Analysis of High-Resolution HapMap of DTNBP1 (Dysbindin) Suggests No Consistency between Reported Common Variant Associations and Schizophrenia (original) (raw)
Abstract
DTNBP1 was first identified as a putative schizophrenia-susceptibility gene in Irish pedigrees, with a report of association to common genetic variation. Several replication studies have reported confirmation of an association to DTNBP1 in independent European samples; however, reported risk alleles and haplotypes appear to differ between studies, and comparison among studies has been confounded because different marker sets were employed by each group. To facilitate evaluation of existing evidence of association and further work, we supplemented the extensive genotype data, available through the International HapMap Project (HapMap), about DTNBP1 by specifically typing all associated single-nucleotide polymorphisms reported in each of the studies of the Centre d'Étude du Polymorphisme Humain (CEPH)–derived HapMap sample (CEU). Using this high-density reference map, we compared the putative disease-associated haplotype from each study and found that the association studies are inconsistent with regard to the identity of the disease-associated haplotype at DTNBP1. Specifically, all five “replication” studies define a positively associated haplotype that is different from the association originally reported. We further demonstrate that, in all six studies, the European-derived populations studied have haplotype patterns and frequencies that are consistent with HapMap CEU samples (and each other). Thus, it is unlikely that population differences are creating the inconsistency of the association studies. Evidence of association is, at present, equivocal and unsatisfactory. The new dense map of the region may be valuable in more-comprehensive follow-up studies.
Schizophrenia (SCZ [MIM 181500]) is a common psychiatric disorder, with a lifetime morbidity risk of 0.72%,1 that presents with psychotic symptoms (delusions and hallucinations), thought disorder, and deficit features described as “negative” symptoms. Although SCZ is highly heritable,2 the genetic etiology is complex, and identification of replicable susceptibility genes has proved difficult.3 A number of SCZ-susceptibility genes have now been reported (see the review by Owen et al.3). One of the most prominent of these genes is the dystrobrevin binding protein 1 gene (DTNBP1 [MIM 607145]) on 6p22.3, which encodes the dysbindin protein. DTNBP1 was first identified as an SCZ-susceptibility gene in SCZ-affected Irish pedigrees.4,5 These data were later reanalyzed, and the association signal was attributed to a haplotype of low frequency (0.058)—defined as G-G-A-A-T-G-C-G on the minus strand of SNPs rs3213207, rs1011313, rs2619528, rs2005976, rs760761, rs2619522, rs1018381, and _rs1474605_—at the locus (see table 4 in the work of van den Oord et al.6). Schwab and colleagues next reported association at DTNBP1 in sib pairs and parents-proband trios from Germany, Hungary, and Israel.7 In that study, the strongest evidence of association came from the most common haplotype across the “associated” region, as determined by van den Oord et al.6 Although not formally tested, the low-frequency haplotype that was associated in the study by van den Oord et al.6 shows evidence of undertransmission in the sample of Schwab et al.7 A series of subsequent studies also purported to replicate association between DTNBP1 and SCZ in family-based8,9 and population-based samples.10–14 In our “Material and Methods” section, we detail the association findings from these studies. To date, these association studies, either individually or in combination, have not identified the causal variant(s) at DTNBP1 that contribute to SCZ risk. However, there is some evidence to suggest that DTNBP1 expression is altered in the brain of patients with SCZ, although no consistent haplotype has been associated with these changes.15,16
A major difficulty in interpretation of the results from _DTNBP1_-association studies is that the same SNPs have not been genotyped in all studies, which precludes direct comparison of risk alleles and haplotypes. Furthermore, where positive findings have been reported using the same SNP, there are instances in which the associated allele differs among samples. This has been attributed to potential differences in the genetic architecture of the sampled populations.
Here, we sought to address these problems by using data generated as part of the International HapMap Project (HapMap).17 To make studies directly comparable, we genotyped all SNPs from the _DTNBP1_-association studies in the CEPH-derived trios employed by HapMap, and we made a composite haplotype map for this locus, to evaluate the similarity of SNP and haplotype frequencies in the populations that were sampled in the DTNBP1 studies. We have considered only data from studies of samples ascertained from populations of European ancestry. This allows appraisal of the degree of similarity of local linkage-disequilibrium (LD) structure at DTNBP1 among the different European samples and determination of whether this is a contributing factor to the differences observed in reported associations.
Material and Methods
SNP Selection for Genotyping and Analysis
SNPs were selected from three sources: previous association studies of DTNBP1, dbSNP, and phase II of HapMap.
Previously Associated SNPs
We identified all SNPs reported in six association studies of DTNBP1 and SCZ that used samples of European-derived ancestry (_n_=31 SNPs).5–7,9,10,12,18 Of those SNPs, 26 were genotyped in our lab, 3 SNPs (rs2619539, rs1474605, and rs1997679) had already been genotyped as part of phase I of HapMap, 1 marker (SNP N) was a rare 1-base insertion/deletion not associated with SCZ in the original study,18 and 1 SNP (SNP O) was rare and also was not associated with SCZ in the original paper.18 Of the associated SNPs that were genotyped in our lab, 12 (rs12524251, rs2005976, rs909706, rs2743852, rs742105, rs16876738, rs13198335, rs13198195, rs2619542, rs2619550, rs2619537, and rs12204704) were not available as part of the HapMap data release 19 (HapMap).
dbSNP
Before the availability of phase II of the HapMap data, we selected an additional 63 SNPs from across the DTNBP1 gene and 10 kb upstream and downstream from those included in dbSNP (see CHIP Bioinformatics Tools Web site). Thus, there was a total of 89 SNPs genotyped. These SNPs were genotyped in the CEPH-derived HapMap (CEU) trio samples (_n_=30 trios) used as part of the HapMap Project (Coriell Institute Cell Repository).
