Mitf regulation of Dia1 controls melanoma proliferation and invasiveness (original) (raw)
Abstract
It is widely held that cells with metastatic properties such as invasiveness and expression of matrix metalloproteinases arise through the stepwise accumulation of genetic lesions arising from genetic instability and “clonal evolution.” By contrast, we show here that in melanomas invasiveness can be regulated epigenetically by the microphthalmia-associated transcription factor, Mitf, via regulation of the DIAPH1 gene encoding the diaphanous-related formin Dia1 that promotes actin polymerization and coordinates the actin cytoskeleton and microtubule networks at the cell periphery. Low Mitf levels lead to down-regulation of Dia1, reorganization of the actin cytoskeleton, and increased ROCK-dependent invasiveness, whereas increased Mitf expression leads to decreased invasiveness. Significantly the regulation of Dia1 by Mitf also controls p27Kip1-degradation such that reduced Mitf levels lead to a p27Kip1-dependent G1 arrest. Thus Mitf, via regulation of Dia1, can both inhibit invasiveness and promote proliferation. The results imply variations in the repertoire of environmental cues that determine Mitf activity will dictate the differentiation, proliferative, and invasive/migratory potential of melanoma cells through a dynamic epigenetic mechanism.
Keywords: Dia1, melanoma, Mitf, actin, p27
The ability of cancer cells to acquire properties of invasiveness and the potential to metastasize is not entirely understood. In the traditional model, genetic instability within a tumor would lead within some cells to the accumulation of genetic and stable epigenetic lesions that are compatible with increased invasiveness and metastatic potential. These genetic lesions and stable epigenetic modifications, such as histone and DNA methylation, would be inherited by the metastatic progeny and dictate alterations in gene expression compatible with metastatic potential (for a review, see Baylin and Ohm 2006). Alternatively, invasive potential could represent a specific epigenetic state that is inherently unstable and nonheritable, being imposed by the microenvironment of the cell; in this model, properties associated with metastasis may be lost once the invasive cell has taken up residence in another location. While considerable resources have been expended on trying to pinpoint genetic mutations that correlate with metastatic potential and, more recently, the resulting stable epigenetic changes, the molecular mechanisms that would underpin a dynamic epigenetic model for cancer metastasis have been relatively little explored.
Some cancer types, such as melanoma, are intrinsically more metastatic than others, and it has been postulated that the ability of melanomas to invade and proliferate may be related to the inherent ability of melanoblasts to migrate from the neural crest and proliferate to populate the epidermis and hair follicles (Vance and Goding 2004; Gupta et al. 2005). Some genes have been identified whose expression appears to correlate with the ability of melanomas to metastasize. The expression of SLUG, for example, can promote metastasis and facilitates the loss of E-cadherin (Gupta et al. 2005), a key molecule that mediates melanocyte adhesion to keratinocytes that is lost when melanomas acquire invasive potential (Haass et al. 2005), while NEDD9, an adaptor protein that interacts with focal adhesion kinase, is frequently overexpressed in metastatic melanoma and promotes invasiveness (Kim et al. 2006). However, despite these advances, and some preliminary investigations into the concept that metastatic melanoma might arise from a dynamic epigenetic event (Seftor et al. 2005), how such an epigenetic mechanism might operate in melanoma to induce the changes in cytoskeletal organization and metalloproteinase expression that accompany the acquisition of invasive potential remains poorly understood.
The Microphthalmia-associated transcription factor Mitf (Steingrimsson et al. 2004) is crucially important for many aspects of melanocyte development and function. It is critically required for survival of melanoblasts (Opdecamp et al. 1997) and postnatal melanocyte stem cells (Nishimura et al. 2005). Mitf is also implicated in promoting differentiation and activating expression of melanogenic enzymes (Steingrimsson et al. 2004) and can induce a G1 cell cycle exit through up-regulation of the p21Cip1 and p16INK4a cyclin-dependent kinase inhibitors (Carreira et al. 2005; Loercher et al. 2005). The anti-proliferative role of Mitf is also underscored by the observation that activated BRAF expression can suppress Mitf expression and that increasing Mitf expression in melanomas leads to decreased proliferation (Wellbrock and Marais 2005). On the other hand, Mitf has also been termed a “lineage survival oncogene,” being amplified >100-fold in some melanomas and required for the proliferation of melanoma cells bearing activated BRAF (Widlund et al. 2002; Du et al. 2004; Garraway et al. 2005). Why Mitf is required for proliferation of melanoma cells is not known, and how Mitf can apparently operate as both a pro- and anti-proliferative factor is an unresolved paradox.
In this report we identify Mitf as a novel regulator of the DIAPH1 gene encoding the diaphanous-related formin Dia1 that controls actin polymerization. Since Dia1 also regulates Skp2, an F-box protein that promotes degradation of p27Kip1, depletion of Mitf in melanoma cells leads to a p27Kip1-dependent G1 arrest and reorganization of the actin cytoskeleton accompanied by increased invasiveness. The results explain why Mitf is required for proliferation of melanomas and suggest that variations in the repertoire of signals that determine Mitf activity will dictate the proliferative and invasive potential of individual cells through a dynamic epigenetic mechanism.
Results
Depletion of Mitf induces a G1 cell cycle arrest
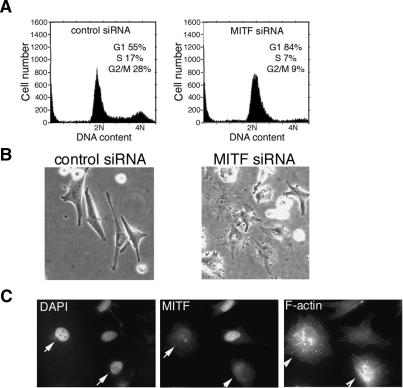
In the course of experiments where we recently demonstrated that elevated Mitf levels induce a p21Cip1-dependent G1 cell cycle arrest (Carreira et al. 2005), we also observed that depletion of Mitf using small interfering RNA (siRNA) led to a G1 cell cycle arrest but no increase in the sub-G1 fraction (Fig. 1A) or PARP cleavage (data not shown), in agreement with previous observations made using other methods that Mitf is necessary for melanoma proliferation (Widlund et al. 2002; Du et al. 2004; Garraway et al. 2005) and that depletion of Mitf does not lead to increased apoptosis (Larribere et al. 2005). Intriguingly, while elevated Mitf induced a more elongated phenotype (Carreira et al. 2005), Mitf-depleted cells appeared more rounded (Fig. 1B), and consistent with this, immunofluorescence assays (Fig. 1C) revealed that Mitf-depleted cells exhibited an F-actin profile that was strikingly different from cells that maintained Mitf expression, although the precise morphology of the cells and their F-actin profile varied from cell to cell, presumably reflecting in part the degree of Mitf depletion and the length of time that cells had been cell cycle arrested. Importantly, depletion of Mitf did not kill the cells as once the effects of the Mitf siRNA treatment declined several days after transfection the cells resumed proliferation.
Figure 1.
Depletion of Mitf induces a G1 cell cycle arrest. (A) Flow cytometry of 501 mel melanoma cells treated with control or Mitf-specific siRNA1. (B) Phase image of control and Mitf-depleted cells. (C) Immunofluorescence assay of cells treated with Mitf-specific siRNA using anti-Mitf antibody or phalloidin to reveal F-actin. Arrows indicate Mitf-depleted cells.
Mitf is required to suppress expression of p27Kip1
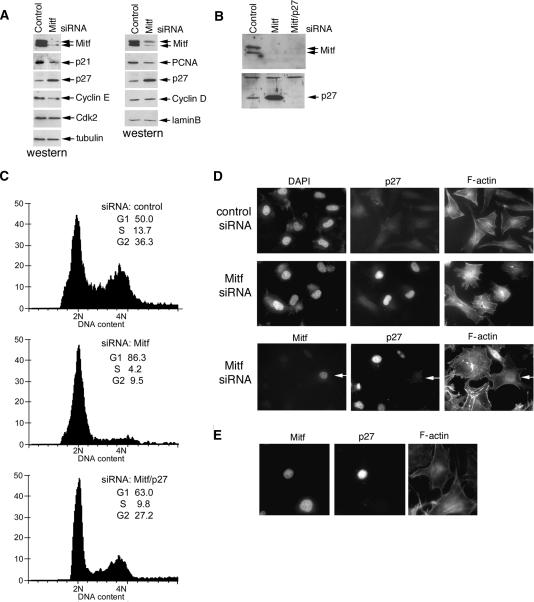
Although Mitf is clearly required for melanoma proliferation, why it is necessary has not been previously established. To understand how Mitf depletion led to a block in G1/S transition, we examined a number of known markers of proliferation and the cell cycle. Western blotting of cells transfected with control or Mitf-specific siRNA (Fig. 2A) revealed that depletion of Mitf led, as expected from our previous work (Carreira et al. 2005), to decreased expression of p21Cip1, but intriguingly also induced expression of the p27Kip1 cyclin-dependent kinase inhibitor. Note that similar results were obtained using a second Mitf siRNA directed against a different region of Mitf (Supplementary Fig. S1). We also observed reduced expression of cyclin E, and PCNA, most likely as an indirect effect of the cell cycle arrest, but no change in cyclin D1 or Cdk2 levels. Tubulin and lamin B were used as loading controls.
