Acm1 Is a Negative Regulator of the Cdh1-Dependent Anaphase-Promoting Complex/Cyclosome in Budding Yeast (original) (raw)
Abstract
Cdh1 is a coactivator of the anaphase-promoting complex/cyclosome (APC/C) and contributes to mitotic exit and G1 maintenance by facilitating the polyubiquitination and subsequent proteolysis of specific substrates. Here, we report that budding yeast Cdh1 is a component of a cell cycle-regulated complex that includes the 14-3-3 homologs Bmh1 and Bmh2 and a previously uncharacterized protein, which we name Acm1 (_A_PC/C_C_dh1 _m_odulator 1). Association of Cdh1 with Bmh1 and Bmh2 requires Acm1, and the Acm1 protein is cell cycle regulated, appearing late in G1 and disappearing in late M. In _acm1_Δ strains, Cdh1 localization to the bud neck and association with two substrates, Clb2 and Hsl1, were strongly enhanced. Several lines of evidence suggest that Acm1 can suppress APC/CCdh1-mediated proteolysis of mitotic cyclins. First, overexpression of Acm1 fully restored viability to cells expressing toxic levels of Cdh1 or a constitutively active Cdh1 mutant lacking inhibitory phosphorylation sites. Second, overexpression of Acm1 was toxic in _sic1_Δ cells. Third, ACM1 deletion exacerbated a low-penetrance elongated-bud phenotype caused by modest overexpression of Cdh1. This bud elongation was independent of the morphogenesis checkpoint, and the combination of _acm1_Δ and _hsl1_Δ resulted in a dramatic enhancement of bud elongation and G2/M delay. Effects on bud elongation were attenuated when Cdh1 was replaced with a mutant lacking the C-terminal IR dipeptide, suggesting that APC/C-dependent proteolysis is required for this phenotype. We propose that Acm1 and Bmh1/Bmh2 constitute a specialized inhibitor of APC/CCdh1.
Ubiquitin-mediated proteolysis of key proteins plays a critical role in regulating cell cycle progression in eukaryotes. This proteolysis is directed primarily by the activity of two large E3 ubiquitin protein ligase complexes, the Skp1/cullin/F-box protein (SCF) complex and the anaphase-promoting complex/cyclosome (APC/C) (57). Important roles for APC/C include triggering the onset of anaphase and facilitating mitotic exit by targeting the anaphase inhibitor securin and mitotic cyclins, respectively, for proteolysis (13, 49, 59). APC/C also helps establish and maintain the G1 state until conditions are appropriate to begin another round of cell division (23, 24).
APC/C is present constitutively during the cell cycle; however, its activity is thought to be limited to the interval from early M through late G1 and is directed to substrate proteins in a highly selective manner. The temporal regulation and substrate specificity of APC/C are attributed to a conserved family of WD40 repeat-containing proteins that includes Cdc20 (also called fizzy) and Cdh1 (fizzy related) (29, 46, 56, 60). These proteins are required to activate APC/C at specific times during the cell cycle. Although the mechanism of activation is a subject of debate, the most widely accepted models propose that Cdc20 family members contribute to substrate recruitment by directly binding to specific substrates, either independently of APC/C or in complex with APC/C (10, 16, 28, 36).
Phosphorylation of Cdh1 is critical for regulating its ability to activate APC/C and thereby restricting activity to a specific period of the cell cycle. APC/CCdh1 is inactivated at the G1/S boundary when cyclin-dependent kinase (CDK) activity phosphorylates Cdh1, preventing it from associating with APC/C (26, 30, 60). In budding yeast, Cdh1 phosphorylation also results in its export from the nucleus to the cytoplasm (25). In late anaphase, Cdc14 phosphatase is activated and dephosphorylates Cdh1, allowing it to reactivate APC/C and promote mitotic exit (26, 55). APC/CCdh1 also triggers the inactivation of APC/CCdc20 by targeting Cdc20 for degradation in late M (50).
An interesting question regarding regulation of Cdh1 is why its level remains relatively high from S through M phase when APC/CCdh1 activity is absent (30, 31, 40). In budding yeast, the Cdh1 protein level appears low when cells are arrested in G1 with the α-factor mating pheromone but is elevated in S phase and M phase (23, 25, 40, 60). In vertebrate systems, Cdh1 is reported to target itself for proteolysis during G1, effectively reducing its level and contributing to its own inactivation (31). Its level then increases during the subsequent S phase and peaks during mitosis. There are no defined functions for Cdh1 during this cell cycle interval when APC/CCdh1 activity is turned off, although one report has implicated Cdh1 in a G2 DNA damage checkpoint response in vertebrate cells (52). Since several reports demonstrate that Cdh1 can interact with its substrates independently of APC/C (9, 10, 28, 38, 47), the issue of its fate following G1 exit is important because it could potentially influence or interfere with the normal function of its substrates during this time. In higher eukaryotes, two proteins that inhibit Cdh1 through physical association, Emi1 (Rca1 in Drosophila) (20, 41) and Mad2B (12, 39), have been described. The biological function of Mad2B is unclear, but evidence suggests that Emi1 and Rca1 are required for proper inactivation of APC/CCdh1 at the end of G1 and in G2, respectively. Emi1 has been proposed to act by inhibiting substrate binding to Cdh1 (41). To date, similar Cdh1-bound inhibitors of APC/C activity have not been found in yeast.
To further understand the fate of budding yeast Cdh1 following its nuclear export and inactivation, we sought to identify proteins that stably associate with it. In this paper, we report the identification of a stable protein complex containing Cdh1, the 14-3-3 homologs Bmh1 and Bmh2, and a previously uncharacterized protein. This complex is strictly cell cycle regulated, appearing in late G1 and disappearing in late M, corresponding to the cell cycle interval in which APC/CCdh1 is inactive. Our results are consistent with this complex acting as a specialized inhibitor of APC/CCdh1 activity and further imply that Cdh1 function might be required under specific conditions during S and/or M phase.
MATERIALS AND METHODS
Reagents.
EZview anti-FLAG M2 and anti-hemagglutinin (HA)-7 affinity resins and anti-FLAG M2 antibody were from Sigma. Anti-HA 12CA5 antibody was from Roche Applied Science. Rabbit anti-Clb2, goat anti-Cdc20, and goat anti-Cdc28 antibodies were from Santa Cruz Biotechnology. Ase1 polyclonal antibody was a generous gift from David Pellman (Harvard University). Fluorescent anti-mouse, anti-rabbit, and anti-goat secondary antibodies for imaging with the LI-COR Odyssey system were from Molecular Probes or Rockland Immunochemicals.
Standard yeast medium was used for cell growth. YPD contained 20 g/liter dextrose, 20 g/liter peptone, and 10 g/liter yeast extract. Selective medium contained 20 g/liter dextrose, 6.7 g/liter yeast nitrogen base, and the appropriate amino acid dropout mixture. For experiments requiring galactose induction, liquid selective medium contained 2% raffinose and agar plates contained 2% galactose in place of dextrose as the carbon source.
Chemicals for cell cycle arrest were α-factor peptide (GenScript Corp.), hydroxyurea (HU; Sigma), and nocodazole (EMD Biosciences). Sytox green dye for flow cytometry was from Molecular Probes. Sequencing grade porcine trypsin was from Promega. The ProteoSilver staining kit used was from Sigma.
Strain and plasmid constructions.
All strains (Table 1) were from the W303 or the BY4741 background. Many experiments were performed in both backgrounds and always yielded similar results. Complete-gene deletions were generated by standard PCR-based approaches (7). Chromosomal alleles encoding amino-terminal 3HA or 3FLAG epitope fusion proteins were generated as previously described (44). Gene deletions were confirmed by PCR. Introduction of epitope tags was confirmed by PCR, DNA sequencing, and Western blotting.
TABLE 1.
Yeast strains used in this study
| Name | Relevant genotype | Source or reference |
|---|---|---|
| W303 background | ||
| YKA150 | MATabar1::URA3 | 21 |
| YKA247 | MATabar1::URA3 acm1::KanMX4 | This study |
| DLY3033 | MATabar1::URA3 cdc15-2 | Daniel Lew, Duke University |
| BY4741 background | ||
| YKA226 | MATa3HA-ACM1 | This study |
| YKA227 | MATabar1::URA3 3HA-ACM1 | This study |
| YKA233 | MATabar1::hisG | This study |
| YKA237 | MATabar1::hisG acm1::URA3 | This study |
| YKA245 | MATabar1::URA3 3HA-ACM1 cdh1::KanMX4 | This study |
| YKA249 | MATabar1::hisG 3HA-BMH1 | This study |
| YKA250 | MATabar1::hisG 3HA-CDH1 | This study |
| YKA252 | MATabar1::hisG 3HA-HSL1 | This study |
| YKA255 | MATabar1::hisG swe1::KanMX4 acm1::URA3 | This study |
| YKA256 | MATabar1::hisG 3HA-CDH1 acm1::KanMX4 | This study |
| YKA257 | MATabar1::hisG 3HA-HSL1 acm1::KanMX4 | This study |
| YKA258 | MATabar1::hisG hsl1::KanMX4 acm1::URA3 | This study |
| YKA259 | MATabar1::hisG clb2::KanMX4 acm1::URA3 | This study |
| YKA274 | MATabar1::hisG 3HA-BMH1 acm1::KanMX4 | This study |
| YKA290 | MATabar1::hisG 3FLAG-CDH1 3HA-ACM1 | This study |
| YKA291 | MATabar1::hisG 3FLAG-CDH1 | This study |
| BY4741 _hsl1_Δ | MATahsl1::KanMX4 | Open Biosystems |
| BY4741 _clb2_Δ | MATaclb2::KanMX4 | Open Biosystems |
| BY4741 _bmh1_Δ | MATabmh1::KanMX4 | Open Biosystems |
| BY4741 _bmh2_Δ | MATabmh2::KanMX4 | Open Biosystems |
| BY4741 _sic1_Δ | MATasic1::KanMX4 | Open Biosystems |
Plasmid pMPY-3xFLAG for construction of strains expressing endogenous proteins with amino-terminal 3FLAG tags was created by replacing the 3HA sequences from pMPY-3xHA (44) with sequences encoding three tandem FLAG epitopes. Oligonucleotides 5′-CTGGAGCTCCACCGCGGTGGCGGCCGCGAATTCTCTGACTACAAAGACCACGACGGTGATTATAAAGATCATGACATC-3′ and 5′-CCCCTCGAGTCTAGAGCGGCCGCACTGAGCCTTGTCATCGTCATCCTTGTAATCGATGTCATGATCTTTATAATCACC-3′ were annealed, and the 3′ ends were extended with Klenow DNA polymerase. The product was digested with NotI and ligated into the NotI sites of pMPY-3xHA to replace the 3HA sequence upstream of URA3. The same product was then digested with EcoRI and XhoI and ligated into the EcoRI/XhoI-treated plasmid generated in the previous step to replace the 3HA sequence downstream of URA3. The same primers used to amplify the integrating DNA from pMPY-3xHA (44) were also used for pMPY-3xFLAG.
