ESEfinder: a web resource to identify exonic splicing enhancers (original) (raw)
Abstract
Point mutations frequently cause genetic diseases by disrupting the correct pattern of pre-mRNA splicing. The effect of a point mutation within a coding sequence is traditionally attributed to the deduced change in the corresponding amino acid. However, some point mutations can have much more severe effects on the structure of the encoded protein, for example when they inactivate an exonic splicing enhancer (ESE), thereby resulting in exon skipping. ESEs also appear to be especially important in exons that normally undergo alternative splicing. Different classes of ESE consensus motifs have been described, but they are not always easily identified. ESEfinder (http://exon.cshl.edu/ESE/) is a web-based resource that facilitates rapid analysis of exon sequences to identify putative ESEs responsive to the human SR proteins SF2/ASF, SC35, SRp40 and SRp55, and to predict whether exonic mutations disrupt such elements.
INTRODUCTION
Accurate and efficient removal of introns from pre-mRNAs is essential to ensure correct gene expression. However, the information content present in the canonical splice signals (5′ splice site, branch site and 3′ splice site) is insufficient to precisely define exons, as a large excess of sequences that conform to these weakly defined consensus elements is present in introns but these sequences are never used (1,2). Additional regulatory _cis_-elements exist in the form of splicing enhancers and silencers (3). These elements become particularly important in the presence of weak splice sites or when alternative splicing is involved. It is estimated that over 60% of human genes undergo alternative splicing (4). Not only is this one of the main mechanisms by which the relatively small number of human genes accounts for the complexity of the proteome, but the generation of different isoforms can be differentially regulated depending on developmental stage, cell type and in response to a wide array of physiological and pathological signals (4,5).
Up to 50% of all point mutations responsible for genetic diseases cause aberrant splicing (3). Such mutations can disrupt splicing by directly inactivating or creating a splice site, by activating a cryptic splice site or by interfering with splicing regulatory elements. Point mutations in the coding regions of genes were traditionally assumed to exert their effects by altering single amino acids in the encoded proteins. However, some of these exonic mutations also affect pre-mRNA splicing. Nonsense, missense and even translationally silent mutations can disrupt exonic splicing enhancers (ESEs) and cause the splicing machinery to skip the mutant exon, with dramatic effects on the structure of the gene product. Since in most cases the effects of mutations are predicted solely based on genomic sequence information, the prevalence of mutations whose primary consequence is aberrant splicing has been substantially underestimated (3).
ESEs are common in both alternative and constitutive exons, where they act as binding sites for Ser/Arg-rich proteins (SR proteins), a family of conserved splicing factors that participate in multiple steps of the splicing pathway (6). SR proteins bind to ESEs through their RNA-binding domain, and promote exon definition by recruiting spliceosomal components via protein–protein interactions mediated by their RS domain and/or by antagonizing the action of nearby splicing silencers. Different SR proteins have different substrate specificities, and multiple classes of ESE consensus motifs have been described (3,6,7).
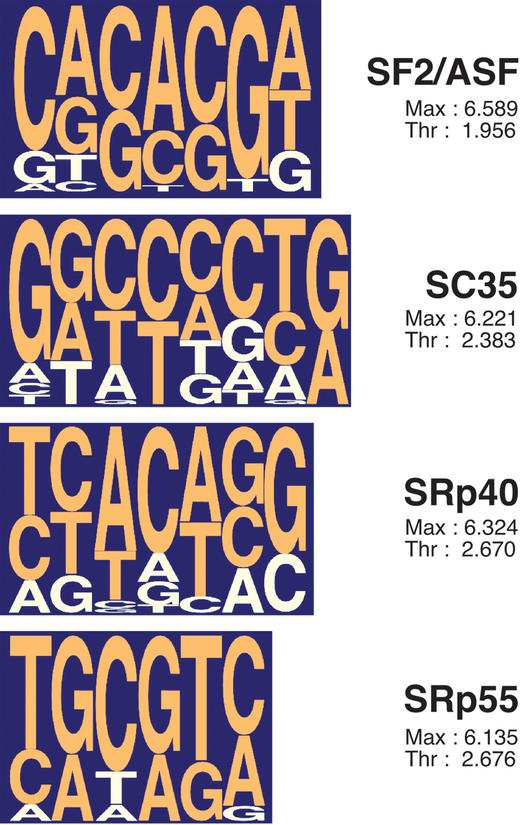
We previously used functional SELEX [Systematic Evolution of Ligands by Exponential enrichment (8)], to identify ESE motifs specific for a subset of SR proteins (9,10). In this approach, a natural enhancer in an IgM minigene was replaced by random 20 nt sequences from an oligonucleotide library. The resulting pool of minigenes was then used to generate pre-mRNA transcripts, which were spliced as a pool in vitro under conditions in which splicing was completely dependent on both an ESE and a recombinant SR protein able to productively recognize this ESE. Spliced mRNAs were gel-purified, amplified and used to rebuild minigene templates, allowing the procedure to be iterated. Specific ESE motifs were thus gradually enriched and eventually cloned, sequenced and individually tested. Using the sequences that resulted from the functional selection procedure, we derived nucleotide-frequency matrices (available on the web site), which define consensus motifs for these SR proteins. The motifs are short (6–8 nt), degenerate and can partially overlap (3) (Fig. 1). Here we describe the implementation of the motif-scoring matrices in a web-based program called ESEfinder (release 2.0: http://exon.cshl.edu/ESE/) which allows scanning of nucleotide sequences to predict putative ESEs responsive to the human SR proteins SF2/ASF, SC35, SRp40 or SRp55. ESEfinder has been freely available for non commercial uses since May 2002, and it has already been used successfully to predict ESEs and/or their disruption in a variety of genes, including ACF (11), BRCA1 (12), BRCA2 (13), FBN1 (14), IGF1 (15), PDHA1 (16), SMN1 (17), SMN2 (17), TNFRSF5 (18), CFTR (19,20) and others.