Figure 2.
Mitf depletion induces a p27Kip1-dependent cell cycle arrest. (A) Western blot of extracts from control or Mitf-depleted 501 mel cells using antibodies specific for the indicated proteins. Note that Mitf appears as two bands corresponding to hyper- and hypophosphorylated forms. (B) Western blot of 501 mel cells treated with control siRNA or siRNA1 specific for Mitf and/or p27Kip1-specific siRNA and probed with the indicated antibodies. (C) Flow cytometry of cells corresponding to those assayed by Western blotting in B. The p27Kip1-specific siRNA alone had no effect on the cell cycle profile (not shown). (D, top panels) Immunofluorescence using anti-p27Kip1 or Phalloidin to stain F-actin on 501 mel melanoma cells transfected with control or Mitf-specific siRNAs as indicated. (Bottom panels) Triple immunofluorescence assay showing p27Kip1 and F-actin expression in Mitf-depleted cells. Arrow indicates a control cell in which Mitf is not depleted. Note that all cells exhibiting Mitf depletion expressed elevated p27Kip1 levels and altered actin. (E) Ectopic overexpression of p27Kip1 does not affect F-actin organization. 501 mel cells were transfected with a p27Kip1 expression vector and subjected to triple immunofluorescence using anti-Mitf, anti-p27Kip1, and phalloidin.
We next tested whether the cell cycle arrest induced by depletion of Mitf was a consequence of elevated p27Kip1 expression. Western blotting of melanoma cells transfected with siRNA specific for Mitf (Fig. 2B) led to decreased Mitf expression, increased p27Kip1 levels, and increased Rb1 hypophosphorylation (data not shown). By contrast, if cells were simultaneously depleted for both Mitf and p27Kip1 (Fig. 2B), the effects the cell cycle were reversed and no G1 arrest was observed (Fig. 2C). The slight increase in the G1 population compared with the control cells in the Mitf and p27Kip1 double siRNA-treated cells most likely represents an inefficient depletion of p27Kip1 in some cells. No significant alteration in the cell cycle profile was observed using p27Kip1 siRNA alone (data not shown).
Immunofluorescence assays on cells treated with control or Mitf-specific siRNA confirmed that Mitf depletion led to increased nuclear p27Kip1 expression and that cells expressing high p27Kip1 also exhibited an alteration in the F-actin organization (Fig. 2D), whereas no change in F-actin was evident in cells with low p27Kip1 levels. The effects of Mitf depletion were not restricted to the 501 mel cells, as similar results were observed on a range of other melanoma cell lines and on normal epidermal melanocytes where Mitf depletion also led to elevated p27Kip1 (Supplementary Fig. S2), although the effects on the actin cytoskeleton were not always so dramatic. However, the altered organization of the F-actin network was not a consequence of elevated p27Kip1, as 501 mel cells not depleted for Mitf and ectopically overexpressing p27Kip1 did not exhibit a noticeably altered F-actin organization (Fig. 2E).
An inverse correlation between Mitf and p27Kip1 expression in vivo
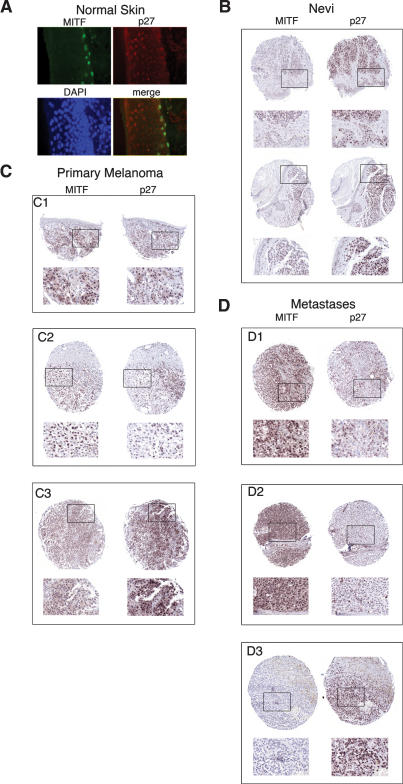
To determine whether the inverse correlation between Mitf expression and p27Kip1 was reproduced in vivo, we examined normal skin, benign nevi, and primary melanomas and metastases for expression of both p27Kip1 and Mitf. In normal skin, immunofluorescence revealed Mitf-positive melanocytes located in the basal layer of the epidermis (Fig. 3A). While differentiating keratinocytes were readily stained for p27Kip1, no significant p27Kip1 expression was observed for the Mitf-positive melanocytes. Immunohistochemistry revealed that nevi, which contain predominantly senescent melanocytes (Michaloglou et al. 2005), were Mitf-negative (Fig. 3B), consistent with the down-regulation of Mitf observed in melanocytes undergoing senescence in culture (Schwahn et al. 2005). By contrast, most nevi stained positive for p27Kip1 expression.
Figure 3.
Expression of Mitf and p27Kip1 in vivo. _(_A) Immunofluorescence assay on normal skin showing expression of Mitf (green), p27Kip1 (red), and DNA (blue). (B) Immunohistochemistry on Nevi using indicated primary antibodies. Boxed areas are shown as enlarged insets below. (C) Immunohistochemistry on primary melanomas using indicated primary antibodies. Indicated insets are shown below each section. (D) Immunohistochemistry on melanoma metastases using indicated primary antibodies. Indicated insets are shown below each section.
In primary melanomas (Fig. 3C), a highly variable staining for both Mitf and p27Kip1 was observed, consistent with the variation in p27Kip1 expression reported previously (Florenes et al. 1998; Alonso et al. 2004). Some melanomas (Fig. 3C, panel C1), exhibited a variable levels of both Mitf and p27Kip1 within the same region of the tumor. By contrast, other melanomas exhibited regions that were positive for Mitf and negative for p27Kip1 (Fig. 3C, panel C2), while other tumors were largely negative for Mitf but expressed a generally high level of p27Kip1 (Fig. 3C, panel C3). Metastases (Fig. 3D) also showed a variable expression between samples, with some metastases being highly Mitf positive but p27Kip1 negative (Fig. 3D, panels D1,D2), whereas others were Mitf negative and highly p27Kip1 positive (Fig. 3D, panel D3). In summary, the results obtained from normal melanocytes, nevi, as well as primary and metastatic melanomas (summarized in Supplementary Table 1) provide substantial evidence for an inverse correlation between Mitf and p27Kip1 expression and indicate that melanomas exhibit significantly different Mitf profiles both between and within individual tumors. However, it is important to stress that while a clear inverse correlation between Mitf and p27Kip1 expression is evident, immunohistochemistry will only give a snapshot of the dynamic events regulating both factors in vivo, and it is not possible to directly relate these data to the subsequent biological behavior of each melanoma examined.
Mitf regulates Dia1 to control actin polymerization and p27Kip1 stability
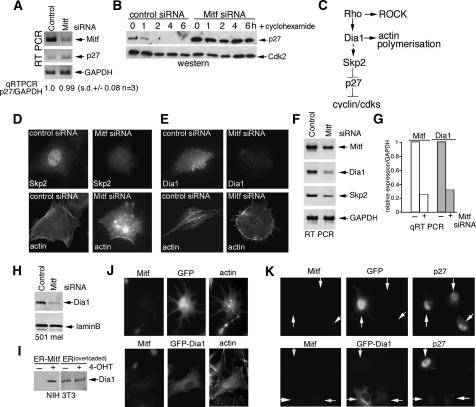
We next investigated the mechanism underlying the increase in p27Kip1 levels in the anticipation that it might yield clues to the mechanism underlying Mitf control of F-actin organization. Although p27Kip1 protein was increased substantially in Mitf-depleted cells, we observed no significant increase in p27Kip1 mRNA compared with GAPDH expression as determined by RT–PCR or quantitative real-time PCR (Fig. 4A), suggesting that p27Kip1 protein stability could be increased by down-regulation of Mitf. To verify that p27Kip1 stability was affected by depletion of Mitf, we treated control and Mitf siRNA transfected cells with cyclohexamide and assayed p27Kip1 levels by Western blotting. The results (Fig. 4B) revealed that whereas cyclohexamide induced a loss p27Kip1 expression within 2 h of treatment, p27Kip1 was stabilized in Mitf-depleted cells.
Figure 4.