The following plasmids are derivatives of centromeric plasmid p415ADH (34). pHLP130 (formerly p415ADH-FLAGCdh1), expressing 3FLAG-CDH1 from the ADH promoter, was constructed previously (21). pHLP117, expressing 3HA-ACM1 from the natural ACM1 promoter, was created by first digesting p415ADH with SacI and XbaI to excise the ADH promoter, treating the remaining vector product with mung bean nuclease to create blunt ends, and religating. Into the SmaI and PstI sites of this vector, we subcloned 3HA-ACM1 with roughly 700 bp of 5′ flanking sequence that had been amplified by PCR from YKA226 genomic DNA. Cell cycle-regulated expression similar to that of endogenous ACM1 was confirmed by Western blotting. To create pHLP114 expressing 3FLAG-CDH1-EGFP from the ADH promoter, we first subcloned the CDH1 coding sequence into the BamHI and SalI sites of pESC-EGFP (a gift from H. Charbonneau) to create the CDH1-EGFP fusion. A HindIII/XhoI fragment from this plasmid was then subcloned into pHLP130. The QuikChange site-directed mutagenesis system (Stratagene) was used to delete the last two codons of CDH1 from pHLP130, creating the 3FLAG-CDH1_Δ_IR allele (pHLP120).
The following are all centromeric galactose-inducible expression plasmids. 3FLAG-ACM1 was amplified by PCR from another construct and subcloned into the XbaI and XhoI sites of p416GAL1 (URA3) to create pHLP112. ACM1 was amplified from yeast genomic DNA with a forward primer containing a single HA epitope sequence and subcloned into the XbaI and XhoI sites of p415GAL1 (LEU2) to create pHLP109. pHLP163 (previously called pNC219-FLAGCdh1), expressing 3FLAG-CDH1 from the GAL1 promoter and carrying the TRP1 selectable marker, was described previously (21). Site-directed mutagenesis was used to create the cdh1-m11 mutant allele in pHLP163. The 3FLAG-cdh1-m11 allele was excised by digestion with XbaI and XmaI and subcloned into the XbaI and XmaI sites of p415GALS (LEU2) to create pHLP154. 3FLAG-Cdh1 was excised from pHLP130 by XbaI and XhoI digestion and subcloned into the XbaI and XhoI sites of p415GAL1 (LEU2) to create pHLP162. All plasmid constructions involving PCR were verified by DNA sequencing of the amplified region.
Cell growth, arrests, and synchronization.
Synchronized cultures were generated by block and release with 5 μg/ml α-factor or 15 μg/ml nocodazole as previously described (2). Arrest was monitored by microscopy until >95% of the cells exhibited the desired morphology. After release, aliquots were removed at the desired time points for flow cytometry and immunoblotting analysis. For immunoblotting, samples from each time point were resuspended at identical cell densities in sodium dodecyl sulfate (SDS) loading dye, boiled for 5 min, disrupted for 5 min by vortexing with glass beads, and cleared by centrifugation.
Cells were arrested in G1 by α-factor treatment (50 μg/liter for bar1 and 5 μg/ml for BAR1 strains), in S with 10 mg/ml HU, in G2/M with 15 μg/ml nocodazole, or in late M (with a cdc15-2 strain) by shifting growth from 25°C to 37°C. Arrests were confirmed by visual inspection of cell morphology by phase-contrast microscopy or by flow cytometry.
For microscopic examination of cell morphology, liquid cultures were grown at 30°C to saturation in selective medium and then diluted to an optical density at 600 nm of 0.05 to 0.1 in YPD. Cultures were allowed to pass through several divisions at 30°C before harvesting at an optical density at 600 nm of 0.6 to 0.8 for analysis by microscopy, flow cytometry, and immunoblotting. For localization experiments, selective medium was used exclusively, except for nocodazole arrest, which requires growth in YPD. Otherwise, the growth procedure was the same.
Immunoaffinity purification of protein complexes.
3FLAG-Cdh1 and 3HA-Acm1 were purified from yeast extracts by the same procedure. Frozen cell pellets were thawed on ice; resuspended in 2 volumes of buffer L (50 mM sodium phosphate [pH 7.5], 400 mM NaCl, 10% glycerol, 0.1% Triton X-100, 0.5 mM dithiothreitol, 50 mM sodium fluoride, 50 mM β-glycerophosphate) supplemented with 1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, 100 μM leupeptin, and 5 mM EDTA; and distributed into 1.5-ml microcentrifuge tubes. Glass beads were added to half of the total liquid volume, and cells were agitated in a Disruptor Genie (Scientific Industries) until greater than 75% of the cells were lysed (typically, 15 to 20 min). Extracts were cleared by centrifugation at 16,000 × g in a microcentrifuge for 30 min, and the contents of each tube were pooled. Anti-FLAG or anti-HA resin was added to the soluble extracts (approximately 1 μl/5 mg protein), and the mixture was incubated on a rotating platform for 2 h at 4°C. Resin was washed four times with 25 ml buffer L. Bound protein was eluted twice with 250 μg/ml 3FLAG peptide or HA peptide (Sigma) in buffer L for 20 min at 30°C. Elutions were combined and separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 4 to 12% gradient gels (Bio-Rad), and proteins were visualized by staining with silver or Coomassie blue.
MS.
SDS-PAGE bands were excised, destained, and subjected to in-gel digestion overnight at 37°C with 20 μg/ml trypsin in fresh 50 mM ammonium bicarbonate. Extracted peptides were analyzed on an Applied Biosystems 4700 matrix-assisted laser desorption ionization-tandem time of flight mass spectrometer in positive reflectron mass spectrometry (MS) and tandem MS modes. Combined MS and tandem MS database searches were performed with GPS Explorer software (Applied Biosystems) and the Mascot search engine (Matrix Science). For each protein identification, a single statistically significant score was obtained from Saccharomyces cerevisiae, with the exception of Bmh1 and Bmh2, which cross-matched because of their 93% sequence identity. However, peptides unique to both Bmh1 and Bmh2 were detected and confirmed by tandem MS.
Co-IP and Western blotting.
Coimmunopurification (Co-IP) experiments were performed essentially as described above for purifications of the Cdh1 complex. For Cdh1-substrate Co-IPs, buffer C (50 mM sodium phosphate [pH 7.5], 100 mM NaCl, 10% glycerol, 0.1% Triton X-100, 5 mM EDTA) was used. For Cdh1-Acm1 and Cdh1-Bmh1 Co-IPs, buffer L was used. The signal from immunoblots was visualized with ECL Plus reagents (GE Healthcare) or directly with a LI-COR Odyssey infrared imaging system. For quantification, immunoblot signals were analyzed either from films by densitometry on an Alpha Innotech Corporation Fluorchem imager or with the Odyssey image analysis software.
Flow cytometry.
A 500-μl volume of mid-log-phase culture was washed with water and fixed in 70% ethanol overnight at 4°C. Cells were washed again, treated with 1 mg/ml RNase A in 50 mM Tris-HCl (pH 8)-15 mM NaCl for at least 2 h at 37°C, treated with 5 mg/ml freshly made pepsin in 0.17% HCl for not more than 20 min, washed with 50 mM Tris-HCl (pH 8), and suspended in 1 μM Sytox green in 50 mM Tris-HCl (pH 8). Cells were briefly sonicated immediately prior to analysis on a Beckman Coulter Cytomics FC 500 flow cytometer.
Microscopy.
Fluorescence and differential interference contrast (DIC) microscope images were acquired on a Leica DMRX HC microscope equipped with a Hamamatsu ORCA ER digital camera and OpenLab software (Improvision Inc.). All images in a given experiment were acquired with identical microscope and camera settings, and any additional image processing was applied to all images with ImageJ or Adobe Photoshop software. The relative intensity of bud neck-specific green fluorescent protein fluorescence signal in raw image files was determined with ImageJ by measuring the average pixel density in an area demarcating the visible bud neck band and subtracting the average pixel density in an equivalent area of adjacent cytoplasm. The resulting values from 20 randomly selected cells that showed visible bud neck-specific fluorescence in wild-type and _acm1_Δ strains were then averaged. Cells were fixed with formaldehyde for all microscopy experiments.
To prepare cells for indirect immunofluorescence, spheroplasts were generated essentially as previously described (8) and spotted on poly-l-lysine-treated slides. After a 15-min incubation at room temperature (RT), slides were submerged first in methanol (−20°C) for 6 min and second in acetone (−20°C) for 30 s and air dried. Samples were incubated in blocking solution (0.1 mg/ml streptavidin and 3% bovine serum albumin [BSA] in phosphate-buffered saline-Tween 20 [PBS-T]) at 4°C for 1 h and then in anti-HA 12CA5 antibody (Roche) diluted 1:500 in PBS-T containing 0.5 mg/ml d-biotin and 3% BSA for 1 h at RT. Slides were washed three times for 5 min with PBS-T and then incubated with biotin-conjugated anti-mouse immunoglobulin G (Vector Laboratories) diluted 1:200 in PBS-T containing 3% BSA at RT for 30 min. Slides were washed again three times for 5 min with PBS-T. Finally, slides were incubated with streptavidin-conjugated Alexa Fluor 488 (Molecular Probes) diluted 1:200 in PBS-T containing 3% BSA for 30 min at RT and washed three times for 5 min with PBS-T.