Phase II HapMap SNPs
Subsequently, all SNPs available from HapMap phase II (HapMap data release 19; phase II of the October 2005 National Center for Biotechnology Information [NCBI] build 34 assembly; dbSNP build 124) across the DTNBP1 gene and 10 kb upstream and downstream were selected. There was a total of 214 SNPs. Of those, 61 were monomorphic, 5 had genotyping call rates <90%, and 1 SNP had significant discrepancies with our genotyping; those 67 SNPs were not included in further analyses.
SNP Genotyping
Genotyping was performed by mass spectrometry as described elsewhere,19 with use of amplification and extension primers designed by Spectro-Designer software (Sequenom). For each SNP, genotype data that met the following quality-control metrics were considered for analysis: (1) >90% of DNA samples attempted for genotyping per SNP were obtained, (2) <1% of chromosomes from parents to probands had Mendelian inheritance errors, (3) parental alleles were in Hardy-Weinberg equilibrium, and (4) a minor-allele frequency of >1%. SNP primer sequences are listed in table 1.
Table 1. .
SNP Amplification and Extension-Primer Sequences
| Amplification Primer(5′→3′) | ||||
|---|---|---|---|---|
| SNP | Marker | Forward | Reverse | Included in HapMap Release #19 |
| 1 | rs1474605 | AGCGGATAACATAGTGTGGTATGTGAGTCC | AGCGGATAACAACCCCTTCCTCTTTGAAGC | Yes |
| 2 | rs11757499 | AGCGGATAACGGGTCAAGTTTAGCCCTAAC | AGCGGATAACGGCTCTGAGTTTCACATATC | Yes |
| 3 | rs1011313 | AGCGGATAACATTCACAGGCTACAGAATGG | AGCGGATAACGCCAAGTTACTGCACACAAG | Yes |
| 4 | rs1988856 | AGCGGATAACCGCCAACTGACTACTACTTC | AGCGGATAACCATTTTCTGCATCCTCCTGG | Yes |
| 5 | rs17470454 | AGCGGATAACCAGGGCTTTTTCTTCCCTAC | AGCGGATAACTTGGAACCTGGAGGGTAATC | Yes |
| 6 | rs1018382 | AGCGGATAACTGTGATCAGATAAGCTCCAG | AGCGGATAACGAACCTTTAGCACGCTGATG | Yes |
| 7 | rs12213676 | AGCGGATAACAGATCTAGGCCAAGGTTTCC | AGCGGATAACCAGCTTCCACATGCTGTTAG | No |
| 8 | rs12204704 | AGCGGATAACGCCAGTGAGGTAAGTAGCAC | AGCGGATAACTCACTGTTTTCATTGCTGGG | No |
| 9 | rs12203173 | AGCGGATAACAAGCAAGGACTGAGCTGATG | AGCGGATAACGTTCTCGATAAATGTTGCCC | Yes |
| 10 | rs2252470 | AGCGGATAACACGCACACACACCACAAAAC | AGCGGATAACGGAGAGCCAGACACTTAAAG | Yes |
| 11 | rs13217513 | AGCGGATAACGTAGTAGCCTAAAAGGTGTC | AGCGGATAACTGTCCAGGTTCCTTTCTGAG | Yes |
| 12 | rs2619553 | AGCGGATAACAGGTGTCAGTTCTTAGAGCC | AGCGGATAACGGGTCCTTGGTTATGGATAG | No |
| 13 | rs9296978 | AGCGGATAACTTGCCATGACTCTTCTTGGG | AGCGGATAACCCGCTCAAACTGTAGACAAG | Yes |
| 14 | rs2743867 | AGCGGATAACGTTGTTTGCTTAATACCACTC | AGCGGATAACGAGACTGCATTTTCTAAACAG | Yes |
| 15 | rs9370822 | AGCGGATAACACTCACACAGTGATGATGGG | AGCGGATAACCGGTTTTGAAAGGAACTGCC | Yes |
| 16 | rs2619535 | AGCGGATAACTAAAACTGTCCTTGCCCACC | AGCGGATAACGCCTAGACTTAATCCTAGAC | Yes |
| 17 | rs7760564 | AGCGGATAACTGAAAGTGCCTCTCAGGAAG | AGCGGATAACCTACTTCATCATCCTCTCGG | Yes |
| 18 | rs2619536 | AGCGGATAACAGAGAGGAACTATGGAGTGC | AGCGGATAACCTCAGTGGCTTTCAATGCAG | Yes |
| 19 | rs2743854 | AGCGGATAACAAGGGAGAGACAAGGCAAAC | AGCGGATAACCCACATATATCCATTGCTGAG | Yes |
| 20 | rs2743548 | AGCGGATAACCCACAAAAAGAAATCTTTGA | AGCGGATAACGCTCCATATGAATTCAACAG | No |
| 21 | rs909706 | AGCGGATAACGTCAAGTCAGTTTCCAAGGG | AGCGGATAACAGATCAGGGTAACCCTAAAC | No |
| 22 | rs9476837 | AGCGGATAACCTGGAAGCACACAGCATTTG | AGCGGATAACAACTTGGATGAACCTGGAGC | Yes |
| 23 | rs9476844 | AGCGGATAACCCTTTCCTAAGCCTAATTCC | AGCGGATAACATCTAATACGCCACAGTGCC | No |
| 24 | rs2743550 | AGCGGATAACGCGGTATAGAAAGAGAATGG | AGCGGATAACGAGTTTCCATAGTGTTCAGTG | Yes |
| 25 | rs4715986 | AGCGGATAACTGTTGGCTACAATATCTTGG | AGCGGATAACGTGGGAAGGTAAAGAGCTTG | No |