Mitf regulates p27Kip1 via Dia1. (A) RT–PCR of mRNA derived from control or Mitf-depleted 501 mel cells using primers specific for the indicated genes. (B) Western blot using anti-p27Kip1 or anti-Cdk2 antibodies of protein from control or Mitf-depleted cells treated with cyclohexamide for the indicated times. (C) Schematic showing how Rho signaling meditates up-regulation of p27Kip1 protein stability via Dia1 and Skp2. (D,E) Immunofluorescence showing actin and Skp2 or Dia1 levels in control or Mitf siRNA-treated 501 mel cells. (F) RT–PCR of mRNA derived from control or Mitf-depleted cells using primers specific for the indicated genes. (G) Real-time quantitative RT–PCR of Mitf and Dia1 mRNA from control or Mitf-depleted 501 mel cells as indicated. (H) Western blot using anti-Dia1 or anti-laminB antibodies of control or Mitf-depleted 501 mel cells. (I) Western blot using anti-Dia1 antibody of NIH 3T3 cells expressing ER-Mitf or ER only ±4-OHT as indicated. (J) Immunofluorescence using anti-Mitf or phalloidin staining on 501 mel cells depleted for Mitf using siRNA1 and transfected with either a GFP or GFP-Dia1 expression vector as indicated. (K) Immunofluorescence assay using anti-Mitf or anti-p27Kip1 antibodies on 501 mel cells depleted for Mitf using siRNA1 and transfected with either a GFP or GFP-Dia1 expression vector as indicated.
The data so far indicate that Mitf depletion induces a G1 arrest by indirectly increasing p27Kip1 protein stability rather than up-regulating transcription of the p27Kip1 gene. Moreover, since ectopic overexpression of p27Kip1 did not affect F-actin organization (Fig. 2E), the deregulation of p27Kip1 expression appeared to lie downstream from the observed effects on F-actin and change in cell shape. The stability of p27Kip1 is controlled by the F-box protein Skp2 (Carrano et al. 1999; Tsvetkov et al. 1999), and indeed, in primary melanomas it has been reported that Skp2 and p27Kip1 expression are inversely correlated (Li et al. 2004). Skp2 expression in turn is regulated by the Diaphanous-related formin Dia1 (Mammoto et al. 2004), a Rho effector that plays a key role in F-actin polymerization (Fig. 4C; Alberts 2002). Our observations led us to speculate that Mitf might act on this pathway by directly regulating Dia1 expression, thereby accounting for the impact of Mitf on both the cell cycle and F-actin organization. Consistent with this, immunofluorescence revealed that Mitf depletion led to a decrease in both Skp2 and Dia1 protein levels (Fig. 4D,E), a result confirmed by RT–PCR showing that Mitf-depleted cells exhibited decreased Dia1 and Skp2 mRNA expression (Fig. 4F). We therefore focused our attention on Dia1 since it lies upstream of Skp2 and confirmed by real-time quantitative PCR that down-regulation of Dia1 mRNA correlated with Mitf depletion (Fig. 4G). The impression gained from the immunofluorescence and RT–PCR assays was substantiated by Western blotting of control or Mitf-depleted cells using anti-Dia1 antibody (Fig. 4H), and importantly, in a 3T3 cell line expressing a tamoxifen (4-OHT)-inducible Mitf fused to the ligand-binding domain to the estrogen receptor (ER) (Carreira et al. 2005), treatment with 4-OHT led to increased expression of Dia1 protein, whereas no effect on Dia1 was observed in control cells expressing ER alone (Fig. 4I; note that to see the basal level of Dia1 in the ER-expressing cells the gel was overloaded in these lanes).
To confirm that the effects of Mitf-depletion on F- actin were indeed a consequence of down-regulation of Dia1, we transfected control or Mitf-depleted cells with either a GFP or GFP-Dia1 expression vector. As expected, ectopic expression of GFP failed to affect the disruption of the F-actin cytoskeleton characteristic of Mitf depletion (Fig. 4J). By contrast, cells expressing GFP-Dia1 reversed the effect of Mitf depletion and exhibited a normal F-actin phenotype, consistent with Dia1 being a major effector of Mitf in regulating F-actin organization.
We also assessed the ability of GFP-Dia1 to restore p27Kip1 expression in cells depleted for Mitf. Thus, cells depleted for Mitf express high levels of nuclear p27Kip1 irrespective of whether GFP is coexpressed or not (Fig. 4K, top panels). By contrast, expression of GFP-Dia1 in Mitf depleted cells led to suppression of p27Kip1 expression (Fig. 4K, bottom panels). Consistent with this, FACS analysis of transfected cells gated for GFP expression revealed that in Mitf depleted cells the expression of GFP-Dia1 increases the S-phase fraction and decreases the proportion of cells in G1 (data not shown), suggesting that Dia1 can contribute to regulating the cell cycle. Together these data indicate that Mitf-mediated regulation of Dia1 expression plays a major role in determining the invasive and proliferative capacity of melanoma cells.
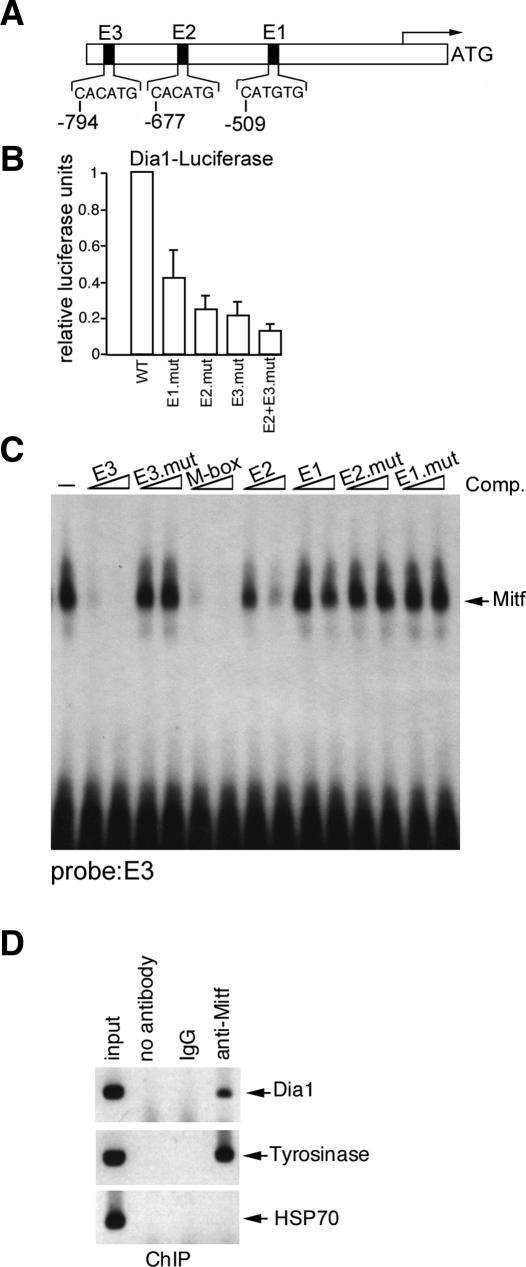
Mitf targets the DIAPH1 promoter
The results suggest that DIAPH1 is a candidate target gene for Mitf and that Dia1 is the molecule through which Mitf would mediate its effects on both the actin cytoskeleton and indirectly through Skp2, on p27Kip1 stability. The effect of Mitf on DIAPH1 mRNA and protein expression was direct. Three potential Mitf-binding sites (CATGTG) were identified in the DIAPH1 promoter that we termed E1-E3 (Fig. 5A). Mutation of these in the context of a DIAPH1 promoter-luciferase reporter revealed that the E2 and E3 elements were critical for DIAPH1 promoter activity in transfected 501 mel cells that contain high levels of endogenous Mitf (Fig. 5B). Moreover in these cells cotransfection with an Mitf expression vector led to activation of the DIAPH1 promoter up to fourfold (data not shown). The fact that higher levels of activation were not achieved in these cells may reflect the high levels of endogenous Mitf present. The ability of Mitf to bind directly the E-boxes present within the DIAPH1 promoter was assessed using band shift assay and a combination of a radiolabeled probe containing the E3 element and bacterially expressed and purified HIS-tagged Mitf. The results (Fig. 5C) indicated that binding by Mitf to the E3 element could be competed by both wild-type (WT) E2 and E3 E-box-containing oligonucleotides, but not by mutated versions, with the E3 element being as efficient as the well-characterized M-box Mitf target motif. Binding of Mitf to the DIAPH1 promoter in vivo was confirmed by chromatin immunoprecipitation (Fig. 5D) using anti-Mitf antibody and the TYROSINASE and HSP70 promoters as positive and negative controls respectively. No immunoprecipitated chromatin was detected using either no antibody or nonspecific IgG as controls.
Figure 5.
Direct regulation of the Dia1 promoter by Mitf. (A) Schematic showing the sequences of the putative Mitf-binding sites E1, E2, and E3 in the Dia1 promoter. (B) Results of luciferase assays in which 100 ng of WT or indicated Dia1 promoter mutants were transfected into 501 mel cells. Results represent the mean of three experiments and SD. (C) Band shift assay showing Mitf binding to a radiolabeled Dia1 E3 probe and competition using 10 or 50 ng or the indicated WT or mutant competitor oligonucleotides. (D) Chromatin immunoprecipitation using anti-Mitf, nonspecific IgG, or no antibody and PCR primers specific for the indicated genes.