Figure 1.
Pictograms (1) representing the functional-SELEX consensus ESE motifs. The height of each letter reflects the frequency of each nucleotide at a given position, after adjusting for background nucleotide composition. At each position, the nucleotides are shown from top to bottom in order of decreasing frequency; orange letters indicate above-background frequencies. For each motif, the threshold value and the highest possible score are provided.
DESCRIPTION
ESEfinder performs searches for putative ESEs in query sequences by using weight matrices corresponding to the motifs for four different human SR proteins. The matrices are based on frequency values derived from the alignment of winner sequences obtained by functional SELEX experiments, adjusted on the basis of the background nucleotide frequency of the initial SELEX library, which was made by chemical synthesis (9,10). We have now developed a user-friendly WWW interface and a representation of the program output is shown in Figure 2.
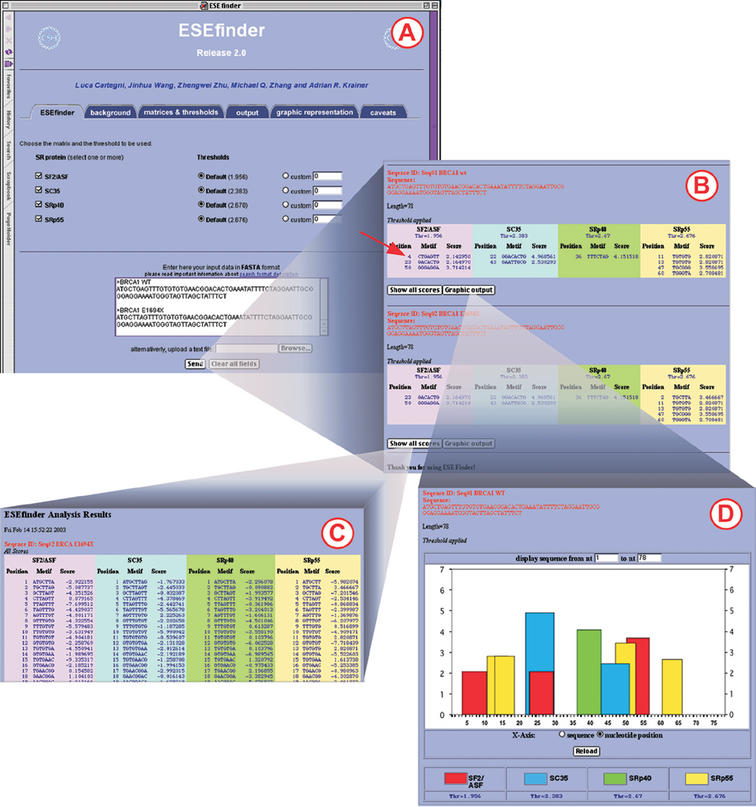
Figure 2.
Example of ESEfinder input and output windows. (A) Input window. Two query sequences, BRCA1 exon 18 and a single point mutation variant (E1694X) are shown. All four matrices and their default threshold values were selected. Additional information is available from the tab links. (B) Output window. High scores, tabulated under each SR protein, are listed. Note that an SF2/ASF high score (arrow) has been abrogated by the mutation. (C) Output window with complete list of scores. (D) Graphic output window. High scores are represented as color-coded bars. The height of each bar indicates the score value, and its width and placement on the _x_-axis represent the length of the motif (6–8 nt) and its position along the sequence.
The query sequences can be directly pasted into the input box or can be uploaded from a text file. Multiple sequences can be analyzed simultaneously, provided that a FASTA-format descriptive line (beginning with ‘>’) precedes them (Fig. 2A). Even though ESEfinder is an RNA analysis tool, only standard DNA notation is accepted (A, C, G and T, not U). The program will ignore any character other than A, C, G and T, including spaces and paragraph breaks. Both upper and lower cases are accepted but the output lines will be in upper case.