| 26 | rs2619533 | AGCGGATAACGAAGATCTTCGTCCTCATTG | AGCGGATAACTTCCACCTCCTCTACCTTTG | No |
| 27 | rs2619538 | AGCGGATAACTCACTGTTTTCATTGCTGGG | AGCGGATAACAGTGAGGTAAGTAGCACAAG | Yes |
| 28 | rs9476860 | AGCGGATAACAGTAAAACCTGGACTGCAAG | AGCGGATAACGTACTAATGAGTAATTTTGAGG | Yes |
| 29 | rs9464795 | AGCGGATAACTAACGGCATGGAGAGGCCTG | AGCGGATAACAGGCTCTCAGGCTTGAGGAC | Yes |
| 30 | rs734129 | AGCGGATAACCCAGGAAGAGGAAAGAACAG | AGCGGATAACGTGGCTCCTTCAATAAATAAG | Yes |
| 31 | rs9464807 | AGCGGATAACGACTCCTTTTCCATCTCCAG | AGCGGATAACCCTAATTCAGTTAGTGCTTTG | Yes |
| 32 | rs9476887 | AGCGGATAACCCGCCGAGGAAAGTAACGA | AGCGGATAACGGGACCTAAGTTACTTTGCG | No |
| 33 | rs9296981 | AGCGGATAACGGGACTATTCTGTACTGGAG | AGCGGATAACGATTACAAACACAAATTATGC | Yes |
| 34 | rs2056942 | AGCGGATAACCTCTCTATTAAAGATTAAGAGC | AGCGGATAACCACCTGAATTGTAAATATTG | Yes |
| 35 | rs2743858 | AGCGGATAACTCCTAAGTATTTTTTGATGC | AGCGGATAACCAGTTGTTTCTATATACTACC | No |
| 36 | rs1474588 | AGCGGATAACAGGGTTGCTGAAGGAAAGAC | AGCGGATAACAAGGAACAAGGAGGGATGAC | Yes |
| 37 | rs2743852 | AGCGGATAACTATAAGGAGCCAGACAAGGG | AGCGGATAACGTGTTCTTAGAAAATTCCAGG | No |
| 38 | rs3213207 | AGCGGATAACGTATTAGGGAACTTTTCTTTG | AGCGGATAACCTACCACTAACAACCAAAAAG | Yes |
| 39 | rs3829893 | AGCGGATAACCTCTACCTCCTCAAAACTCG | AGCGGATAACGAGGATTCTGACTTTTGAGG | Yes |
| 40 | rs742106 | AGCGGATAACCAAGGAGCAGACTCAAATGG | AGCGGATAACCCGGTAACTTTGGTGAGTTG | Yes |
| 41 | rs1018381 | AGCGGATAACGTAAATGAAACGTCATGCAGG | AGCGGATAACGAGTACTACAATGACTGCTG | Yes |
| 42 | rs742105 | AGCGGATAACCTCACTGCACCTTCAACCTC | AGCGGATAACGTGCATACCTGTAGTCCAAG | No |
| 43 | rs760761 | AGCGGATAACGGTCTTTTTAGATATAACATC | AGCGGATAACTTGACCAAGTCCATTGTGTC | Yes |
| 44 | rs12525702 | AGCGGATAACACCAGGTTTTAGGCACAAAG | AGCGGATAACAATCTCTACTGAGTAGAGGG | Yes |
| 45 | rs1047631 | AGCGGATAACGTTTACCGTCCTCACACTTT | AGCGGATAACGCCAGGTTGTTTTATAGAGG | Yes |
| 46 | rs2619528 | AGCGGATAACGGTACAGAGTTTCCATTTTGC | AGCGGATAACCATTCTTAAGCTTAGTAGTGC | Yes |
| 47 | rs885773 | AGCGGATAACACTGTTGCCTTCAGAACCAG | AGCGGATAACATTCAAACAGGCACTAGCCC | No |
| 48 | rs2619522 | AGCGGATAACGCTCTTATGTCTACCTTTCC | AGCGGATAACAATAGCTGGCAGAAGCAGTG | Yes |
| 49 | rs16876738 | AGCGGATAACAATTACACCAAACCCTGCCC | AGCGGATAACCAGCAAATCTGAGTAAGTCC | No |
| 50 | rs760666 | AGCGGATAACCCAGTGAGTACTCGTCATTC | AGCGGATAACAATAAGTACACTAAGGTGGG | Yes |
| 51 | rs2619537 | AGCGGATAACGAAGAACTGTCTGTGTTCCC | AGCGGATAACCTGGAACCTCCTTCTCTTTC | No |
| 52 | rs12527496 | AGCGGATAACAGACTTCCTTTCGTAAAGCC | AGCGGATAACCTACCACTAACAACCAAAAAG | Yes |
| 53 | rs12524251 | AGCGGATAACCAAAGGAAGTGAGGCTGAAG | AGCGGATAACTATCTGCTTAAGCCATCAGC | No |
| 54 | SNP_H-Cardiffa | AGCGGATAACGTTCCCTAATACATTTAGAA | AGCGGATAACGCCAGTTTCCTCAAAATTCC | No |
| 55 | rs2005976 | AGCGGATAACTGTCAGTCTTCAGGGAAACG | AGCGGATAACCAAAGTGCTGGGATTATAGG | No |
Of the 89 SNPs we genotyped, 36 were not included in the subsequent analyses (minor-allele frequency <1% [_n_=26]; genotyping call rate <90% [_n_=5]; Hardy-Weinberg violation [_n_=4]). One further SNP (rs1988856) was eliminated, since 35% of the genotypes differed from those in HapMap. (None of the 26 SNPs from the literature failed.) Of the remaining 53 SNPs, the average genotyping success rate was 98.9%. There were only two SNPs with genotyping success rates between 90% and 95%. Since a number of these SNPs were genotyped independently by HapMap, we were able to compare genotypes, to estimate a concordance rate of our genotyping with that of HapMap. A total of 3,059 SNP genotypes in 36 SNPs were available for comparison, in which there were 5 discordant genotypes (0.163%).