Mitf depletion increases invasiveness and expression of MMP-2
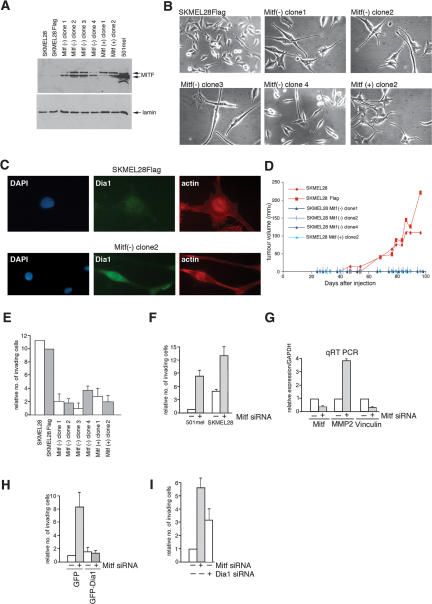
The results reveal an ability of Mitf to regulate F-actin and the cell cycle through Dia1 and p27Kip1. Since the organization of the actin cytoskeleton is a key determinant of cell motility and potential to metastasize (Vasiliev 2004) we took an invasive melanoma cell line SKMel28 and asked whether increasing Mitf would affect its behavior in vivo. To do this we stably transfected SKMel28 cells with an Mitf expression vector, isolated clones and verified by Western blotting that the derived clones expressed increased levels of Mitf (Fig. 6A). The Mitf expressing clones had a more elongated morphology compared with the control SKMel28 cells (Fig. 6B), and consistent with the results of several laboratories, increased Mitf expression reduced the proliferation rate of the SKMel28 cells in culture (data not shown). Results from immunofluorescence assays obtained using the Mitf(−) clone 2 that were also representative of the other SKMel28 cell lines ectopically expressing Mitf revealed that ectopic Mitf expression led to expression of higher levels of Dia1 and a reorganization of the actin cytoskeleton (Fig. 6C; data not shown). Of the clones isolated, three ectopically expressing the Mitf(−) isoform and one the Mitf(+) isoform, which have been shown previously to have slightly different effects on cell cycle progression (Bismuth et al. 2005), were used for subcutaneous injection into nude mice together with the parental SKMel28 cells and a control cell line transfected with an empty expression vector. Strikingly, while the parental SKMel28 cells and those transfected with an empty vector readily formed tumors, no tumors were observed with any of the four cell lines that expressed increased levels of Mitf (Fig. 6D), a result reminiscent of those from Selzer et al. (2002) in which ectopic expression of Mitf led to decreased tumor growth in a xenograft model for melanoma tumorigenesis.
Figure 6.
Mitf depletion induces a hyperinvasive phenotype. (A) SKMEL28 cells were stably transfected with Mitf expression vectors for the Mitf(+) and Mitf(−) isoforms, clones were isolated, and expression of Mitf was determined by Western blotting. (B) Phase images showing that ectopic expression of Mitf leads to a more elongated morphology. (C) Immunofluorescence assay using indicated antibodies of control SKMel28-Flag cells or the Mitf(−)-expressing clone 2. Increased Dia1 expression was also observed using other Mitf-expressing SKMel28 cell lines (not shown). (D) Tumor formation in nude mice of SKMel28 cells or indicated derivatives ectopically expressing Mitf or containing an empty vector. (E) SKMel28 cells or indicated derivatives were assessed using a Matrigel assay for their invasive potential. Relative number of invasive cells in this assay means relative to the number of cells that invade using SKMel28-Flag control set to a value of 10. (F) Invasiveness of parental and Mitf-depleted 501 mel and SKmel28 cell lines determined using the Matrigel assay. Depletion was achieved using siRNA1, although similar results were obtained using a second independent Mitf siRNA2 (not shown). Quantification shows the mean fold difference from five independent assays. Relative number of invasive cells in this assay and all remaining assays presented means relative to the number of cells that invade using control siRNA in 501 mel cells set to a value of 1. (G) Quantitative RT–PCR of control or Mitf-depleted (siRNA1) 501 mel cells assayed using the indicated primers. (H) GFP or GFP-Dia1 expression vectors were used to transfect 501 mel cells treated with control of Mitf-specific siRNA and fluorescent cells were assayed for their ability to invade using a Matrigel assay. The results shown are normalized for the number of transfected cells. The number of cells invading is relative to that of the control siRNA transfected with GFP. (I) 501 mel cells were treated with control, Mitf, or Dia1-specific siRNA1 as indicated and assayed for invasiveness using the Matrigel assay. Relative number of invading cells refers to the number of cells invading using the control siRNA.
Given this result and the consequent difficulty of assessing metastasis in vivo, we used the Matrigel assay to determine the potential of Mitf to regulate invasiveness. Consistent with the results obtained in vivo, increasing Mitf expression in all SKMel28 derivatives strongly inhibited the ability of the SKMel28 cells to invade using this assay (Fig. 6E). By contrast, siRNA-mediated depletion of Mitf led to an eightfold increase in invasiveness compared with control siRNA treated cells in the normally noninvasive 501 mel cell line (Fig. 6F), and a two- to threefold increase in the already invasive SKMel28 cells. Thus increasing or decreasing Mitf levels can reduce or enhance invasiveness respectively.
In cancers, including melanoma, invasiveness is frequently characterized by increased metalloprotease expression (Hendrix et al. 2003; Hofmann et al. 2005) and alterations in cell surface molecules that mediate cell-cell adhesion (Haass et al. 2005). Consistent with Mitf depletion inducing an invasive phenotype, cells treated with Mitf-specific siRNA exhibited a substantial increase in matrix metalloprotease MMP-2 expression and a decrease in vinculin (Fig. 6G). Although we have not examined the mechanism underlying the changes in MMP2 expression, these data provide further support for the notion that Mitf regulates the invasive potential of melanoma cells.
To confirm that loss of Dia1 expression was a key event in the increased invasiveness of Mitf-depleted melanoma cells, we depleted Mitf using siRNA and transfected the cells with expression vectors for either GFP or GFP-Dia1 and determined the proportion of GFP or GFP-Dia1 positive cells passing through the membrane in the Matrigel assay. Consistent with Mitf-mediated regulation of Dia1 playing a key role in controlling invasive potential, expression of GFP-Dia1, but not GFP alone, abrogated the increased invasiveness associated with Mitf depletion (Fig. 6H). In the converse experiment, siRNA-mediated depletion of Dia1 also resulted in increased invasive potential to ∼60% of that observed on Mitf depletion (Fig. 6I).
Mitf depletion leads to ROCK-dependent invasiveness
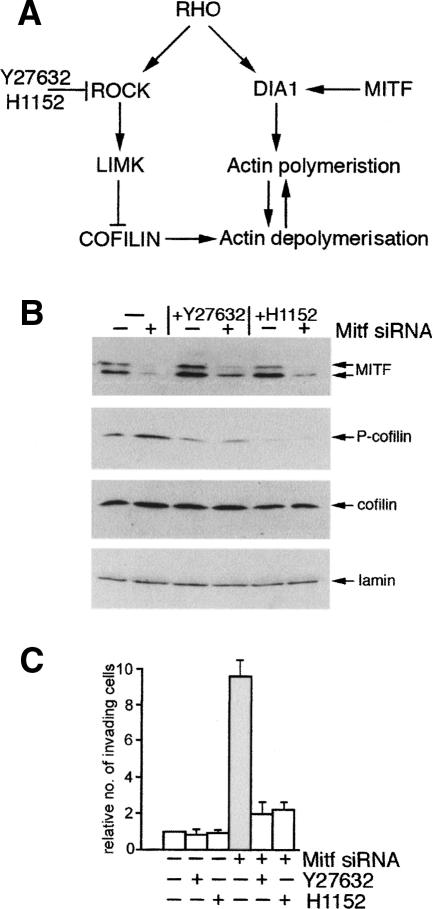
The actin cytoskeleton, and consequently cell migration, motility and invasiveness, are controlled by the balance between the activities of the Rho family GTPases, Rho, CDC42 and Rac (Jaffe and Hall 2005). Since Rac, which promotes actin polymerization at the cell periphery, can be stimulated by Dia1 (Tsuji et al. 2002), the regulation of Dia1 by Mitf will affect the balance between Rac and Rho signaling. It will also affect the relative activities of the two Rho effector pathways that operate through Dia1 on the one hand, and the ROCK kinases on the other, which can act antagonistically on the formation of adherens junctions (Sahai and Marshall 2002). Since Mitf specifically affects Dia1 expression, it can potentially lead to an imbalance between the Dia1 and ROCK signaling pathways downstream from Rho (Fig. 7A).