RESULTS
Cdh1 stably interacts with three other proteins.
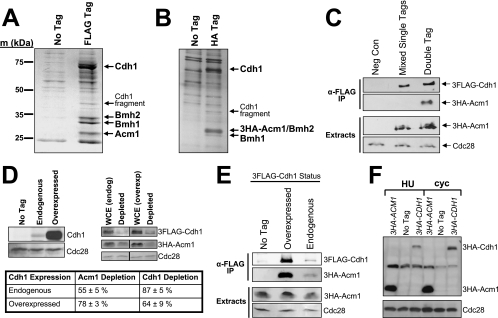
We used immunoaffinity purification and MS to identify proteins associated with Cdh1 during cell cycle stages when APC/CCdh1 activity is thought to be absent. 3FLAG epitope-tagged Cdh1 was purified from yeast extracts with anti-FLAG affinity resin, and eluted proteins were visualized by SDS-PAGE (Fig. 1A). Three additional protein bands were detected specifically in the presence of 3FLAG-Cdh1 and were identified by peptide mass fingerprinting and tandem MS. Two of them, Bmh1 and Bmh2, are homologs of the highly conserved 14-3-3 family. The third protein is the product of a previously uncharacterized open reading frame, YPL267W, which we have named Acm1 (_A_PC/C_C_dh1 _M_odulator 1). We also used anti-HA affinity resin to purify endogenous 3HA epitope-tagged Acm1 and any bound proteins (Fig. 1B). Cdh1, Bmh1, and Bmh2 were identified by MS, suggesting that Cdh1 associates with Acm1 and the 14-3-3 proteins in a single complex. However, our data do not distinguish between Bmh1 and Bmh2 existing together in the complex as a heterodimer or individually, forming distinct subcomplexes. We observed the complex by using cells arrested in late M by temperature shift in a cdc15-2 strain (Fig. 1A), in S by HU treatment (Fig. 1B and 2D), and in asynchronous cultures (not shown). The same interacting proteins were observed when endogenous Cdh1 (not shown) or Cdh1 overproduced from the ADH promoter was purified and when the 3FLAG tag was at the N terminus or C terminus (not shown) of Cdh1. The complex is highly salt stable since the purifications were performed at 500 mM Na+.
FIG. 1.
Cdh1 is a component of a multiprotein complex. (A) 3FLAG-Cdh1 expressed from the ADH promoter on a CEN plasmid (pHLP130) was purified from a cdc15-2 strain (DLY3033) arrested in late M at 37°C. Eluted proteins were analyzed by SDS-PAGE and Coomassie blue staining and compared to a control preparation from cells lacking the 3FLAG-Cdh1 expression plasmid. Bands specific to the 3FLAG-Cdh1 sample were excised and analyzed by MS. Identified proteins are labeled with arrows. m, markers. (B) Endogenous 3HA-Acm1 was purified from strain YKA226 arrested in S phase with HU and analyzed by SDS-PAGE and silver staining. Bands present in the preparation of 3HA-Acm1 but not in a control preparation from cells lacking the 3HA epitope tag (BY4741) were excised and analyzed by MS. Note that 3HA-Acm1 and Bmh2 comigrate on this gel but were still both unambiguously identified. (C) Western analysis of endogenous 3HA-Acm1 copurifying with endogenous 3FLAG-Cdh1 from yeast extracts derived either from two mixed cultures (YKA227 and YKA291), each individually expressing one of the tagged proteins (mixed single tags) or from a single strain (YKA290) expressing both tagged proteins (double tag). The control (Neg Con) sample is BY4741 lacking the epitope tags. Cdc28 is a loading control. IP, immunoprecipitate. (D) Western blotting was used to quantify the amount of Cdh1 bound to Acm1 in HU-arrested cells. Whole-cell extracts (WCE) containing endogenous 3HA-Acm1 and either endogenous (YKA290) or overexpressed (pHLP130 in YKA245) 3FLAG-Cdh1 were depleted of 3HA-Acm1 with anti-HA resin. The difference in level of endogenous and overexpressed 3FLAG-Cdh1 is illustrated in the immunoblot on the left. Codepletion of Cdh1, indicating the amount bound to 3HA-Acm1, was then measured by anti-FLAG immunoblotting (illustrated on the right) with a LI-COR Odyssey imager. Cdc28 served as a loading control and was used to normalize the 3HA-Acm1 and 3FLAG-Cdh1 levels in each sample. Different exposures are shown for endogenous and overexpressed 3FLAG-Cdh1 because of the large difference in protein level. The table shows the average depletion of 3HA-Acm1 and codepletion of endogenous and overexpressed 3FLAG-Cdh1 in three independent experiments with standard deviations. (E) Overexpressed (pHLP130 in YKA245) and endogenous (YKA290) 3FLAG-Cdh1 were immunopurified from extracts of HU-arrested cells expressing 3HA-Acm1. The amount of copurified 3HA-Acm1 was then monitored by anti-HA immunoblotting. Band intensities were quantified with the LI-COR imager. The ratio of overexpressed to endogenous immunopurified 3FLAG-Cdh1 was 11 ± 4, and the corresponding ratio of copurified 3HA-Acm1 was 12 ± 2 (three trials). The control strain in the No Tag lane was YKA227. (F) Expression of endogenous 3HA-Acm1 (YKA227) and 3HA-Cdh1 (YKA250) was compared by anti-HA immunoblotting in HU-arrested S-phase cells (HU) and asynchronous log-phase cells (cyc). Cdc28 was a loading control, and the control strain in the No Tag lane was YKA233.
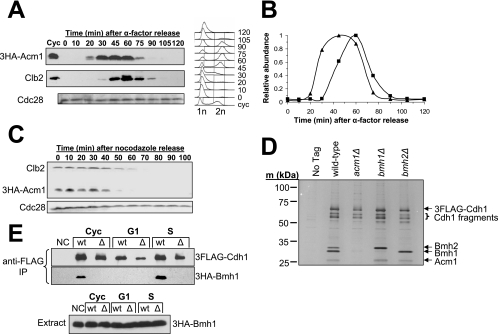
FIG. 2.
The Cdh1 complex is regulated via cell cycle-dependent expression of ACM1. (A) Western analysis of the 3HA-Acm1 level in strain YKA226 synchronized by α-factor block and release. After 45 min, α-factor was added back to rearrest in the following G1 phase. The blot was reprobed with anti-Clb2 for comparison and anti-Cdc28 as a loading control. cyc, cycling asynchronous culture. Samples from each time point were also analyzed by flow cytometry to monitor cell cycle progression. n, genomic DNA content. (B) A quantitative comparison of 3HA-Acm1 (▴) and Clb2 (▪) levels obtained from the data in panel A by densitometry. The peak level of each protein was normalized to 1. (C) 3HA-Acm1 expressed from its natural promoter on a single-copy plasmid (pHLP117) in strain YKA247 was monitored by Western blotting in a synchronized culture released from a nocodazole arrest into medium containing α-factor. Immunoblotting of Clb2 was included for comparison. Cell cycle progression was also monitored by flow cytometry (not shown). (D) 3FLAG-Cdh1 expressed from the ADH promoter on a CEN plasmid (pHLP130) was purified from the indicated BY4741 yeast strains arrested in S phase with HU. The presence or absence of the other complex components was monitored by SDS-PAGE and silver staining and confirmed by MS. No Tag, wild-type with empty plasmid. (E) Co-IP of endogenous 3HA-Bmh1 with 3FLAG-Cdh1 expressed from pHLP130 in YKA249 and YKA274 cells containing (wt) or lacking (Δ) ACM1, respectively. Anti-FLAG resin was used for immunopurification, and copurification of Bmh1 was measured by anti-HA immunoblotting. Cyc, asynchronous log-phase cells; G1, α-factor arrest; S, HU arrest; NC, negative control lacking the 3FLAG-Cdh1 expression plasmid; IP, immunoprecipitate.
The Cdh1 protein complex forms in vivo.
14-3-3 proteins are highly expressed and interact with numerous proteins (54). We were initially concerned that the complex might be an artifact of the cell extracts and would not normally exist in vivo. To test this, we used a method reported previously to confirm in vivo formation of a Cdc20-CCT chaperonin complex (11). Side-by-side Co-IPs of endogenous 3HA-Acm1 with endogenous 3FLAG-Cdh1 were performed with either a doubly tagged strain or a mixture of two singly tagged strains. In the latter sample, the two tagged proteins can only interact after cell lysis, whereas in the doubly tagged strain the two tagged proteins have the opportunity to associate inside the cell prior to lysis. Figure 1C clearly shows that the interaction between 3HA-Acm1 and 3FLAG-Cdh1 is observed exclusively when using the doubly tagged strain, providing strong evidence that the Cdh1 complex forms in a physiological context in vivo. We monitored the Acm1-Cdh1 interaction because, as shown in Fig. 2D and E, association of Bmh1 and Bmh2 with Cdh1 is dependent on Acm1 and therefore Acm1 binding is a suitable indicator of full complex assembly.
Acm1, Bmh1, and Bmh2 were observed in roughly stoichiometric amounts in our preparations of 3FLAG-Cdh1 overexpressed from the ADH promoter (Fig. 1A), suggesting that Cdh1 might exist exclusively in complex-bound form at specific cell cycle stages. We quantified the relative amounts of endogenous and overexpressed 3FLAG-Cdh1 present in S-phase whole-cell extracts prior to and following depletion of endogenous 3HA-Acm1 with anti-HA resin (Fig. 1D). 3HA-Acm1 depletion reduced the level of endogenous 3FLAG-Cdh1 by an average of 87%. When 3FLAG-Cdh1 was overproduced with the ADH promoter (a roughly 20-fold increase), we still observed a 64% reduction following depletion of endogenous 3HA-Acm1. We also observed a proportionate increase in 3HA-Acm1 copurified with overexpressed 3FLAG-Cdh1 compared to endogenous 3FLAG-Cdh1 (Fig. 1E). Not surprisingly, immunoblot analysis revealed that endogenous 3HA-Acm1 is expressed at a much higher level than endogenous 3HA-Cdh1 in both asynchronous and HU-arrested cells (Fig. 1F). Together, these results suggest that Cdh1 is probably stoichiometrically associated with the complex, at least during S phase, and that Acm1, Bmh1, and Bmh2 are not limiting factors in maintaining Cdh1 in a complexed state.
