Conditional Suppression of Cellular Genes: Lentivirus Vector-Mediated Drug-Inducible RNA Interference (original) (raw)
Abstract
RNA interference has emerged as a powerful technique to downregulate the expression of specific genes in cells and in animals, thus opening new perspectives in fields ranging from developmental genetics to molecular therapeutics. Here, we describe a method that significantly expands the potential of RNA interference by permitting the conditional suppression of genes in mammalian cells. Within a lentivirus vector background, we subjected the polymerase III promoter-dependent production of small interfering RNAs to doxycycline-controllable transcriptional repression. The resulting system can achieve the highly efficient and completely drug-inducible knockdown of cellular genes. As lentivirus vectors can stably transduce a wide variety of targets both in vitro and in vivo and can be used to generate transgenic animals, the present system should have broad applications.
The externally controllable expression of exogenous cDNAs can be readily obtained in cells or in animals owing to techniques pioneered more than a decade ago (2, 8). Recently, it was demonstrated that the knockdown of endogenous genes could be achieved by RNA interference, and plasmid- or viral vector-based delivery systems for the stable expression of small interfering RNAs (siRNAs) were rapidly created (1, 3, 5, 7, 9, 17). In many situations, however, it is desirable to suppress genes in a regulated fashion, for instance, to study cellular factors that play essential roles during differentiation or development. On the basis of this premise, we created a lentivirus vector-based system for drug-inducible production of siRNAs in stably transduced mammalian cells.
MATERIALS AND METHODS
Vector construction.
Vectors were constructed by using standard cloning procedures. The pSUPER and pSUPER-p53 constructs were described previously (5). pSUPER-siGFP was provided by F. Iseni (Geneva, Switzerland), and pSUPER-siLamin was a gift from R. Oggi (Lausanne, Switzerland). pLV-H was constructed by inserting the H1 promoter from pSUPER into the 3′ long terminal repeat (LTR) of pWPXL (http://www.tronolab.unige.ch/). To construct pLV-TH, the tetO cassette was excised from pUHD13-3 (obtained from H. Bujard, Heidelberg, Germany) and cloned into pLV-H, upstream of the H1 promoter. Finally, the H1 promoter cassette in pLV-H and pLV-TH was replaced by the H1-siRNA cassette excised from pSUPER-siRNA, generating pLV-H/siRNA and pLV-TH/siRNA, respectively. The sequence encoding tTR-KRAB (kindly provided by P. Lorenz and H.-J. Thiesen, Rostock, Germany) was cloned into pWPXL, replacing the green fluorescent protein (GFP) marker (pLV-tTR-KRAB), or as part of a bicistronic unit also encoding Discosoma sp. Red, using the encephalomyocarditis virus 5′ internal ribosome entry site.
The lentivirus vectors described here are available upon request (www.tronolab.unige.ch/).
Cell culture and transduction with lentivirus vectors.
The 293T, HeLa, and MCF-7 cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. All recombinant lentiviruses were produced by transient transfection of 293T cells according to standard protocols (21). Briefly, subconfluent 293T cells were cotransfected with 20 μg of a plasmid vector, 15 μg of pCMV-ΔR8.91, and 5 μg of pMD2G-VSVG by calcium phosphate precipitation. After 16 h medium was changed, and recombinant lentivirus vectors were harvested 24 h later.
To analyze the regulation of GFP, a HeLa cell clone carrying a single copy of the WPXL-GFP provirus (HeLa-GFP) was used. For transduction, HeLa-GFP, MCF-7, or HeLa cells were plated on 24-well plate (20 × 104 cells/well), and after 16 h medium containing recombinant lentivirus vectors was added. Following 16 h of incubation, the cells were washed and split, and doxycycline (DOX) was added to half of the transduced cells at a final concentration of 5 μg/ml. Five days later the cells were harvested and analyzed by fluorescence-activated cell sorting (FACS).
Western blotting.