Figure 7.
Reduced Mitf expression leads to a ROCK-dependent invasive phenotype. (A) Pathways downstream from Rho. (B) Western blot using indicated antibodies of control and Mitf-depleted 501 mel cells grown in the presence or absence of the ROCK inhibitors Y27632 and H1152 at 10 μM and 115 nM, respectively. (C) Invasiveness of 501 mel cells treated with indicated inhibitors, and control or Mitf-specific siRNA as indicated.
To determine whether depletion of Mitf, and therefore Dia1, would affect ROCK signaling, we examined the phosphorylation status of cofilin, an actin depolymerising factor. Cofilin is inhibited by phosphorylation by LIM-Kinase (Arber et al. 1998; Yang et al. 1998), which is in turn activated by ROCK. As such, activation of the ROCK pathway leads to phosphorylation of cofilin and stabilization of actin filaments. In cells transfected with the control siRNA there is a constitutive level of phosphorylated cofilin, but consistent with ROCK signaling being elevated in Mitf-depleted cells, the amount of phosphorylated, and therefore inactivated, cofilin was reproducibly increased between two- and fourfold on Mitf depletion, with the levels of phospho-cofilin being decreased by treatment with either of two ROCK inhibitors, Y27632 or H1152 (Fig. 7B). This result is entirely consistent with the phenotype of the Mitf depleted cells (Fig. 1C) that show high levels of polymerized actin that is not coordinated by Dia1 with the cell periphery.
In some melanoma cells Rho–ROCK signaling is required for a form of invasion of three-dimensional matrices that is characterised by a membrane-blebbing morphology (Sahai and Marshall 2003; Wilkinson et al. 2005), more akin to an amoeboid form of cell invasion than the mesenchymal elongated form of migration (Friedl 2004). Since the rounded cell morphology apparent on Mitf depletion is reminiscent of the ROCK-dependent invasion observed previously, we asked whether treatment of Mitf-depleted cells with the ROCK inhibitors Y27632 or H1152 affected the increased ability of Mitf-depleted cells to invade. The results obtained (Fig. 7C) revealed that in the presence of either ROCK inhibitor, Mitf depletion failed to increase significantly the numbers of invasive cells in the Matrigel assay. However, as expected, inhibition of ROCK did not rescue the actin phenotype associated with decreased Mitf expression (data not shown), consistent with the fact that while the inhibitors might block the ROCK-dependent actin-myosin contractility necessary for invasion, they cannot restore the Dia1-mediated coordination of the actin and microtubule networks at the cell periphery.
Taken together these results reinforce the notion that Mitf activity can suppress the invasive potential of melanoma cells, most likely through altering Dia1 and ROCK activity downstream from Rho and by repressing metalloproteinase expression.
Discussion
In this study we have identified a mechanism by which Mitf is required for melanoma proliferation: Regulation of the diaphanous-related formin Dia1 enables Mitf to coordinate control of actin polymerization with cell cycle progression via regulation of p27Kip1 stability. The ability of Mitf to regulate cell shape and invasiveness clearly has important implications both for our understanding of melanoma proliferation and metastasis as well as melanoblast migration from the neural crest.
The malignant transformation of a cell can be accomplished by the acquisition of pro-proliferative signals coupled to the suppression of senescence. In melanomas the pro-proliferative signal is usually achieved by gain of function mutations in BRAF (Davies et al. 2002) or NRAS (Kraemer 2003) while senescence, found in nevi (Michaloglou et al. 2005), is bypassed by inactivation of the Rb1 pathway, frequently by inactivating the INK4a locus (Chin et al. 1998b). However, what is less clear is how cells in the primary lesion localized to the skin might go on to progress rapidly to a highly invasive vertical growth phase melanoma with the potential to metastasize. Although it is evident that the acquisition of genomic lesions may contribute in some cases to melanoma progression, NEDD9 being clear example (Kim et al. 2006), the high metastatic potential of melanoma might favor the model of cancer progression highlighted by Bernards and Weinberg (2002) in which the same mutations that lead to the initial acquisition of enhanced proliferative and anti-senescence properties would also tend to be prometastatic. Our results would indicate that in melanoma cells one of the determinants of invasive potential is the activity of Mitf. Low Mitf activity leads to a p27Kip1-dependent cell cycle arrest, reorganization of the actin cytoskeleton compatible with increased invasiveness arising through reduced Dia1 expression, increased expression of matrix metalloproteinases and reduced vinculin levels. Thus elevated p27Kip1 expression may mark melanoma cells with metastatic potential, a notion consistent with the observation that invasive primary melanomas are characterized by high p27Kip1 levels (Bales et al. 1999).
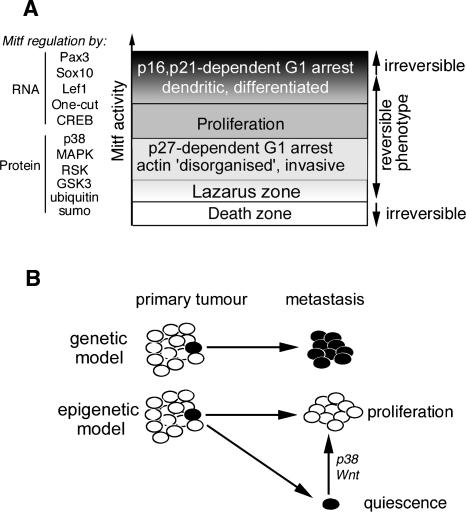
Since elevated Mitf activity is associated with a p16INK4a- (Loercher et al. 2005) and p21Cip1-dependent (Carreira et al. 2005) G1 arrest and increased expression of differentiation markers, rather than acting as a simple on-off switch, Mitf appears to act as a rheostat (Fig. 8A), integrating a wide variety of signals regulating its expression and activity to yield a variable output that translates into differential biological phenotypes. The biological outputs regulated by Mitf range from apoptosis in the absence of Mitf in normal melanocytes/melanoblasts, p27Kip1-mediated G1 arrest with invasive properties at low levels, proliferation at intermediate levels, and, at the highest levels of Mitf activity, differentiation. Within this range, terminal differentiation at one end, or cell death at the other would be irreversible events, but everything between would be reversible and dynamic. Thus for melanomas to proliferate, Mitf activity must be maintained within a window compatible with proliferation, high enough to suppress p27Kip1 expression through regulation of Dia1, but not so high that differentiation might occur. We note that as p16INK4a is likely to be inactivated by mutation or promoter methylation, and p21Cip1 suppressed by Tbx2 (Prince et al. 2004; Vance et al. 2005) in melanomas, terminal differentiation may not be an option, and apoptosis prevented by loss of APAF1 for example (Soengas et al. 2001). Moreover, our view is that although the rheostat model provides a framework for understanding the role of Mitf in melanoma progression and melanocyte development and reconciles the pro- and anti-proliferative roles of Mitf, understanding precisely what we mean by “Mitf activity” will represent a major challenge. Implicit in the model is the idea that post-translational modifications will modulate Mitf DNA-binding specificity to determine whether it will regulate a class of genes implicated in differentiation versus a second class required for proliferation, as well as its capacity to activate or possibly repress gene expression. Only once these molecular switches are identified and superimposed on the rheostat model presented will we obtain a more three-dimensional view of the role of Mitf in development and disease.
Figure 8.
An epigenetic model for melanoma metastasis. (A) Model describing Mitf function in cell cycle control in melanoma cells. Mitf activity is regulated by a wide range of factors that act at the RNA and protein levels. Low levels of Mitf are required for survival as invasive G1 arrested cells with low Dia1, high p27KIP1, and disorganized actin; moderate Mitf activity promotes actin polymerization and suppresses p27KIP1expression leading to proliferation; and elevated Mitf leads to differentiation-associated G1-arrested cells with high levels of p16INK4A and p21CIP1. Activation of NRAS or BRAF would strongly down-regulate Mitf expression. (B) Epigenetic versus genetic models for metastasis. The traditional view is that accumulation of genetic lesions leads to a cell acquiring metastatic potential that will be retained in progeny within a metastasis. In contrast, the epigenetic model states that variations in the tumor microenvironment may lead to the acquisition of metastatic potential, (e.g., by down-regulating Mitf), but that once in a different environment at the site of metastasis, a reversal of the epigenetic change (e.g., up-regulation of Mitf) may lead to renewed proliferation. Alternatively, if the metastatic cell ends up in a different environment, long-term quiescence may ensue. A change in the status of Mitf—for example, by activation of the Wnt pathway or the p38 stress signaling pathway—might then lead to a resumption of proliferation and recurrence of disease many years after initial relapse.