Production of a Joined Data Set
Genotypes from the 53 SNPs were joined with genotypes from phase II of HapMap. When a SNP was genotyped in our lab, it was used in the final analysis, to avoid errors in strandedness. A total of 166 SNPs were used to build the LD map and to determine tagging SNPs (tSNPs) for the entire region, with use of Haploview version 3.32,20 and the Tagger implementation therein.
Identification of Associated SNPs or Haplotypes in Published Studies of SCZ and DTNBP1
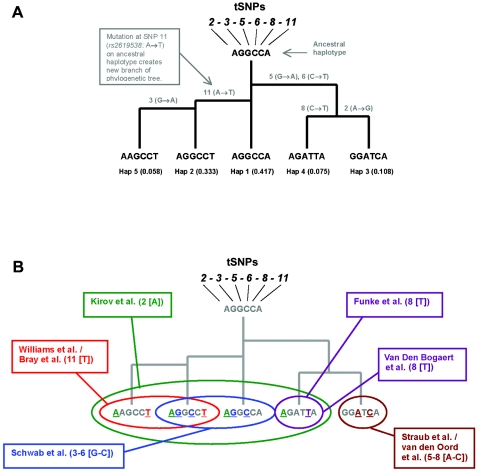
For this study, we concentrated on association studies of DTNBP1 and SCZ in European-derived samples (table 2). Within each study, we identified the single-marker or multimarker haplotype result that best captured the association signal in each sample. We paid particular attention to determining which SNPs tagged the associated haplotypes reported in the original study; this would later simplify the task of amalgamating independent findings for a common analysis. Across the six studies, only 11 SNPs were required to define all associated alleles or haplotypes (table 3). Associated haplotypes from each of the studies are shown in bold type in table 3, with tSNPs identified by shading. These tSNPs are shown in figure 1. For each of the studies, we identified the strongest evidence of association for the following alleles or haplotypes: Kirov et al.9 (study A) found strongest association, in their sample, with allele A of SNP 2 (rs3213207). The associated haplotype reported by Williams et al.18 (study B) in their United Kingdom and Ireland sample was later redefined by Bray et al.21 as A-A-T at SNPs 1-2-11. This haplotype can effectively be tagged by allele T at SNP 11 (rs2619538). Schwab et al.7 (study C) reported their strongest finding from the A-G-G-C-T-C haplotype at SNPs 2-3-4-6-7-8. This haplotype can be tagged by the G-C haplotype from SNPs 3–6 (rs1011313–rs760761). The data from the original study by Straub et al.5 (study D) was later reanalyzed by van den Oord et al.,6 who identified the strongest evidence of association as coming from the G-G-A-A-T-G-C-G haplotype at SNPs 2-3-4-5-6-7-8-9. This haplotype can be tagged by the A-C haplotype from SNPs 5–8 (rs2005976–rs1018381). Van Den Bogaert et al.10 (study E) reported their strongest finding with the A-G-A-T-T haplotype from SNPs 2-3-5-6-8. This haplotype can be tagged by allele T at SNP 8 (rs1018381). Funke et al.12 (study F) reported their strongest finding with the G-A-G-T-G-T-G haplotype from SNPs 2-3-4-5-6-7-9. This haplotype can be tagged by allele T at SNP 8 (rs1018381). Haplotype frequencies for table 3 in the CEU were calculated using Haploview.20 For these haplotype frequencies, the 95% CIs in the CEU were calculated using the method of Clopper and Pearson.22 A phylogenetic tree was derived for DTNBP1. A simple series of mutations, excluding back mutations, homoplasy (the same mutation occurring twice), or recombination explained the likely evolution of the haplotypes at this locus.
Table 2. .
Studies Reporting Association of DTNBP1 with SCZ in European-Derived Samples
| Study | Reference | Sample Size and Type | Sample Origin |
|---|---|---|---|
| A | Kirov et al.9 | 488 Parents-proband trios | Bulgaria |
| B | Williams et al.18; Bray et al.24 | 708 Cases and 711 controls | United Kingdom and Ireland |
| C | Schwab et al.7 | 78 Sib-pair families and 125 triads (affected proband and parents) | Germany, Hungary, and Israel |
| D | Straub et al.4,5; van den Oord et al.6 | 268 Multiplex families (1,405 individuals genotyped) | Ireland |
| E | Van Den Bogaert et al.10 | 418 German cases and 285 German controls; 142 Swedish cases and 272 Swedish controls; 294 Polish cases and 113 Polish controls | Germany, Sweden, and Poland |
| F | Funke at al.12 | 258 White cases and 467 white controls | United States |
Table 3. .
