Overexpression of groESL in Clostridium acetobutylicum Results in Increased Solvent Production and Tolerance, Prolonged Metabolism, and Changes in the Cell's Transcriptional Program (original) (raw)
Abstract
DNA array and Western analyses were used to examine the effects of groESL overexpression and host-plasmid interactions on solvent production in Clostridium acetobutylicum ATCC 824. Strain 824(pGROE1) was created to overexpress the groESL operon genes from a clostridial thiolase promoter. The growth of 824(pGROE1) was inhibited up to 85% less by a butanol challenge than that of the control strain, 824(pSOS95del). Overexpression of groESL resulted in increased final solvent titers 40% and 33% higher than those of the wild type and plasmid control strains, respectively. Active metabolism lasted two and one half times longer in 824(pGROE1) than in the wild type. Transcriptional analysis of 824(pGROE1) revealed increased expression of motility and chemotaxis genes and a decrease in the expression of the other major stress response genes. Decreased expression of the dnaKJ operon upon overexpression of groESL suggests that groESL functions as a modulator of the CIRCE regulon, which is shown here to include the hsp90 gene. Analysis of the plasmid control strain 824(pSOS95del) revealed complex host-plasmid interactions relative to the wild-type strain, resulting in prolonged biphasic growth and metabolism. Decreased expression of four DNA gyrases resulted in differential expression of many key primary metabolism genes. The ftsA and ftsZ genes were expressed at higher levels in 824(pSOS95del), revealing an altered cell division and sporulation pattern. Both transcriptional and Western analyses revealed elevated stress protein expression in the plasmid-carrying strain.
The toxic nature of solvents on bacteria is a major limiting factor in the production of chemicals by fermentation or whole-cell biotransformations (18). The toxic effects of solvents have been widely studied in gram-negative bacteria (17, 18, 27). Accumulation of organic solvents has been shown to permeabilize the cell membrane, resulting in a passive flux of ATP, protons, ions, and macromolecules such as RNA and proteins (36). The flux of ions dissipates the proton motive force and affects the proton gradient (ΔpH) and electrochemical potential (ΔΨ), thereby diminishing the energy status of the cell. Solvents may also disrupt the function of embedded membrane proteins and drastically alter membrane fluidity (4, 35).
Most organisms with demonstrated ability to tolerate otherwise toxic solvent levels have cellular adaptations which effectively suppress solvent effects on the membrane through changes in membrane composition (18). Such specific alterations have primarily been demonstrated in Pseudomonas strains. Another class of solvent-tolerant bacteria includes those with an efflux system which actively decreases the concentration of toxic solvents within the cell (27). A third mechanism, similar to that of antibiotic resistance, is degradation of the toxic substance to a less toxic product (10). Finally, toxic solvents have been shown to induce known stress (heat shock) proteins (HSPs) (38). Growth has been shown to be the most sensitive cellular activity, while glycolysis is more resistant to the effects of solvents (17).
Clostridium acetobutylicum ATCC 824 is capable of producing the solvents butanol and acetone. Butanol toxicity in C. acetobutylicum is quite severe and has been attributed to its chaotropic effect on the cell membrane (4, 44). High butanol concentrations have been shown to inhibit nutrient transport, glucose uptake, and membrane-bound ATPase activity (4). The cellular energy status is also severely affected: intracellular pH and ATP concentrations decrease, and the membrane ΔpH and ΔΨ are abolished. C. acetobutylicum fermentations rarely produce more than 13 g/liter, a level generally considered the toxic limit (19). However, we have generated several recombinant strains able to accumulate up to 18.5 g of butanol/liter without any specific membrane alteration or selection (13, 14). This suggests that the membrane model is not sufficient to explain butanol toxicity. Moderate increases in butanol concentration have been shown to elicit a response similar to heat shock (38). However, the role of stress proteins (HSPs) in solvent tolerance has yet to be demonstrated.
The stress response provides cells with a homeostatic mechanism to survive a variety of environmental stresses (24). HSPs, also called molecular chaperones, play an essential role in the synthesis, transport, and folding of proteins. Class I heat shock genes in Bacillus subtilis include the dnaK and groESL operons, whose expression involves the _cis_-active inverted repeat CIRCE (controlling inverted repeat of chaperone expression) (23). Under normal physiological conditions, class I HSPs are responsible for folding newly translated proteins, translocation of proteins across membranes, disassembly of oligomeric protein structures, and proteolytic degradation of unstable proteins. Under heat shock or stress conditions, they primarily serve to prevent aggregation and assist in protein folding.
GroEL forms a large cage with two rings of seven subunits surrounding a central cavity. Misfolded proteins bind to the hydrophobic lining of the cavity and are released upon binding of GroES and hydrolysis of ATP (28). In contrast, DnaK is characterized by a “brick-shaped” domain, with a hole through which bound peptides run. The peptides are in a completely extended conformation and are also released by hydrolysis of ATP. Class I heat shock operons are negatively regulated by the HrcA repressor protein, which interacts with the CIRCE element. The dnaK and groESL operon structures in C. acetobutylicum are nearly identical to those in B. subtilis (1). It has been proposed that regulation of the class I heat shock genes in C. acetobutylicum is very similar to that in B. subtilis.
Host-plasmid interactions have been widely reported in the literature (2, 30, 45). The presence of a plasmid has been shown to change growth rate, ribosome content, heat shock protein levels, and host enzymatic activities (2). The prevailing theory regarding host-plasmid interactions is that plasmids represent a metabolic burden (30). This idea has been accepted based on the large amounts of experimental data consistent with this theory. However, recent evidence suggests that the complex interactions between host and plasmid are a result of alterations to the cellular regulatory status. The ability to perform large-scale transcriptional analysis with DNA arrays may help elucidate the extent and nature of host-plasmid interactions. Plasmid-containing strains of C. acetobutylicum have been shown to produce higher levels of solvents and lower level of acids than wild-type cells in controlled pH 4.5 batch fermentations (45). The observed effect was independent of the plasmid DNA sequence. Less of an effect was observed in higher-pH fermentations (pH 5.5).
The first aim of this study was to test the hypothesis that overexpression of groESL will increase butanol tolerance and allow accumulation of increased solvent titers. A multicopy replicative plasmid was constructed to overexpress the groESL operon genes from the clostridial thiolase promoter. The corresponding control plasmid was used in the second aim, to better understand the observed host-plasmid interactions. A combination of phenotypic, large-scale transcriptional and Western analyses were used to this end. The results show that overexpression of groESL provides resistance to butanol and that host-plasmid interactions are the result of an altered regulatory state accompanied by a general stress response. In addition, new insights into the regulation of heat shock protein expression were gained.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or referenceb |
|---|---|---|
| Strains | ||
| E. coli ER2275 | hsdR mcrA recA1 endA1 | NEB |
| E. coli Top 10 | hsdR mcrA recA1 endA1 | Invitrogen |
| C. acetobutylicum ATCC 824 | Wild-type strain | ATCC |
| Plasmids | ||
| pAN1 | p15A ori Cmr φ3T I | 22 |
| pSOS95 | ColE1 pIM13 Apr MLSr P-thl ctfA/B adc | 42 |
| pSOS95del | ColE1 pIM13 Apr MLSr P-thl | This study |
| pGROE1 | ColE1 pIM13 Apr MLSr P-thl groES groEL | This study |
Analytical methods.
Cell growth was determined by measuring the absorbance at 600 nm (_A_600) with a Beckman (Fullerton, Calif.) DU64 spectrophotometer. Samples were diluted with the appropriate medium to maintain absorbance readings below 0.40. Supernatants from the bioreactor samples were analyzed for acetate, butyrate, acetone, butanol, ethanol, acetoin, and glucose levels with a Waters (Milford, Mass.) high-pressure liquid chromatography system (5). A Bio-Rad (Hercules, Calif.) Aminex HPX-87H ion exchange column (7.8 by 300 mm) with a Cation H guard column (4.6 by 30 mm) at 15°C was used with a mobile phase of 0.05 mM sulfuric acid flowing at 0.50 ml/min. A refractive index (RI) detector (Waters) was used at 30°C for signal detection.
Growth conditions and maintenance.
Escherichia coli strains were grown aerobically in liquid Luria-Bertani (LB) medium at 220 rpm and 37°C and on agar-solidified LB at 37°C unless otherwise noted. The medium was supplemented with the appropriate antibiotics: ampicillin at 100 μg/ml and chloramphenicol at 35 μg/ml. Frozen stocks were prepared from overnight cultures and stored in LB plus 15% glycerol at −85°C. C. acetobutylicum strains were grown in an anaerobic chamber (Forma Scientific, Marietta, Ohio) at 37°C. Liquid cultures were grown in clostridia growth medium (CGM) (46), and colonies were obtained from agar-solidified 2× YTG (46). Cells from a single colony, at least 5 days old, were used to inoculate liquid cultures, which were then heat shocked at 70 to 80°C for 8 min before growth at 37°C. Frozen stocks were prepared from cells at an _A_600 of 0.8 to 1.0 and stored in CGM plus 20% glycerol at −85°C.
