Proposal for Standardization of Optimized Mycobacterial Interspersed Repetitive Unit-Variable-Number Tandem Repeat Typing of Mycobacterium tuberculosis (original) (raw)
Abstract
Molecular typing based on 12 loci containing variable numbers of tandem repeats of mycobacterial interspersed repetitive units (MIRU-VNTRs) has been adopted in combination with spoligotyping as the basis for large-scale, high-throughput genotyping of Mycobacterium tuberculosis. However, even the combination of these two methods is still less discriminatory than IS_6110_ fingerprinting. Here, we define an optimized set of MIRU-VNTR loci with a significantly higher discriminatory power. The resolution and the stability/robustness of 29 loci were analyzed, using a total of 824 tubercle bacillus isolates, including representatives of the main lineages identified worldwide so far. Five loci were excluded for lack of robustness and/or stability in serial isolates or isolates from epidemiologically linked patients. The use of the 24 remaining loci increased the number of types by 40%—and by 23% in combination with spoligotyping—among isolates from cosmopolitan origins, compared to those obtained with the original set of 12 loci. Consequently, the clustering rate was decreased by fourfold—by threefold in combination with spoligotyping—under the same conditions. A discriminatory subset of 15 loci with the highest evolutionary rates was then defined that concentrated 96% of the total resolution obtained with the full 24-locus set. Its predictive value for evaluating M. tuberculosis transmission was found to be equal to that of IS_6110_ restriction fragment length polymorphism typing, as shown in a companion population-based study. This 15-locus system is therefore proposed as the new standard for routine epidemiological discrimination of M. tuberculosis isolates and the 24-locus system as a high-resolution tool for phylogenetic studies.
The genotyping of Mycobacterium tuberculosis isolates contributes to tuberculosis (TB) control by, e.g., indicating possible epidemiological links between TB patients, detecting (un)suspected outbreaks and laboratory cross-contamination, and distinguishing exogenous reinfection from endogenous reactivation in relapse cases. For these purposes, IS_6110_ restriction fragment length polymorphism (RFLP) typing (48) has been used as the gold standard method for more than a decade. However, this method is labor-intensive, requires weeks for culturing the isolates and subsequent DNA purification, and suffers from problems of interpretability and portability of the complex banding patterns. In addition, it provides insufficient discrimination among isolates with low (<6) IS_6110_ copy numbers, a problem that is only partly overcome by using PCR-based spoligotyping as a secondary method (6).
Genotyping based on variable numbers of tandem repeats (VNTRs) of different classes of interspersed genetic elements named mycobacterial interspersed repetitive units (MIRUs) (12, 25, 32, 36, 40, 43, 44) is increasingly used to solve these problems. This method relies on PCR amplification of multiple loci using primers specific for the flanking regions of each repeat locus and on the determination of the sizes of the amplicons, which reflect the numbers of the targeted MIRU-VNTR copies. MIRU-VNTR typing is technically flexible, as sizing can be done using capillary (1, 24) or gel (28) electrophoresis or nondenaturing high-performance liquid chromatography (8). It is considerably faster than IS_6110_-RFLP typing, is applicable to crude DNA extracts from early mycobacterial cultures, and has been adapted to high-throughput conditions (1, 24, 42). Moreover, the results are expressed as numerical codes and are therefore very easy to compare and exchange.
Among different sets of MIRU-VNTR loci described for typing M. tuberculosis isolates (12, 14, 18, 21, 25-27, 32, 33, 36, 39, 45), a system based on 12 loci (28, 44) is currently the most widely used and has been integrated in TB control systems on a national scale in, e.g., the United States (1, 6). Based on pilot studies with limited numbers of isolates, the discriminatory power of this set approached that of IS_6110_-RFLP typing to discriminate epidemiologically unrelated cases (28, 42), while the genotypes based on this set are stable among isolates from epidemiologically linked cases (4, 16, 24, 34). A recent population-based study indicated that the use of this system as a first-line method in combination with spoligotyping provides adequate discrimination in most cases for large-scale, prospective genotyping of M. tuberculosis in the United States. However, IS_6110_ fingerprinting is still required as an additional method to type the clustered isolates in a number of cases, when contact investigation or demographic or epidemiological data do not provide independent clues to the existence or the absence of links between patients (6).
Alternative sets of MIRU-VNTR loci have been suggested to further improve the discrimination of unrelated isolates, compared to that provided by this 12-locus system (18, 21, 25, 32, 33, 36, 39, 45, 49). However, the collections of isolates studied were restricted to small samples of local origin and/or included only Mycobacterium bovis or representatives of only one or two of the defined M. tuberculosis lineages. The overall technical robustness and the clonal stability of the individual MIRU-VNTR loci in the sets tested were not assessed. Furthermore, none of these studies were nonselected, population-based studies, and contact tracing data were not available, making it impossible to establish the predictive value of the various sets for studying ongoing M. tuberculosis transmission at the population level.
Here, we have investigated the resolution power and the clonal stability and technical applicability for molecular epidemiological typing of 29 MIRU-VNTR loci, comprising our original 12 loci and most of the other exploitable loci disclosed so far. These parameters were tested on 824 tubercle bacillus isolates, including worldwide representatives of the main M. tuberculosis lineages identified by the use of a diversity of genetic markers, as well as multiple groups of epidemiologically linked or clonal isolates. On this basis, an optimized set of 24 loci was defined, including a highly discriminatory subset of 15 loci for specific first-line epidemiological investigations. The predictive value of this optimized set for evaluating M. tuberculosis transmission was evaluated in a companion population-based study (M. Cardoso-Oelemann, R. Diel, V. Vatin, W. Haas, S. Rüsch-Gerdes, C. Locht, S. Niemann, and P. Supply, submitted for publication) and compared to that of IS_6110_-RFLP typing.
MATERIALS AND METHODS
Strains.
A total of 824 tubercle bacillus isolates were used in this study. A first set comprised 529 isolates from cosmopolitan origins representing the main M. tuberculosis genetic lineages (n = 417) and other members of the M. tuberculosis complex (see Table S1 in the supplemental material). This set included the standardized collection of 90 M. tuberculosis complex isolates from 38 countries described in reference 23; 100 isolates from seven countries/territories described in reference 41; 132 isolates from the National Reference Center for Mycobacteria, Borstel, Germany (including 53 isolates from Ghana [31], 43 from Germany and 20 from Uganda [30], 14 from Kazakhstan [17], and 2 from the ATCC), 136 isolates from patients mostly of foreign origins residing in the Brussels region, Belgium; 36 reference variants of the M. tuberculosis Beijing/W lineage collected at the Public Health Research Institute, New Jersey (3, 22); and 35 isolates from East Africa representing eight clonal groups, corresponding to extant representatives of the progenitor species of M. tuberculosis, proposed to be named Mycobacterium prototuberculosis (15).
A second set assembled at the National Institute for Public Health and the Environment, The Netherlands, included 61 single-colony cultures obtained from 13 different M. tuberculosis complex strains, which were previously used in the study described in reference 7, as well as a collection of 110 isolates, representing 52 different groups of related strains from different countries. The latter collection comprised pairs or groups of serial M. tuberculosis isolates obtained from 42 individual patients from Belgium (n = 1), Denmark (n = 12), Estonia (n = 4), The Netherlands (n = 17), the United States (n = 7), and Vietnam (n = 1), as well as 10 groups of clustered isolates obtained from different patients from The Netherlands linked by contact tracing, with one or two human-to-human transmissions (K. Kremer, R. Warren, K. Brudey, R. Skuce, C. Gutierrez, T. Lillebaek, E. Fair, C. Arnold, G. Saunders, and D. van Soolingen, Abstr. EU Concerted Action Project Meet., Prague, Czech Republic, p. 58, 2003).
A third set included isolates from 125 patients in 42 IS_6110_-polymorphic GC-rich sequence (PGRS) RFLP strain clusters, assigned to different transmission groups (TGs) according to the likelihood of epidemiological linkage between patients after thorough contact tracing and epidemiological analysis. These clusters were identified in a population-based study conducted in the province of North Holland, The Netherlands, from July 1998 to July 2000 (46, 47).