SNPs Reported at DTNBP1 That Contribute to Finding of Association in at Least One Study[Note]
| SNP Allele | Haplotype Frequency in | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | CEU (95% CI) | Original Study |
| dbSNP ID | rs1047631 | rs3213207 | rs1011313 | rs2619528 | rs2005976 | rs760761 | rs2619522 | rs1018381 | rs1474605 | rs909706 | rs2619538 | |
| Alternative name | … | P1635 | P1325 | P1765 | P1757 | P1320 | P1763 | P1578 | P1792 | P1583 | SNP A | |
| Chromosome 6 location | 15631080 | 15736081 | 15741411 | 15757808 | 15758781 | 15759111 | 15761628 | 15765049 | 15766191 | 15768850 | 15773188 | |
| Allelesa | A/G | A/G | A/G | A/G | A/G | C/T | G/T | C/T | A/G | A/G | A/T | |
| Study: | ||||||||||||
| A | .881 (.812–.935) | .895b | ||||||||||
| A (Kirov et al.9) | G | .119 (.065–.188) | .105 | |||||||||
| A | A | A | .398 (.312–.493) | .438c | ||||||||
| A | A | T | .392 (.304–.485) | .404 | ||||||||
| G | G | A | .108 (.059–.178) | .079 | ||||||||
| B (Bray et al.24) | G | A | A | .093 (.047–.158) | .043 | |||||||
| A | G | G | C | T | C | .75 (.663–.824) | .738d | |||||
| G | G | A | T | G | C | .108 (.059–.178) | .090 | |||||
| A | G | A | T | G | T | .075 (.035–.138) | .071 | |||||
| C (Schwab et al.7) | A | A | G | C | T | C | .058 (.024–.116) | .061 | ||||
| A | G | G | G | C | T | C | A | .750 (.663–.824) | .733e | |||
| G | G | A | A | T | G | C | G | .108 (.059–.178) | .058 | |||
| A | G | A | A | T | G | T | G | .075 (.035–.138) | .06 | |||
| A | A | G | G | C | T | C | A | .058 (.024–.116) | .071 | |||
| D (van den Oord et al.6) | G | A | G | G | T | T | C | G | .008 (.000–.046) | .015 | ||
| A | G | G | C | C | .750 (.663–.824) | .753f | ||||||
| G | G | A | T | C | .108 (.059–.178) | .094 | ||||||
| A | G | A | T | T | .075 (.035–.138) | .031 | ||||||
| E (Van Den Bogaert et al.10) | A | A | G | C | C | .058 (.024–.116) | .060 | |||||
| A | G | G | C | T | C | G | .450 (.359–.543) | .499c | ||||
| A | G | G | C | T | C | A | .300 (.220–.390) | .266 | ||||
| G | G | A | T | G | C | G | .110 (.059–.178) | .082 | ||||
| A | G | A | T | G | T | G | .075 (.035–.138) | .071 | ||||
| F (Funke et al.12) | A | A | A | C | T | C | A | .058 (.024–.116) | .065 |
Figure 1. .
A, Phylogenetic tree detailing the likely evolution of the five common haplotypes derived from tSNPs 2, 3, 5, 6, 8, and 11 (see table 3) in the CEU sample. Haplotype frequencies are shown at the bottom of the tree. Mutational events are detailed on the horizontal lines of the tree. The ancestral haplotype remains the most common haplotype in the CEU sample. Hap = haplotype. B, Each associated allele or haplotype from the six association studies of DTNBP1 and SCZ, mapped onto the phylogenetic tree. tSNPs are shown in parentheses, and haplotypes are shown in brackets.
Structure of LD at DTNBP1
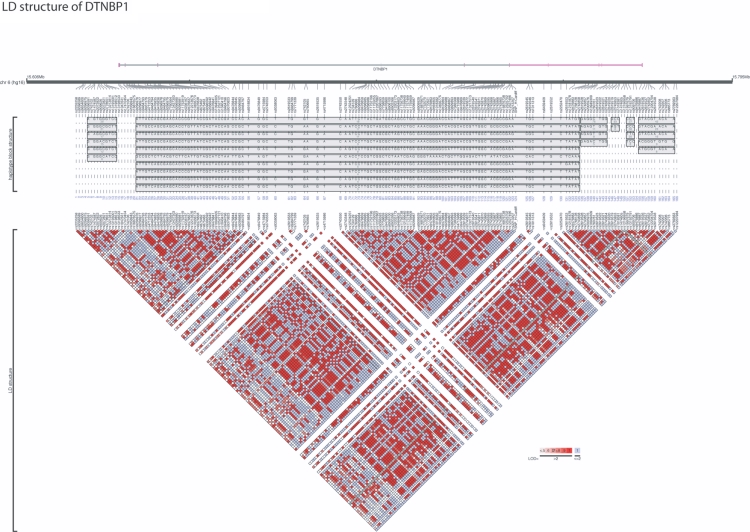
With use of the joined set of markers consisting of the three original categories, an LD map was constructed using Haploview.20 Markers were used to construct an LD map from the 172 SNPs that were present in allele frequencies >1%. A map of haplotypes present at frequency >2% is shown in figure 2. Graphic representation of the LD structure (fig. 2) was created by LocusView version 2.0. To construct a common set of markers that could be used to investigate the full haplotype structure of DTNBP1, we used the program Tagger23 (Haploview). Tagger employs both pairwise and effective (multimarker) haplotype predictors to capture alleles of interest. We used an _r_2 threshold of 0.8, a LOD score of 3.0, and the “pairwise” option for tSNP selection. A list of tSNPs is shown in table 4.
Figure 2. .
LD and haplotype-block structure of the DTNBP1 gene. Gene structure is seen, with vertical lines indicating exons. Markers are displayed relative to gene location. SNPs and their positions are given in table 4. LD structure (_D′_’) between marker pairs is indicated by the colored matrices. Haplotype blocks spanning the DTNBP1 gene are shown according to the method of Gabriel et al.25 Genomic positions are according to the NCBI Build 34, hg16 human genome assembly. The figure was generated using LocusView version 2.0.
Table 4. .