DNA isolation, manipulation, and transformation.
Isolation of plasmid DNA from E. coli was performed by standard protocols (33). Large-scale plasmid DNA preparations were performed with the QIAFilter plasmid maxi kit (Qiagen, Inc., Valencia, Calif.). Plasmid and chromosomal DNAs from C. acetobutylicum were isolated as previously described (15, 42). E. coli transformations were carried out with One Shot Top10 Chemically Competent cells (Invitrogen, Carlsbad, Calif.) according to the manufacturer's protocol, except outgrowth was done at room temperature. C. acetobutylicum transformations were performed as previously described (22). All cloning enzymes were used according to the supplier's protocols (New England Biolabs, Beverly, Mass.; MBI Fermentas, Hanover, Md.; Amersham Pharmacia Biotech, Piscataway, N.J.). DNA was purified with a GFX PCR DNA and gel band purification kit from Amersham Pharmacia Biotech. PCRs were performed with rTth DNA polymerase XL (Applied Biosystems, Foster City, Calif.) in Invitrogen's PCR buffer B (10 mM MgCl2, pH 8.5).
Plasmid constructs.
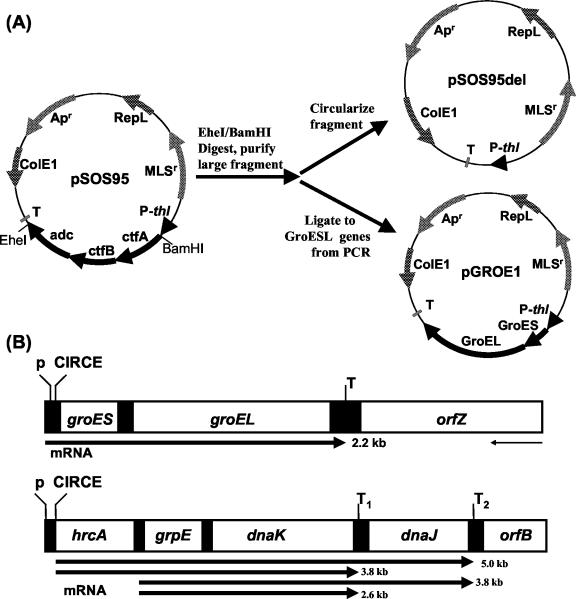
Plasmid pGROE1 was created to overexpress the groESL operon genes (groES and groEL) under control of the clostridial thiolase promoter, while pSOS95del is the corresponding control plasmid. Construction of pSOS95del and pGROE1 is outlined in Fig. 1. Plasmid pSOS95 (42) was digested with _Ehe_I, purified with a GFX DNA purification column, and subsequently digested with _Bam_HI. The large fragment (5.0 kb) was isolated with a GFX gel band purification column and blunt ended with Klenow fragment. Religation of the large fragment resulted in the formation of pSOS95del. For pGROE1, the _gro_ESL operon minus its putative promoter region was amplified by PCR with primers UP-GROE (5′-GCCAAAATTAAGTTTATACTAAAAG-3′) and DN-GROE (5′-AATGCACTCTTATTACATTAATC-3′), with chromosomal DNA as a template. The groESL insert (2,144 bp) was ligated to the linear, blunt-ended pSOS95del fragment, and the ligation mix was transformed into E. coli Top10 chemically competent cells at room temperature. Transformations at 37°C failed to produce transformants. Ampicillin-resistant colonies were grown at room temperature, and the resulting plasmid DNA was screened by digestion with _Pvu_II and _Hin_dIII.
FIG. 1.
Construction of pSOS95del and pGROE1 (A) and structure of groESL and dnaKJ operons of C. acetobutylicum (B) (adapted from Bahl et al. [1]). The location and direction of relevant genes are indicated with arrows. Relevant restriction sites are shown. Abbreviations: Apr, ampicillin resistance; MLSr, macrolide-lincosamide-streptogramin B resistance; RepL, pIM13 origin of replication; ColE1, ColE1 origin of replication; P-thl, clostridial thiolase promoter; T, terminator; ctfA, CoAT subunit A gene; ctfB, CoAT subunit B gene; adc, acetoacetate decarboxylase gene; p, promoter.
Bioreactor experiments.
Large-scale fermentations were carried out in Biostat M (B. Braun, Allentown, Pa.) and BioFlo II (New Brunswick Scientific, Edison, N.J.) bioreactors with 1.5-liter and 4.0-liter working volumes, respectively. The reactor medium (CGM) was supplemented with 75 μg of clarithromycin (Abbott Labs, Abbott Park, Ill.) per ml and 0.15% antifoam C (Sigma Chemical Co., St. Louis, Mo.). Preculture tubes containing 10 ml of CGM were inoculated with heat-shocked colonies, grown to an _A_600 of 0.6, and used to inoculate flasks containing CGM equal to 10% of the reactor working volume. When the flasks reached an _A_600 of 0.20, the flasks were used to inoculate the bioreactors (1:10 dilution). Reactor pH was controlled at a low-end value of 5.0 with 6 N ammonium hydroxide. When glucose levels reached half their initial concentration, 3.5 M glucose was fed to return the glucose concentration to preinoculum levels. Cell samples from throughout the fermentation were plated on 2× YTG plates, with and without erythromycin, to ensure the presence of the plasmid. One round of replica plating was also performed for further verification. Additionally, plasmid DNA preparations and digests were used to verify the presence of the plasmid. Finally, the presence of the pSOL1 megaplasmid (7) was verified by monitoring amylase activity on 2× YTGMA plates (15, 32).
Metabolic flux analysis.
Metabolite concentration data from the bioreactor experiments were analyzed with a metabolic flux analysis program developed by Desai et al. (8). Fluxes calculated with this method typically have errors of less than 10% (14).
Crude cell extracts.
Cell pellets from 2 to 8 ml of culture were collected by centrifugation at 16,000 × g for 5 min, washed with sterile water, and spun again for 2 min. The cell pellets were immediately frozen at −85°C until extracts were prepared. After thawing on ice, 150 μl of sodium dodecyl sulfate (SDS) buffer (50 mM Tris [pH 6.8], 2% SDS, 10% glycerol, and 5% β-mercaptoethanol) was added, and the mixture was boiled for 3 min. The samples were spun at 16,000 × g for 3 min, and the supernatant was collected, aliquoted, and stored at −85°C. Total protein concentrations were determined with an RC DC protein assay kit (Bio-Rad) with bovine serum albumin as a standard, with a standard error of the mean of less than 10%.
Western blot analysis.
For electrophoresis of proteins, 10 μg of total protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Ready-gels (12% polyacrylamide, 4% stacking) from Bio-Rad Laboratories. After electrophoresis, proteins were transferred to 0.2-μm Immun-Blot polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked with Tris-buffered saline plus Tween 20 (TBST)-4% ECL blocking agent (Amersham Pharmacia Biotech) before hybridization. The blocked membrane was incubated with either rabbit anti-GroEL polyclonal antibody (E. coli GroEL as immunogen; StressGen, Victoria, Canada), rabbit anti-HSP70 polyclonal antibody (E. coli DnaK as immunogen; Upstate Biotechnology, Lake Placid, N.Y.), rabbit anti-acetoacetate decarboxylase (AADC) antiserum (25), or sheep anti-coenzyme A transferase (CoAT) antiserum (6) at dilutions of 1:2,000, 1:1,000, 1:4,000, and 1:10,000, respectively. Goat anti-rabbit immunoglobulin G for GroEL, DnaK, and AADC immunoblots or donkey anti-sheep immunoglobulin G for CoAT immunoblots conjugated with horseradish peroxidase (1:10,000 dilution) were used as a secondary antibody (Sigma). All antibody dilutions were prepared in TBST-4% ECL blocking agent. The conjugated antibodies were fluorescently reacted with an ECL+ kit (Amersham Pharmacia Biotech) and detected with a Storm 860 Imager (Molecular Dynamics, Sunnyvale, Calif.).
RNA sampling, isolation, and purification.
Cell pellets from 5 to 15 ml of culture were collected by centrifugation at 5,000 × g for 10 min at 4°C. The pellets were resuspended in 200 μl of SET buffer (25% sucrose, 50 mM Tris [pH 8], 50 mM EDTA [pH 8]) with 20 mg of lysozyme per ml, and the samples were incubated at 37°C for 5 min. Cold Trizol reagent (1 ml; Invitrogen) was added, and the samples were vortexed for 30 s. The Trizol samples were immediately frozen at −85°C, and the RNA was purified within 1 month to avoid degradation.