Acm1 protein is cell cycle regulated.
We predicted that if this Cdh1-bound complex plays a role in regulating APC/CCdh1 activity during the cell cycle, then the complex itself must be regulated in a cell cycle-dependent manner. Microarray analysis of yeast gene expression revealed a late G1 transcriptional peak for ACM1 (YPL267W), and the promoter region of ACM1 contains SCB and MCB sequence elements characteristic of numerous genes induced in late G1 (51). Therefore, we monitored 3HA-Acm1 protein by Western blotting in synchronously growing cultures released either from pheromone-induced G1 arrest or nocodazole-induced M arrest (Fig. 2A to C). Acm1 is absent from G1 cells. It then appears very quickly after G1 release, just prior to initiation of DNA replication, as evidenced by flow cytometry. Acm1 disappears again late in mitosis around the same time as, or slightly earlier than, the major mitotic cyclin Clb2. Likewise, following release from a nocodazole arrest, Acm1 disappeared in late mitosis slightly earlier than Clb2.
Cdh1-Bmh1/Bmh2 association is Acm1 dependent.
To begin to define the determinants for assembly and regulation of the Cdh1 complex, we purified 3FLAG-Cdh1 from strains harboring deletions of the ACM1, BMH1, and BMH2 genes (Fig. 2D). Strikingly, Bmh1 and Bmh2 were not detected in 3FLAG-Cdh1 preparations from cells lacking Acm1. In contrast, absence of Bmh1 or Bmh2 had no effect on the association of the other three complex components, suggesting that either is sufficient for complex formation. To further confirm the dependence of the Cdh1-Bmh1 interaction on Acm1, we monitored Co-IP of 3HA-Bmh1 with 3FLAG-Cdh1 using asynchronous G1- and S-phase cultures (Fig. 2E). As expected, the Cdh1-Bmh1 interaction was not detected in G1 cells or in the absence of Acm1. This was true when we used either our typical complex purification buffer containing 500 mM Na+ (Fig. 2E) or a more physiological buffer containing 150 mM Na+ (not shown). Although we cannot rule out the possibility that absence of Acm1 weakens a direct Cdh1-Bmh1/Bmh2 association enough that it is lost during our purification procedure, these results suggest that Acm1 mediates the association of Cdh1 with Bmh1 and Bmh2.
Cdh1 bud neck localization is enhanced in cells lacking Acm1.
A common function of 14-3-3 proteins is to control cellular localization of their binding targets, for example, by sequestering them in the cytoplasm (35). Since Cdh1 is exported to the cytoplasm at the end of G1 when it is inactivated (25), we hypothesized that association with Bmh1 and Bmh2 might be required to maintain cytoplasmic localization of Cdh1 during its inactive period. To directly test this, we mimicked the experimental design of Jaquenoud et al. (25) by expressing Cdh1 as a C-terminal enhanced green fluorescent protein (EGFP) fusion (in our case, from the ADH promoter on a single-copy plasmid) and monitoring its intracellular localization in wild-type and _acm1_Δ cells at different cell cycle stages by fluorescence microscopy. In wild-type cells, 3FLAG-Cdh1-EGFP localized to the nuclei of unbudded cells in an asynchronous population and pheromone-arrested cells and was primarily dispersed throughout the cytoplasm in cells with large buds or in cells arrested in M phase with nocodazole (Fig. 3A and B), consistent with the previous results (25). In _acm1_Δ cells, 3FLAG-Cdh1-EGFP also localized to the nucleus in G1 and was found dispersed throughout the cytoplasm in large budded cells (Fig. 3A and B). Therefore, we conclude that Acm1/Bmh1/Bmh2 is not required to sequester Cdh1 in the cytoplasm following its inactivation.
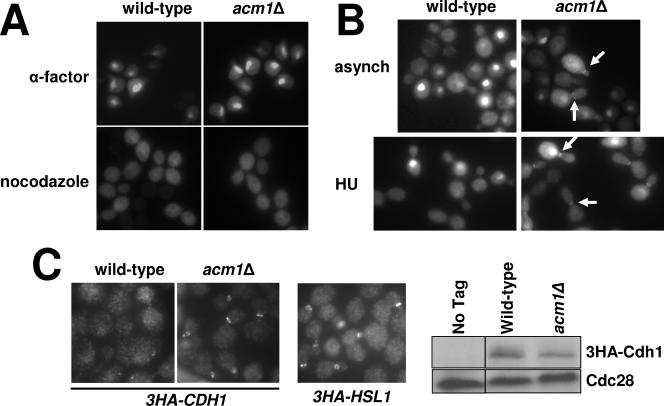
FIG. 3.
Effects of the Cdh1 complex on cellular localization of Cdh1. (A) 3FLAG-Cdh1-EGFP expressed from the ADH promoter (pHLP114) was visualized with a fluorescence microscope in fixed wild-type (YKA150) or _acm1_Δ (YKA247) cells arrested in either G1 with α-factor or in M with nocodazole. (B) Analysis of bud neck localization of 3FLAG-Cdh1-EGFP in wild-type (YKA150) versus _acm1_Δ (YKA247) cells in asynchronous (asynch) and HU-treated cultures. Arrows indicate examples of bud neck localization of 3FLAG-Cdh1-EGFP in the _acm1_Δ strain. (C) Localization of endogenous 3HA-Cdh1 in asynchronous wild-type (YKA250) and _acm1_Δ (YKA256) cells was visualized by indirect immunofluorescence by a signal amplification strategy described in Materials and Methods. Bud neck localization of 3HA-Hsl1 in strain YKA252 was used as a positive control. The immunoblot on the right illustrates the relative 3HA-Cdh1 levels in the wild-type and _acm1_Δ strains. Cdc28 was a loading control. All three images were obtained with a 100× oil immersion objective.
Cdh1 was also shown previously to localize to the bud neck (25). In our analysis of Cdh1 nuclear versus cytoplasmic localization, we observed a dramatic difference in the frequency and intensity of bud neck staining in _acm1_Δ cells compared to the isogenic wild-type strain (Fig. 3B). In an asynchronous culture, bud neck staining was observed in both small- and large-budded cells and was also observed in cells arrested in S with HU and to a lesser extent in cells arrested in M with nocodazole. We detected bud neck-specific EGFP signal in only 14% of budded wild-type cells in an asynchronous culture. In contrast, bud neck fluorescence was detected in 59% of budded _acm1_Δ cells (n = 100). After cell cycle arrest with HU, bud neck-specific EGFP signal was detected in 6% of wild-type cells and 51% of _acm1_Δ cells (n = 200). Furthermore, the detectable bud neck-specific fluorescence was sevenfold more intense, on average, in _acm1_Δ cells than in wild-type cells. The average cytoplasmic fluorescence intensity differed by less than 10%, and immunoblot assays confirmed equivalent expression of 3FLAG-Cdh1-EGFP in the wild-type and _acm1_Δ strains (not shown).
To rule out the possibility that bud neck localization was an artifact of Cdh1 overexpression or use of a C-terminal tag, we used indirect immunofluorescence to visualize endogenous 3HA-Cdh1 localization in wild-type and _acm1_Δ cells (Fig. 3C). A signal amplification strategy (see Materials and Methods) was required to detect endogenous 3HA-Cdh1 fluorescence. Consistent with the EGFP experiments, 3HA-Cdh1 localization to the bud neck was only detected in _acm1_Δ cells. We conclude that binding of Acm1 and Bmh1/Bmh2 can prevent localization of Cdh1 to the bud neck.
Specific Cdh1-substrate interactions are enhanced in the absence of Acm1.
Several APC/CCdh1 substrates localize to the bud neck, including the kinases Hsl1 and Cdc5 (4, 6) and the mitotic cyclin Clb2 (5, 22). It is possible that localization of Cdh1 to the bud neck in an _acm1_Δ strain was observed because of enhanced association with one or more of its substrates and that the complex with Acm1 and Bmh1/Bmh2 functions to block substrate binding. To test this hypothesis, we compared the interaction between overexpressed 3FLAG-Cdh1 and several of its substrates in _acm1_Δ cells and wild-type cells by Co-IP. The amounts of Clb2 and 3HA-Hsl1 that copurified with 3FLAG-Cdh1 using asynchronous cultures was between two- and fourfold higher in the absence of Acm1 after normalization to 3FLAG-Cdh1 (Fig. 4A and B). Using extracts from asynchronous cultures could be problematic because the proteolytic machinery capable of degrading both Acm1 and the APC/CCdh1 substrates is likely active. Therefore, we repeated these experiments after arresting cells in S phase by treatment with HU. The results were striking. The amounts of Clb2 and 3HA-Hsl1 that copurified with 3FLAG-Cdh1 increased by 25-fold and more than 100-fold, respectively, in the absence of Acm1.
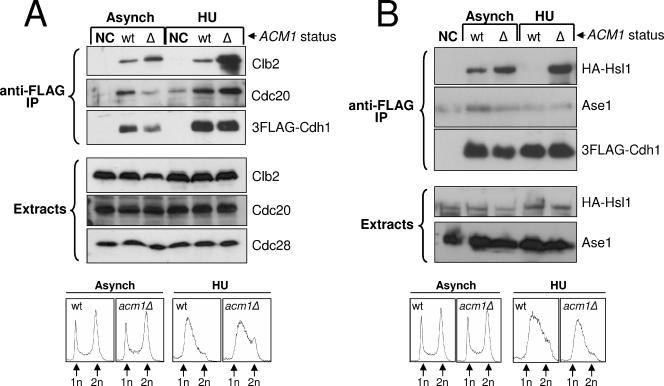
FIG. 4.