Consistent with the notion discussed by Bernards and Weinberg (2002) that prometastatic genes could in fact be those oncogenes that are activated early in the transformation process, gain of function mutations in BRAF that are acquired early in melanomagenesis (Pollock et al. 2003) strongly down-regulate Mitf expression, at least in melanocytes (Wellbrock and Marais 2005) and will therefore give a predisposition to metastasis, though they may not by themselves be sufficient. Superimposed on the prometastatic background conferred by activated BRAF-mediated regulation of Mitf expression and stability would be the effects of the many additional signals and transcription factors that up- or down-regulate Mitf expression and activity. These would include Wnt/β-catenin, p38, GSK3 (Steingrimsson et al. 2004), sumoylation (Miller et al. 2005; Murakami and Arnheiter 2005), various receptor tyrosine kinases (Chin et al. 1998a; Easty and Bennett 2000), competition with USF (Corre and Galibert 2005) and many others. Each of these regulatory events would be affected either by genetic mutations, such as activating mutations in β-catenin (Rubinfeld et al. 1997) or amplification of the MITF gene (Garraway et al. 2005), or by autocrine, paracrine or other external signals arising from the tumor microenvironment through interactions with other melanoma cells, keratinocytes, fibroblasts, etc. (Halaban et al. 1998; Haass et al. 2005). Our view is that signaling by many of the multiple signaling pathways that modulate Mitf activity will be significantly affected by the tumor microenvironment, and thereby, in the context of activating mutations in BRAF or NRAS, will yield melanoma metastases.
One conclusion from this dynamic epigenetic model (Fig. 8B) would be that metastatic melanoma cells may frequently be genetically identical to their nonmetastatic parents. Moreover, it is extremely important to distinguish between metastatic cells, which may represent a very small minority within the primary tumor, and a metastasis derived from it. The metastasis will only be detected once the pioneer metastatic cell has arrived in a location where its environment is compatible with proliferation, and consequently metastatic cells and detectable metastases are likely to have different gene expression profiles. Since proliferation and metastatic potential may both be determined by the effects of the microenvironment superimposed on BRAF mutations, making sense of gene expression profiles in an attempt to identify a metastasis signature may not be simple (Hoek et al. 2006).
Although it is widely believed that within a primary tumor, those cells with high metastatic capacity would also be highly proliferating, our results raise the possibility that the opposite may be true; that the more invasive fraction will be dividing slowly, expressing low levels of Mitf and high levels of p27Kip1. This conclusion is consistent with observations that in vitro proliferation rates of uveal melanoma cells are not necessarily representative of their metastatic potential (Marshall et al. 2004), as well as recent results based on gene array studies that indicated that melanomas will contain highly proliferating cells with low metastatic potential as well slow dividing and highly metastatic cells (Hoek et al. 2006). Since current therapies are designed to target dividing cells, the cohort of melanoma cells with low levels of Mitf may be resistant to treatment. As such, rather than inhibiting Mitf expression as proposed previously (Garraway et al. 2005), a more effective approach may in fact be to elevate Mitf expression and induce proliferation in the fraction of cells with low Mitf and high metastatic potential to render them sensitive to chemotherapy.
Finally, although we have restricted our analysis in this study to melanoma cells, the observation that Mitf may regulate actin dynamics is also likely to be significant for melanocyte function. In melanocytes, Mitf regulates the expression of genes required for melanosome biogenesis (Steingrimsson et al. 2004). Once melanosomes have matured into pigment producing organelles they are transported to the tips of melanocyte dendrites and transferred to surrounding keratinocytes. For this process to work efficiently, melanosome biogenesis, actin and microtubule-dependent transport, and dendrite formation should be coordinated (Scott 2002). The ability of Mitf to play a key role in each step of the process would provide a simple mechanism to coordinate the response of melanocytes to physiological stimuli provided by keratinocytes in response to UV irradiation that necessitate increased melanosome production (Hirobe 2005).
In summary, Mitf appears to lie right at the heart of the melanocyte, coordinating survival, cell cycle entry and exit, cytoskeletal organization, melanosome assembly and transport, differentiation and migration/metastasis. As such, understanding Mitf regulation and function may well be the key to achieving one of the major aims of cancer research, an effective melanoma therapy.
Materials and methods
Generation and growth of cell lines and tumor formation assay
All cell lines were grown as described previously (Carreira et al. 2005). Human epidermal foreskin melanocytes derived from healthy children were supplied by Gentaur. siRNA-mediated down-regulation of Mitf was achieved as described (Prince et al. 2004) using the following Mitf and Dia1-specific sequences: Mitf siRNA1, 5′-r(AGCAGUACCUUUCUACCAC)d(TT)-3′; Mitf siRNA2, 5′-r(CUCCUGUCCAGCCAACCUU)d(TT)-3′; and Dia1 siRNA1, 5′-r(AAGGAGAGCUCUAAGUCUGCC)d (TT)-3′. The control siRNA sequence used was 5′-r(UUCUCC GAACGUGUCACGU)d(TT)-3′.
RT–PCR was performed as described (Carreira et al. 2005) using primers specific for Mitf (5′-ATGCTGCAAATGCTAG AATAA-3′ and 5′-CAATCAGGTTGTGATTGTCC-3′), p27Kip1 (5′-CGTGCGAGTGTCTAACGGGAGC-3′ and 5′-TGCGTGT CCTCAGAGTTAGCC-3′), Dia1 (5′-GAGCCCAGATGAGCT GCCCTCGG-3′ and 5′-ATGTCCAATAAGGAGGCCAAGC C-3′), Skp2 (5′-CTCCAGGAGATTCCAGACCTGAG-3′ and 5′-CTCCAGGAGATTCCAGACCTGAG-3′), or G3PDH (5′-CCAACTGCTTAGCCCCCCTGGCCAAG-3′ and 5′-CTCCT TGGAGGCCATGTAGGCCATG-3′).
Following siRNA treatment for 3 d, cells were assayed by Western blotting, immunofluorescence, or RT–PCR. Luciferase assays were performed as described (Carreira et al. 2000) and assayed 2 d post-transfection.
For the SKMel28 cell lines overexpressing Mitf, puromycin-selectable vectors expressing either Flag epitope-tagged Mitf(+) or Mitf(−) isoforms were transfected into SKMel28 cells and individual clones were isolated following puromycin selection. Positive clones were detected by Western blotting and used either for the Matrigel invasion assay or for the tumor formation assay, in which 100,000 parental, control, or Mitf-expressing cells were injected subcutaneously into 6- to 8-wk-old female athymic nude mice (nu/nu Balb/c, Charles River strain) and tumor growth was monitored. Tumor size was assessed by caliper measurements every 2–3 d, and volume was calculated by the formula length × width × width/2. Mice were weighed every week. For ethical reasons, the animals were sacrificed when their tumors reached 400 mm3. All experiments were approved by the Local Committee on Ethics of Animal Experimentation, Orsay, France.
Tissue samples, tissue microarray (TMA), and immunohistochemistry
Formalin-fixed and paraffin-embedded tissue specimens for TMA construction were obtained from Istituto Europeo di Oncologia. TMAs were prepared as described previously (Kononen et al. 1998), Briefly, two representative tumor areas (diameter, 0.6 mm) from each sample, identified previously on H&E-stained sections, were removed from the donor blocks and deposited on the recipient block using a custom-built precision instrument (Tissue Arrayer, Beecher Instruments). Serial sections (3 μm) of the resulting recipient block were cut, mounted on glass slides, and processed for immunohistochemistry with anti-Mitf (1/250) and anti-p27 (1/300) (Dako) monoclonal antibodies.
Matrigel invasion assay
Quantitative invasion assays were performed using a cell invasion assay kit (Chemicon) according to the manufacturer’s instructions. 501 melanoma cells were transfected with control or Mitf-specific siRNA1. Forty-eight hours post-transfection, the cells were trypsinized and seeded at 1.5 × 105 in 300 μL of RPMI without serum onto the inserts. FBS was added to the lower chamber to act as a chemoattractant. After 16-h incubation, the noninvading cells and ECMs were removed gently by cotton swab, and the invading cells, present on the lower side of the chamber, were stained, air dried, and photographed. The invading cells were counted under the microscope. The ROCK inhibitors Y27632 and H1152 were used at 10 μM and 115 nM, respectively.
Flow cytometry
Cells were harvested and fixed as a single cell suspension in −20°C methanol. Cells were recovered by centrifugation and then stained with propidium iodide and subjected to analysis using a Beckman Coulter EpicsXL flow cytometer.
Western blot analysis
Western blotting on whole-cell extracts was performed as described (Prince et al. 2004). The primary antibodies used were anti-Mitf mouse monoclonal (C5, Neomarkers), rabbit polyclonal anti-p21Cip1 (Santa Cruz Biotechnology), goat polyclonal anti-p27Kip1 (Santa Cruz Biotechnology), rabbit polyclonal anti-cyclinE (Santa Cruz Biotechnology), goat polyclonal anti-CDK2 (Santa Cruz Biotechnology), mouse monoclonal anti-PCNA (Santa Cruz Biotechnology), mouse monoclonal anti-cyclin D1 (Santa Cruz Biotechnology), and goat polyclonal anti-Dia1 (Santa Cruz Biotechnology).