For isolation and purification, 300 μl of chloroform was added to 1 ml of the Trizol-treated samples (quickly thawed at 37°C), vortexed, and allowed to stand for 2 min at room temperature. The samples were centrifuged at 12,000 × g for 15 min at 4°C, and the aqueous phase was transferred to a fresh tube. Then 0.5 ml of isopropanol was added, and the samples were allowed to stand for 3 min and then centrifuged at 12,000 × g for 10 min at 4°C. The resulting pellet was washed with 75% ethanol and spun at 8,000 × g for 4 min at 4°C. The pellet was washed with an Li/Mg buffer (0.38 M LiCl2 [pH 8], 4.7 mM MgCl2, 50% isopropanol), followed by another ethanol wash. The RNA was finally resuspended in RNase-free water and quantitated with a UV spectrophotometer. Samples were stored at −85°C.
cDNA microarrays.
cDNA microarrays with spots representing 1,019 open reading frames, approximately one-fourth of the C. acetobutylicum genome, were printed (39) with the TIGR protocol (16). A complete list of genes included on the arrays, along with complete expression profile data, is available at http://www.chem-eng.northwestern.edu/Faculty/papou.html. For array fabrication, PCR primers were designed to amplify gene fragments with an average size of approximately 470 bp so that nonspecific hybridization is minimized. The resulting PCR products were dried in a vacuum centrifuge, dissolved in a 35% dimethyl sulfoxide solution, and spotted at least in triplicate on Corning (Corning, N.Y.) CMT-GAPS glass microarray slides with a BioRobotics (Woburn, Mass.) MicroGrid II DNA arrayer. Most genes involved in solventogenesis and sporulation are represented by as many as 12 spots. The slides were UV cross-linked (Stratagene cross-linker) and baked at 80°C for 2 h.
cDNA labeling and hybridization.
Labeled cDNA was synthesized by random hexamer-primed reverse transcription reactions in the presence of indocarbocyanine (Cy3)-dUTP or indodicarbocyanine (Cy5)-dUTP with Moloney murine leukemia virus reverse transcriptase. Forty micrograms of RNA was mixed with 160 μg of random hexamer primers (Roche, Indianapolis, Ind.), heated to 75°C for 7 min, cooled to 37°C for 1 min, and allowed to sit at room temperature for 10 min. To this sample were added unlabeled deoxynucleoside triphosphates (0.60 mM dATP, 0.15 mM dTTP, and 0.40 mM dGTP and dCTP), either indocarbocyanine- or indodicarbocyanine-labeled dUTP (Amersham), 200 U of Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI), 5× Moloney murine leukemia virus buffer, and 0.50 μl of SUPERaseIn (Ambion, Austin, Tex.) to a final volume of 25 μl. The samples were incubated at 37°C for 2 h, with the addition of 200 more units of Moloney murine leukemia virus reverse transcriptase 1 h into the incubation. The RNA was degraded by addition of 0.5 N NaOH (30 mM final concentration) and incubation at 70°C for 10 min. The mixture was cooled on ice and neutralized by adding 0.5 N HCl (30 mM final concentration) and 400 μl of Tris-EDTA (TE; 50 mM Tris, 50 mM EDTA, pH 8).
The labeled probe was purified by centrifugation in a Microcon-30 centrifugal filter device and washed once with 250 μl of TE (pH 8). The final volume of each probe was adjusted to 10 μl with TE buffer. Five microliters of oppositely labeled probes was mixed, 1 μl of sonicated salmon sperm DNA (10 mg/ml) was added, and the mixture was denatured at 95°C for 3 min. An equal volume of 2× hybridization buffer (10× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 50% formamide, 0.2% SDS) was added, and the sample was placed on the array under a HybriSlip (22 by 22 mm; Grace Biolabs, Bend, Oreg.). The slides were hybridized overnight at 44°C in Corning hybridization chambers with 100 μl of 10× SSC to maintain humidity. The hybridized arrays were analyzed with a GSI Lumonics scanner and ScanArray software (PerkinElmer Life Sciences, Boston, Mass.). Spot intensities were quantitated with QuantArray Microarray analysis software (PerkinElmer Life Sciences).
Microarray data analysis.
All array data were subjected to a filtering criterion as previously described (39). The data were normalized, and genes which showed significant differences in expression levels were determined with the normalization and gene selection method described by Yang et al. (48). Average linkage hierarchical clustering was performed with Cluster (9). Self-organizing map analysis (37) was performed with GeneCluster 2.0 (Whitehead Institute for Biomedical Research, Cambridge, Mass.). Gene clusters were visualized in Tree View (9).
Quantitative reverse transcription-PCR.
Random hexamer-primed reverse transcription reactions were carried out for 2 h at 37°C with 5 μg of RNA, 2.5 μg of random hexamers, 1.0 μl of SUPERaseIn, 0.60 mM each dATP and dTTP, 0.40 mM each dGTP and dCTP, and 200 units of Moloney murine leukemia virus reverse transcriptase in a total volume of 25 μl. Each PCR contained 0.5 μl of reverse transcription reaction product, 1× SYBR Green PCR master mix (Applied Biosystems), and 0.8 μM gene-specific primers. PCRs were run on an ABI Prism 7700 sequence detection system with the following program: 10 min at 95°C, followed by 40 amplification cycles of 95°C for 45 s, 58°C for 45 s, and 72°C for 1 min. All reactions were performed at least six times. All genes were normalized to the pullulanase gene, which was determined to be expressed at a constant level in all three strains based on the rank invariant method of Tseng et al. (40).
Butanol challenge experiments.
Preculture tubes were grown as they were for the bioreactor experiments and used to inoculate 400-ml static flasks. Each flask was allowed to grow to an _A_600 of 1.0 ± 0.05 and then split into four 80-ml closed static flasks. One flask was allowed to grow without butanol addition, and butanol was added to the remaining three flasks at 0.25%, 0.50%, and 0.75% (vol/vol), respectively. Growth was monitored into the early stationary phase. Growth inhibition for the butanol-challenged cultures was calculated relative to the respective unchallenged culture at each time point by subtracting the _A_600 of the challenged culture from the _A_600 of the unchallenged culture and dividing by the _A_600 of the unchallenged culture. An integral analysis was used to calculate early and overall average inhibition rates by calculating the area under each curve with the trapezoidal rule and dividing by time (8). The integral analysis was then used to calculate the percent growth inhibition of 824(pGROE1) relative to 824(pSOS95del) at each level of butanol challenge by taking a ratio of percent inhibition for 824(pGROE1) to percent inhibition of 824(pSOS95del) and multiplying by 100.
RESULTS
Verification of groES and groEL overexpression.
Quantitative reverse transcription-PCR was performed to verify increased transcription of the groES and groEL genes in 824(pGROE1) relative to wild-type C. acetobutylicum ATCC 824 and the plasmid control strain 824(pSOS95del). A rank invariant method based on that of Tseng et al. (40) was used to determine that the pullulanase gene has constant gene expression across all strains and samples. The pullulanase gene was subsequently used to normalize all quantitative reverse transcription-PCR results. Both transcripts were present at levels 6 to 15 times higher than in the wild-type and plasmid control strains during exponential growth and into the early stationary phase. Transcriptional analysis with cDNA microarrays comparing 824(pGROE1) to 824(pSOS95del) revealed increased transcript levels of 1.8- to 4.5-fold and 2.1- to 8.9-fold for groES and groEL, respectively. It is well known that DNA arrays typically underestimate differences relative to Northern analysis and quantitative reverse transcription-PCR (48).
Fermentation characteristics.
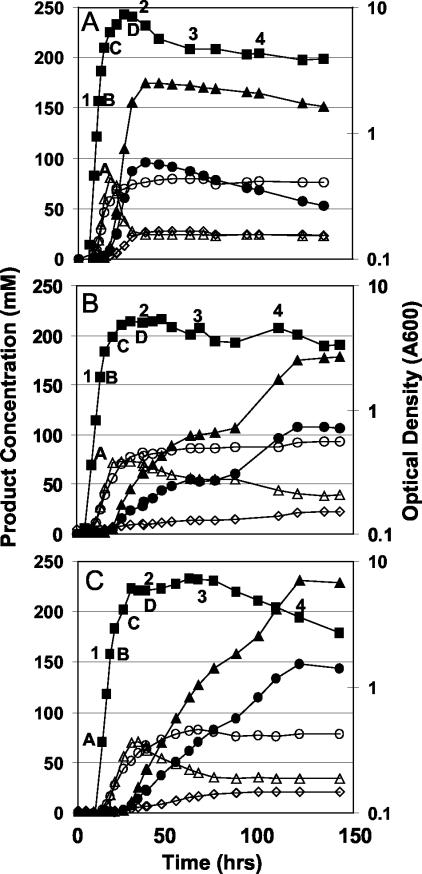
Replicate pH-controlled glucose-fed batch fermentations with the wild-type, 824(pSOS95del), and 824(pGROE1) strains resulted in significantly different final solvent concentrations and product formation profiles (Fig. 2). Strain 824(pGROE1) produced 148 mM acetone and 231 mM butanol, an increase of 38% and 30%, respectively, relative to the plasmid control strain and 54% and 32%, respectively, relative to the wild-type strain. Final ethanol titers were slightly lower in the 824(pSOS95del) and 824(pGROE1) strains (23 mM and 21 mM, respectively) compared to the wild-type strain (28 mM). Acetate levels showed no statistical difference between strains, while butyrate levels were slightly lower in the two recombinant strains (73 mM and 70 mM) compared to the wild type (80 mM). The doubling times for the two recombinant strains were nearly identical (2.01 and 1.99 h), both exhibiting slower exponential growth than the wild-type strain (1.24 h). 824(pGROE1) grew to higher optical densities (_A_600 of 7.18) than the 824(pSOS95del) control strain (_A_600 of 5.38), but was slightly lower than the wild-type strain (_A_600 of 8.88).