Cell extracts were prepared in radioimmunoprecipitation assay lysis buffer (25 mM Tris [pH 7.5], 1% Triton X-100, 0.5% sodium deoxycholate, 5 mM EDTA, 150 mM NaCl) containing a cocktail of protease inhibitors (Sigma). The protein samples (10 μg) were separated on sodium dodecyl sulfate-4 to 20% gradient polyacrylamide gels, electroblotted to polyvinylidene difluoride membranes (Perkin-Elmer), and exposed to antibodies against p53 (Santa Cruz Biotechnology), lamin A/C (Santa Cruz Biotechnology), GFP (Clontech), and actin (Calbiochem). Antibodies conjugated with horseradish peroxidase (Amersham) and enhanced chemiluminescence (Amersham) were used for detection.
FACS analysis.
Harvested HeLa-GFP cells transduced with lentivirus vectors carrying ΔNGFR cDNA were incubated with monoclonal antibody specific for human nerve growth factor receptor NGFR (Becton Dickinson PharMingen) labeled with phycoerythrin, washed twice, and analyzed with a FACSscan (Becton Dickinson) for green (GFP) and red (NGFR-phycoerythrin) fluorescence. MCF-7 and HeLa cells cotransduced with LV-THsi/p53 or LV-THsi/lamin and pLV-tTR-KRAB-Red and cultured in presence or absence of DOX were harvested and analyzed with a FACSscan for green and red (dsRed) fluorescence.
Immunofluorescence.
MCF-7 and HeLa cells cotransduced with LV-THsi/p53 or LV-THsi/lamin and pLV-tTR-KRAB-Red and cultured for 5 days in the presence or absence of DOX were fixed with methanol (10 min, −20°C), blocked with phosphate-buffered saline-1% bovine serum albumin, and stained with antibodies against p53 (Santa Cruz Biotechnology) or lamin A/C (Santa Cruz Biotechnology), using secondary antibodies conjugated with Alexa 633 (Molecular Probes) for detection. Images were acquired by using three-color confocal microscopy (LSM 510; Carl Zeiss) and analyzed with Zeiss software.
RESULTS AND DISCUSSION
We took advantage of a tetracycline-controlled hybrid protein, tTR-KRAB, in which the tetracycline repressor (tTR) from Escherichia coli Tn_10_ is fused to the KRAB domain of human Kox1 (6, 8). KRAB is an approximately 75-amino-acid transcriptional repression module found in many zinc finger-containing proteins, which can suppress, in an orientation-independent manner, both polymerase II- and polymerase III-mediated transcription within a distance of up to 3 kb from its binding site, presumably by triggering the formation of heterochromatin (4, 6, 12, 14, 19). When linked to the DNA-binding domain of tTR, KRAB can modulate transcription from an integrated promoter juxtaposed with tet operator (tetO) sequences (6). In the absence of DOX, tTR-KRAB binds specifically to tetO and suppresses the activity of the nearby promoter. Conversely, in the presence of DOX, tTR-KRAB is sequestered away from tetO, thus permitting gene expression (6).
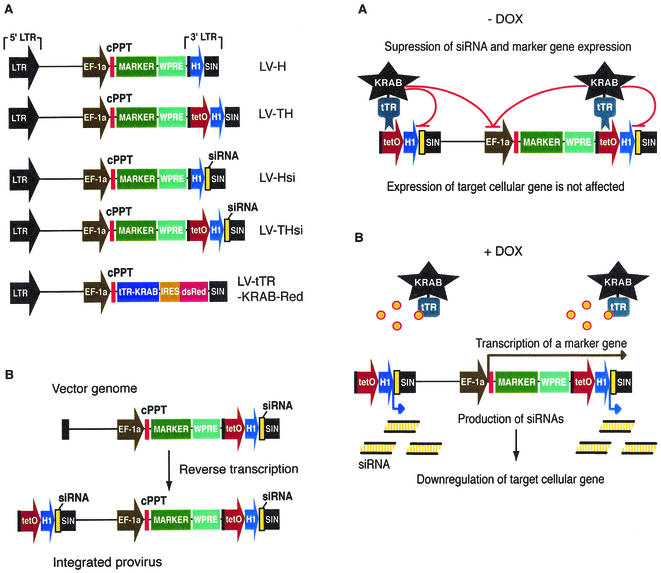