The Cdh1 complex can inhibit association of Cdh1 with specific substrates. (A) 3FLAG-Cdh1 expressed from pHLP130 was purified from mid-log-phase (Asynch) and HU-arrested cultures with anti-FLAG resin. Copurification of APC/C substrates Clb2 and Cdc20 with 3FLAG-Cdh1 from _acm1_Δ (Δ) and isogenic wild-type (wt) strains (YKA237 and YKA233) was probed by Western blotting with anti-Clb2 or anti-Cdc20 antibodies. (B) The same experiment as in panel A was performed with _acm1_Δ and isogenic wild-type strains expressing a 3HA-HSL1 allele (YKA257 and YKA252). Copurification of 3HA-Hsl1 and Ase1 with 3FLAG-Cdh1 was probed by Western blotting with anti-HA or anti-Ase1 antibodies. In both panels, NC indicates the negative control lacking the plasmid expressing 3FLAG-Cdh1. Extract immunoblots demonstrate that roughly equal amounts of substrate were present in all starting samples. Cdc28 was a loading control. Flow cytometry analysis (bottom of each panel) demonstrates that the cell cycle distribution of _acm1_Δ and wild-type cells is not significantly different and confirms the HU arrests. n, genomic DNA content. Asynch, asynchronous; IP, immunoprecipitate.
Surprisingly, when we probed the same Co-IP samples for two additional Cdh1 substrates, Ase1 and Cdc20, the results were very different (Fig. 4A and B). Cdc20 association with 3FLAG-Cdh1 did not increase in the absence of Acm1 in an asynchronous culture and was enhanced less than threefold in the absence of Acm1 when cells were treated with HU. Ase1 association with 3FLAG-Cdh1 was undetectable above the nonspecific background, although the protein produced a very strong signal in the cell extracts, even in the absence of Acm1. These results suggest that there may be inherent differences in the way Clb2 and Hsl1 interact with Cdh1 compared to other substrates. We also observed a similar enhanced association with Clb2 in _acm1_Δ cells expressing endogenous 3HA-Cdh1 (not shown), ruling out the possibility that these results are artifacts of Cdh1 overexpression.
Acm1 can suppress APC/CCdh1 activity in vivo.
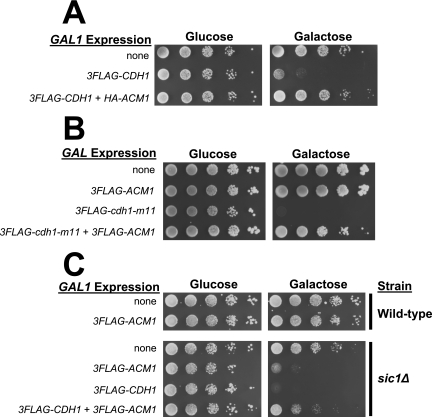
Because Cdh1 is complexed with Acm1, Bmh1, and Bmh2 during the same cell cycle interval in which it is thought to be inactive and because the complex can block association of Cdh1 with certain substrates, including Clb2, we performed a series of biological experiments to determine if Acm1 can inhibit APC/CCdh1 activity in vivo. High-level overexpression of CDH1 from a GAL promoter results in a G2 cell cycle arrest that has been attributed to constitutive APC/C activation and depletion of mitotic cyclins needed to drive mitotic entry (46, 56). We reproduced this toxic effect of high-level Cdh1 overexpression with a single-copy plasmid expressing 3FLAG-Cdh1 from the GAL1 promoter (Fig. 5A). We predicted that if Acm1 acts as an inhibitor of APC/CCdh1, then co-overexpression of Acm1 might restore viability to these cells. Indeed, co-overexpression of HA-Acm1 from the GAL1 promoter on a single-copy plasmid restored wild-type viability to cells expressing toxic levels of Cdh1 (Fig. 5A). Second, we tested if Acm1 overexpression could suppress the toxicity of the Cdh1-m11 mutant (60) that lacks inhibitory CDK phosphorylation sites. Expression of 3FLAG-Cdh1-m11 at a much lower level from the weak GALS promoter caused inviability on galactose medium (Fig. 5B). Overexpression of 3FLAG-Acm1 restored near-normal viability to these cells. Third, since cells lacking both Cdh1 and Sic1 are inviable (46, 56), we hypothesized that overexpression of Acm1 might result in constitutive APC/CCdh1 inhibition that would lead to low viability in a _sic1_Δ background. We observed a severe reduction in viability of _sic1_Δ but not isogenic wild-type cells overexpressing 3FLAG-Acm1 from the GAL1 promoter on a single-copy plasmid (Fig. 5C). This effect was Cdh1 dependent because substantial viability was restored by co-overexpression of 3FLAG-Cdh1. Taken together, these results strongly support the conclusion that Acm1/Bmh1/Bmh2 is an inhibitor of APC/CCdh1-mediated proteolysis of mitotic cyclins.
FIG. 5.
Acm1 suppresses APC/CCdh1 activity in vivo. (A) Liquid YKA150 cultures harboring empty control plasmids or single-copy GAL1 promoter-driven expression plasmids (pHLP163 for 3FLAG-CDH1 and pHLP109 for HA-ACM1) were grown in selective raffinose medium, and 10-fold serial dilutions were spotted and grown on selective medium containing either glucose or galactose as the carbon source. (B) Same experiment as in panel A with pHLP154 expressing 3FLAG-cdh1-m11 from the attenuated GALS promoter and pHLP112 expressing 3FLAG-ACM1 from the GAL1 promoter. (C) BY4741 (wild-type) and _sic1_Δ cells containing empty control plasmids or single-copy GAL1 promoter-driven expression plasmids (pHLP112 for 3FLAG-ACM1 and pHLP162 for 3FLAG-Cdh1) were grown and processed as in panel A.
Deletion of ACM1 enhances the elongated-bud phenotype of cells overexpressing Cdh1.
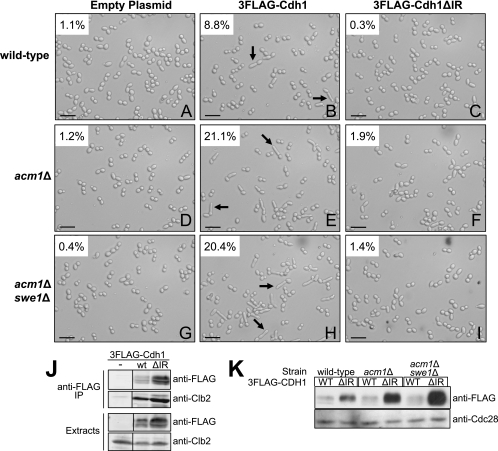
While studying Cdh1 localization, we noticed that many _acm1_Δ cells had elongated buds. Since elongated buds are a hallmark of insufficient mitotic CDK activity (18, 42) and can be generated by defects in regulators of a bud morphogenesis checkpoint (27), including Hsl1, we decided to explore this phenotype in more detail. Overexpression of 3FLAG-Cdh1 from the ADH promoter on a single-copy plasmid resulted in a low frequency of highly elongated buds (8.8%) following treatment with HU (Fig. 6A, compare to panel B). A bud was considered elongated when its length was at least twice the width of the mother cell, a criterion defined in a previous study (32). Deletion of ACM1 greatly enhanced the penetrance of this elongated-bud phenotype (21.1% elongated; Fig. 6B, compare to panel E). In addition, the average ratio of bud length to mother cell width in cells meeting the criteria for an elongated bud increased substantially (2.80 for _acm1_Δ versus 2.39 for wild-type). Elongated buds resulting from defects in regulators of the morphogenesis checkpoint can typically be suppressed by deletion of SWE1, encoding a CDK inhibitory kinase (27). SWE1 deletion had no significant effect on bud elongation associated with Cdh1 overexpression in _acm1_Δ cells (20.4% elongated; Fig. 6E, compare to panel H), demonstrating that this phenotype is independent of the morphogenesis checkpoint. To determine if this phenotype requires APC/CCdh1-dependent proteolysis, we took advantage of a recently described Cdh1 mutant lacking the last two amino acids (Cdh1ΔIR) that is defective in APC/C activation but still able to interact with substrates (10, 37, 58). We confirmed that Cdh1ΔIR associated with Clb2, similar to Cdh1 (Fig. 6J), and bound the full Acm1/Bmh1/Bmh2 complex (not shown), demonstrating that the mutation did not globally affect protein folding or structure. Overexpressed 3FLAG-Cdh1ΔIR did not elicit an elongated-bud phenotype in wild-type or _acm1_Δ cells (Fig. 6C, F, and I) despite being present at a significantly higher level than overexpressed wild-type 3FLAG-Cdh1 (Fig. 6K). This observation suggests that hyperpolarized bud growth promoted by Cdh1 in the absence of Acm1 requires the ability to activate APC/C-dependent proteolysis and further supports the conclusion that Acm1 is a negative regulator of mitotic cyclin proteolysis.
FIG. 6.
Deletion of ACM1 in cells moderately overexpressing Cdh1 results in hyperpolarized growth independent of the morphogenesis checkpoint. (A to I) Cells from HU-treated cultures of BY4741 (wild-type), YKA237 (_acm1_Δ), and YKA255 (_acm1_Δ _swe1_Δ) harboring an empty plasmid or a plasmid overexpressing the 3FLAG-Cdh1 (pHLP130) or the 3FLAG-Cdh1ΔIR mutant (pHLP120) from the ADH promoter were visualized by DIC microscopy. Black arrows indicate examples of hyperpolarized buds. The percentage of cells containing an elongated bud is shown in each panel and was determined by manual inspection of approximately 500 cells. The scale bar equals 32 μm. A 40× objective lens was used. (J) Co-IP of Clb2 with 3FLAG-Cdh1 (wild type [wt]) and 3FLAG-Cdh1ΔIR (ΔIR) from HU-arrested YKA237 cells. IP, immunoprecipitate. (K) Immunoblotting of extracts from strains shown in panels A to I with anti-FLAG antibody to visualize overexpressed 3FLAG-Cdh1 and 3FLAG-Cdh1ΔIR. Cdc28 was a loading control. WT, wild type.
ACM1 and HSL1 interact genetically.