FIG. 2.
Product concentration and optical density profiles for controlled pH (>5.0) batch fermentations with the wild-type (A), 824(pSOS95del) (B), and 824(pGROE1) (C) strains. Profiles shown represent the averages of two (for the recombinant strains) or three (for the wild type) fermentations. Samples used for array analysis are labeled A through D. Samples used for Western analysis are labeled 1 through 4. Symbols: ▪, _A_600; ○, acetate; •, acetone; ▴, butanol; ▵, butyrate; ⋄, ethanol.
The growth and product formation patterns of strain 824(pSOS95del) were characterized by two distinct phases, the first lasting approximately 75 h and the second approximately 60 h. This pattern has been observed in many large-scale fermentations with this and other plasmid control strains (e.g., 824[pIMP1]; unpublished data) and is likely due to host-plasmid interactions (45). Strain 824(pGROE1) exhibited a much less pronounced two-phase product formation pattern in both replicate fermentations, with active metabolism (glucose uptake and product formation) lasting more than 110 h.
To eliminate the possibility of plasmid instability, cell samples were taken throughout the course of the fermentation and plated in triplicate on 2× YTG plates both with and without erythromycin so that at least 30 colonies were present. The plates were allowed to grow overnight, and the number of colonies on the plates with and without erythromycin was counted and compared with a t test statistic (P = 0.05). Replica plating from plates containing no erythromycin to plates containing erythromycin further verified plasmid stability even in the absence of an external selective pressure. Finally, plasmid preparations and digestions were performed to verify the presence of the desired plasmid.
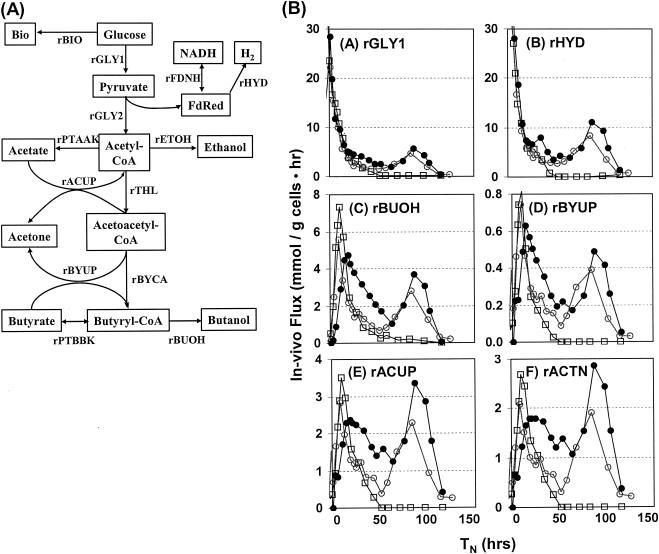
Metabolic flux analysis.
An examination of specific in vivo fluxes for a number of key metabolic reactions provides an excellent portrait of differences in carbon flow between strains while accounting for differences in cell densities. The specific in vivo fluxes illustrated drastic differences between the wild-type, 824(pSOS95del), and 824(pGROE1) strains (Fig. 3). Strain 824(pGROE1) exhibited elevated glucose utilization (rGLY1, Fig. 3A) and acetyl-coenzyme A utilization (profile very similar to rGLY1; data not shown) relative to the control strain. 824(pGROE1) also produced more hydrogen (rHYD, Fig. 3B) during the later stages of fermentation. Strain 824(pGROE1) had higher in vivo butanol formation fluxes (rBUOH, Fig. 3C) than strain 824(pSOS95del). The acetate and butyrate uptake rates (rBYUP and rACUP, respectively; Fig. 3D and 3E, respectively) were also higher in strain 824(pGROE1), which resulted in increased acetone formation (rACTN; Fig. 3F).
FIG. 3.
C. acetobutylicum primary metabolic pathways (A) and corresponding in vivo fluxes and time course profiles of several metabolic fluxes in cultures of the wild-type (□), 824(pSOS95del) (○), and 824(pGROE1) (•) strains (B). Profiles shown represent the averages of two (for the recombinant strains) or three (for the wild type) fermentations. rACTN = rBYUP + rACUP. TN represents normalized time such that TN = 0 h at _A_600 = 1.0. Abbreviations: Bio, biomass; FdRed, reduced ferrodoxin; rACUP, acetate uptake flux; rBIO, bimass formation flux; rBUOH, butanol formation flux; rBYCA, butyryl-CoA formation flux; rBYUP, butyrate uptake flux; rETOH, ethanol formation flux; rFDNH and rHYD, electron transport fluxes; rGLY1 and rGLY2, pyruvate and acetyl-CoA formation fluxes, respectively; rPTAAK, acetate formation flux; rPTBBK, butyrate formation flux; rTHL, acetoacetyl-CoA formation flux.
Acetone is formed through the uptake of either acetate or butyrate. The increases in both butyrate uptake and butyrate formation (data not shown) were relatively small in comparison to the increases in acetate uptake and acetate formation (data not shown). For strain 824(pGROE1), acetate uptake appeared to play a larger role in increased acetone production than butyrate uptake. For both recombinant strains, the fluxes for acetate and butyrate uptake as well as for acetone and butanol formation exhibited a distinct bimodal pattern similar to those for product formation. The second phase was more pronounced for 824(pGROE1) in terms of specific in vivo fluxes as a result of lower optical density near the end of the fermentation relative to 824(pSOS95del). The recombinant strains also exhibited metabolic activity and solvent production over a period two and a half times longer than that of the wild type (115 h and 45 h, respectively).
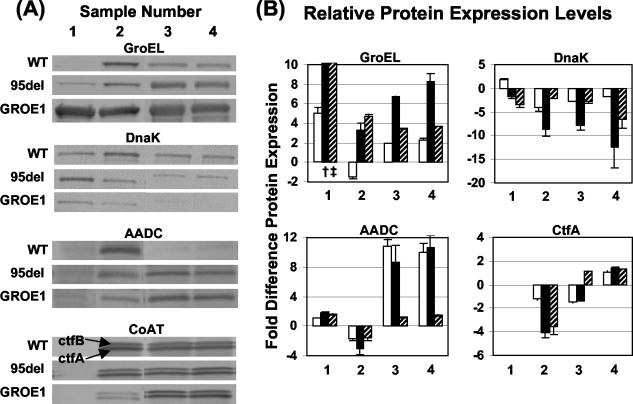
Western analysis of GroEL and DnaK expression.
Antibodies against the GroEL and DnaK stress (chaperone) proteins from E. coli have been shown to cross-react with the GroEL and DnaK proteins of C. acetobutylicum (26). An anti-GroEL antibody from E. coli was used to verify that increased transcription in 824(pGROE1) resulted in increased protein levels (Fig. 4). During exponential growth (sample 1), GroEL levels in 824(pGROE1) were 180 ± 45 and 37 ± 13 times higher than in the wild-type and 824(pSOS95del) strain, respectively, and at least 3.3 ± 0.9 times higher during the transition through stationary phase (samples 2 to 4; Fig. 4). GroEL levels in 824(pSOS95del) were elevated relative to wild-type levels during mid-exponential growth (sample 1) and during stationary phase (samples 3 and 4), suggesting that the presence of the control plasmid imposes a stress on C. acetobutylicum. Sample 2 corresponds to late solventogenesis in the wild-type strain and early solventogenesis in the recombinant strains, as evidenced by the product profiles (Fig. 2) and Western analysis of AADC (a key enzyme in solvent production; Fig. 4). The same wild-type sample also had increased DnaK expression relative to the other wild-type samples. This is further evidence that solventogenesis in C. acetobutylicum correlates with an increase in HSP production (26). DnaK expression levels in 824(pGROE1) were up to 12 ± 4 times lower than in the wild type and up to 6.6 ± 1.8 times lower than in 824(pSOS95del).
FIG. 4.
Western blots for GroEL, DnaK, AADC, and CoAT (A) and their relative protein expression levels between the wild type (WT), 824(pSOS95del), and 824(pGROE1) (B). Reported values are the averages of two gels (± standard error of the mean). All samples used to calculate protein expression ratios were run on the same gel. No quantifiable bands were present in sample 1 on the CtfA or CtfB blot. The relative protein expression levels for CtfB were nearly identical to those shown for CtfA. A negative difference represents higher expression for the strain in the denominator of the ratio. Symbols: open bars, 824(pSOS95del)/wild type; black bars, 824(pGROE1)/wild type; hatched bars, 824(pGROE1)/824(pSOS95del); †, 180 ± 45 times higher; ‡, 37 ± 13 times higher.
