T. Wingfield, and C. C. Hyde. 1999. The crystal structure of the SIV gp41 ectodomain at 1.47 Å resolution. J. Struct. Biol. 126**:**131-144. [DOI] [PubMed] [Google Scholar]
- 80.Yao, Q., and R. W. Compans. 1996. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 223**:**103-112. [DOI] [PubMed] [Google Scholar]
- 81.Yoo, D. W., M. D. Parker, and L. A. Babiuk. 1991. The S2 subunit of the spike glycoprotein of bovine coronavirus mediates membrane fusion in insect cells. Virology 180**:**395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Young, J. K., R. P. Hicks, G. E. Wright, and T. G. Morrison. 1997. Analysis of a peptide inhibitor of paramyxovirus (NDV) fusion using biological assays, NMR, and molecular modeling. Virology 238**:**291-304. [DOI] [PubMed] [Google Scholar]
- 83.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97**:**14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]