The user selects which matrices will be used, up to all four matrices simultaneously. For each matrix, the output is provided as a series of scores calculated in 1 nt increments. In the initial output window (Fig. 2B), only the ‘hits’ or ‘high-score motifs’ are displayed, giving the position of the first nucleotide, the sequence of the motif match, and the calculated score. A score is considered a high score when it is greater than the threshold value defined in the input page. Any score can be chosen as the cutoff value by selecting the ‘custom’ button and typing the desired value in the box. We suggest that for most routine analyses, users select the ‘default’ threshold values, above which we consider a score for a given sequence to be potentially significant. Our default threshold values are defined as the median of the highest scores for each sequence in a set of 30 randomly chosen 20 nt sequences (from the starting pool used for functional SELEX experiments). Such values are currently set as follows: SF2/ASF, 1.956; SC35, 2.383; SRp40, 2.670; SRp55, 2.676. Any refinements or updates will be incorporated as they become available. From the output window, the complete set of scores for the input sequence can be selected (Fig. 2C).
To facilitate the interpretation of the results and to standardize their representation, we implemented a graphic output of the query that is accessible from the output page (Fig. 2D). The query (exonic) sequence is reproduced along the _x_-axis. The presence of a high-score motif (above the selected threshold) is indicated by the color-coded bars. The height of the bars represents the motif scores, whereas their width indicates the length and position (6–8 nt).
DISCUSSION
ESEfinder allows for the identification of putative ESEs and one of its most useful applications is the correct interpretation of the effects of disease-associated point mutations or polymorphisms. We have previously shown that ESEs predicted by this matrix-based approach tend to cluster in regions where natural enhancers have been experimentally mapped and are more frequent in exons than in introns (9,10). In a database of 50 human point mutations known to cause in vivo exon skipping, the majority reduced or eliminated at least one predicted ESE (12). Considering that we can currently search for putative ESEs using matrices for just four SR proteins, it is likely that a large fraction of skipping-associated mutations do indeed cause ESE disruption, and that a higher predictive value will be obtained when matrices for other relevant splicing factors become available. A computational approach (RESCUE-ESE) was recently described (7), in which putative ESE motifs are identified by comparing the frequency of hexamers in exons surrounded by ‘weak’ versus ‘strong’ splice sites. Several hexamer families enriched in the weak exons, which likely depend on enhancers for correct expression, were identified, and some of these overlap with the motifs defined by ESEfinder.
The ESEfinder matrices have been used to show that disruption of ESEs recognized by various SR proteins cause exon skipping in several genes (11–18). In some contexts, ESEfinder appears to be remarkably accurate. For example, using a _BRCA1_-derived three-exon minigene system, which is very responsive to point mutations within a critical ESE, we showed that when multiple SF2/ASF-dependent ESEs were substituted for each other or mutated, there was a strong correlation between exon-inclusion efficiency and the matrix scores (12,17). Furthermore, ESEfinder was used in combination with mutational analysis, in vitro and in vivo splicing, and site-specific UV-crosslinking experiments to demonstrate that the translationally silent, single-nucleotide difference between SMN1 and SMN2 disrupts an ESE, which in SMN1 is directly recognized by splicing factor SF2/ASF (17). The disruption of the SF2/ASF-dependent ESE causes inefficient SMN2 exon 7 inclusion. In the absence of SMN1, SMN2 is unable to produce enough full-length SMN protein, thus resulting in a spinal muscular atrophy phenotype. Finally, we exploited the degeneracy of the consensus motif, and used ESEfinder to design a second-site suppressor mutation that reconstituted the high-score motif and fully restored exon 7 inclusion in the SMN2 context in vivo and in vitro, as predicted (17). More than a dozen wild-type and mutant SF2/ASF heptamer motifs were tested in the SMN and BRCA1 systems (12,17). All of the motifs that maintained a high-score promoted exon inclusion in a manner roughly proportional to the motif score, even though, because of the degeneracy of the consensus motif, some of them did not share a single nucleotide. All of the motifs with below-threshold scores resulted in reduced levels of exon inclusion.
It should be emphasized, however, that the presence of a high-score motif in a sequence does not necessarily identify that sequence as a functional ESE, and that, in general, there is not a very strict quantitative correlation between numerical scores and ESE activity. Until stronger predictive algorithms are available, direct experimental evidence will remain necessary before safely concluding that a particular sequence can act as an ESE in its natural context. Conversely, the lack of a high-score motif does not imply that no ESEs are present. Several important variables, such as the local sequence context, the splice-site strengths, the position of the ESE along the exon and the presence of silencer elements, are likely to play a significant role in ESE activity. Furthermore, even mutations that abrogate genuine ESEs might not always exert a noticeable effect, because of the presence of redundant ESEs nearby. Finally, it should be noted that our matrices were defined in a mammalian system and reflect the sequence specificity of the human SR proteins. Their relevance to other species depends on the extent of conservation of each SR protein.