Western analysis of solvent formation proteins AADC and CoAT.
The proteins AADC and CoAT catalyze the reactions required for acetone formation. CoAT converts acetoacetyl-coenzyme A to acetoacetate by transferring the coenzyme A to acetate or butyrate, resulting in the formation of acetyl-coenzyme A and butyryl-coenzyme A, respectively. The acetoacetate is then converted to acetone by AADC. All three strains, as expected, had very low levels of AADC expression and no measurable CoAT expression during exponential growth (sample 1, Fig. 4; CtfA is shown graphically for CoAT and is nearly identical to CtfB). Wild-type expression of AADC was 1.8 ± 0.2 and 3.0 ± 0.7 times higher in sample 2 relative to 824(pSOS95del) and 824(pGROE1), respectively. Similarly, wild-type expression of CoAT (CtfA and CtfB) was slightly higher (1.2 ± 0.1 and 1.3 ± 0.1 times higher for CtfA and CtfB, respectively) in 824(pSOS95del). This correlates well with an earlier and initially more rapid rate of solvent production in the wild-type strain.
Into the stationary phase, there were only trace levels of AADC in the wild type, while 824(pSOS95del) and 824(pGROE1) had relatively high levels of AADC expression (at least eight times higher than in the wild type). There was no statistical difference (P < 0.80) in the amounts of AADC in 824(pSOS95del) and 824(pGROE1) during the stationary phase. CoAT was present in amounts easily visualized on Western blots in all three strains during the stationary phase, with 1.4 ± 0.1 times less in the two recombinant strains in sample 3 and 1.2 ± 0.1 times more in sample 4. The lack of AADC during the stationary phase in the wild-type strain suggests that AADC is not stable under typical fermentation conditions or is not being transcribed, while both of the CoAT subunits are more stable than the AADC protein or continue to be transcribed and translated.
Transcriptional analysis of host-plasmid interactions.
Transcriptional analysis of the plasmid control strain 824(pSOS95del) versus the wild type was performed to better understand the observed host-plasmid interactions. Samples from four time points, ranging from exponential through early stationary phase (Fig. 2), were analyzed on eight DNA arrays. All duplicate arrays were hybridized with reverse-labeled samples (e.g., wild-type-Cy3/824[pSOS95del]-Cy5 and wild-type-Cy5/824[pSOS95del]-Cy3). The resulting array data were normalized, and the genes identified as differentially expressed at the 95% significance level (48) were further analyzed by average-linkage hierarchical clustering (9) and self-organizing map analysis (37).
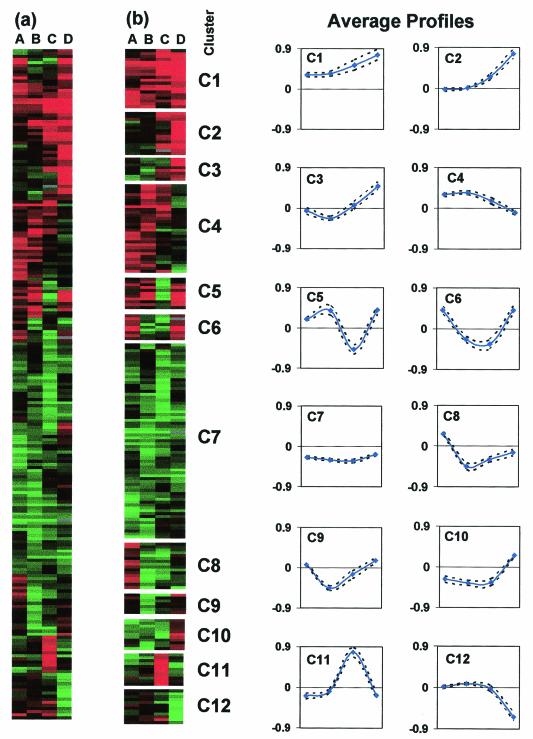
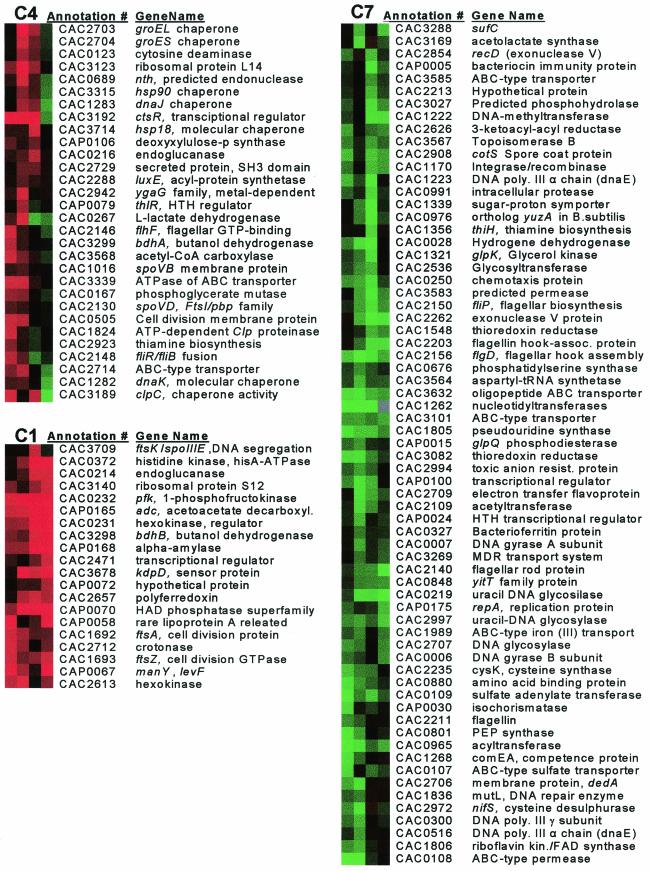
A total of 217 genes were organized by hierarchical clustering (Fig. 5a) and into 12 clusters with self-organizing maps (Fig. 5b). Cluster C4 contained a large number of stress-related genes (Fig. 6) whose expression was higher in 824(pSOS95del) through the transition phase (samples A to C), but higher in the wild type nearer the onset of solventogenesis (sample D). This further suggests that the presence of the control plasmid represents a cellular stress, as in E. coli (2). Stress genes in cluster C4 include groES and groEL (CAC2704 and CAC2703), dnaK and dnaJ (CAC1282 and CAC1283), hsp18 (CAC3714), and hsp90 (CAC3315). Also included are clpC (CAC3189) and clpA (CAC1842), members of the class III hsp100 heat shock family, and an associated regulator of class III heat shock proteins, ctsR (CAC3192). The mRNA and protein expression levels for groEL appeared to be directly correlated when microarray and Western samples taken at the same time were compared (B and D with 1 and 2, respectively).
FIG. 5.
Expression profile of differentially expressed genes for the wild type versus 824(pSOS95del). Hierarchical gene clustering (a) and self-organizing map analysis with graphs showing average profiles of the logarithm of expression ratios (824[pSOS95del]/wild type) (b) of the 217 differentially expressed genes. Red indicates higher expression in 824(pSOS95del), and green indicates higher expression in the wild type. Samples A to D correspond to those indicated on Fig. 2.
FIG. 6.
Wild type versus 824(pSOS95del) clusters C1, C4, and C7 arranged by hierarchical clustering. Genes in cluster C1 are expressed at a higher level in 824(pSOS95del), while those in C7 are expressed at a higher level in the wild type. Cluster C4 contains many stress response genes. Abbreviations: FAD, flavin adenine dinucleotide; MDR, multiple-drug resistance; HTH, helix-turn-helix; HAD, haloacid dehydrogenase; poly.III, polymerase III; PEP, phosphoenolpyruvate; kin., kinase.
The largest gene cluster (C7) was composed of genes having lower expression in 824(pSOS95del) relative to the wild type (Fig. 6). The genes in clusters C8 and C10 also had lower expression in 824(pSOS95del) except at the first and last time points, respectively. These three clusters contained 19 genes (16 in cluster C7) related to DNA topology, replication, modification, recombination, and competence. Included are four DNA gyrases (CAC0006, CAC0007, CAC2262, and CAC3567), four DNA polymerase III genes (CAC0125, CAC0300, CAC0515, and CAC1223), four recombination genes (CAC1296, CAC1170, CAC2854, and CAP0095), and three DNA glycosylases (CAC0219, CAC2707, and CAC2997). The same DNA gyrases were shown to have lower expression in M5, an asporogenous, non-solvent-producing strain (7), relative to the wild type during exponential growth (39).
The genes in the glycolytic pathway were also expressed at a lower level in 824(pSOS95del) except for hexokinase, which had increased expression. This suggests that DNA topology plays a major role in the regulation of carbon metabolism in C. acetobutylicum. Also included in these three clusters were 13 genes related to motility and chemotaxis, representing all four predicted motility and chemotaxis operons (39), and two glycosyltransferases (CAC2522 and CAC2536). This pattern of gene expression agrees with previous microscopic observations of plasmid control strains (e.g., 824[pIMP1]), which revealed a phenotype characterized by increased sporulation and decreased motility (15). The same motility and chemotaxis gene expression patterns were observed in M5 (39), suggesting that DNA topology may also play a role in regulating motility.