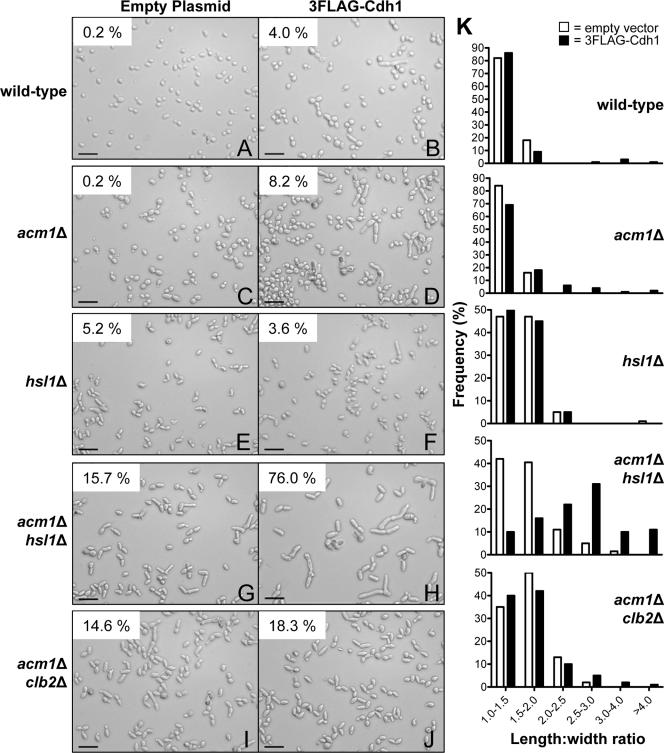
We next tested whether a genetic interaction might exist between ACM1 and the bud morphogenesis checkpoint that would influence bud elongation and cell cycle progression by synergistically downregulating mitotic CDK activity. We chose to examine Hsl1, which helps relieve the morphogenesis checkpoint by inactivating Swe1. We monitored cell elongation in asynchronous cultures and compared cells containing either the same 3FLAG-Cdh1 overexpression construct used above or an empty vector. In an _acm1_Δ strain overexpressing Cdh1, 8.2% of the cells were elongated (Fig. 7D). _hsl1_Δ cells exhibited a mild bud elongation phenotype (5.2%) that has been reported previously in some strain backgrounds (33). This phenotype was not enhanced by overexpression of 3FLAG-Cdh1 (compare Fig. 7E and F). Strikingly, the combination of _hsl1_Δ, _acm1_Δ, and 3FLAG-Cdh1 overexpression resulted in a dramatic increase in the severity of bud elongation (Fig. 7H and K). Three-quarters of these cells met the elongation criteria and, in general, they were noticeably larger than those of the other strains analyzed in this experiment. Liquid cultures grew very slowly compared to the wild-type and single-deletion strains, and cells had a greater tendency to clump (data not shown). Consistent with the HU-treated cultures shown in Fig. 6, the effects of Cdh1 overexpression were diminished when the 3FLAG-Cdh1ΔIR mutant was used (data not shown). A _clb2_Δ strain also exhibited a mild bud elongation phenotype, similar to _hsl1_Δ cells and consistent with previous reports (42) (data not shown). Combining _acm1_Δ and 3FLAG-Cdh1 overexpression with _clb2_Δ did not enhance the severity of bud elongation to the same extent as it did with _hsl1_Δ (Fig. 7J and K). So although HSL1 and CLB2 are both components of the morphogenesis checkpoint, their genetic interactions with ACM1 are different. Surprisingly, given the apparent requirement for APC/C activation by Cdh1 in the experiments described above and the previous reports of Clb2 depletion in cells overexpressing toxic levels of Cdh1, we did not observe a large reduction in Clb2 level in cells exhibiting the most severe bud elongation (data not shown), suggesting that Acm1 may act to protect a specific subpopulation of Clb2 from APC/CCdh1-mediated proteolysis (see Discussion).
FIG. 7.
ACM1 interacts genetically with HSL1. (A to J) Cells from asynchronous mid-log-phase cultures of BY4741 with the indicated genotypes and either pHLP130 expressing 3FLAG-Cdh1 from the ADH promoter or an empty plasmid control were visualized by DIC microscopy. Scale bar, 32 μm. A 40× objective lens was used. The percentage of elongated cells is indicated in the top left corner of each panel (n = >500 for all panels except H, where n = 300) and was obtained from images compiled from three independent experiments. Cells were counted as elongated if their length was at least twice their width. (K) Frequency distributions of length/width ratio for the cells pictured in panels A to J obtained from careful measurements of at least 100 randomly selected cells each. Open bars represent cells with the empty-plasmid control, and solid bars represent cells with the 3FLAG-Cdh1 expression plasmid.
Effects of _acm1_Δ on cells expressing endogenous Cdh1.
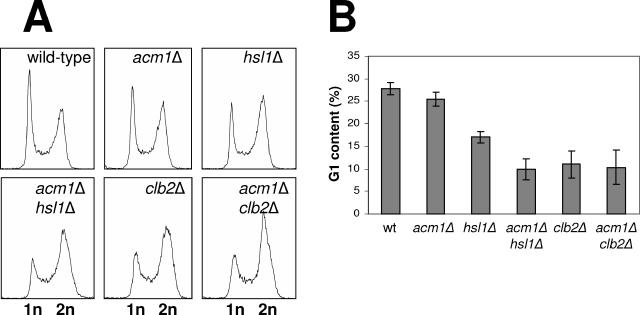
Even with endogenous Cdh1 expression, _acm1_Δ _hsl1_Δ cells exhibited a more pronounced polarized growth phenotype than either single-deletion strain (Fig. 7K and G, compare to panels E and C), although not nearly to the same extent as with overexpressed Cdh1. To further explore a possible cell cycle defect associated with _acm1_Δ at the endogenous Cdh1 level, we examined cell cycle distribution in a panel of yeast strains by flow cytometry. The cell cycle profile of _acm1_Δ cells was very similar to that of wild-type cells (Fig. 8). _hsl1_Δ cells exhibited a mild G2/M delay. The _acm1_Δ _hsl1_Δ double-deletion strain had a much more pronounced G2/M delay than the _hsl1_Δ single-deletion mutant. This synergistic effect was of a magnitude similar to that of the delay observed in a _clb2_Δ strain. Moreover, the G2/M delay in _acm1_Δ _hsl1_Δ double-deletion cells was the same as in the _clb2_Δ single-mutant strain. These results closely mirror the bud elongation results in Fig. 7, and together they suggest that Acm1 and Hsl1 are independent positive regulators of mitotic CDK.
FIG. 8.
Cell cycle distribution in _acm1_Δ _hsl1_Δ cells expressing endogenous Cdh1. (A) Yeast strains (BY4741 background) with the indicated genotypes were grown to mid-log phase and analyzed by flow cytometry. n = genomic DNA content. (B) Data from panel A were analyzed with ModFit LT software to determine the percentages of cells in G1, S, and G2/M. Percent G1 content is plotted for each strain. Data represent the average of three independent cultures, and error bars are standard deviations. wt, wild type.
ACM1 is conserved in budding yeast species.
Cdh1 and 14-3-3 proteins are highly conserved in eukaryotes, prompting us to search for homologs of Acm1 in other eukaryotic organisms. Standard BLAST searches did not reveal any related proteins. However, iterative PSI-BLAST searching revealed apparent orthologs in several other budding yeasts (order Saccharomycetales), including species such as Candida albicans and Debaryomyces hansenii that are only distantly related to S. cerevisiae within this group (see Fig. S1 in the supplemental material). The fission yeast Schizosaccharomyces pombe does not appear to have an Acm1 homolog, and we did not find any homologs in any non-yeast species. The most highly conserved sequence elements in Acm1 and its homologs include several consensus CDK recognition sites (S/T-P-X-K/R), primarily concentrated near the amino terminus. We have identified a number of prominent phosphorylation sites, including several at these consensus CDK sequences, by MS (D. Jeong and M. Hall, unpublished data). These results and the evolutionary conservation of the consensus CDK recognition sequences suggest that Acm1 is likely regulated by CDK phosphorylation. In addition, Acm1 has several sequences that match degrons common to APC/C substrates including three D box sequences (R-x-x-L) and a KEN box. We are currently testing if Acm1 proteolysis is APC/C dependent.
DISCUSSION
A cell cycle-regulated complex associates with Cdh1.
We have identified a protein complex associated with the APC/C coactivator Cdh1 and demonstrated that this complex is a negative regulator of APC/CCdh1 function in vivo. In addition to Cdh1, the complex contains the budding yeast 14-3-3 homologs Bmh1 and Bmh2 and a previously uncharacterized protein, which we named Acm1. Acm1 appears to be the critical subunit for regulating assembly and dissociation of the Cdh1 complex. Acm1 is cell cycle regulated, appearing in late G1 and disappearing in late mitosis, and is required for stable interaction between Cdh1 and the 14-3-3 proteins. As a result, the Cdh1-Bmh1/Bmh2 interaction is restricted to the cell cycle interval in which Acm1 is expressed. In S-phase-arrested cells, the majority of Cdh1 is associated with this complex.
Knowledge of YPL267W (ACM1) prior to this study was limited to a number of large-scale genomic and proteomic screens. In addition to the global gene expression data already mentioned, the product of YPL267W was identified as an in vitro substrate of Clb2-Cdc28 in a proteomic screen for targets of the mitotic CDK in budding yeast (53). Alignment of Acm1 with orthologs in other budding yeast species suggests that CDK phosphorylation is likely conserved. We are currently studying the function of Acm1 phosphorylation sites in regulating the Cdh1 complex. The product of YPL267W (ACM1) was also identified as a binding partner of the cyclins Cln2 and Clb3 in another proteomic study (3).
The 14-3-3 family is highly conserved in eukaryotes. Bmh1 and Bmh2 share over 90% sequence identity with each other and 70% identity with the human epsilon 14-3-3 isoform (19). 14-3-3 proteins function primarily as homo- and heterodimers, interact with numerous proteins involved in diverse biological processes, and exhibit a strong preference for phosphorylated binding targets (54). Neither BMH1 nor BMH2 is an essential gene, but loss of both is lethal in most strain backgrounds. In many processes, they perform redundant functions. We predict that Bmh1 and Bmh2 homo- or heterodimers bind to one or more phosphorylation sites on Acm1, and we are currently investigating this possibility.
Bud neck localization and substrate binding.