The development and refinement of reliable prediction tools for auxiliary splicing elements will have important implications for our ability to accurately identify the exon/intron structures of genes and predict their expression profile, to correctly interpret the effects of point mutations and/or polymorphisms, and to assess phenotypic risk.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the many users that sent us useful comments and suggestions which have been incorporated in the current release. We thank Xavier Roca for comments on the manuscript and Gengxin Chen for assistance. This work was supported by NIH grants GM42699 to A.R.K. and CA88351 and HG01696 to M.Q.Z.
REFERENCES
- 1.Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to messenger RNAs by the spliceosome. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds) The RNA World II, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560. [Google Scholar]
- 2.Sun H. and Chasin,L.A. (2000) Multiple splicing defects in an intronic false exon. Mol. Cell. Biol., 20, 6414–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartegni L., Chew,S.L. and Krainer,A.R. (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet., 3, 285–298. [DOI] [PubMed] [Google Scholar]
- 4.Maniatis T. and Tasic,B. (2002) Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature, 418, 236–243. [DOI] [PubMed] [Google Scholar]
- 5.Ladd A.N. and Cooper,T.A. (2002) Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol., 3, reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graveley B.R. (2000) Sorting out the complexity of SR protein functions. RNA, 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fairbrother W.G., Yeh,R.F., Sharp,P.A. and Burge,C.B. (2002) Predictive identification of exonic splicing enhancers in human genes. Science, 297, 1007–1013. [DOI] [PubMed] [Google Scholar]
- 8.Tuerk C. and Gold,L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 9.Liu H.X., Zhang,M. and Krainer,A.R. (1998) Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev., 12, 1998–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H.X., Chew,S.L., Cartegni,L., Zhang,M.Q. and Krainer,A.R. (2000) Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol., 20, 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dance G.S., Sowden,M.P., Cartegni,L., Cooper,E., Krainer,A.R. and Smith,H.C. (2002) Two proteins essential for apolipoprotein B mRNA editing are expressed from a single gene through alternative splicing. J. Biol. Chem., 277, 12703–12709. [DOI] [PubMed] [Google Scholar]
- 12.Liu H.X., Cartegni,L., Zhang,M.Q. and Krainer,A.R. (2001) A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nature Genet., 27, 55–58. [DOI] [PubMed] [Google Scholar]
- 13.Fackenthal J.D., Cartegni,L., Krainer,A.R. and Olopade,O.L. (2002) BRCA2 T2722R is a deleterious allele that causes exon skipping. Am. J. Hum. Genet., 71, 625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caputi M., Kendzior,R.J.,Jr and Beemon,K.L. (2002) A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes Dev., 16, 1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith P.J., Spurrell,E.L., Coakley,J., Hinds,C.J., Ross,R.J.M., Krainer,A.R. and Chew,S.L. (2002) An exonic splicing enhancer in human IGF-I pre-mRNA mediates recognition of alternative exon 5 by the serine-arginine protein splicing factor-2/alternative splicing factor. Endocrinology, 143, 146–154. [DOI] [PubMed] [Google Scholar]
- 16.Mine M., Brivet,M., Touati,G., Grabowski,P.J., Abitbol,M. and Marsac,C. (2003) Splicing error in E1 alpha PDH mRNA caused by novel intronic mutation responsible for lactic acidosis and mental retardation. J. Biol. Chem., 278, 11768–11772. [DOI] [PubMed] [Google Scholar]
- 17.Cartegni L. and Krainer,A.R. (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nature Genet., 30, 377–384. [DOI] [PubMed] [Google Scholar]
- 18.Ferrari S., Giliani,S., Insalaco,A., Al-Ghonaium,A., Soresina,A.R., Loubser,M., Avanzini,M.A., Marconi,M., Badolato,R., Ugazio,A.G. et al. (2001) Mutations of CD40 gene cause an autosomal recessive form of immunodeficiency with hyper IgM. Proc. Natl Acad. Sci. USA, 98, 12614–12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pagani F., Buratti,E., Stuani,C. and Baralle,F.E. (2003) Missense, nonsense and neutral mutations define juxtaposed regulatory elements of splicing in CFTR Exon 9. J. Biol. Chem., PMID: 12732620. [DOI] [PubMed] [Google Scholar]
- 20.Pagani F., Stuani,C., Tzetis,M., Kanavakis,E., Efthymiadou,A., Doudounakis,S., Casals,T. and Baralle,F.E. (2003) New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet., 12, 1111–1120. [DOI] [PubMed] [Google Scholar]