SNPs and tSNPs in LD Map
| Figure 2 Reference | SNP | Hg16 Position | tSNP |
|---|---|---|---|
| 1 | rs2235258 | 15621461 | Yes |
| 2 | rs9396589 | 15621723 | |
| 3 | rs9654600 | 15622092 | |
| 4 | rs9296975 | 15623486 | Yes |
| 5 | rs9396591 | 15624264 | |
| 6 | rs742102 | 15624612 | Yes |
| 7 | rs1474587 | 15624688 | Yes |
| 8 | rs2072822 | 15625638 | Yes |
| 9 | rs909626 | 15625663 | Yes |
| 10 | rs2072821 | 15626015 | |
| 11 | rs3778651 | 15626511 | |
| 12 | rs13213814 | 15627355 | |
| 13 | rs13195001 | 15627382 | |
| 14 | rs13198512 | 15627864 | Yes |
| 15 | rs13198533 | 15627893 | |
| 16 | rs1047631 | 15631080 | Yes |
| 17 | rs17470454 | 15631427 | Yes |
| 18 | rs742106 | 15632459 | Yes |
| 19 | rs2056943 | 15632542 | Yes |
| 20 | rs16876575 | 15633136 | |
| 21 | rs9296976 | 15633432 | |
| 22 | rs9296977 | 15633471 | |
| 23 | rs9464793 | 15633842 | |
| 24 | rs6937379 | 15633976 | |
| 25 | rs4712253 | 15634396 | Yes |
| 26 | rs9464794 | 15634758 | |
| 27 | rs9476835 | 15635176 | |
| 28 | rs9476836 | 15635291 | |
| 29 | rs9296978 | 15636346 | |
| 30 | rs9464795 | 15638143 | |
| 31 | rs9296979 | 15638763 | |
| 32 | rs7760564 | 15638887 | Yes |
| 33 | rs13437303 | 15639583 | |
| 34 | rs9296980 | 15640573 | Yes |
| 35 | rs9296981 | 15640879 | |
| 36 | rs11753919 | 15641712 | |
| 37 | rs4236167 | 15641930 | |
| 38 | rs9476837 | 15642249 | |
| 39 | rs12213676 | 15644761 | |
| 40 | rs10456773 | 15645304 | |
| 41 | rs9476838 | 15646186 | |
| 42 | rs875462 | 15646415 | |
| 43 | rs875463 | 15646664 | |
| 44 | rs9396592 | 15646989 | |
| 45 | rs2056942 | 15650277 | |
| 46 | rs9476841 | 15651323 | |
| 47 | rs10949305 | 15653842 | |
| 48 | rs12527121 | 15654192 | Yes |
| 49 | rs1040410 | 15655455 | |
| 50 | rs9464796 | 15657743 | |
| 51 | rs9370823 | 15658637 | Yes |
| 52 | rs9476844 | 15661998 | |
| 53 | rs9476845 | 15662074 | |
| 54 | rs9296983 | 15663405 | |
| 55 | rs9464797 | 15664265 | |
| 56 | rs6918834 | 15665818 | |
| 57 | rs9476849 | 15668562 | |
| 58 | rs4715984 | 15669870 | |
| 59 | rs2743553 | 15670729 | |
| 60 | rs9358063 | 15673010 | |
| 61 | rs2619533 | 15676872 | |
| 62 | rs9464799 | 15677538 | |
| 63 | rs7771339 | 15677985 | Yes |
| 64 | rs742105 | 15681053 | |
| 65 | rs760665 | 15681329 | |
| 66 | rs2619535 | 15684274 | |
| 67 | rs4715986 | 15686104 | |
| 68 | rs2743550 | 15690386 | |
| 69 | rs2743548 | 15691803 | |
| 70 | rs12524251 | 15694111 | |
| 71 | rs734129 | 15696990 | |
| 72 | rs760666 | 15697100 | |
| 73 | rs9296984 | 15697285 | |
| 74 | rs9476859 | 15697387 | |
| 75 | rs9296985 | 15697994 | |
| 76 | rs9296986 | 15698067 | |
| 77 | rs11752196 | 15698631 | |
| 78 | rs11755055 | 15698664 | |
| 79 | rs12207867 | 15699482 | |
| 80 | rs9396593 | 15700538 | |
| 81 | rs7758659 | 15701219 | |
| 82 | rs9476860 | 15702040 | |
| 83 | rs6906100 | 15702998 | |
| 84 | rs6903266 | 15704971 | |
| 85 | rs12203173 | 15705548 | |
| 86 | rs9464803 | 15705716 | |
| 87 | rs11756738 | 15706140 | |
| 88 | rs12199640 | 15706859 | Yes |
| 89 | rs11757499 | 15707709 | |
| 90 | rs11759609 | 15707878 | |
| 91 | rs9296987 | 15708794 | |
| 92 | rs6909929 | 15712434 | |
| 93 | rs16876671 | 15712574 | |
| 94 | rs7752070 | 15712898 | Yes |
| 95 | rs7770921 | 15713093 | |
| 96 | rs9476863 | 15713948 | |
| 97 | rs9464805 | 15714212 | |
| 98 | rs1018382 | 15715876 | |
| 99 | rs10456775 | 15717091 | |
| 100 | rs9476864 | 15719806 | |
| 101 | rs9296988 | 15720490 | |
| 102 | rs9464807 | 15722896 | |
| 103 | rs3829893 | 15723616 | Yes |
| 104 | rs7383568 | 15725911 | |
| 105 | rs4715988 | 15726088 | Yes |
| 106 | rs9476869 | 15726501 | |
| 107 | rs2619539 | 15728834 | |
| 108 | rs2743872 | 15730592 | |
| 109 | rs13217513 | 15731386 | |
| 110 | rs2619540 | 15731803 | |
| 111 | rs2619541 | 15731919 | |
| 112 | rs2743871 | 15732565 | |
| 113 | rs2743870 | 15732741 | |
| 114 | rs2743869 | 15733463 | |
| 115 | rs9396595 | 15733478 | Yes |
| 116 | rs2743868 | 15733787 | |
| 117 | rs2743867 | 15733865 | |
| 118 | rs2743866 | 15734194 | |
| 119 | rs2743865 | 15734282 | |
| 120 | rs16876738 | 15735532 | Yes |
| 121 | rs12525702 | 15735750 | Yes |
| 122 | rs12527496 | 15736060 | |
| 123 | rs3213207 | 15736081 | Yes |
| 124 | SNP_H-Cardiffa | 15736141 | Yes |
| 125 | rs2619545 | 15740720 | |
| 126 | rs2619546 | 15740792 | |
| 127 | rs1011313 | 15741411 | Yes |
| 128 | rs6459409 | 15744832 | |
| 129 | rs2619552 | 15746774 | |
| 130 | rs2252470 | 15749400 | |
| 131 | rs2619553 | 15750885 | |
| 132 | rs12181878 | 15753025 | |
| 133 | rs9476883 | 15755970 | |
| 134 | rs7768128 | 15756679 | |
| 135 | rs2743858 | 15756820 | |
| 136 | rs2619528 | 15757808 | Yes |
| 137 | rs2743857 | 15758475 | |
| 138 | rs2005976 | 15758781 | Yes |
| 139 | rs10949309 | 15758863 | |
| 140 | rs760761 | 15759111 | Yes |
| 141 | rs2619523 | 15761412 | |
| 142 | rs2619522 | 15761628 | Yes |
| 143 | rs2619521 | 15762440 | |
| 144 | rs2743854 | 15763248 | |
| 145 | rs2619520 | 15764134 | |
| 146 | rs2619519 | 15764181 | |
| 147 | rs2743853 | 15764769 | |
| 148 | rs1018381 | 15765049 | Yes |