One of the many reported functions of 14-3-3 proteins is the cytoplasmic sequestration of binding targets. Such regulation of cellular localization by the 14-3-3 family has been described for several proteins involved in cell cycle control (54). Our results suggest that Bmh1 and Bmh2 are not required for cytoplasmic localization of Cdh1. Instead, we found that the complex prevents localization of Cdh1 to the bud neck (Fig. 3). Several Cdh1 substrates also localize to the bud neck, including Clb2 and Hsl1, and we found that association of Cdh1 with these two substrates was strongly enhanced in the absence of Acm1 in S-phase-arrested cells. Cdh1 currently has no known function at the bud neck. It is possible that Cdh1 transiently localizes to the bud neck at a specific time during normal cell cycle progression or in response to cell cycle perturbations and that Acm1/Bmh1/Bmh2 regulates its localization and function there. Alternatively, it is possible that Acm1/Bmh1/Bmh2 merely acts to prevent Cdh1 localization to the bud neck during the cell cycle interval in which Cdh1 is located in the cytoplasm. Because the Acm1 protein may be specific to budding yeast species (based on our similarity searches), one can speculate that it might regulate a Cdh1-dependent function unique to budding yeast.
Based on the enhanced association of Cdh1 with Clb2 and Hsl1 in the absence of Acm1, we predict that Acm1 acts, at least in part, by preventing binding of specific substrates to Cdh1 similar to the mechanism demonstrated for the vertebrate Cdh1 and Cdc20 inhibitor Emi1 (41). The stable stoichiometric interaction between Acm1 and Cdh1 (Fig. 1) is consistent with this possibility. In addition, we found that expression of just the WD40 region of Cdh1, which has been shown to be the site of substrate binding (28), was sufficient for assembly of the intact complex with Acm1, Bmh1, and Bmh2 (our unpublished observations). A roughly 40-kDa Cdh1 proteolytic fragment that copurifies with 3HA-Acm1 (labeled in Fig. 1B) comprises the intact C-terminal WD40 domain. In vitro experiments will ultimately be required to study how this complex affects substrate binding to Cdh1.
Acm1 can negatively regulate APC/CCdh1 function in vivo.
We have presented several lines of evidence that strongly suggest that Acm1 is able to inhibit APC/CCdh1-dependent proteolysis of mitotic cyclins in vivo. Overexpression of Acm1 restored viability to cells expressing toxic levels of Cdh1 and the Cdh1-m11 mutant lacking inhibitory CDK phosphorylation sites. Lethality following high-level Cdh1 overexpression or in the presence of Cdh1-m11 has previously been attributed to inappropriate APC/C-mediated proteolysis of Clb2 (46, 56, 60), and we therefore conclude that rescue of this lethality by Acm1 indicates suppression of APC/CCdh1-mediated proteolysis of Clb2. Importantly, in the absence of CDK inhibition of Cdh1, Acm1 is capable of fulfilling the role of APC/CCdh1 inhibitor to allow continued cell cycle progression, suggesting overlapping or partly redundant functions. Synthetic lethality of _cdh1_Δ with _sic1_Δ has previously been attributed to an inability of cells to inactivate Clb2-Cdc28 activity at mitotic exit (46, 56), and our finding that Acm1 overexpression is toxic in a _sic1_Δ strain provides further evidence that Acm1 can suppress APC/CCdh1-dependent proteolysis of Clb2. Finally, this conclusion is supported by our observations that (i) _acm1_Δ exacerbates another phenotype (hyperpolarized growth) associated with deficient mitotic CDK activity in an APC/CCdh1 activity-dependent manner and (ii) ACM1 interacts genetically with HSL1, which encodes a positive regulator of mitotic CDK, resulting in synergistic hyperpolarization and G2/M cell cycle delay in the absence of both.
Whether Acm1/Bmh1/Bmh2 represents a general negative regulator of APC/CCdh1-catalyzed ubiquitination remains to be seen. In light of the data reported here, we favor a model in which Acm1 acts as a specialized inhibitor of mitotic cyclin proteolysis. This is consistent with our observation that single deletion of ACM1 to prevent formation of the complex has no significant effect on cell cycle progression and growth (Fig. 8 and data not shown). Thus, the complex does not appear to be required for regulation of APC/CCdh1 activity during standard laboratory growth but perhaps performs a more specialized function related to cell cycle progression under specific conditions. Interestingly, Surana and colleagues recently reported that in yeast unable to activate mitotic CDK by dephosphorylation of Cdc28 tyrosine 19, certain substrates of APC/CCdh1 remain sensitive to proteolysis whereas Clb2 is stable (14). Perhaps Acm1 contributes to this apparent difference in APC/CCdh1 substrate stability.
It is also apparent from our studies that absence of Acm1 does not result in massive depletion of Clb2, even when Cdh1 is overexpressed from the ADH promoter. This is in contrast to previous studies that demonstrated rapid loss of Clb2 following high-level galactose-induced Cdh1 expression (46, 56). We expected to find that Clb2 was lost or severely depleted in the _acm1_Δ _hsl1_Δ cells that showed the most severe hyperpolarization and growth defect (Fig. 7). Surprisingly, the Clb2 level was relatively high in these cells (not shown). This suggests that under these experimental conditions Cdh1 may be targeting only a subpopulation of Clb2 for proteolysis in the absence of Acm1 (e.g., the population located at the bud neck or in the cytoplasm) and this fraction of total cellular Clb2 is important for promoting isotropic bud growth and mitotic entry. It is important to note that Cdh1 is primarily cytoplasmic outside G1 phase and thus the nuclear population of Clb2 could be protected from Cdh1 during this time. Although the formal possibility exists that Cdh1 is simply interfering with Clb2-Cdc28 function by inappropriately binding to Clb2, our experiments with the cdh1_Δ_IR allele argue against it. The fact that the phenotypes observed when wild-type Cdh1 is overexpressed are greatly diminished with Cdh1ΔIR, which binds substrates but is unable to activate APC/C (10, 37, 58), suggests that APC/C-mediated proteolysis is required.
The significance of the enhanced interaction between Cdh1 and Hsl1 in _acm1_Δ cells is not clear. Inappropriate Hsl1 degradation in the absence of Acm1 seems unlikely because it should result in stabilization of Swe1, which would contribute to the elongated-bud phenotype. Instead, the effects we observed on bud elongation were clearly SWE1 independent. Perhaps the effects on Hsl1 are minimal and secondary compared to those on Clb2 and do not perturb the regulation of Swe1 enough to influence bud morphology. Like Clb2, Hsl1 was not significantly depleted in _acm1_Δ cells overexpressing Cdh1 (not shown).
What is the biological function of Acm1?
It is unclear why Cdh1 is not eliminated from cells after its G1 function is complete, especially since a convenient mechanism seems to already be in place to do so. Sic1 protein plays a redundant role with Cdh1 in promoting mitotic exit by stoichiometrically inhibiting mitotic CDK activity (48). Sic1 is phosphorylated on multiple sites in a G1 CDK-dependent manner in the following cell cycle (45), resulting in its recognition and polyubiquitination by the SCFCdc4 ubiquitin ligase (17) and consequent proteolysis. Cdh1 is inactivated around the same time as Sic1, also by multisite CDK phosphorylation (26, 60). However, it is not eliminated by proteolysis. Instead, it is exported to the cytoplasm and its level stabilizes (23, 25, 60). A similar expression pattern is found in higher eukaryotic cells (31). This persistence of Cdh1 after the G1/S transition suggests that it might be required for an as-yet-unidentified function during S, G2, and/or early M. Perhaps the association with Acm1, Bmh1, and Bmh2 stabilizes Cdh1 and maintains it in an inactive state following G1 exit such that APC/CCdh1 activity can be activated in response to a specific signal.
What type of signal would be expected to activate APC/CCdh1 in S phase or early M? The elongated-bud phenotypes observed when we overexpress Cdh1 in an _acm1_Δ background are reminiscent of the hyperpolarized cell morphology seen in pseudohyphal yeast. The switch from the yeast form to filamentous growth is believed to involve down-regulation of mitotic CDK activity that allows an extended polarized growth period, resulting in the highly elongated cells found in pseudohyphae (1). In fact, it has been proposed that a distinct inhibitor of Clb2-Cdc28 activity might exist to elicit this response (1). Components of the morphogenesis checkpoint, including Hsl1 and Swe1, are involved in controlling the switch to filamentous growth (15), and a role for APC/CCdh1 in this process is conceivable. Moreover, Bmh1 and Bmh2 have been implicated in the filamentous growth response as regulators of the Ste20 protein (43). Another possible function for Cdh1 could be down-regulation of Clb-Cdc28 activity in response to a checkpoint signal that occurs in S, G2, or early M and that contributes to cell cycle arrest. Other possibilities exist as well, including effects of Cdh1 on substrates other than Clb2. We are currently testing a variety of possibilities in an effort to define the biological significance of Acm1 and its association with Cdh1 and the biochemical mechanism by which it inhibits APC/CCdh1.
Supplementary Material
[Supplemental material]
Acknowledgments
This work was supported by an award from the American Heart Association to M.C.H. and by an American Cancer Society Institutional Research Grant to the Purdue Cancer Center.
We thank David Pellman for providing the Ase1 antibody. We thank members of the Purdue University Cytology Labs for assistance with flow cytometry analyses. We thank Ann Kirchmaier and Harry Charbonneau for providing reagents. We are also grateful to Harry Charbonneau for allowing us to use the Leica fluorescence microscope and for providing assistance with the acquisition and analysis of images. We thank Sandra Rossie for suggesting and providing reagents for the indirect immunofluorescence procedure with signal amplification. Finally, we thank Harry Charbonneau and Joe Ogas for helpful comments on the manuscript and during the course of this study.
Footnotes
▿
Published ahead of print on 9 October 2006.
§
Journal paper 17975 from the Purdue University Agricultural Experiment Station.