Cluster C1 represents genes with higher expression in 824(pSOS95del) (Fig. 6). Both adc and bdhB were present despite the fact that solvent production in 824(pSOS95del) was delayed and initially slower than in the wild type. However, Western analysis indicates (Fig. 4) increased AADC expression at the corresponding early time point (sample 1) and lower AADC levels at the later time point (sample 2) in 824(pSOS95del). Several key metabolic genes are in this cluster, including hexokinase (CAC2613) and crotonase (CAC2712). ftsA and ftsZ, which form a predicted bicistronic operon (CAC1692 and CAC1693) (39), also belong to this cluster. Along with ftsK (also present, CAC3709), these genes play critical roles in cell division and sporulation in many prokaryotes (39), including B. subtilis (11, 20). Cluster C1 also contains ribosomal protein S12. Two additional ribosomal proteins (CAC2847 and CAC3123) are in clusters C4 and C2 (also higher in 824[pSOS95del]). The presence of a plasmid has been shown to alter levels of ribosome components within a cell (2). Cluster C2 also contains three genes from a predicted operon central in the regulation of sporulation, sigF, anti-sigF, and anti-anti-sigF (CAC2306 to CAC2308). Several other sporulation-specific genes showed altered gene expression, including spoVB, spoVD, spoIIIE, a spoIIIE ATPase, cotS, spoT, and spo0J. Finally, cluster C11 contained a gene encoding a special sigma factor located on the megaplasmid (CAP0126).
Transcriptional analysis of groESL overexpression.
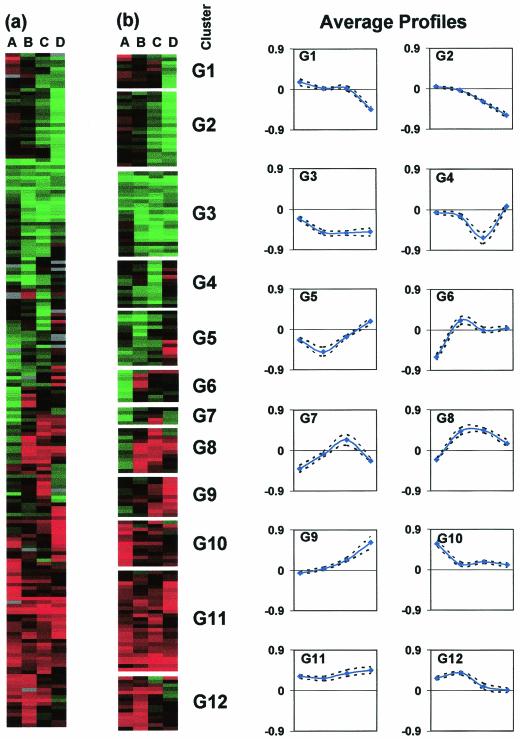
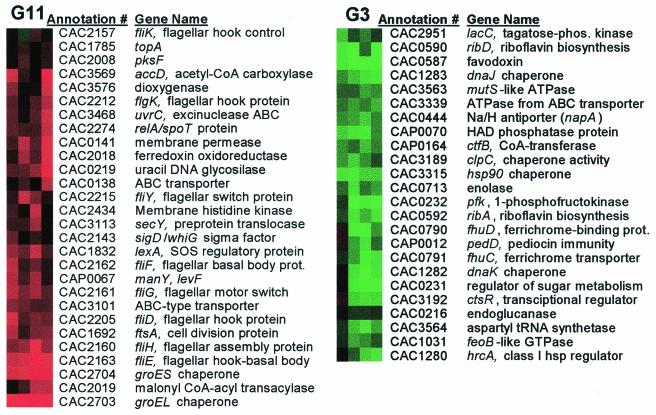
Transcriptional analysis of the _groESL_-overexpressing strain 824(pGROE1) relative to the plasmid control strain 824(pSOS95del) identified 174 genes as differentially expressed (with the same criteria as above). The 174 differentially expressed genes were organized by hierarchical clustering (Fig. 7a) and into 12 clusters with self-organizing maps (Fig. 7b). The genes for groES and groEL were in cluster G11 (Fig. 8), which represents genes with higher expression in 824(pGROE1) relative to 824(pSOS95del). Cluster G11 also contains seven motility genes which fall into one of the four operons discussed above. This pattern of gene expression suggests that overexpression of groES and groEL shifts expression levels of the motility-related genes back toward wild-type levels.
FIG. 7.
Expression profile of differentially expressed genes for 824(pSOS95del) versus 824(pGROE1). Hierarchical gene clustering (a) and self-organizing map analysis with graphs showing average profiles of the logarithm of expression ratios (824[pSOS95del]/wild type) (b) of the 174 differentially expressed genes. Red indicates higher expression in 824(pGROE1), and green indicates higher expression in 824(pSOS95del). Samples A to D correspond to those indicated on Fig. 2.
FIG. 8.
824(pSOS95del) versus 824(pGROE1) clusters G3 and G11 arranged by hierarchical clustering. Genes in cluster G11 are expressed at a higher level in 824(pGROE1), while those in G3 are expressed at a higher level in 824(pSOS95del).
It is interesting that only 2 of the 19 genes related to DNA topology, replication, modification, recombination, and competence were differentially regulated in 824(pGROE1) relative to 824(pSOS95del): a uracil DNA glycosylase (CAC0219) in cluster G11 and a recO recombination protein homolog (CAC1296) in cluster G6. ftsA, however, was expressed at an even higher level in 824(pGROE1) than in 824(pSOS95del), which itself had higher expression than the wild type. The other major HSP genes were in clusters G2 and G3, with lower relative expression levels in 824(pGROE1). Cluster G3 (Fig. 8) included hrcA (CAC1280), dnaK, dnaJ, hsp90, clpC, and ctsR, while hsp18 and a dnaK homolog (CAC0472) were in cluster G2. The downregulation of the dnaKJ operon genes (CAC1280 to CAC1284) upon overexpression of groESL further supports the idea that groEL acts as a negative modulator of class I heat shock genes through stabilization of the HrcA protein.
The CIRCE element sequence (TTAGCACTC-N9-GAGTGCTAA) was used to search the entire intergenic region of the C. acetobutylicum genome, and four exact matches were found. In addition to the previously reported elements in front of the groESL and dnaKJ operons, an additional sequence was found upstream of hsp90 (CAC3315), which also falls within cluster G3. hsp90 has also been clustered with the groESL and dnaKJ operon genes in other sets of microarray experiments (39, 41). This suggests that hsp90 is regulated in the same manner as the groESL and dnaKJ operons. Key solvent formation genes also showed early decreased expression in 824(pGROE1), including ctfB (cluster G3; CAP0164), adc (cluster G2; CAP0165), and bdhA (cluster G5; CAC3299). Additionally, several glycolytic genes had lower expression in 824(pGROE1), including triosephosphate isomerase (CAC0711), enolase (CAC0713), and pfk (CAC0232).
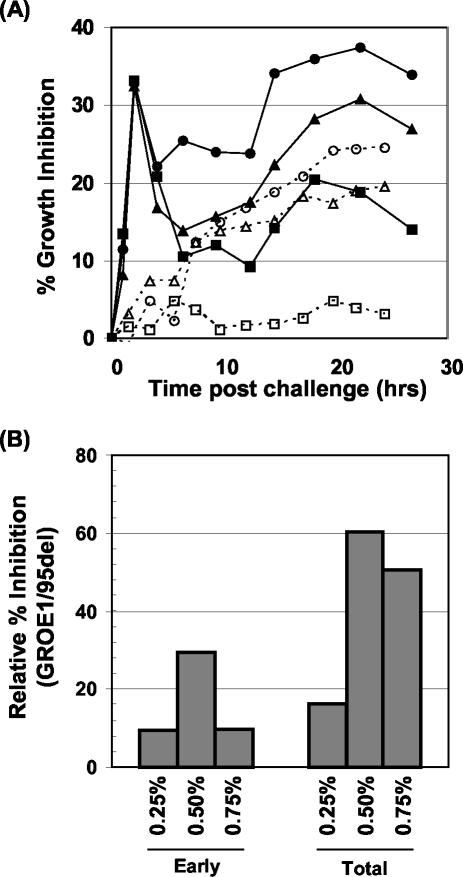
824(pGROE1) growth displays increased tolerance to butanol addition.