Immunofluorescence microscopy
Immunofluorescence was performed as described (Carreira et al. 2005), using 1:100 dilution of monoclonal anti-Mitf (NeoMarkers), anti-p27Kip1 (Santa Cruz Biotechnology), anti-Dia1 (Santa Cruz Biotechnology), and anti-SKP2 (Zymed) antibodies and with 1:100 dilution of appropriate secondary antibodies (Vector Laboratories). F-Actin was visualized by phalloidin TRITC labeled (Sigma).
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed as described previously for Mitf (Carreira et al. 2005) using 10 μg anti-Mitf mouse monoclonal antibody (C5, Neomarkers) or 10 μg nonspecific IgG (Bio-Rad). The DNA recovered was subjected to PCR amplification before analysis by agarose gel electrophoresis. The primers used for the PCRs were the human Dia1 promoter region (5′-CTTGGGCAAGTCACTTCAACTCTC-3′ and 5′-GACAGGTTTTCCAAGAGAATGCAG-3′), the HSP70 promoter (5′-CCTCCAGTGAATCCCAGAAGACTCT-3′ and 5′-TGGGACAACGGGAGTCACTCTC-3′), and the Tyrosinase promoter (5′-GCTCTATTCCTGACACTACCTCTC-3′ and 5′-CAAGGTCTGCAGGAACTGGCTAATTG-3′).
DNA-binding assays
Electrophoretic mobility shift assays were performed using purified HIS-tagged Mitf and 32P-labeled oligonucleotide Dia1E3 probe as described previously (Carreira et al. 2005). The sequences for probe and competitor DNA were Dia1 E3(−805/−780) (5′-GCAGAATCTTATCACATGGGACTGTGG-3′), Dia1 E2(−698/−673) (5′-CTGTTTAGGCACATGTTGGCA ATAAT-3′), Dia1 E1(−520/−493) (5′-TATAATTTTTTTCAT GTGTTCCATGGGT-3′), M-box (5′-CAGTGGGGAGGGAGT CATGTGCTGCCTAGT-3′), E3.mut (5′-GCAGAATCTATCA CcTaGGACTGTGG-3′), E2.mut (5′-CTGTTTAGGCACtTGgT GGCAATAAT-3′), and E1.mut (5′-TATAATTTTTTcCAaGT GCTCCATGGGT-3′). Lowercase indicates mutated residues.
Luciferase assays
The human Dia1 promoter extending between −1831 and +32 was cloned into pGL3 using appropriate primers. The E-box elements were mutated by site-directed mutagenesis using the Strategene QuikChange system as follows, where lowercase indicates the mutated residues: E3.mut (5′-TCTatctcgAgGGA-3′), E2.mut (5′-AGGCtCgaGTTG-3′), E1.mut (5′-TTTTCac GaGTTC-3′), and mutations confirmed by sequencing. For the luciferase assays, 501 mel cells were transfected with 100 ng WT or mutated reporter and assayed 48-h post-transfection.
Acknowledgments
We thank S. Narumiya for the GFP-Dia1 expression vector and Richard Treisman and Erk Sahai for other Dia1 and ROCK vectors. This work was supported by Marie Curie Cancer Care and the Association for International Cancer Research (S.C., J.G., and C.G.), and K.H. was supported by the Swiss National Foundation (Grant no. 310040-103671/1). L.D. and L.L. were supported by the Ligue Nationale Contre le Cancer.
Footnotes
References
- Alberts, A.S. Diaphanous-related Formin homology proteins. Curr. Biol. 2002;12:R796. doi: 10.1016/s0960-9822(02)01309-x. [DOI] [PubMed] [Google Scholar]
- Alonso, S.R., Ortiz, P., Pollan, M., Perez-Gomez, B., Sanchez, L., Acuna, M.J., Pajares, R., Martinez-Tello, F.J., Hortelano, C.M., Piris, M.A., et al. Progression in cutaneous malignant melanoma is associated with distinct expression profiles: A tissue microarray-based study. Am. J. Pathol. 2004;164:193–203. doi: 10.1016/s0002-9440(10)63110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber, S., Barbayannis, F.A., Hanser, H., Schneider, C., Stanyon, C.A., Bernard, O., Caroni, P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- Bales, E.S., Dietrich, C., Bandyopadhyay, D., Schwahn, D.J., Xu, W., Didenko, V., Leiss, P., Conrad, N., Pereira-Smith, O., Orengo, I., et al. High levels of expression of p27KIP1 and cyclin E in invasive primary malignant melanomas. J. Invest. Dermatol. 1999;113:1039–1046. doi: 10.1046/j.1523-1747.1999.00812.x. [DOI] [PubMed] [Google Scholar]
- Baylin, S.B., Ohm, J.E. Epigenetic gene silencing in cancer—A mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Bernards, R., Weinberg, R.A. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- Bismuth, K., Maric, D., Arnheiter, H. MITF and cell proliferation: The role of alternative splice forms. Pigment Cell Res. 2005;18:349–359. doi: 10.1111/j.1600-0749.2005.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano, A.C., Eytan, E., Hershko, A., Pagano, M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Carreira, S., Liu, B., Goding, C.R. The gene encoding the T-box transcription factor Tbx2 is a target for the microphthalmia-associated transcription factor in melanocytes. J. Biol. Chem. 2000;275:21920–21927. doi: 10.1074/jbc.M000035200. [DOI] [PubMed] [Google Scholar]
- Carreira, S., Goodall, J., Aksan, I., La Rocca, S.A., Galibert, M.D., Denat, L., Larue, L., Goding, C.R. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- Chin, L., Merlino, G., Depinho, R.A. Malignant melanoma: Modern black plague and genetic black box. Genes & Dev. 1998a;12:3467–3481. doi: 10.1101/gad.12.22.3467. [DOI] [PubMed] [Google Scholar]
- Chin, L., Pomerantz, J., DePinho, R.A. The INK4a/ARF tumor supressor: One gene —two products—two pathways. Trends Biochem. Sci. 1998b;23:291–296. doi: 10.1016/s0968-0004(98)01236-5. [DOI] [PubMed] [Google Scholar]
- Corre, S., Galibert, M.D. Upstream stimulating factors: Highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- Davies, H., Bignell, G.R., Cox, C., Stephens, P., Edkins, S., Clegg, S., Teague, J., Woffendin, H., Garnett, M.J., Bottomley, W., et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Du, J., Widlund, H.R., Horstmann, M.A., Ramaswamy, S., Ross, K., Huber, W.E., Nishimura, E.K., Golub, T.R., Fisher, D.E. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Easty, D.J., Bennett, D.C. Protein tyrosine kinases in malignant melanoma. Melanoma Res. 2000;10:401–411. doi: 10.1097/00008390-200010000-00001. [DOI] [PubMed] [Google Scholar]
- Florenes, V.A., Maelandsmo, G.M., Kerbel, R.S., Slingerland, J.M., Nesland, J.M., Holm, R. Protein expression of the cell-cycle inhibitor p27Kip1 in malignant melanoma: Inverse correlation with disease-free survival. Am. J. Pathol. 1998;153:305–312. doi: 10.1016/S0002-9440(10)65572-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl, P. Prespecification and plasticity: Shifting mechanisms of cell migration. Curr. Opin. Cell Biol. 2004;16:14–23. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Garraway, L.A., Widlund, H.R., Rubin, M.A., Getz, G., Berger, A.J., Ramaswamy, S., Beroukhim, R., Milner, D.A., Granter, S.R., Du, J., et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Gupta, P.B., Kuperwasser, C., Brunet, J.P., Ramaswamy, S., Kuo, W.L., Gray, J.W., Naber, S.P., Weinberg, R.A. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat. Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass, N.K., Smalley, K.S., Li, L., Herlyn, M. Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res. 2005;18:150–159. doi: 10.1111/j.1600-0749.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- Halaban, R., Miglarese, M.R., Smicun, Y., Puig, S. Melanomas, from the cell cycle point of view. Int. J. Mol. Med. 1998;1:419–425. doi: 10.3892/ijmm.1.2.419. [DOI] [PubMed] [Google Scholar]
- Hendrix, M.J., Seftor, E.A., Kirschmann, D.A., Quaranta, V., Seftor, R.E. Remodeling of the microenvironment by aggressive melanoma tumor cells. Ann. N. Y. Acad. Sci. 2003;995:151–161. doi: 10.1111/j.1749-6632.2003.tb03218.x. [DOI] [PubMed] [Google Scholar]
- Hirobe, T. Role of keratinocyte-derived factors involved in regulating the proliferation and differentiation of mammalian epidermal melanocytes. Pigment Cell Res. 2005;18:2–12. doi: 10.1111/j.1600-0749.2004.00198.x. [DOI] [PubMed] [Google Scholar]