MIRU-VNTR genotyping.
The isolates were genotyped by PCR amplification of the original 12 MIRU-VNTR loci as described in reference 44 and 17 other loci containing VNTRs of other interspersed sequences (25, 32, 36, 40, 44). They are collectively referred to as MIRU-VNTR in this study. The primers and the conditions for their amplification, their standardized designation, and correspondence with alias designations are described in Tables 1 and 2. Most analyses were performed using multiplex PCR, Rox-labeled MapMarker 1000 size standard (Bioventures), and gel (ABI 377) or capillary electrophoresis-based (ABI 3730-XL) sequencers at the Institut Pasteur de Lille, except for PCR fragments with sizes above 1,000 bp, which were also analyzed by electrophoresis at 150 V for 6 h 30 min using 25-cm gels and 1.2% Ultra Pure electrophoresis-grade agarose (Gibco BRL). Subsets of isolates or loci were analyzed at the Institut Pasteur de Bruxelles and at the National Reference Center for Mycobacteria, Germany, or at Queen's University of Belfast, Northern Ireland, using 3100-Avant capillary electrophoresis-based sequencers or agarose gel electrophoresis, respectively. Sizing of the PCR fragments and assignment of the various VNTR alleles were done using customized GeneScan and Genotyper or Genemapper software packages (PE Applied Biosystems). The reproducibility and accuracy of sizing and the size offsets, which correct differences in relative migration between the size standard and the amplicons depending on the locus and the polymer used for capillary electrophoresis, were checked and standardized among the different laboratories by analyzing selected PCR fragments amplified from M. tuberculosis H37Rv and other reference isolates, as described previously (1, 42).
TABLE 1.
Locus designations and PCR primer sequences used in this study for the 24-locus set
| Set and multiplexa | Locus | Alias(es) | Repeat unit length (bp)b | PCR primer pairs (5′ to 3′, with labeling indicated)c |
|---|---|---|---|---|
| Discriminatory | ||||
| Mix 1 | 580 | MIRU 4; ETR D | 77 | GCGCGAGAGCCCGAACTGC (FAM) |
| GCGCAGCAGAAACGCCAGC | ||||
| 2996 | MIRU 26 | 51 | TAGGTCTACCGTCGAAATCTGTGAC | |
| CATAGGCGACCAGGCGAATAG (VIC) | ||||
| 802 | MIRU 40 | 54 | GGGTTGCTGGATGACAACGTGT (NED) | |
| GGGTGATCTCGGCGAAATCAGATA | ||||
| Mix 2 | 960 | MIRU 10 | 53 | GTTCTTGACCAACTGCAGTCGTCC |
| GCCACCTTGGTGATCAGCTACCT (FAM) | ||||
| 1644 | MIRU 16 | 53 | TCGGTGATCGGGTCCAGTCCAAGTA | |
| CCCGTCGTGCAGCCCTGGTAC (VIC) | ||||
| 3192 | MIRU 31; ETR E | 53 | ACTGATTGGCTTCATACGGCTTTA | |
| GTGCCGACGTGGTCTTGAT (NED) | ||||
| Mix 3 | 424 | Mtub04 | 51 | CTTGGCCGGCATCAAGCGCATTATT |
| GGCAGCAGAGCCCGGGATTCTTC (FAM) | ||||
| 577 | ETR C | 58 | CGAGAGTGGCAGTGGCGGTTATCT (VIC) | |
| AATGACTTGAACGCGCAAATTGTGA | ||||
| 2165 | ETR A | 75 | AAATCGGTCCCATCACCTTCTTAT (NED) | |
| CGAAGCCTGGGGTGCCCGCGATTT | ||||
| Mix 4 | 2401 | Mtub30 | 58 | CTTGAAGCCCCGGTCTCATCTGT (FAM) |
| ACTTGAACCCCCACGCCCATTAGTA | ||||
| 3690 | Mtub39 | 58 | CGGTGGAGGCGATGAACGTCTTC (VIC) | |
| TAGAGCGGCACGGGGGAAAGCTTAG | ||||
| 4156 | QUB-4156 | 59 | TGACCACGGATTGCTCTAGT | |
| GCCGGCGTCCATGTT (NED) | ||||
| Mix 5 | 2163b | QUB-11b | 69 | CGTAAGGGGGATGCGGGAAATAGG |
| CGAAGTGAATGGTGGCAT (FAM) | ||||
| 1955 | Mtub21 | 57 | AGATCCCAGTTGTCGTCGTC (VIC) | |
| CAACATCGCCTGGTTCTGTA | ||||
| 4052 | QUB-26 | 111 | AACGCTCAGCTGTCGGAT (NED) | |
| CGGCCGTGCCGGCCAGGTCCTTCCCGAT | ||||
| Auxiliary | ||||
| Mix 6 | 154 | MIRU 2 | 53 | TGGACTTGCAGCAATGGACCAACT |
| TACTCGGACGCCGGCTCAAAAT (FAM) | ||||
| 2531 | MIRU 23 | 53 | CTGTCGATGGCCGCAACAAAACG (VIC) | |
| AGCTCAACGGGTTCGCCCTTTTGTC | ||||
| 4348 | MIRU 39 | 53 | CGCATCGACAAACTGGAGCCAAAC | |
| CGGAAACGTCTACGCCCCACACAT (NED) | ||||
| Mix 7 | 2059 | MIRU 20 | 77 | TCGGAGAGATGCCCTTCGAGTTAG (FAM) |
| GGAGACCGCGACCAGGTACTTGTA | ||||
| 2687 | MIRU 24 | 54 | CGACCAAGATGTGCAGGAATACAT | |
| GGGCGAGTTGAGCTCACAGAA (VIC) | ||||
| 3007 | MIRU 27; QUB-5 | 53 | TCGAAAGCCTCTGCGTGCCAGTAA | |
| GCGATGTGAGCGTGCCACTCAA (NED) | ||||
| Mix 8 | 2347 | Mtub29 | 57 | GCCAGCCGCCGTGCATAAACCT (FAM) |
| AGCCACCCGGTGTGCCTTGTATGAC | ||||
| 2461 | ETR B | 57 | ATGGCCACCCGATACCGCTTCAGT (VIC) | |
| CGACGGGCCATCTTGGATCAGCTAC | ||||
| 3171 | Mtub34 | 54 | GGTGCGCACCTGCTCCAGATAA (NED) | |
| GGCTCTCATTGCTGGAGGGTTGTAC |
For analysis on automated sequencers, PCR mixtures were prepared as follows, using 96-well plates and the HotStartTaq DNA polymerase kit (QIAGEN, Hilden, Germany). Two nanograms of DNA was added to a final volume of 20 μl containing 0.08 μl of DNA polymerase (0.4 U); 4 μl of Q-solution; 0.2 mM each of dATP, dCTP, dGTP, and dTTP (Pharmacia, Uppsala, Sweden); 2 μl of PCR buffer; 1.5 to 3.0 mM MgCl2; 0.4 μM of each unlabeled oligonucleotide; and from 0.04 to 0.4 μM of labeled oligonucleotide (Tables 1 and 2). The Multiplex PCR kit (QIAGEN, Hilden, Germany) was specifically used for mix 5 (Table 1) to avoid pronounced stutter peaks observed with large alleles of QUB-26. Therefore, 2 ng of DNA was added to a to a final volume of 20 μl containing 10 μl of PCR Master Mix; 1 μl dimethyl sulfoxide; and 0.08, 0.28, and 1 μM of each unlabeled and labeled oligonucleotide for loci 2163a, 1955, and QUB-26, respectively. For analysis using agarose gel electrophoresis, PCRs were performed and analyzed as described previously (32). As additional controls for VNTR 3232 and 3336, oligonucleotides and PCR conditions described in reference 32 were used. Negative controls consisted of the PCR performed in the absence of mycobacterial DNA.