We used human immunodeficiency virus type 1-derived lentivirus vectors (designated LV) as delivery vehicles because we aimed for a system that would be easy to apply to a broad variety of cellular targets, both ex vivo (cell lines, primary cells including stem cells, fertilized oocytes, and blastocysts) and in vivo (e.g., brain and liver) (9, 13, 15, 16-18, 20), and because _tetO_-linked transcriptional units are repressed by tTR-KRAB only when integrated in the genome. The tTR-KRAB cDNA was expressed from the ubiquitously active EF1-α promoter as part of a bicistronic transcript also producing the dsRed marker (Fig. 1A, LV-tTR-KRAB). The regulated siRNA vectors were constructed by inserting a _tetO_-H1 promoter-siRNA cassette into the U3 region of the 3′ LTR of a self-inactivating lentivirus vector (Fig. 1A, LV-THsi). During reverse transcription, the vector RNA 3′ U3 region serves as the template for the synthesis of its 5′ DNA homologue, so that the _tetO_-H1-siRNA cassette is duplicated in the integrated provirus (Fig. 1B). We chose this double-copy configuration to obtain higher rates of siRNA synthesis. Sequences encoding siRNA hairpin precursors were designed as described previously (5). Control vectors carried either a constitutively active H1-siRNA cassette (LV-Hsi) or the H1- or _tetO_-H1 transcriptional elements without downstream siRNA-coding sequence (LV-H and LV-TH, respectively). All siRNA and control vectors also carried a marker gene downstream of an internal EF1-α promoter. We predicted (Fig. 2A) that cells cotransduced with LV-THsi and LV-tTR-KRAB would normally express the gene targeted by the siRNA when maintained in the absence of DOX, owing to tTR-KRAB-mediated suppression of siRNA synthesis. In contrast, addition of the drug would relieve this inhibition and allow for target gene downregulation (Fig. 2B). Expression of the internal marker gene would also be subjected to conditional tTR-KRAB repression, thus providing an internal monitoring device.
FIG. 1.
(Left panels) A lentivirus vector-based system for conditional gene suppression with DOX-inducible siRNAs. (A) Schematic drawing of lentivirus vector plasmids used in this work. Cassettes consisting of the H1 promoter without (LV-H) or with (pLV-TH) the upstream tetO sequence, H1-siRNA (LV-Hsi), and _tetO_-H1-siRNA (LV-THsi) were cloned in the 3′ U3 region of pWPXL (http://www.tronolab.unige.ch/). All of the vectors contain an internal marker cDNA under transcriptional control of the EF-1α promoter. (B) Double-copy design of siRNA lentivirus vectors. During reverse transcription, the U3 region of the 5′ LTR is synthesized by using its 3′ homologue as a template, which results in a duplication of the siRNA cassette in the provirus integrated in the genome of transduced cells.
FIG.2.
(Right panels) Mode of action of the DOX-controllable transrepressor. (A) In the absence of DOX, tTR-KRAB binds to tetO and suppresses H1-mediated siRNA transcription, thus allowing normal expression of the cellular target gene (on). (B) In the presence of DOX, tTR-KRAB cannot bind to tetO and hence siRNAs are produced, leading to downregulation of their target (off). The internal marker contained in the siRNA vectors provides an inverse monitoring device, as it is on in the presence of DOX and off in its absence.