- Hoek, K.S., Schlegel, N.C., Brafford, P., Sucker, A., Ugurel, S., Kumar, R., Weber, B.L., Nathanson, K.L., Phillips, D.J., Herlyn, M., et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Hofmann, U.B., Houben, R., Brocker, E.B., Becker, J.C. Role of matrix metalloproteinases in melanoma cell invasion. Biochimie. 2005;87:307–314. doi: 10.1016/j.biochi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Jaffe, A.B., Hall, A. RHO GTPASES: Biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Kim, M., Gans, J.D., Nogueira, C., Wang, A., Paik, J.H., Feng, B., Brennan, C., Hahn, W.C., Cordon-Cardo, C., Wagner, S.N., et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Kononen, J., Bubendorf, L., Kallioniemi, A., Barlund, M., Schraml, P., Leighton, S., Torhorst, J., Mihatsch, M.J., Sauter, G., Kallioniemi, O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- Kraemer, K.H. NRAS hypermutability in familial melanoma with CDKN2A mutations—Cause and effect? J. Natl. Cancer Inst. 2003;95:768–769. doi: 10.1093/jnci/95.11.768. [DOI] [PubMed] [Google Scholar]
- Larribere, L., Hilmi, C., Khaled, M., Gaggioli, C., Bille, K., Auberger, P., Ortonne, J.P., Ballotti, R., Bertolotto, C. The cleavage of microphthalmia-associated transcription factor, MITF, by caspases plays an essential role in melanocyte and melanoma cell apoptosis. Genes & Dev. 2005;19:1980–1985. doi: 10.1101/gad.335905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Murphy, M., Ross, J., Sheehan, C., Carlson, J.A. Skp2 and p27kip1 expression in melanocytic nevi and melanoma: An inverse relationship. J. Cutan. Pathol. 2004;31:633–642. doi: 10.1111/j.0303-6987.2004.00243.x. [DOI] [PubMed] [Google Scholar]
- Loercher, A.E., Tank, E.M., Delston, R.B., Harbour, J.W. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J. Cell Biol. 2005;168:35–40. doi: 10.1083/jcb.200410115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto, A., Huang, S., Moore, K., Oh, P., Ingber, D.E. Role of RhoA, mDia, and ROCK in cell shape-dependent control of the Skp2–p27kip1 pathway and the G1/S transition. J. Biol. Chem. 2004;279:26323–26330. doi: 10.1074/jbc.M402725200. [DOI] [PubMed] [Google Scholar]
- Marshall, J.C., Caissie, A.L., Callejo, S.A., Antecka, E., Burnier, M.N., Jr. Cell proliferation profile of five human uveal melanoma cell lines of different metastatic potential. Pathobiology. 2004;71:241–245. doi: 10.1159/000080057. [DOI] [PubMed] [Google Scholar]
- Michaloglou, C., Vredeveld, L.C., Soengas, M.S., Denoyelle, C., Kuilman, T., van der Horst, C.M., Majoor, D.M., Shay, J.W., Mooi, W.J., Peeper, D.S. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Miller, A.J., Levy, C., Davis, I.J., Razin, E., Fisher, D.E. Sumoylation of MITF and its related family members TFE3 and TFEB. J. Biol. Chem. 2005;280:146–155. doi: 10.1074/jbc.M411757200. [DOI] [PubMed] [Google Scholar]
- Murakami, H., Arnheiter, H. Sumoylation modulates transcriptional activity of MITF in a promoter-specific manner. Pigment Cell Res. 2005;18:265–277. doi: 10.1111/j.1600-0749.2005.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, E.K., Granter, S.R., Fisher, D.E. Mechanisms of hair graying: Incomplete melanocyte stem cell maintenance in the niche. Science. 2005;307:720–724. doi: 10.1126/science.1099593. [DOI] [PubMed] [Google Scholar]
- Opdecamp, K., Nakayama, A., Nguyen, M.T., Hodgkinson, C.A., Pavan, W.J., Arnheiter, H. Melanocyte development in vivo and in neural crest cell cultures: Crucial dependence on the Mitf basic-helix–loop–helix-zipper transcription factor. Development. 1997;124:2377–2386. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- Pollock, P.M., Harper, U.L., Hansen, K.S., Yudt, L.M., Stark, M., Robbins, C.M., Moses, T.Y., Hostetter, G., Wagner, U., Kakareka, J., et al. High frequency of BRAF mutations in nevi. Nat. Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- Prince, S., Carreira, S., Vance, K.W., Abrahams, A., Goding, C.R. Tbx2 directly represses the expression of the p21(WAF1) cyclin-dependent kinase inhibitor. Cancer Res. 2004;64:1669–1674. doi: 10.1158/0008-5472.can-03-3286. [DOI] [PubMed] [Google Scholar]
- Rubinfeld, B., Robbins, P., El-Gamil, M., Albert, I., Porfiri, E., Polakis, P. Stabilization of β-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- Sahai, E., Marshall, C.J. ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat. Cell Biol. 2002;4:408–415. doi: 10.1038/ncb796. [DOI] [PubMed] [Google Scholar]
- Sahai, E., Marshall, C.J. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- Schwahn, D.J., Timchenko, N.A., Shibahara, S., Medrano, E.E. Dynamic regulation of the human dopachrome tautomerase promoter by MITF, ER-α and chromatin remodelers during proliferation and senescence of human melanocytes. Pigment Cell Res. 2005;18:203–213. doi: 10.1111/j.1600-0749.2005.00229.x. [DOI] [PubMed] [Google Scholar]
- Scott, G. Rac and rho: The story behind melanocyte dendrite formation. Pigment Cell Res. 2002;15:322–330. doi: 10.1034/j.1600-0749.2002.02056.x. [DOI] [PubMed] [Google Scholar]
- Seftor, E.A., Brown, K.M., Chin, L., Kirschmann, D.A., Wheaton, W.W., Protopopov, A., Feng, B., Balagurunathan, Y., Trent, J.M., Nickoloff, B.J., et al. Epigenetic transdifferentiation of normal melanocytes by a metastatic melanoma microenvironment. Cancer Res. 2005;65:10164–10169. doi: 10.1158/0008-5472.CAN-05-2497. [DOI] [PubMed] [Google Scholar]
- Selzer, E., Wacheck, V., Lucas, T., Heere-Ress, E., Wu, M., Weilbaecher, K.N., Schlegel, W., Valent, P., Wrba, F., Pehamberger, H., et al. The melanocyte-specific isoform of the microphthalmia transcription factor affects the phenotype of human melanoma. Cancer Res. 2002;62:2098–2103. [PubMed] [Google Scholar]
- Soengas, M.S., Capodieci, P., Polsky, D., Mora, J., Esteller, M., Opitz-Araya, X., McCombie, R., Herman, J.G., Gerald, W.L., Lazebnik, Y.A., et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- Steingrimsson, E., Copeland, N.G., Jenkins, N.A. Melanocytes and the Microphthalmia Transcription Factor Network. Annu. Rev. Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Tsuji, T., Ishizaki, T., Okamoto, M., Higashida, C., Kimura, K., Furuyashiki, T., Arakawa, Y., Birge, R.B., Nakamoto, T., Hirai, H., et al. ROCK and mDia1 antagonize in Rho-dependent Rac activation in Swiss 3T3 fibroblasts. J. Cell Biol. 2002;157:819–830. doi: 10.1083/jcb.200112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov, L.M., Yeh, K.H., Lee, S.J., Sun, H., Zhang, H. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- Vance, K.W., Goding, C.R. The transcription network regulating melanocyte development and melanoma. Pigment Cell Res. 2004;17:318–325. doi: 10.1111/j.1600-0749.2004.00164.x. [DOI] [PubMed] [Google Scholar]
- Vance, K.W., Carreira, S., Brosch, G., Goding, C.R. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 2005;65:2260–2268. doi: 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- Vasiliev, J.M. Cytoskeletal mechanisms responsible for invasive migration of neoplastic cells. Int. J. Dev. Biol. 2004;48:425–439. doi: 10.1387/ijdb.041806jv. [DOI] [PubMed] [Google Scholar]
- Wellbrock, C., Marais, R. Elevated expression of MITF counteracts B-RAF-stimulated melanocyte and melanoma cell proliferation. J. Cell Biol. 2005;170:703–708. doi: 10.1083/jcb.200505059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund, H.R., Horstmann, M.A., Price, E.R., Cui, J., Lessnick, S.L., Wu, M., He, X., Fisher, D.E. β-catenin-induced melanoma growth requires the downstream target Microphthalmia-associated transcription factor. J. Cell Biol. 2002;158:1079–1087. doi: 10.1083/jcb.200202049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, S., Paterson, H.F., Marshall, C.J. Cdc42–MRCK and Rho–ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat. Cell Biol. 2005;7:255–261. doi: 10.1038/ncb1230. [DOI] [PubMed] [Google Scholar]
- Yang, N., Higuchi, O., Ohashi, K., Nagata, K., Wada, A., Kangawa, K., Nishida, E., Mizuno, K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]