TABLE 2.
Locus designations and PCR conditions for excluded locia
| Locus | Alias | VNTR length (bp) | PCR primer pairs (5′ to 3′, with labeling indicated)b |
|---|---|---|---|
| 1895 | QUB-1895 | 57 | GTGAGCAGGCCCAGCAGACT (NED) |
| CCACGAAATGTTCAAACACCTCAAT | |||
| 1982 | QUB-18 | 78 | CCGGAATCTGCAATGGCGGCAAATTAAAAG |
| TGATCTGACTCTGCCGCCGCTGCAAATA (NED) | |||
| 2163a | QUB-11a | 69 | CCCATCCCGCTTAGCACATTCGTA |
| TTCAGGGGGGATCCGGGA (FAM) | |||
| 3232 | QUB-3232 | 56/57 | TGCCGCCATGTTTCATCAGGATTAA |
| GCAGACGTCGTGCTCATCGATACA (FAM) | |||
| 3336 | QUB-3336 | 59 | AAACAGCACACCGGTGATTTT (VIC) |
| TTCTACGACTTCGCAACCAAGTATC |
Results for exact tandem repeat (ETR) A and the 12 MIRU-VNTR loci for the two collections of the first set of isolates were from references 23, 41, and 42. The results of the analysis of 12 MIRU-VNTR loci for the third set of isolates were from reference 47, while results for MIRU-VNTR loci of M. prototuberculosis isolates were from reference 15.
Spoligotyping.
Spoligotyping was performed according to the previously described method (19). Spoligotype families were assigned as described in reference 10.
Allelic diversity, genetic distance, and clustering analysis.
The MIRU-VNTR allelic diversity (h) at a given locus was calculated as h = 1 − Σ_xi_2 [(n/n − 1)], where xi is the frequency of the _i_th allele at the locus and n is the number of isolates. Minimum spanning tree analysis was performed using Bionumerics (Applied Maths, Belgium) and a categorical coefficient. The priority rule was to first link types that had the highest number of single-locus variants (SLVs). To enable easier detection of locus variations between existing types, creation of hypothetical types was not allowed. The frequencies of SLVs, double-locus variants (DLVs), and triple-locus variants (TLVs) for the different MIRU-VNTR loci were calculated using alleles detected in the different complexes defined by MIRU-VNTR relationships and corroborated by consistent spoligotype family designations. The phylogenetic consistency of the Ghana and Uganda designation (with two subfamilies in the latter case) was supported by independent analysis of genomic deletions (T. Wirth et al., unpublished). The clustering rate was defined as (nc − c)/n, where n is the total number of cases in the sample, c is the number of genotypes represented by at least two cases, and nc is the total number of cases in clusters of two or more patients (38).
RESULTS
Robustness and variability of MIRU-VNTR loci in a standardized collection.
A standardized collection of 90 M. tuberculosis complex isolates from 38 countries (23) was used to perform a first screen of the relative variability and robustness of a set of 29 MIRU-VNTR loci (Tables 1 and 2), including the 12 original MIRU-VNTR (43, 44) and three nonredundant ETR loci (12) that had already been analyzed in this collection (23, 42). Among the 29 loci, five (3232, 3336, 2163a, QUB-1895, and QUB-18) showed a number of problems and were thus subsequently excluded from the final selection.
The problems of these five loci consisted of the absence of PCR products, PCR products that were difficult to interpret, and amplification of multiple alleles, as detailed below. No allele could be amplified from locus 2163a of four isolates despite several attempts. Repeated PCR assays were regularly required to obtain interpretable single amplicons from large alleles of locus QUB-18, even after simplex PCR. For loci 3232 and 3336 two distinct alleles were simultaneously detected in a total of 10 isolates and for locus QUB-1895 in one isolate. These patterns were clearly different from the stutter peaks frequently observed with some VNTRs, as described previously (42). These double alleles were also detected when different DNA extracts, PCR conditions, and primers were used but were not observed for any of the other loci using the same DNA from these isolates, ruling out contamination by foreign DNA or mixed infections. In addition, for locus 3336 a range of intermediate alleles, corresponding to inclusion of repeats with apparent sizes of 5 to 25 bp instead of the 56 bp expected for a full-size repeat, were detected on sequencers, complicating the interpretation of the respective VNTR patterns. When loci 3232, 3336, and 1895 were analyzed by two different laboratories by using the automated and/or the manual techniques, 35 allelic discordances were found for the 90 isolates, of which 17 were for locus 3232, 13 for 3336, and 4 for 1895. In contrast, a single discordance and no discordance were found for MIRU 26 and ETR B, respectively, added as controls. The discordances resulted from differential amplification or interpretation of dual alleles, in addition to simpler sizing problems (see Note S1 in the supplemental material for details).
Finally, QUB-3232, 3336, and 2163a appeared hypervariable compared to the other loci, as seen from the respective numbers of alleles detected (21, 18, and 14), maximal numbers of repeats (25, 21, and 29), and allelic diversities (0.92, 0.91, and 0.87) (Table 3), which can explain part of the problems encountered.
TABLE 3.
Parameters of locus variability among geographically diverse isolates
| Isolate group and locusa | Subsetd | Allelic diversity | Allele no. | No. of repeats (range) |
|---|---|---|---|---|
| 90 isolatesb | ||||
| QUB-2163b | 1 | 0.84 | 10 | 2-11 |
| QUB-26 | 1 | 0.83 | 9 | 2-12 |
| MIRU 10 | 1 | 0.79 | 7 | 2-8 |
| ETR A | 1 | 0.78 | 9 | 2-10 |
| VNTR 1955 | 1 | 0.74 | 10 | 1-14 |
| MIRU 26 | 1 | 0.72 | 8 | 1-8 |
| MIRU 40 | 1 | 0.71 | 7 | 1-8 |
| ETR C | 1 | 0.71 | 5 | 2-6 |
| MIRU 31 | 1 | 0.7 | 6 | 1-6 |
| QUB-4156 | 1 | 0.7 | 5 | 0-4 |
| MIRU 23 | 2 | 0.68 | 9 | 1-11 |
| VNTR 0424 | 1 | 0.66 | 8 | 0-8 |
| VNTR 3690 | 1 | 0.64 | 7 | 1-8 |
| VNTR 2401 | 1 | 0.61 | 4 | 1-4 |
| ETR B | 2 | 0.58 | 5 | 1-5 |
| MIRU 04 | 1 | 0.55 | 9 | 1-6 |
| VNTR 2347 | 2 | 0.52 | 4 | 2-5 |
| MIRU 16 | 1 | 0.48 | 4 | 1-4 |
| MIRU 39 | 2 | 0.41 | 4 | 1-4 |
| MIRU 24 | 2 | 0.39 | 3 | 1-6 |
| VNTR 3171 | 2 | 0.35 | 5 | 1-5 |
| MIRU 20 | 2 | 0.2 | 2 | 1-2 |
| MIRU 27 | 2 | 0.2 | 3 | 2-4 |
| MIRU 02 | 2 | 0.1 | 3 | 1-3 |
| QUB-3232 | Excluded | 0.92 | 21 | 0-24 |
| QUB-3336 | Excluded | 0.91 | 18 | 2-21 |
| QUB-18 | Excluded | 0.87 | 12 | 2-16 |
| QUB-2163a | Excluded | 0.87 | 14 | 1-29 |
| QUB-1895 | Excluded | 0.65 | 6 | 1-7 |
| 494 isolatesc | ||||
| QUB-26 | 1 | 0.84 | ||
| QUB-2163b | 1 | 0.82 | ||
| VNTR 1955 | 1 | 0.76 | ||
| MIRU 26 | 1 | 0.75 | ||
| ETR A | 1 | 0.75 | ||
| MIRU 10 | 1 | 0.74 | ||
| MIRU 40 | 1 | 0.73 | ||
| MIRU 31 | 1 | 0.72 | ||
| VNTR 0424 | 1 | 0.71 | ||
| ETR C | 1 | 0.69 | ||
| VNTR 3690 | 1 | 0.69 | ||
| QUB-4156 | 1 | 0.67 | ||
| MIRU 23 | 2 | 0.65 | ||
| VNTR 2401 | 1 | 0.62 | ||
| MIRU 16 | 1 | 0.53 | ||
| VNTR 2347 | 1 | 0.48 | ||
| MIRU 39 | 2 | 0.45 | ||
| ETR B | 2 | 0.44 | ||
| MIRU 04 | 1 | 0.38 | ||
| MIRU 24 | 2 | 0.35 | ||
| MIRU 20 | 2 | 0.30 | ||
| VNTR 3171 | 2 | 0.27 | ||
| MIRU 27 | 2 | 0.25 | ||
| MIRU 02 | 2 | 0.16 |
The remaining 24 retained loci could all be amplified readily, except for QUB-2163b, from which the corresponding amplicon could not be obtained from one isolate with a T1 spoligotype, and from two exceptional M. bovis variants isolated from oryxes.