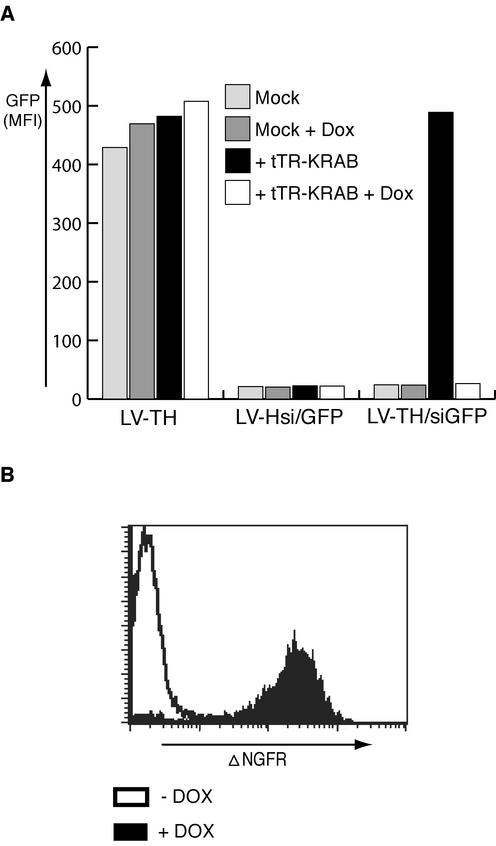
In a first series of experiments, we probed the ability of this system to regulate the production of GFP in HeLa cells stably expressing this fluorophore (Fig. 3A). Vectors were used at a multiplicity of infection of 10 to ensure good rates of (co)transduction. HeLa-GFP cells transduced with the empty LV-TH vector remained strongly GFP positive irrespective of their culture conditions. In contrast, cells transduced with the constitutively active LV-Hsi/GFP vector exhibited a strong downregulation of the marker. In cells transduced with the controllable LV-THsi/GFP vector, GFP expression was observed only in the presence of tTR-KRAB and in the absence of DOX (Fig. 3A). Correspondingly, in the absence of drug, tTR-KRAB suppressed the expression of the vector's ΔNGFR internal reporter gene (Fig. 3B). As expected, the tTR-KRAB-mediated suppression of siRNA production was equally efficient whether tetO was inserted in the sense or antisense orientation and upstream or downstream of the H1 promoter (data not shown).
FIG. 3.
Regulation of GFP expression by using DOX-inducible siRNA. (A) HeLa cells carrying a single copy of a lentivirus vector expressing GFP from the EF-1α promoter (HeLa-GFP) were transduced with a control lentivirus vector (LV-TH) or with vectors producing a GFP-specific siRNA in a constitutive (LV-Hsi) or regulated (LV-THsi) manner, with or without LV-tTR-KRAB (lacking the internal ribosome entry site-dsRed cassette) and/or DOX as indicated. A truncated form of NGFR (ΔNGFR) served as an internal reporter in the siRNA vectors. (B) Conditional expression of the internal marker gene. HeLa-GFP cells dually transduced with LV-THsi/GFP and LV-tTR-KRAB were maintained in the presence or absence of DOX before FACS analysis with a monoclonal antibody specific for the extracellular domain of NGFR.
Next, we tested our system for the regulation of truly endogenous genes. We the chose p53 and lamin genes as targets because highly effective siRNAs directed against these genes were previously identified and well characterized (5, 7). MCF-7 breast cancer cells were used as substrates for p53 downregulation studies (Fig. 4, left panels). Cells cotransduced with LV-tTR-KRAB and LV-THsi/p53 produced wild-type levels of p53 when cultured in the absence of DOX, indicating full repression of siRNA synthesis (lower blot, lane 7). This repression was mediated by tTR-KRAB, since p53 was undetectable in cells transduced only with LV-THsi/p53, whether or not DOX was present in the culture medium (upper blot, lanes 7 and 8). In contrast, addition of the drug to the dually transduced cells resulted in rates of p53 downmodulation as robust as those observed in cells containing the constitutively active LV-Hsi/p53 vector (compare lane 8 in the lower blot with lanes 5 and 6 in both blots). Similar results were obtained for lamin in HeLa cells transduced with the corresponding siRNA lentivirus vectors (Fig. 4, right panels). It is noteworthy that in both settings the drug-induced production of the siRNAs, and hence the suppression of the p53 or lamin target gene, correlated with the expression of the lentivirus vector internal GFP marker, whether examined by Western blotting (Fig. 4) or by FACS or confocal microscopy (data not shown).
FIG. 4.
Regulation of endogenous genes by using DOX-inducible siRNAs. Left panels, downmodulation of p53. MCF-7 cells were infected with the indicated lentivirus vectors as described in Materials and Methods. Western blotting was performed with monoclonal antibodies against p53, GFP, or actin (as a control). Right panels, downmodulation of lamin A/C. The same experiment as for the left panels was performed with HeLa cells, using lamin-specific siRNA vectors and antibodies.