To test whether overexpression of groESL imparts resistance to butanol in the growth phase, 824(pSOS95del) and 824(pGROE1) cultures were subject to various levels of butanol challenge (up to 0.75%, vol/vol) during mid-exponential growth (_A_60 of 1.0), and the level of growth inhibition was determined relative to the unchallenged cultures (Fig. 9A). Growth of 824(pSOS95del) was initially inhibited approximately 33% at all three levels of butanol challenge (2 h postchallenge), while 824(pGROE1) was inhibited less than 5% at 0.25% and 0.50% butanol and 7% at 0.75% butanol. The initial response to growth inhibition appeared to be dose independent. The relative inhibition of all three 824(pSOS95del) cultures fell briefly at approximately 6 h postchallenge and then increased in a butanol dose-dependent manner (Fig. 9A). Growth inhibition of 824(pGROE1) was less than 5% with 0.25% butanol.
FIG. 9.
Butanol challenge experiments. (A) Growth inhibition profiles for 824(pSOS95del) (solid symbols) and 824(pGROE1) (open symbols) at various levels of butanol challenge: squares, 0.25% (vol/vol); triangles, 0.50% (vol/vol); circles, 0.75% (vol/vol). (B) Relative inhibition of 824(pGROE1) to 824(pSOS95del) for the early (0 to 6 h) and total course of butanol challenge.
Integral analysis (8) was used to compare growth inhibition during the early stages of butanol challenge (up to 6 h) and overall (total) growth inhibition. Inhibition of 824(pGROE1) was an average of 16% that of 824(pSOS95del) during the early stage of butanol challenge. Over the course of the culture, 824(pGROE1) inhibition was only 15% that of 824(pSOS95del) at 0.25% butanol, similar to the initial inhibition level. At 0.50% and 0.75% butanol, inhibition of 824(pGROE1) was 60% and 50%, respectively, that of 824(pSOS95del).
DISCUSSION
824(pGROE1) was created in an effort to assess the effects of groESL overexpression on metabolism and solvent production in C. acetobutylicum. Overexpression of the groESL operon was verified by three methods: two at the transcriptional level (DNA arrays and quantitative reverse transcription-PCR) and one at the protein level (Western analysis). Overexpression resulted in increased final solvent titers relative to both the wild-type (40% higher) and the plasmid control 824(pSOS95del) strain (33% higher). Increased acetone production in 824(pGROE1) relative to 824(pSOS95del) was achieved primarily through increased acetate uptake rather than butyrate uptake. Increased acetone production in 824(pGROE1) relative to the control strain was observed despite the fact that AADC and CoAT protein levels were not significantly higher during the stationary phase. Increased levels of GroES and GroEL may hold the AADC and CoAT proteins in a more active state, resulting in increased acetone production. Alternatively, overexpression of groESL may increase the level of metabolic intermediates available for acetone formation by stabilizing the proteins involved in their formation.
Increased butanol titers similar to those observed in 824(pGROE1) have been achieved through overexpression of the alcohol-aldehyde dehydrogenase gene (aad) (13, 14). However, aad is likely too large (96 kDa) to be stabilized by the GroESL machinery, which has a size limit of approximately 56 kDa in most bacterial species (28). This suggests that increased solvent production is a result of a broader stabilization of the biosynthetic machinery. This is supported by the fact that cells overexpressing groESL were better able to grow in the presence of butanol. In addition, analysis of specific in vivo fluxes related to carbon metabolism (glucose and hydrogen) and solvent formation (butanol and acetone) revealed that overall metabolic activity in 824(pGROE1) was significantly higher than that in 824(pSOS95del). Increased levels of GroES and GroEL may also help to overcome the stress associated with the presence of a plasmid, resulting in initially higher cell densities and a motility/chemotaxis gene expression pattern which is more like that of the wild-type strain. Enhanced sporulation of the 824(pSOS95del) strain may result in lower cell densities and lost solvent productivity.
Several conclusions with regard to the regulation of HSPs in C. acetobutylicum can be drawn. A CIRCE element was found upstream of the hsp90 start site, and its transcriptional expression pattern was identical to that of the dnaKJ operon in this and several previous studies (39, 41). Therefore, hsp90 is likely part of the CIRCE heat shock regulon. The decrease in expression of the dnaKJ and hsp90 operon genes at the transcriptional level along with decreased DnaK protein levels suggests that groESL acts as a major modulator of the CIRCE heat shock regulon, as in B. subtilis (23). In B. subtilis, GroEL has been shown to stabilize the heat shock repressor protein HrcA (23, 29), which is known to negatively regulate both the dnaKJ and groESL operons through binding of the CIRCE element (Fig. 1B) (34). The dnaKJ and groESL operons in C. acetobutylicum have the same organization as in B. subtilis, including an HrcA homolog (CAC1280) and CIRCE elements. It is likely that GroEL acts as a negative modulator of HSPs in C. acetobutylicum. DnaK has been shown to play a role in HSP regulation in other bacteria, such as Lactococcus lactis (21). However, a dnaK insertion mutation in B. subtilis had no effect on the expression of other heat shock proteins (23).
Previous in vitro studies have demonstrated that DnaK prevents aggregation of HrcA in C. acetobutylicum (31), but its in vivo role has not yet been demonstrated. Efforts to overexpress the dnaKJ operon with pSOS95del as the expression vector have proven difficult. Transformant colonies transferred to liquid cultures failed to grow beyond the exponential phase, an effect also observed in E. coli (3). Decreased viability of cultures overexpressing DnaK has been attributed to a defect in cell septation and can be suppressed by overproduction of FtsZ (3). Lower expression of DnaK results in the same phenotype but does not appear to have a negative effect on the growth of 824(pGROE1) relative to 824(pSOS95del), possibly as a result of increased ftsZ expression.
One of the most widely accepted views on host-plasmid interactions is that the presence of a plasmid represents a metabolic burden and a cellular stress (30). The presence of a plasmid has been shown to result in increased levels of HSPs and altered levels of carbon metabolism genes (2). The expression of HSPs in 824(pSOS95del) was elevated relative to the wild type except at the onset of solventogenesis. Recent evidence suggests that host-plasmid interactions are related to alteration of the cellular regulatory status (30).
The degree of differential gene expression in 824(pSOS95del) relative to the wild type would indicate that there is a complex interaction between the host and the plasmid in C. acetobutylicum. An altered motility and chemotaxis program was observed in the plasmid control strain, along with differential expression of sporulation-specific regulatory factors. Perhaps most interesting is the differential regulation of many genes related to DNA topology. It has been suggested that regulation of nearly all bacterial operons is sensitive to DNA supercoiling (12). In C. acetobutylicum, high concentrations of gyrase inhibitors have been shown to increase the transcription of groESL, hsp18, adc, and aad while lowering the transcription of ptb, thlA, and ctfAB (43, 47). High gyrase inhibitor concentrations are required to decrease negative supercoiling because intermediate gyrase inhibitor levels have been shown to increase supercoiling. Increased expression of groESL, hsp18, and adc in 824(pGROE1) is consistent with these findings, while expression of thlA was actually higher in 824(pSOS95del). ptb, aad, and ctfA/B were not found to be differentially expressed in 824(pSOS95del). It is possible that ptb, aad, and ctfA/B were differentially expressed in the previous studies (43, 47) due to the very high levels of gyrase inhibitors used to study the effects of DNA topology.
The same DNA gyrases which were differentially expressed in 824(pSOS95del) were also shown to have lower expression in M5 (7) relative to the wild type during exponential growth (39). The genes in the glycolytic pathway, except for hexokinase, were also expressed at a lower level in M5. This same gene expression pattern, including the opposite expression of hexokinase, was also observed in 824(pSOS95del). These findings suggest that DNA topology plays a major role in regulation of primary metabolism and that host-plasmid interactions in C. acetobutylicum are the result of a drastically altered regulatory state. This may help explain the prolonged (115 versus 45 h), biphasic growth pattern observed in the two recombinant strains compared to the wild type. For example, DnaK and DnaJ have been shown to play a role in DNA replication and cell division (49). Defects in cell division due to DnaK over- or underexpression can be reduced through overexpression of ftsZ (3). Expression of ftsZ is higher in 824(pSOS95del) than in the wild type and even higher in 824(pGROE1). The additional increase in ftsZ expression in 824(pGROE1) may result in a growth pattern that is more similar to that of the wild type. ftsZ is also involved in regulation of sporulation and cell motility, a program also more like the wild-type program in 824(pGROE1) than in 824(pSOS95del).
The complex global response to the presence of a plasmid and overexpression of groESL makes it impossible to assign a single gene expression pattern to a specific phenotype. However, the wealth of information generated by the combination of phenotypic, Western, and microarray analyses can be used to pose new hypotheses for future testing.
Acknowledgments
This work was supported by grant BES-9911231 from the National Science Foundation and grant R828562 from the Environmental Protection Agency.
We acknowledge use of the Keck Biophysics Facility and the Center for Genetic Medicine facilities at Northwestern University. We thank J. S. Takahashi's lab for use of the ABI Prism 7700 sequence detection system and Abbott Laboratories for the donation of clarithromycin.