By using the 24 remaining MIRU-VNTR loci, 89 distinct genotypes were distinguished for the 90 isolates included in this standardized collection. The only remaining cluster was composed of the clonal M. tuberculosis strains H37Rv and H37Ra, which were also undistinguishable by spoligotyping. Even when the five problematic loci were considered, these two strains remained clustered. By testing multiple combinations of markers within the set of 24 robust loci, we found that the maximal resolution of 89 types in this collection could already be achieved using a minimal group of nine markers (MIRU 04, 10, 16, 26, and 40 and VNTR 0577, 2163b, 2165, and 4052).
Stability of MIRU-VNTRs among epidemiologically linked or serial patient isolates and single-colony cultures.
The clonal stability of the individual MIRU-VNTR loci was evaluated using a total of 171 isolates, including a standardized set of 52 different groups of epidemiologically linked or serial isolates from six different countries and 13 groups of 2 to 10 single-colony cultures obtained from different M. tuberculosis complex strains (Table 4).
TABLE 4.
MIRU-VNTR stability among 65 groups of clonal M. tuberculosis complex isolates
| Locus group and locus | No. of changes in: | |
|---|---|---|
| Epidemiologically linked and serial patient isolates (n = 110; 52 groups) | Single-colony cultures (n = 61; 13 groups) | |
| Includedc | ||
| QUB-26 | 2 | NDf |
| VNTR 3690 | 1 | 1 |
| MIRU 10 | 1 | 0 |
| MIRU 40 | 1 | 0 |
| VNTR 2163b | 1 | ND |
| All others | 0 | 0a |
| Excluded | ||
| QUB-3232 | 3-7b | ND |
| QUB-3336 | 4-0b | ND |
| QUB-18 | 1e | 1 |
| QUB-2163a | 0 | ND |
| QUB-1895 | 0d | 0d |
Among the 24 robust loci, only seven changes were detected in total: six within the epidemiologically linked group and one within the single-colony group. They affected once MIRU 10 and 40 and VNTR 2163b and twice VNTR 3690 and QUB-26. Six of these changes consisted of SLVs within groups, while a single DLV involving MIRU 10 and VNTR 2163b was observed between two serial isolates from an individual patient.
In contrast, at least eight changes were assigned in total for the five excluded loci among the epidemiologically linked or serial isolates and one among the single-colony cultures. Two laboratories analyzed independently loci VNTR 3232 and 3336 from the 52 epidemiologically linked or serial isolate groups and scored for these loci three and four, and seven and zero, changes, respectively. Some changes were scored as uncertain, as the patterns were irreproducible or repeatedly complex with two or even three alleles, and there was no correspondence between any changes scored as certain or possible by the two laboratories for these two loci. A total of 17 discordant results were noticed among the alleles assigned by the two laboratories in the 52 groups. Finally, noninterpretable long amplicons were obtained for locus 1895 of nine epidemiologically linked, serial isolate, and single-colony culture groups (see Note S2 in the supplemental material for details), and locus QUB-18 could not be amplified from one isolate.
MIRU-VNTR variability in an extended collection of isolates from cosmopolitan origins.
In order to analyze the relative evolutionary rate and the resolution power of various marker sets in a sample that is more representative of the worldwide diversity of M. tuberculosis, the above-described standardized collection of 90 M. tuberculosis complex isolates was extended with 404 additional isolates to include a total of 494 isolates from widespread origins, representing the main M. tuberculosis spoligotype prototypes identified so far and the other, more rarely encountered members of the complex. The analysis was restricted to the 24 robust loci in combination with spoligotyping.
The 24 loci resolved 446 types among the 494 isolates (Fig. 1 and 2); 420 isolates had a unique profile, while the remaining 74 isolates were in 26 clusters. All but one of the MIRU-VNTR clusters were composed of two or three isolates of the same spoligotype family. The single larger cluster included 10 epidemiologically related multidrug-resistant (MDR) isolates from the Beijing/W family. Only eight additional types among the 494 isolates were identified when spoligotyping was combined with the 24-locus MIRU-VNTR typing, bringing the total number of types up to 454 by combining the two methods. The values and the overall hierarchies of the allelic diversities among the 24 loci were similar between the small standardized collection and the extended one (Table 3), including 404 additional isolates.
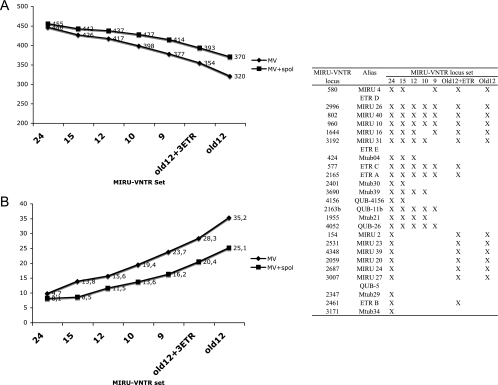
FIG. 1.
Discrimination by MIRU-VNTR typing alone or in combination with spoligotyping in a collection of isolates from cosmopolitan origins. The number of types distinguished among 494 isolates of the M. tuberculosis complex (A) and the corresponding clustering rates (B) obtained by different combinations of markers are shown on the y axes. Because of their particular population structure characterized by homogeneous clonal groups with horizontal gene transfers among them (15), M. prototuberculosis isolates were not considered for this analysis. Diamonds and squares correspond to values obtained using MIRU-VNTR (MV) alone and in combination with spoligotyping (spol), respectively. The composition of the different MIRU-VNTR sets is given in the inset table; 24, full set of 24 robust loci; 15, 15-locus discriminatory subset; Old12, original 12 MIRU-VNTR loci; 9, minimal set resulting in maximal resolution in the standardized collection of 90 isolates (see text).
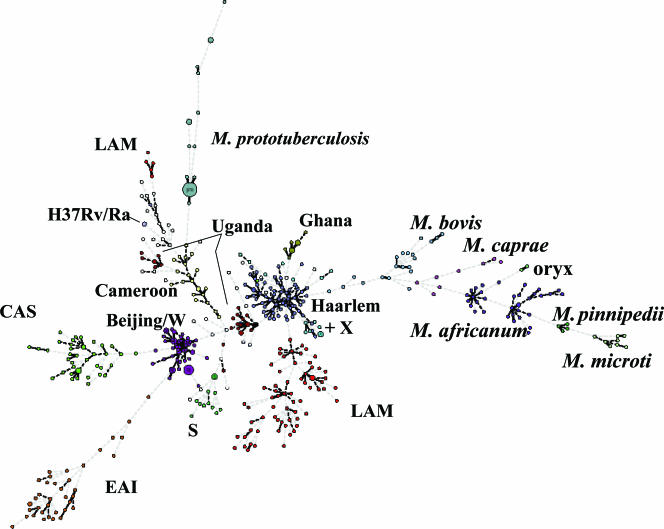
FIG. 2.