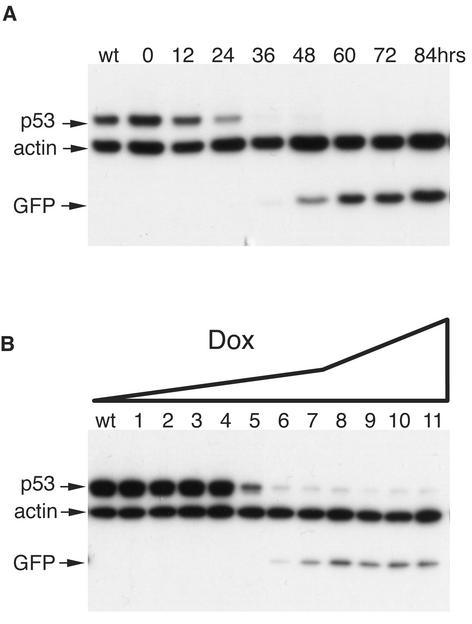
Taken together, these results indicate that the tTR-KRAB-regulated, lentivirus vector-mediated delivery of siRNAs allows for the controllable suppression of cellular genes both with a high degree of efficacy and without significant leakiness. To complete the characterization of this system, we defined its kinetics and DOX dose responsiveness (Fig. 5). We chose p53 as target for these analyses because the half-life of this protein is relatively short, around 12 h. In MCF-7 cells dually transduced with the LV-THsi/p53 and LV-tTR-KRAB vectors, p53 steady-state levels started to decrease as early as 12 h after addition of 5 μg of DOX per ml to the culture medium and became undetectable by Western blotting within 36 h (Fig. 5A). This suggests that RNA interference was fully effective in less than 24 h, implying that the DOX-mediated sequestration of tTR-KRAB rapidly unleashes high rates of siRNA production from the integrated H1 promoters. A dose-response analysis further revealed an extreme sensitivity to DOX control, while pointing to the possibility of some tuning of the gene suppression. Indeed, whereas p53 downregulation was already apparent at the low DOX concentration of 0.004 μg/ml, full-blown suppression was achieved only at a dose of 0.25 μg/ml (Fig. 5B). The anti-p53 siRNA used in this experiment being very efficient, a greater range of DOX concentrations may allow for a modulation of the degree of gene knockdown with siRNAs of lower specific activity.
FIG.5.
Kinetics and dose responsiveness of DOX-inducible RNA interference. (A) MCF-7 cells were cotransduced with LV-THsi/p53 and LV-tTR-KRAB as described in Materials and Methods. Five days later, DOX was added at a concentration of 5 μg/ml. Cells were harvested just before DOX treatment (lane 0) and then at indicated time points. wt, nontransduced cells. Whole-cell extracts were analyzed by Western blotting with p53-specific antibodies. (B) At 5 days posttransduction as described for panel A, cells were placed in medium containing the following concentrations of DOX (in micrograms per milliliter): 0 (lane 1), 0.0005 (lane 2), 0.001 (lane 3), 0.002 (lane 4), 0.004 (lane 5), 0.008 (lane 6), 0.016 (lane 7), 0.063 (lane 8), 0.25 (lane 9), 1 (lane 10), and 5 (lane 11). wt, nontransduced cells. Western blot analyses of whole-cell extracts were performed after another 5 days.
In summary, we provide a system for the conditional suppression of genes in mammalian cells. The versatility of its mode of delivery suggests very broad uses, as lentivirus vectors can transduce a wide range of targets, including stem cells, and can be used for generating transgenic animals from several species. In the latter setting, the system described here should offer significant advantages over currently available conditional knockout techniques, among which are its reversibility and simplicity of use. While the lentivirus vector-mediated delivery of drug-inducible RNA interference may thus be of particular interest for the study genes involved in development and differentiation, it is likely to be useful in many other areas of biology as well.
Acknowledgments
We thank P. Lorenz and H.-J. Thiesen for tTR-KRAB cDNA, F. Iseni for pSUPER-siGFP, R. Oggi for pSUPER-siLamin, S. Vianin for technical assistance, and M.-O. Sauvain, other members of our laboratory, and J. Szulc for helpful discussions.