| 149 | rs1474605 | 15766191 | Yes |
| 150 | rs12196958 | 15766584 | |
| 151 | rs1997679 | 15766884 | Yes |
| 152 | rs13192791 | 15767518 | |
| 153 | rs909706 | 15768850 | Yes |
| 154 | rs9476886 | 15769440 | Yes |
| 155 | rs9476887 | 15770870 | Yes |
| 156 | rs2619536 | 15771826 | |
| 157 | rs2619537 | 15772392 | |
| 158 | rs2743852 | 15772743 | |
| 159 | rs12204704 | 15773184 | Yes |
| 160 | rs2619538 | 15773188 | Yes |
| 161 | rs742208 | 15776640 | |
| 162 | rs742207 | 15776846 | |
| 163 | rs742206 | 15777277 | |
| 164 | rs885773 | 15777465 | Yes |
| 165 | rs2769563 | 15779710 | Yes |
| 166 | rs12207984 | 15780272 | Yes |
Results
Comparison of the CEU Allele and Haplotype Frequencies for All Associated SNPs in the Literature
The sample size and population origin for association studies of DTNBP1 are presented in table 2. There are six studies of the association of DTNBP1 and SCZ that have used samples of European ancestry. In general, there was not a common set of SNPs genotyped uniformly across all studies, which makes it impossible to make direct comparisons of allele frequencies between or among the studies. This literature contains 11 SNPs that contribute to the finding of either a SNP or haplotype association in at least one study (table 3). The allele frequencies of these 11 SNPs were determined in the CEU samples that were used as part of the International HapMap Project. Across all six studies of DTNBP1 and SCZ, the haplotypes reported in each study sample are present in the CEU sample and with broadly similar frequencies. This suggests that the LD structure is very similar across all study samples at this locus.
Mapping of Associated Markers onto a Common Framework Map
It was apparent that a smaller set of markers could be used to tag the associated haplotype in each study. For each study, table 3 shows the haplotypes in bold type, and the tSNP(s) for each study are shaded. With the defined associated alleles or haplotypes from each sample, we then mapped each of these associated alleles or haplotypes onto the CEU sample as a reference, to examine all of the studies together. For this analysis, we concentrated on the tSNPs from each study, as defined above (SNPs 2, 3, 5, 6, 7, and 11). From these six SNPs, we identified five common haplotypes in the CEU trios. A simple phylogenetic tree explains the likely evolution of these haplotypes from an ancestral haplotype. Figure 1_A_ displays this tree and identifies the five common CEU haplotypes and their respective frequencies. The ancestral haplotype remains the most common haplotype.
Comparison of Association Results through Mapping onto a Common Framework Map
We were then able to map the associated allele or haplotype from each study onto the phylogenetic tree (fig. 1_B_). The associated allele from the study of Kirov et al.9 (allele A of SNP 2) maps onto haplotypes 1, 2, 4, and 5 in the CEU data. The allele that tags the associated haplotype in the studies of Williams et al.11,24 (allele T at SNP 11) maps onto haplotypes 2 and 5. The associated haplotype from the study of Schwab et al.7 (captured by the haplotype G-C at SNPs 3–6) maps onto haplotypes 1 and 2. The associated haplotype from the study of van den Oord et al.6 (captured by the haplotype A-C at SNPs 5–8) maps onto haplotype 3. The strongest association signals from the studies of both Van Den Bogaert et al.10 and Funke et al.12 are tagged by allele T at SNP 8; this maps onto haplotype 4.
Construction of a Dense LD Map of DTNBP1
We concentrated our examination on the 160-kb region on chromosome 6 (nucleotide positions 15621461–15780272) that contains the DTNBP1 gene (fig. 2), including 10–15 kb upstream and downstream. With use of our joined data set, there are 166 genotyped SNPs in this region that have a minor-allele frequency of >1%, for an average density of 1 SNP per 962 bases. The largest gap between SNPs is 4.58 kb. There are 10 gaps >3 kb. To provide a visualization of this region, we used the block definition that defines strong LD as in the work of Gabriel et al.,25 and we found that this region comprises six blocks. Most of the blocks are small; however, there is one large block, spanning 123 kb, that includes exons 4–7. The LD between pairs of blocks is very high; for example, it is 0.89 between blocks 1 and 2 and 1.00 between blocks 2 and 3, 3 and 4, and 4 and 5. We used the program Tagger (Haploview) to select a reduced set of tSNPs for this region. Tagger (Haploview) does not explicitly use haplotype blocks for tagging; rather, it selects a reduced set of SNPs on the basis of its ability to represent variation present at the larger set of SNPs under preset conditions. Because the 11 SNPs reported in the present study have already been used by many researchers, Tagger (Haploview) has included all of these SNPs. With use of pairwise tagging, a total of 42 SNPs are needed to capture 100% of alleles with _r_2>0.8 (mean _r_2=0.975) across the region covering DTNBP1 (table 4).