Minimum spanning tree based on MIRU-VNTR relationships among tubercle bacilli. Circles correspond to the different types identified by the set of 24 loci among 494 M. tuberculosis complex isolates from cosmopolitan origins and 35 M. prototuberculosis isolates (as reference) and are proportional to the number of clustered isolates with an identical MIRU-VNTR type. The corresponding species names and spoligotype family designations (except T types) are indicated. Linkages by a single, double, or triple locus variation are boldfaced. EAI, East African-Indian; LAM, Latin American-Mediterranean; CAS, Central Asian; S, S spoligotype family; X, X spoligotype family.
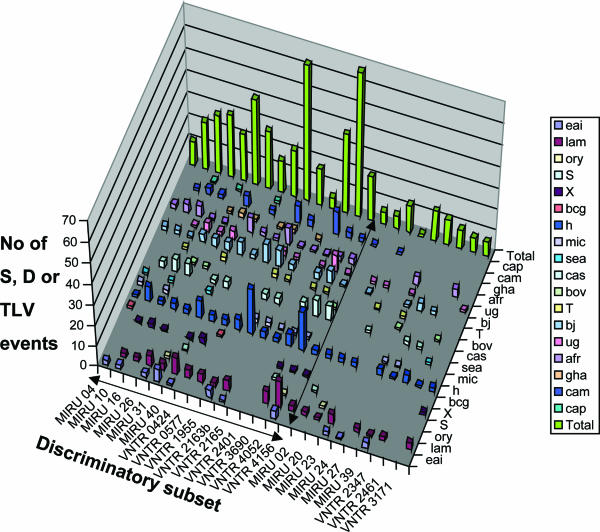
The relative evolutionary rates of the loci were analyzed by calculating the frequency of their involvement in SLVs, DLVs, or TLVs among isolates (Fig. 3). The genetic relationships were analyzed on the basis of the 24 MIRU-VNTR loci using the minimum spanning tree algorithm and compared with independent spoligotype assignations. Consistently, virtually all of the SLVs, DLVs, and TLVs were found among isolates within defined spoligotype families. As expected, the loci most frequently involved in variations among these genetically closely related isolates, such as QUB-2163b and QUB-26, were generally those with the highest allelic diversities. However, other loci with relatively high allelic diversities, such as MIRU 23, were rarely involved in SLVs, DLVs, or TLVs, indicating moderate evolutionary rates within families but rather separate distributions of distinct alleles among different families. At the other extreme, MIRU 02, MIRU 24, and VNTR 3171, the loci with the lowest allelic diversities, were never found as the single locus varying between isolates of the same (or different) families, i.e., they were not involved in any SLVs (not shown), indicating generally low evolutionary rates in the different families. In accordance, these loci could be withdrawn from the 24-locus set without any loss of resolution in this collection.
FIG. 3.
Distribution of single, double, or triple-locus variations in 24 MIRU-VNTR loci among isolates from cosmopolitan origins. Events detected among 494 isolates from widespread geographic origins of the M. tuberculosis complex are shown. Because of their particular population structure, characterized by homogeneous clonal groups with horizontal gene transfers among them (15), M. prototuberculosis isolates were not considered for this analysis. eai, East African-Indian; lam, Latin American-Mediterranean; ory, M. bovis from oryxes; S, S spoligotype family; X, X spoligotype family; bcg, M. bovis BCG; h, Haarlem; mic, Microbacterium microti; sea, M. bovis from seals; cas, Central Asian; bov, classical M. bovis; T, T spoligotypes; bj, Beijing (including W); ug, Uganda; afr, M. africanum; gha, Ghana; cam, Cameroon; cap, Mycobacterium caprae.
Based on these analyses, a discriminatory subset of 15 loci was selected to retain the markers that showed both high allelic diversities and the most frequent involvement in SLVs, DLVs, or TLVs across the different lineages tested, with a cutoff generally set to at least 15 events for the whole collection (Fig. 3). This subset includes the minimal group of nine loci providing the maximal resolution of the isolates in the standardized collection, as described above. The use of this 15-locus subset alone or in combination with spoligotyping resulted in only marginal loss of resolution and increase of clustering rate, as it discriminated 425 or 441 types among the 494 isolates, respectively (Fig. 1). The increase in clustering was evenly distributed among the different lineages (not shown). Substraction of up to five additional loci (MIRU 04 and 10 and VNTR 2401, 0424, and 4156) with relatively lower frequencies of SLVs, DLVs, or TLVs resulted in further minor but increasingly larger losses of resolution, only partly compensated for by the additional use of spoligotyping. In contrast, the use of the 12 original MIRU-VNTR loci alone or in combination with the ETR loci or of the minimal group of nine markers described above and providing maximal resolution in the small standardized collection resulted in stronger decreases of resolution, even if combined with spoligotyping. Similarly, the use of the original 12 MIRU-VNTR loci alone or in combination with spoligotyping resulted in four- to threefold-higher clustering rates than those of the 24- or the 15-locus set alone or in combination with spoligotyping.
Analysis of IS_6110_-RFLP clusters from a population-based study with the discriminatory subset of 15 MIRU-VNTR loci.
The few changes observed in some of the 24 robust loci in the serial isolates and epidemiologically linked and single-colony culture groups described above were all confined to markers composing the 15-locus discriminatory subset. The epidemiological relevance of these changes was therefore assessed by examining isolates from 40 high-copy-number (≥6) and two low-copy-number IS_6110_-PGRS RFLP clusters of a previously published population-based study (46, 47). Based on thorough epidemiological analysis these clusters had been classified into groups with proven epidemiological links (TG1) and proven contacts after DNA fingerprint data had become available (TG2), likely contacts (TG3), or no epidemiological links (TG4).
Among the 24 IS_6110_-PGRS RFLP clusters of epidemiologically linked patients, comprising 57 isolates from TG1 and TG2, only two (clusters 8 and 13) were subdivided by changes in the 15-locus subset (Fig. 4). In both cases, the changes consisted of an SLV in one clustered isolate, affecting locus ETR C or QUB-2163b, respectively.
FIG. 4.
IS_6110_ RFLP, spoligotype, and MIRU-VNTR patterns of the M. tuberculosis isolates from 125 patients in 42 IS_6110_-PGRS clusters, as assigned to four transmission groups (for explanation of transmission groups, see text). Designations of MIRU-VNTR loci are given according to the position (in kilobase pairs) on the M. tuberculosis H37Rv chromosome. Alias designations are in parentheses. Spoligo, spoligotyping. Results of IS_6110_-RFLP, spoligotyping, and MIRU-VNTR loci from VNTR 424 to 4156 were taken from reference 47; results for VNTR 2165 (ETR A), VNTR 1955, 2163b (QUB-11b), and 4052 (QUB-26) are from this study. Differences in MIRU-VNTR patterns among IS_6110_-PGRS RFLP clustered isolates are boxed. For IS_6110_-PGRS RFLP cluster 37, the isolate of the patient from TG3 is compared with the isolate of the first patient from TG4, and they thus differ by an SLV in MIRU-VNTR locus 4052. Likewise, for IS_6110_-PGRS RFLP cluster 38, the isolates of the two patients of category TG3 differ by a four-locus variation (in loci 2165, 1955, 2163b, and 4052).
Of the 23 IS_6110_-PGRS clusters comprising in total 54 isolates from category TG3 patients, six clusters (23, 27, 32, 34, 37, and 38) were subdivided by the 15-locus subset. In clusters 23, 27, 34, and 37, the changes consisted again of an SLV in one clustered isolate, affecting VNTR 424, QUB-26, MIRU 26, and QUB-26, respectively. For the two remaining clusters (clusters 32 and 38), a DLV and a four-locus change were detected between two clustered isolates, respectively. It is noteworthy that these two clusters, as well as cluster 34, were characterized by IS_6110_-PGRS profiles with only five, seven, or five IS_6110_ copies, respectively.
Finally, of the seven IS_6110_-PGRS clusters of isolates from TG4, six (85.7%) were split by differences in two to six loci of the 15-locus MIRU-VNTR subset.