REFERENCES
- 1.Bahl, H., H. Muller, S. Behrens, H. Joseph, and F. Narberhaus. 1995. Expression of heat shock genes in Clostridium acetobutylicum. FEMS Microbiol. Rev. 17**:**341-348. [DOI] [PubMed] [Google Scholar]
- 2.Birnbaum, S., and J. E. Bailey. 1991. Plasmid presence changes the relative levels of many host-cell proteins and ribosome components in recombinant Escherichia coli. Biotechnol. Bioeng. 37**:**736-745. [DOI] [PubMed] [Google Scholar]
- 3.Blum, P., J. Ory, J. Bauernfeind, and J. Krska. 1992. Physiological consequences of DnaK and DnaJ overproduction in Escherichia coli. J. Bacteriol. 174**:**7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowles, L. K., and W. L. Ellefson. 1985. Effects of butanol on Clostridium acetobutylicum. Appl. Environ. Microbiol. 50**:**1165-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buday, Z., J. C. Linden, and M. N. Karim. 1990. Improved acetone-butanol fermentation analysis with subambient HPLC column temperature. Enzyme Microb. Technol. 12**:**24-27. [Google Scholar]
- 6.Cary, J. W., D. J. Petersen, E. T. Papoutsakis, and G. N. Bennett. 1990. Cloning and expression of Clostridium acetobutylicum ATCC 824 acetoacetyl-coenzyme A:acetate/butyrate:coenzyme A transferase in Escherichia coli. Appl. Environ. Microbiol. 56**:**1576-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornillot, E., R. V. Nair, E. T. Papoutsakis, and P. Soucaille. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 179**:**5442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai, R. P., L. K. Nielsen, and E. T. Papoutsakis. 1999. Stoichiometric modeling of Clostridium acetobutylicum fermentations with non-linear constraints. J. Biotechnol. 71**:**191-205. [DOI] [PubMed] [Google Scholar]
- 9.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95**:**14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrante, A. A., J. Augliera, K. Lewis, and A. M. Klibanov. 1995. Cloning of an organic solvent-resistance gene in Escherichia coli: the unexpected role of alkylhydroperoxide reductase. Proc. Natl. Acad. Sci. USA 92**:**7617-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feucht, A., I. Lucet, M. D. Yudkin, and J. Errington. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40**:**115-125. [DOI] [PubMed] [Google Scholar]
- 12.Gellert, M. 1981. DNA topoisomerases. Annu. Rev. Biochem. 50**:**879-910. [DOI] [PubMed] [Google Scholar]
- 13.Harris, L. M., L. Blank, R. P. Desai, N. E. Welker, and E. T. Papoutsakis. 2001. Fermentation characterization and flux analysis of recombinant strains of Clostridium acetobutylicum with an inactivated solR gene. J. Ind. Microbiol. Biol. 27**:**322-328. [DOI] [PubMed] [Google Scholar]
- 14.Harris, L. M., R. P. Desai, N. E. Welker, and E. T. Papoutsakis. 2000. Characterization of recombinant strains of the C_lostridium acetobutylicum_ butyrate kinase inactivation mutant: need for new phenomenological models for solventogenesis and butanol inhibition? Biotechnol. Bioeng. 67**:**1-11. [PubMed] [Google Scholar]
- 15.Harris, L. M., N. E. Welker, and E. T. Papoutsakis. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184**:**3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29**:**548-562. [DOI] [PubMed] [Google Scholar]
- 17.Ingram, L. O. 1990. Ethanol tolerance in bacteria. Crit. Rev. Biotechnol. 9**:**305-319. [DOI] [PubMed] [Google Scholar]
- 18.Isken, S., and J. A. de Bont. 1998. Bacteria tolerant to organic solvents. Extremophiles 2**:**229-238. [DOI] [PubMed] [Google Scholar]
- 19.Jones, D. T., and D. R. Woods. 1986. Acetone-butanol fermentation revisited. Microbiol. Rev. 50**:**484-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kemp, J. T., A. Driks, and R. Losick. 2002. FtsA mutants of Bacillus subtilis impaired in sporulation. J. Bacteriol. 184**:**3856-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch, B., M. Kilstrup, F. K. Vogensen, and K. Hammer. 1998. Induced levels of heat shock proteins in a dnaK mutant of Lactococcus lactis. J. Bacteriol. 180**:**3873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mermelstein, L. D., and E. T. Papoutsakis. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage φ 51 TI methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59**:**1077-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the Circe heat shock regulon of Bacillus subtilis. EMBO J. 16**:**4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morimoto, R. I., A. Tissieres, and C. Georgopoulos. 1994. Stress proteins in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Petersen, D. J., and G. N. Bennett. 1990. Purification of acetoacetate decarboxylase from Clostridium acetobutylicum ATCC 824 and cloning of the acetoacetate decarboxylase gene in Escherichia coli. Appl. Environ. Microbiol. 56**:**3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pich, A., F. Narberhaus, and H. Bahl. 1990. Induction of heat shock proteins during initiation of solvent formation in Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 33**:**697-704. [Google Scholar]
- 27.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-Gonzalez, A. Rojas, W. Teran, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56**:**743-768. [DOI] [PubMed] [Google Scholar]
- 28.Ranson, N. A., H. E. White, and H. R. Saibil. 1998. Chaperonins. Biochem. J. 333**:**233-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reischl, S., T. Wiegert, and W. Schumann. 2002. Isolation and analysis of mutant alleles of the Bacillus subtilis HrcA repressor with reduced dependency on GroE function. J. Biol. Chem. 277**:**32659-32667. [DOI] [PubMed] [Google Scholar]
- 30.Ricci, J. C. D., and M. E. Hernandez. 2000. Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 20**:**79-108. [DOI] [PubMed] [Google Scholar]
- 31.Rungeling, E., T. Laufen, and H. Bahl. 1999. Functional characterization of the chaperones DnaK, DnaJ, and GrpE from Clostridium acetobutylicum. FEMS Microbiol. Lett. 170**:**119-123. [DOI] [PubMed] [Google Scholar]
- 32.Sabathe, F., C. Croux, E. Cornillot, and P. Soucaille. 2002. amyP, a reporter gene to study strain degeneration in Clostridium acetobutylicum ATCC 824. FEMS Microbiol. Lett. 210**:**93-98. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Schulz, A., and W. Schumann. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178**:**1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sikkema, J., J. A. de Bont, and B. Poolman. 1994. Interactions of cyclic hydrocarbons with biological membranes. J. Biol. Chem. 269**:**8022-8028. [PubMed] [Google Scholar]
- 36.Sikkema, J., J. A. de Bont, and B. Poolman. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59**:**201-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamayo, P., D. Slonim, J. Mesirov, Q. Zhu, S. Kitareewan, E. Dmitrovsky, E. S. Lander, and T. R. Golub. 1999. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA 96**:**2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terracciano, J. S., and E. R. Kashket. 1986. Intracellular conditions required for the initiation of solvent production by Clostridium acetobutylicum. Appl. Environ. Microbiol. 52**:**86-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomas, C. A., K. V. Alsaker, H. P. J. Bonarius, W. T. Hendriksen, H. Yang, J. A. Beamish, C. J. Parades, and E. T. Papoutsakis. 2003. DNA-array based transcriptional analysis of asporogenous, nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5. J. Bacteriol. **185:**4539-4547. [DOI] [PMC free article] [PubMed]
- 40.Tseng, G. C., M. K. Oh, L. Rohlin, J. C. Liao, and W. H. Wong. 2001. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res. 29**:**2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tummala, S. B., S. G. Junne, and E. T. Papoutsakis. 2003. Antisense RNA downregulation of coenzyme A transferase combined with alcohol/aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J. Bacteriol. **185:**3644-3653. [DOI] [PMC free article] [PubMed]
- 42.Tummala, S. B., N. E. Welker, and E. T. Papoutsakis. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65**:**3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ullmann, S., A. Kuhn, and P. Durre. 1996. DNA topology and gene expression in Clostridium acetobutylicum: Implications for the regulation of solventogenesis. Biotechnol. Lett. 18**:**1413-1418. [Google Scholar]
- 44.Vollherbst, S. K., J. A. Sands, and B. S. Montenecourt. 1984. Effect of butanol on lipid composition and fluidity of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 47**:**193-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walter, K. A., L. D. Mermelstein, and E. T. Papoutsakis. 1994. Host-plasmid interactions in recombinant strains of Clostridium acetobutylicum ATCC 824. FEMS Microbiol. Lett. 123**:**335-342. [Google Scholar]
- 46.Wiesenborn, D. P., F. B. Rudolph, and E. T. Papoutsakis. 1988. Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl. Environ. Microbiol. 54**:**2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong, J., and G. N. Bennett. 1996. The effect of novobiocin on solvent production by Clostridium acetobutylicum. J. Ind. Microbiol. 16**:**354-359. [DOI] [PubMed] [Google Scholar]
- 48.Yang, H., H. Haddad, C. Tomas, K. Alsaker, and E. T. Papoutsakis. 2003. A segmental nearest neighbor normalization and gene identification method gives superior results for DNA-array analysis. Proc. Natl. Acad. Sci. USA 100**:**1122-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zylicz, M., D. Ang, K. Liberek, and C. Georgopoulos. 1989. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 8**:**1601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]