REFERENCES
- 1.Ahn, S. H., A. Acurio, and S. J. Kron. 1999. Regulation of G2/M progression by the STE mitogen-activated protein kinase pathway in budding yeast filamentous growth. Mol. Biol. Cell 10**:**3301-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amon, A. 2002. Synchronization procedures, p. 457-467. In C. Guthrie and G. R. Fink (ed.), Guide to yeast genetics and molecular and cell biology, vol. 351. Academic Press, San Diego, Calif. [Google Scholar]
- 3.Archambault, V., E. J. Chang, B. J. Drapkin, F. R. Cross, B. T. Chait, and M. P. Rout. 2004. Targeted proteomic study of the cyclin-Cdk module. Mol. Cell 14**:**699-711. [DOI] [PubMed] [Google Scholar]
- 4.Asano, S., J. E. Park, K. Sakchaisri, L. R. Yu, S. Song, P. Supavilai, T. D. Veenstra, and K. S. Lee. 2005. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J. 24**:**2194-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailly, E., S. Cabantous, D. Sondaz, A. Bernadac, and M. N. Simon. 2003. Differential cellular localization among mitotic cyclins from Saccharomyces cerevisiae: a new role for the axial budding protein Bud3 in targeting Clb2 to the mother-bud neck. J. Cell Sci. 116**:**4119-4130. [DOI] [PubMed] [Google Scholar]
- 6.Barral, Y., M. Parra, S. Bidlingmaier, and M. Snyder. 1999. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13**:**176-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14**:**115-132. [DOI] [PubMed] [Google Scholar]
- 8.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Burton, J. L., and M. J. Solomon. 2001. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 15**:**2381-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton, J. L., V. Tsakraklides, and M. J. Solomon. 2005. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol. Cell 18**:**533-542. [DOI] [PubMed] [Google Scholar]
- 11.Camasses, A., A. Bogdanova, A. Shevchenko, and W. Zachariae. 2003. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell 12**:**87-100. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J., and G. Fang. 2001. MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev. 15**:**1765-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen-Fix, O., J. M. Peters, M. W. Kirschner, and D. Koshland. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10**:**3081-3093. [DOI] [PubMed] [Google Scholar]
- 14.Crasta, K., P. Huang, G. Morgan, M. Winey, and U. Surana. 2006. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 25**:**2551-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgington, N. P., M. J. Blacketer, T. A. Bierwagen, and A. M. Myers. 1999. Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol. Cell. Biol. 19**:**1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eytan, E., Y. Moshe, I. Braunstein, and A. Hershko. 2006. Roles of the anaphase-promoting complex/cyclosome and of its activator Cdc20 in functional substrate binding. Proc. Natl. Acad. Sci. USA 103**:**2081-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldman, R. M., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91**:**221-230. [DOI] [PubMed] [Google Scholar]
- 18.Fitch, I., C. Dahmann, U. Surana, A. Amon, K. Nasmyth, L. Goetsch, B. Byers, and B. Futcher. 1992. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 3**:**805-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelperin, D., J. Weigle, K. Nelson, P. Roseboom, K. Irie, K. Matsumoto, and S. Lemmon. 1995. 14-3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92**:**11539-11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosskortenhaus, R., and F. Sprenger. 2002. Rca1 inhibits APC-Cdh1(Fzr) and is required to prevent cyclin degradation in G2. Dev. Cell 2**:**29-40. [DOI] [PubMed] [Google Scholar]
- 21.Hall, M. C., E. N. Warren, and C. H. Borchers. 2004. Multi-kinase phosphorylation of the APC/C activator Cdh1 revealed by mass spectrometry. Cell Cycle 3**:**1278-1284. [DOI] [PubMed] [Google Scholar]
- 22.Hood, J. K., W. W. Hwang, and P. A. Silver. 2001. The Saccharomyces cerevisiae cyclin Clb2p is targeted to multiple subcellular locations by cis- and trans-acting determinants. J. Cell Sci. 114**:**589-597. [DOI] [PubMed] [Google Scholar]
- 23.Huang, J. N., I. Park, E. Ellingson, L. E. Littlepage, and D. Pellman. 2001. Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J. Cell Biol. 154**:**85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irniger, S., and K. Nasmyth. 1997. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J. Cell Sci. 110(Pt. 13)**:**1523-1531. [DOI] [PubMed] [Google Scholar]
- 25.Jaquenoud, M., F. van Drogen, and M. Peter. 2002. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C(Cdh1). EMBO J. 21**:**6515-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaspersen, S. L., J. F. Charles, and D. O. Morgan. 1999. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol. 9**:**227-236. [DOI] [PubMed] [Google Scholar]
- 27.Kellogg, D. R. 2003. Wee1-dependent mechanisms required for coordination of cell growth and cell division. J. Cell Sci. 116**:**4883-4890. [DOI] [PubMed] [Google Scholar]
- 28.Kraft, C., H. C. Vodermaier, S. Maurer-Stroh, F. Eisenhaber, and J. M. Peters. 2005. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol. Cell 18**:**543-553. [DOI] [PubMed] [Google Scholar]
- 29.Kramer, E. R., C. Gieffers, G. Holzl, M. Hengstschlager, and J. M. Peters. 1998. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr. Biol. 8**:**1207-1210. [DOI] [PubMed] [Google Scholar]
- 30.Kramer, E. R., N. Scheuringer, A. V. Podtelejnikov, M. Mann, and J. M. Peters. 2000. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell 11**:**1555-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Listovsky, T., Y. S. Oren, Y. Yudkovsky, H. M. Mahbubani, A. M. Weiss, M. Lebendiker, and M. Brandeis. 2004. Mammalian Cdh1/Fzr mediates its own degradation. EMBO J. 23**:**1619-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longtine, M. S., C. L. Theesfeld, J. N. McMillan, E. Weaver, J. R. Pringle, and D. J. Lew. 2000. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20**:**4049-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma, X. J., Q. Lu, and M. Grunstein. 1996. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10**:**1327-1340. [DOI] [PubMed] [Google Scholar]
- 34.Mumberg, D., R. Müller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156**:**119-122. [DOI] [PubMed] [Google Scholar]
- 35.Muslin, A. J., and H. Xing. 2000. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 12**:**703-709. [DOI] [PubMed] [Google Scholar]
- 36.Passmore, L. A., and D. Barford. 2005. Coactivator functions in a stoichiometric complex with anaphase-promoting complex/cyclosome to mediate substrate recognition. EMBO Rep. 6**:**873-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Passmore, L. A., E. A. McCormack, S. W. Au, A. Paul, K. R. Willison, J. W. Harper, and D. Barford. 2003. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 22**:**786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfleger, C. M., E. Lee, and M. W. Kirschner. 2001. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 15**:**2396-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfleger, C. M., A. Salic, E. Lee, and M. W. Kirschner. 2001. Inhibition of Cdh1-APC by the MAD2-related protein MAD2L2: a novel mechanism for regulating Cdh1. Genes Dev. 15**:**1759-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prinz, S., E. S. Hwang, R. Visintin, and A. Amon. 1998. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr. Biol. 8**:**750-760. [DOI] [PubMed] [Google Scholar]
- 41.Reimann, J. D., B. E. Gardner, F. Margottin-Goguet, and P. K. Jackson. 2001. Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes Dev. 15**:**3278-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richardson, H., D. J. Lew, M. Henze, K. Sugimoto, and S. I. Reed. 1992. Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 6**:**2021-2034. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, R. L., H. U. Mosch, and G. R. Fink. 1997. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 89**:**1055-1065. [DOI] [PubMed] [Google Scholar]
- 44.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11**:**1265-1274. [DOI] [PubMed] [Google Scholar]
- 45.Schneider, B. L., Q. H. Yang, and A. B. Futcher. 1996. Linkage of replication to start by the Cdk inhibitor Sic1. Science 272**:**560-562. [DOI] [PubMed] [Google Scholar]
- 46.Schwab, M., A. S. Lutum, and W. Seufert. 1997. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90**:**683-693. [DOI] [PubMed] [Google Scholar]
- 47.Schwab, M., M. Neutzner, D. Mocker, and W. Seufert. 2001. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 20**:**5165-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwob, E., T. Bohm, M. D. Mendenhall, and K. Nasmyth. 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79**:**233-244. [DOI] [PubMed] [Google Scholar]
- 49.Shirayama, M., A. Toth, M. Galova, and K. Nasmyth. 1999. APCCdc20 promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 402**:**203-207. [DOI] [PubMed] [Google Scholar]
- 50.Shirayama, M., W. Zachariae, R. Ciosk, and K. Nasmyth. 1998. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17**:**1336-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9**:**3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sudo, T., Y. Ota, S. Kotani, M. Nakao, Y. Takami, S. Takeda, and H. Saya. 2001. Activation of Cdh1-dependent APC is required for G1 cell cycle arrest and DNA damage-induced G2 checkpoint in vertebrate cells. EMBO J. 20**:**6499-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ubersax, J. A., E. L. Woodbury, P. N. Quang, M. Paraz, J. D. Blethrow, K. Shah, K. M. Shokat, and D. O. Morgan. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425**:**859-864. [DOI] [PubMed] [Google Scholar]
- 54.van Hemert, M. J., H. Y. Steensma, and G. P. H. van Heusden. 2001. 14-3-3 proteins: key regulators of cell division, signaling and apoptosis. Bioessays 23**:**936-946. [DOI] [PubMed] [Google Scholar]
- 55.Visintin, R., K. Craig, E. S. Hwang, S. Prinz, M. Tyers, and A. Amon. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2**:**709-718. [DOI] [PubMed] [Google Scholar]
- 56.Visintin, R., S. Prinz, and A. Amon. 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278**:**460-463. [DOI] [PubMed] [Google Scholar]
- 57.Vodermaier, H. C. 2004. APC/C and SCF: controlling each other and the cell cycle. Curr. Biol. 14**:**R787-R796. [DOI] [PubMed] [Google Scholar]
- 58.Vodermaier, H. C., C. Gieffers, S. Maurer-Stroh, F. Eisenhaber, and J. M. Peters. 2003. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr. Biol. 13**:**1459-1468. [DOI] [PubMed] [Google Scholar]
- 59.Wäsch, R., and F. R. Cross. 2002. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418**:**556-562. [DOI] [PubMed] [Google Scholar]
- 60.Zachariae, W., M. Schwab, K. Nasmyth, and W. Seufert. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282**:**1721-1724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]