DISCUSSION
MIRU-VNTR typing based on 12 loci in combination with spoligotyping has been adopted as a robust basis for large-scale real-time typing of M. tuberculosis isolates in the United States, after validation in a model population (6). Although the resolution provided by this method has proven to be adequate in many situations, a significant proportion of unrelated isolates remains falsely clustered (6, 35). The properties of ideal molecular markers represent a compromise between (i) sufficient variability to distinguish unrelated strains and (ii) satisfactory clonal stability to reliably identify isolates from the same strains and trace transmission chains, as well as robustness to be applicable to a wide range of strains. To meet these criteria, we have now defined an optimized set of 24 MIRU-VNTR markers, including the original 12-locus set, by analyzing a total of 824 tubercle bacillus isolates from 52 different countries or overseas territories.
In a standardized collection (23) including 90 isolates from 38 countries, the set of the 24 selected MIRU-VNTR loci reached nearly the maximal resolution, with 89 distinct types. This represents the highest discrimination achieved among all typing methods tested so far in this collection and a significant gain compared to the 78, 81, 82, and 84 types obtained with the 12 original loci, the 12 original locus markers combined with three ETR loci, five QUB loci, and IS_6110_-RFLP, respectively (for details see references 21, 23, and 42). The only clustered strains were H37Rv and H37Ra, which were derived from a single M. tuberculosis strain in the 1930s and kept thereafter as laboratory strains.
In an extended collection of 494 isolates from cosmopolitan origins, the use of the 24-locus MIRU-VNTR set increased the number of types by 40%, compared to those obtained with the original 12-locus set, respectively. When spoligotyping was used as an additional method, the gain in the number of types was of 23% with the set of 24 loci compared to the original 12-locus set. This lower relative gain in resolution power observed after the addition of spoligotyping consistently reflects the much lower frequency of subdivision by spoligotyping of clusters defined by the 24 loci, compared to subdivision by spoligotyping of clusters defined by the original 12 loci. The clustering rates were consequently decreased by fourfold—and by threefold in combination with spoligotyping—when the 24-locus MIRU-VNTR set was used instead of the original 12-locus set. Virtually all of the remaining clusters were composed of two to three isolates which were quite evenly distributed within the different spoligotype families, indicating that this 24-locus set discriminates quite evenly and well in the primary M. tuberculosis lineages. The single larger cluster included 10 MDR isolates from the Beijing/W family with subtle differences in complex IS_6110_ fingerprints. However, the isolates of this cluster also share the same distinctive mutations in rpoB, rpsL, embB, and katG and are known to be part of consecutive epidemic waves in New York City, N.Y., originating from a single MDR clone (29). Some other, smaller clusters were also known to correspond to outbreaks and/or to include isolates with identical IS_6110_ fingerprints (data not shown). The values and the overall hierarchies of the allelic diversities among the 24 loci were broadly similar between the smaller standardized collection of 90 M. tuberculosis complex isolates (23) and the extended one including 404 additional isolates from widespread origins, despite marked changes in composition and sizes. This observation indicates a good preservation of the relative performances of the individual loci in these different strain populations, although clear differences of diversities were obvious among different lineages for some loci (e.g., allelic diversity of ETR B was 0.74 in East African-Indian isolates, compared to 0 in Central Asian and Beijing isolates).
Therefore, we have analyzed the relative evolutionary rates of the 24 loci by measuring the frequency of their involvements in single- to triple-locus variations among well-characterized isolates. Such an approach, similar to the BURST approach (9), is less sensitive to sampling bias and to problems of saturation (i.e., ancient changes being obscured by more recent ones) than simple analyses of allelic diversities or reductive analysis among diverse strains. If the combined MIRU-VNTR markers are not subject to convergence (independent evolution to the same state), SLVs, DLVs, or TLVs should inherently occur among closely related isolates and thus, on a probabilistic basis, involve loci with the highest evolutionary rates. This assumption was confirmed by the observation that virtually all of the SLVs, DLVs, and TLVs were found among isolates within phylogenetically consistent spoligotype families (i.e., excluding the T family and apparently rare outliers in families such as the Latin American-Mediterranean family; see reference 11). In general a very good concordance between MIRU-VNTR groupings and these spoligotype families was observed (Fig. 2), which also indicates that the selected 24-locus set is highly informative and, not surprisingly, better than the smaller set of the 12 original loci (11, 13, 42) for phylogenetic identification of M. tuberculosis complex lineages. As expected, the loci the most frequently involved in SLVs, DLVs, or TLVs generally showed the highest overall allelic diversities. The converse situation did not necessarily occur, as found for some loci (e.g., MIRU 23), indicating a lower evolutionary rate but rather separate distributions of distinct alleles among different families.
Consistently, a discriminatory subset gathering the 15 robust MIRU-VNTR markers most frequently involved in SLVs, DLVs, or TLVs distributed across the different families concentrated almost all of the resolution (96%, or 98% in combination with spoligotyping) provided by the full set of 24 loci. The nine less-variable remaining robust loci thus provide auxiliary discrimination, consistent with their exceptional involvement in SLVs in spite of sometimes relatively high allelic diversities in some lineages (e.g., ETR B, see above). The usefulness of these less-variable ancillary loci thus mostly resides in their collective contribution to more accurate phylogenetic identification of clones from the different lineages, when used in addition to the other 15 loci (Fig. 2). When some loci of the 15-locus subset were removed from the analysis, the marginal gain obtained with secondary use of spoligotyping gradually increased, but this gain gradually compensated less for the deficit in resolution, as expected. In agreement with higher evolutionary rates, the few clonal changes observed among the 24 robust loci in the epidemiologically linked, serial isolate, and single-colony culture groups were all confined to some markers composing this 15-locus discriminatory subset. Further in accordance with higher evolutionary rates, this subset of 15 MIRU-VNTR loci was able to split six out of the seven clusters of patients from a population-based study for which no epidemiological link could be established and which had isolates with identical IS_6110_ high-copy-number RFLP fingerprints. This situation likely corresponds to the most stringent conditions to analyze the resolution power in M. tuberculosis. It is noteworthy that cases of subdivision of false clusters based on high-copy-number IS_6110_ RFLP fingerprints will in general probably remain a minority, as another population-based study showed that in most cases the clustering on the basis of IS_6110_ RFLP typing and that on the basis of advanced MIRU-VNTR typing are in agreement (Cardoso-Oelemann et al., submitted). However, our data suggest that optimized MIRU-VNTR typing, especially when combined with spoligotyping, may be overall more accurate for cluster analysis than IS_6110_ RFLP typing (47; Cardoso-Oelemann et al., submitted).
Among the seven changes observed in this 15-locus subset over 123 possible independent events among clones within the epidemiologically linked, serial isolate, and single-colony culture groups, six differences (4.9%) involved only an SLV, and a single one involved a DLV (0.8%). The changes in these SLV cases most likely reflect rare and stochastic MIRU-VNTR mutation events and inherent genetic drift in clonal populations originating from recent transmission (34, 47). However, in the single DLV case, a single-band difference was also observed between the IS_6110_ RFLP fingerprints of the two serial isolates from the elderly, possibly foreign-born patient involved (data not shown); this distinction concordantly seen with IS_6110_ RFLP and two independent MIRU-VNTR loci casts some suspicion on the clonal origin of these two isolates (i.e., this patient might have been infected independently by two closely related strains). Likewise, only two differences (6.1%) involving only an SLV were observed over 33 possible independent events in the population-based clusters including patients with a proven epidemiological link (TG1 and TG2). As expected, a slightly higher proportion of differences were observed in the clusters with only a likely link (TG3). In that case, four changes (12.9% of possible independent events) involved an SLV, while two involved a DLV and a four-locus change, respectively. However, the molecular definition of the two latter clusters was based on RFLP profiles with relatively low IS_6110_ copy numbers, known to be poorly discriminated in spite of the additional use of PGRS fingerprinting and spoligotyping. Therefore, the MIRU-VNTR results cast doubt on the likelihood of the links and thus of the clonal transmission suggested by contact tracing for these two TG3 clusters. Hence, if cautiously only the groups with proven epidemiological links are considered, the chances of erroneous exclusion from a cluster of an isolate showing an SLV or DLV in the 15-locus subset are estimated to be only 5 to 6% or less than 1% (if any; see above), respectively. The probability of incorrect exclusion when mutations involve more than two loci or a locus from the rest of the 24-locus set is close to zero, since no such event was observed among clonal isolates. In contrast to a suggestion from a recent report (35) but in accordance with others studying the 12 original loci (4, 6, 16, 24, 34), these results thus fully support the stability of MIRU-VNTR types among epidemiologically related isolates and thus the use of MIRU-VNTR changes for reliable exclusion of a link (termed “sensitivity of typing” in reference 35) when ongoing transmission is tracked.