This work was supported by the Swiss National Science Foundation under the auspices of the National Center for Competence in Research Frontiers in Genetics program and by the Institut Clayton de la Recherche.
REFERENCES
- 1.Abbas-Terki, T., W. Blanco-Bose, N. Deglon, W. Pralong, and P. Aebischer. 2002. Lentiviral-mediated RNA interference. Hum. Gene Ther. 13**:**2197-2201. [DOI] [PubMed] [Google Scholar]
- 2.Agha-Mohammadi, S., and M. T. Lotze. 2000. Regulatable systems: applications in gene therapy and replicating viruses J. Clin. Investig. 105**:**1177-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, G. M., and R. Medzhitov. 2002. Retroviral delivery of small interfering RNA into primary cells. Proc. Natl. Acad. Sci. USA 99**:**14943-14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellefroid, E. J., D. A. Poncelet, P. J. Lecocq, O. Revelant, and J. A. Martial. 1991. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc. Natl. Acad. Sci. USA 88**:**3608-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., R. Bernards, and R. A. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296**:**550-553. [DOI] [PubMed] [Google Scholar]
- 6.Deuschle, U., W. K. Meyer, and H.-J. Thiesen. 1995. Tetracycline-reversible silencing of eukaryotic promoters. Mol. Cell. Biol. 15**:**1907-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411**:**494-498. [DOI] [PubMed] [Google Scholar]
- 8.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89**:**5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacque, J. M., K. Triques, and M. Stevenson. Modulation of HIV-1 replication by RNA interference. Nature **418:**435-438. [DOI] [PMC free article] [PubMed]
- 10.Kafri, T., U. Blomer, D. A. Peterson, F. H. Gage, and I. M. Verma. 1997. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat. Genet. 17**:**314-317. [DOI] [PubMed] [Google Scholar]
- 11.Lois, C., E. J. Hong, S. Pease, E. J. Brown, and D. Baltimore. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295**:**868-872. [DOI] [PubMed] [Google Scholar]
- 12.Margolin, J. F., J. R. Friedman, W. K. Meyer, H. Vissing, H.-J. Thiesen, and F. J. Rauscher III. 1994. Kruppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. USA 91**:**4509-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyoshi, H., K. A. Smith, D. E. Mosier, I. M. Verma, and B. E. Torbett. 1999. Transduction of human CD34+ cells that mediate long-term engraftment of NOD/SCID mice by HIV vectors. Science 283**:**682-686. [DOI] [PubMed] [Google Scholar]
- 14.Moosmann, P., O. Georgiev, H.-J. Thiesen, M. Hagmann, and W. Schaffner. 1997. Silencing of RNA polymerases II and III-dependent transcription by the KRAB protein domain of KOX1, a Kruppel-type zinc finger factor. Biol. Chem. 378**:**669-677. [DOI] [PubMed] [Google Scholar]
- 15.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272**:**263-267. [DOI] [PubMed] [Google Scholar]
- 16.Pfeifer, A., M. Ikawa, Y. Dayn, and I. M. Verma. 2002. Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc. Natl. Acad. Sci. USA 99**:**2140-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin, X. F., D. S. An, I. S. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100**:**183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubinson, D. A., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, M. Zhang, M. T. McManus, F. B. Gertler, M. L. Scott, and L. Van Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33**:**401-406. [DOI] [PubMed] [Google Scholar]
- 19.Senatore, B., A. Cafier, I. Di Marino, M. Rosati, P. P. Di Nocera, and G. Grimaldi. 1999. A variety of RNA polymerases II and III-dependent promoter classes is repressed by factors containing the Kruppel-associated/finger preceding box of zinc finger proteins. Gene 234**:**381-394. [DOI] [PubMed] [Google Scholar]
- 20.Tiscornia, G., O. Singer, M. Ikawa, and I. M. Verma. 2003. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad. Sci. USA 100**:**1844-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15**:**871-875. [DOI] [PubMed] [Google Scholar]