Acceptable technical problems were occasionally encountered with some of the 24 markers, including relatively frequent—but still interpretable—stutter peaks (e.g., with QUB-26) and exceptional PCR failures for some infrequent genotypes (i.e., with locus 2163b in some Mycobacterium africanum isolates or isolates from oryxes). In such a case, the genotypes can still reliably be compared based on the 23 (or 14) remaining loci.
In contrast to the 24-locus MIRU-VNTR set, five other loci out of a total of 29 tested failed to acceptably fulfill the criteria of sufficient variability yet required stability and robustness as defined above. Three of them, i.e., QUB-3232, 3336, and 2163a, were hypervariable with respect to both allelic diversity among unrelated isolates and frequency of changes among epidemiologically linked and serial patient isolates. Some repeatedly yielded double alleles in several isolates, regardless of the experimental conditions and/or were subject to frequent PCR failures or uninterpretable PCR patterns, resulting in numerous discordances or uncertainties between results obtained in independent laboratories or within the same laboratory (see Note S3 in the supplemental material for details) (20). We believe that even hypothetical technical improvements will at best only partially solve these problems, and the inclusion of these markers in a routine panel for general screening such as proposed elsewhere (18, 45) would reduce the overall reproducibility to an unacceptable rate. Future studies will determine if such hypervariable markers could nevertheless retain some utility in very well controlled conditions for dissection of long transmission chains. It is noteworthy that these markers excluded for M. tuberculosis were not subject to such problems when specifically applied to M. bovis (except QUB-3336), probably because of generally lower repeat numbers in M. bovis isolates. Therefore, panels including some of these markers might be specifically proposed for bovine tubercle bacilli, in order to cope with the particularly high genetic homogeneity reported among these isolates in previous studies (2, 37).
A few additional M. tuberculosis VNTR loci (25, 39) were not included in this study. Among them, VNTR 3820, proposed in another panel of markers (39), displayed features characteristic of hypervariable loci (large repeat and allele numbers and high allelic diversity), which we predicted to lead to the problems described above. As far as the other loci we suspect them to be unlikely to have a significant impact on the discriminatory power, as judged from the limited data available.
In conclusion, we have defined an optimized set of 24 MIRU-VNTR loci, including a discriminatory subset of 15 loci to be used as first-line molecular typing tools. These sets significantly and reliably improve the resolution for molecular epidemiological studies compared to the original set of 12 MIRU-VNTR loci. However, not all 15 or 24 loci are necessarily required to define all the unique isolates in any given situation. Depending on the cases, such as for the standardized collection of 90 isolates (23), nine loci from the 15-locus set can be already sufficient. As a general guideline, loci with the highest evolutionary rates within this set may be applied in the first place, possibly depending on the lineage(s) known to be prevalent in the area of interest (5) (Fig. 3 shows the loci that are the most frequently involved in SLVs, DLVs, or TLVs in the different lineages). In other cases, the SLV distribution and the resolution of the 15-locus set compared with that of the 24-locus set in the worldwide collection indicate that secondary typing with a few ancillary loci may provide additional discrimination of particular genotypes in some specific lineages. Spoligotyping also yielded additional albeit marginal resolution in this study. Therefore, its combined use with MIRU-VNTR typing may be useful, especially as a quick and convenient independent control. Overall, however, the optimized MIRU-VNTR set described here is predicted to be generally effective for M. tuberculosis typing in many settings, as it has been defined based on numerous representatives of the principal worldwide M. tuberculosis lineages. Its predictive value for evaluating M. tuberculosis transmission was found to be equal to that of IS_6110_-RFLP typing in a companion population-based study. Therefore, we propose the 15-locus (for epidemiological studies) and 24-locus (more for phylogenetic studies) sets as a basis for standardized MIRU-VNTR typing of M. tuberculosis. The introduction of this more advanced method provides the possibility of maintaining and reinforcing international communication in the field of TB molecular epidemiology, already facilitated by the recognition of IS_6110_-RFLP as the gold standard since the early 1990s. This situation is almost unique among the infectious diseases.
Supplementary Material
[Supplemental material]
Acknowledgments
Anne-Laure Bañuls is acknowledged for discussions at initial stages of the work. Philip Suffys is gratefully thanked for his support to M.C.-O.
The work was supported by INSERM, the Institut Pasteur de Lille, a grant from the Lille Génopole, and the European Community (grant QLK2-CT-2000-00630). Parts of this work were also supported by the German Ministry of Health and the Robert Koch Institute, Berlin, Germany. E.S. held a Poste Vert from the INSERM. M.C.-O. held a fellowship from the CAPES; P.S. is a Chercheur du Centre National de Recherche Scientifique.
Footnotes
▿
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Allix, C., P. Supply, and M. Fauville-Dufaux. 2004. Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin. Infect. Dis. 39**:**783-789. [DOI] [PubMed] [Google Scholar]
- 2.Allix, C., K. Walravens, C. Saegerman, J. Godfroid, P. Supply, and M. Fauville-Dufaux. 2006. Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS_6110_ restriction fragment length polymorphism analysis and spoligotyping. J. Clin. Microbiol. 44**:**1951-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10**:**45-52. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood, K. S., J. N. Wolfe, and A. M. Kabani. 2004. Application of mycobacterial interspersed repetitive unit typing to Manitoba tuberculosis cases: can restriction fragment length polymorphism be forgotten? J. Clin. Microbiol. 42**:**5001-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Guttierez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, M. L. Ho, C. Martin, C. Martin, I. Mokrousov, O. Narvskaia, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. G. Shemyakin, U. B. Singh, A. Somoskovi, R. A. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. M. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6**:**23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowan, L. S., L. Diem, T. Monson, P. Wand, D. Temporado, T. V. Oemig, and J. T. Crawford. 2005. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J. Clin. Microbiol. 43**:**688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer, A. S., K. Kremer, M. W. Borgdorff, P. E. de Haas, H. F. Heersma, and D. van Soolingen. 2000. Genetic heterogeneity in Mycobacterium tuberculosis isolates reflected in IS_6110_ restriction fragment length polymorphism patterns as low-intensity bands. J. Clin. Microbiol. 38**:**4478-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, J. T., P. M. Hawkey, E. G. Smith, K. A. Boese, R. E. Warren, and G. Hong. 2004. Automated high-throughput mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis strains by a combination of PCR and nondenaturing high-performance liquid chromatography. J. Clin. Microbiol. 42**:**4175-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55**:**561-590. [DOI] [PubMed] [Google Scholar]
- 10.Filliol, I., J. R. Driscoll, D. Van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. D. Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, G. Kallenius, E. Kassa-Kelembho, T. Koivula, H. M. Ly, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. De Waard, C. Sola, and N. Rastogi. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg. Infect. Dis. 8**:**1347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188**:**759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144**:**1189-1196. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, A., T. Brown, L. Baker, and F. Drobniewski. 2005. Can 15-locus mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis provide insight into the evolution of Mycobacterium tuberculosis? Appl. Environ. Microbiol. 71**:**8207-8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goyal, M., D. Young, Y. Zhang, P. A. Jenkins, and R. J. Shaw. 1994. PCR amplification of variable sequence upstream of katG gene to subdivide strains of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 32**:**3070-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez, M. C., S. Brisse, R. Brosch, M. Fabre, B. Omais, M. Marmiesse, P. Supply, and V. Vincent. 2005. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 1**:**e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawkey, P. M., E. G. Smith, J. T. Evans, P. Monk, G. Bryan, H. H. Mohamed, M. Bardhan, and R. N. Pugh. 2003. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS_6110_-based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J. Clin. Microbiol. 41**:**3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillemann, D., T. Kubica, R. Agzamova, B. Venera, S. Rusch-Gerdes, and S. Niemann. 2005. Rifampicin and isoniazid resistance mutations in Mycobacterium tuberculosis strains isolated from patients in Kazakhstan. Int. J. Tuberc. Lung Dis. 9**:**1161-1167. [PubMed] [Google Scholar]
- 18.Kam, K. M., C. W. Yip, L. W. Tse, K. L. Leung, K. L. Wong, W. M. Ko, and W. S. Wong. 2006. Optimization of variable number tandem repeat typing set for differentiating Mycobacterium tuberculosis strains in the Beijing family. FEMS Microbiol. Lett. 256**:**258-265. [DOI] [PubMed] [Google Scholar]
- 19.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35**:**907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kremer, K., C. Arnold, A. Cataldi, M. C. Gutierrez, W. H. Haas, S. Panaiotov, R. A. Skuce, P. Supply, A. G. van der Zanden, and D. van Soolingen. 2005. Discriminatory power and reproducibility of novel DNA typing methods for Mycobacterium tuberculosis complex strains. J. Clin. Microbiol. 43**:**5628-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kremer, K., B. K. Au, P. C. Yip, R. Skuce, P. Supply, K. M. Kam, and D. van Soolingen. 2005. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS_6110_ restriction fragment length polymorphism typing and spoligotyping. J. Clin. Microbiol. 43**:**314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kremer, K., J. R. Glynn, T. Lillebaek, S. Niemann, N. E. Kurepina, B. N. Kreiswirth, P. J. Bifani, and D. van Soolingen. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 42**:**4040-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37**:**2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwara, A., R. Schiro, L. S. Cowan, N. E. Hyslop, M. F. Wiser, S. Roahen Harrison, P. Kissinger, L. Diem, and J. T. Crawford. 2003. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 41**:**2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Fleche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2**:**37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magdalena, J., P. Supply, and C. Locht. 1998. Specific differentiation between Mycobacterium bovis BCG and virulent strains of the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 36**:**2471-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magdalena, J., A. Vachee, P. Supply, and C. Locht. 1998. Identification of a new DNA region specific for members of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 36**:**937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98**:**1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munsiff, S. S., B. Nivin, G. Sacajiu, B. Mathema, P. Bifani, and B. N. Kreiswirth. 2003. Persistence of a highly resistant strain of tuberculosis in New York City during 1990-1999. J. Infect. Dis. 188**:**356-363. [DOI] [PubMed] [Google Scholar]
- 30.Niemann, S., T. Kubica, F. C. Bange, O. Adjei, E. N. Browne, M. A. Chinbuah, R. Diel, J. Gyapong, R. D. Horstmann, M. L. Joloba, C. G. Meyer, R. D. Mugerwa, A. Okwera, I. Osei, E. Owusu-Darbo, S. K. Schwander, and S. Rusch-Gerdes. 2004. The species Mycobacterium africanum in the light of new molecular markers. J. Clin. Microbiol. 42**:**3958-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owusu-Dabo, E., O. Adjei, C. G. Meyer, R. D. Horstmann, A. Enimil, T. F. Kruppa, F. Bonsu, E. N. L. Browne, M. A. Chinbuah, I. Osei, J. Gyapong, C. Berberich, T. Kubica, S. Niemann, and S. Rüsch-Gerdes. 2006. Mycobacterium tuberculosis drug resistance, Ghana. Emerg. Infect. Dis. 12**:**1171-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roring, S., A. Scott, D. Brittain, I. Walker, G. Hewinson, S. Neill, and R. Skuce. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40**:**2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roring, S., A. N. Scott, R. G. Hewinson, S. D. Neill, and R. A. Skuce. 2004. Evaluation of variable number tandem repeat (VNTR) loci in molecular typing of Mycobacterium bovis isolates from Ireland. Vet. Microbiol. 101**:**65-73. [DOI] [PubMed] [Google Scholar]
- 34.Savine, E., R. M. Warren, G. D. van der Spuy, N. Beyers, P. D. van Helden, C. Locht, and P. Supply. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 40**:**4561-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott, A. N., D. Menzies, T. N. Tannenbaum, L. Thibert, R. Kozak, L. Joseph, K. Schwartzman, and M. A. Behr. 2005. Sensitivities and specificities of spoligotyping and mycobacterial interspersed repetitive unit-variable-number tandem repeat typing methods for studying molecular epidemiology of tuberculosis. J. Clin. Microbiol. 43**:**89-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148**:**519-528. [DOI] [PubMed] [Google Scholar]
- 37.Skuce, R. A., S. W. McDowell, T. R. Mallon, B. Luke, E. L. Breadon, P. L. Lagan, C. M. McCormick, S. H. McBride, and J. M. Pollock. 2005. Discrimination of isolates of Mycobacterium bovis in Northern Ireland on the basis of variable numbers of tandem repeats (VNTRs). Vet. Rec. 157**:**501-504. [DOI] [PubMed] [Google Scholar]
- 38.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330**:**1703-1709. [DOI] [PubMed] [Google Scholar]
- 39.Smittipat, N., P. Billamas, M. Palittapongarnpim, A. Thong-On, M. M. Temu, P. Thanakijcharoen, O. Karnkawinpong, and P. Palittapongarnpim. 2005. Polymorphism of variable-number tandem repeats at multiple loci in Mycobacterium tuberculosis. J. Clin. Microbiol. 43**:**5034-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smittipat, N., and P. Palittapongarnpim. 2000. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuber. Lung Dis. 80**:**69-74. [DOI] [PubMed] [Google Scholar]
- 41.Sola, C., I. Filliol, E. Legrand, S. Lesjean, C. Locht, P. Supply, and N. Rastogi. 2003. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect. Genet. Evol. 3**:**125-133. [DOI] [PubMed] [Google Scholar]
- 42.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39**:**3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26**:**991-1003. [DOI] [PubMed] [Google Scholar]
- 44.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36**:**762-771. [DOI] [PubMed] [Google Scholar]
- 45.Surikova, O. V., D. S. Voitech, G. Kuzmicheva, S. I. Tatkov, I. V. Mokrousov, O. V. Narvskaya, M. A. Rot, D. van Soolingen, and M. L. Filipenko. 2005. Efficient differentiation of Mycobacterium tuberculosis strains of the W-Beijing family from Russia using highly polymorphic VNTR loci. Eur. J. Epidemiol. 20**:**963-974. [DOI] [PubMed] [Google Scholar]
- 46.van Deutekom, H., S. P. Hoijng, P. E. de Haas, M. W. Langendam, A. Horsman, D. van Soolingen, and R. A. Coutinho. 2004. Clustered tuberculosis cases: do they represent recent transmission and can they be detected earlier? Am. J. Respir. Crit. Care Med. 169**:**806-810. [DOI] [PubMed] [Google Scholar]
- 47.van Deutekom, H., P. Supply, P. E. de Haas, E. Willery, S. P. Hoijng, C. Locht, R. A. Coutinho, and D. van Soolingen. 2005. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J. Clin. Microbiol. 43**:**4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31**:**406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren, R. M., T. C. Victor, E. M. Streicher, M. Richardson, G. D. van der Spuy, R. Johnson, V. N. Chihota, C. Locht, P. Supply, and P. D. van Helden. 2004. Clonal expansion of a globally disseminated lineage of Mycobacterium tuberculosis with low IS_6110_ copy numbers. J. Clin. Microbiol. 42**:**5774-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]