Discussion
In the human genome, >9 million SNPs have been reported,26 which yields an abundance of markers to use to scan the human genome for disease mutations. Since many of these SNPs have a high degree of allelic association with each other, a reduced set of variation can be used to capture, or tag, genetic variation across a particular gene. However, since many tSNPs are equivalent for this purpose, individual investigators have not always chosen the same markers, so the same small representative set of SNPs has not been used consistently in all studies of a given gene. This is the case for DTNBP1, for which studies have reported association with SCZ in samples of European-derived ancestry.5–7,9,10,12,18 We focused on these studies and used the CEU trios from HapMap, so that alleles and haplotypes could be mapped in samples of similar ancestry. Using this map, we evaluated the evidence in support of DTNBP1 as an SCZ-susceptibility gene and found that evidence for DTNBP1 is equivocal.
For each of the six SCZ-association studies of DTNBP1, the SNPs or haplotypes are of similar frequency in the association samples and in the CEU sample—this suggests that each European-derived sample is genetically similar and that population stratification cannot explain differences in published results. It was not possible to study this topic in greater detail, because LD measurements were not routinely published for all analyzed SNPs in each study. When each DTNBP1 study result is reduced to a tSNP or haplotype that defines the strongest association signal studied in that sample, it is possible to map all results onto the common haplotypes that are defined by the same tSNPs in the CEU sample. The simple phylogenetic tree explains the evolution of these haplotypes. The phylogeny of haplotypes was examined by van den Oord et al.6 Although our phylogenetic tree was not determined by computer software, it is essentially identical to the phylogram of haplotypes from Irish pedigrees that is presented in figure 1 of the work by van den Oord et al.6 In both their study and ours, the ancestral haplotype is the most common haplotype (van den Oord and colleagues6 did not genotype rs2619538 [SNP 11], which differentiates haplotypes 1 and 2 in our study). However, there are two low-frequency (<2%) haplotypes in the Irish sample that were not associated with SCZ that are not detected in the CEU sample. Haplotype frequencies are very similar in both our studies and the study by van den Oord et al.6 Frequencies differ by no more than 1.5%, with the exception of the Irish-associated haplotype (haplotype 2 in the work of van den Oord et al.6), which has a frequency of 5.8% in the Irish families, compared with a frequency of 10.8% in the CEU sample (haplotype 3 in the present study).
We have demonstrated that each common DTNBP1 haplotype is tagged by the association signal of at least one study, which implies that there is not one common causal variant that is contributing to SCZ risk at the DTNBP1 locus. There are a number of caveats to be aware of in interpreting the results of the present study. First, this study does not represent a true meta-analysis, since we did not have access to raw genotypes from each completed study of DTNBP1. We could rely only on the published findings of each study, and we concentrated specifically on only one association result from each study. Second, it is possible that only some of the studies have produced false-positive results but that there is some agreement among studies. This is not something that can be evaluated given the current data. Third, differences in environmental factors, ascertainment schemes, and diagnosis could lead to different results. Furthermore, the samples were of varying size, type, and degrees of statistical power as replication samples. Our study does not rule out the possibility of an appreciable number of rare variants that have arisen on multiple common backgrounds. However, the spectrum of rare penetrant variation would not be expected to result in highly significant associations with multiple common haplotypes. Therefore, in the absence of a common causal variant and without raw genotypic and phenotypic data from these large samples, it is impossible to tease out subtle influence on the SCZ phenotype of genetic variants at DTNBP1.
All studies (European-derived populations) had allele or haplotype frequencies compatible with the HapMap CEU sample. The present study has shown the utility of HapMap in successfully relating association studies that have used diverse marker sets and, furthermore, emphasizes the utility of testing a common set of SNPs, to enable direct comparisons between or among association studies. Because we find that all of the association samples of European-derived ancestry have a similar genetic structure, the conflicting results among studies cannot simply be attributed to population differences. This calls into question the interpretation of the replication studies at this locus. Although a large number of studies have reported association of DTNBP1 with SCZ, it is important to unambiguously confirm this association and to identify the risk alleles. We have produced a dense genetic map and a list of tSNPs of the region that can now be used for large-scale association studies, to help determine how DTNBP1 contributes to SCZ susceptibility.
Acknowledgments
We are grateful to Andrew Kirby for merging our genotyping data with HapMap data. We also thank Tracey Petryshen, Jinbo Fan, Annette Taberner, Lauren Weiss, and Paul de Bakker, for enlightening discussion and suggestions. We thank the International HapMap Project for providing access to raw genotype data. D.W.M. was supported by a fellowship from the Health Research Board of Ireland. Funding for this work was provided by The Heinz C. Prechter Fund for Manic Depression.
Web Resources
The URLs for data presented herein are as follows:
- CHIP Bioinformatics Tools, http://snpper.chip.org/
- Coriell Institute Cell Repository, http://ccr.coriell.org/nigms/products/hapmap.html
- dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/
- Haploview, http://www.broad.mit.edu/mpg/haploview/ (for Tagger)
- International HapMap Project, http://www.hapmap.org/
- LocusView, http://www.broad.mit.edu/mpg/locusview/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SCZ and DTNBP1)
References
- 1.Saha S, Chant D, Welham J, McGrath J (2005) A systematic review of the prevalence of schizophrenia. PLoS Med 2:e141 10.1371/journal.pmed.0020141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardno AG, Gottesman, II (2000) Twin studies of schizophrenia: from bow-and-arrow concordances to Star Wars Mx and functional genomics. Am J Med Genet 97:12–17 [DOI] [PubMed] [Google Scholar]
- 3.Owen MJ, Craddock N, O’Donovan MC (2005) Schizophrenia: genes at last? Trends Genet 21:518–525 10.1016/j.tig.2005.06.011 [DOI] [PubMed] [Google Scholar]
- 4.Straub RE, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, O’Neill FA, Walsh D, Kendler KS (2002) Genome-wide scans of three independent sets of 90 Irish multiplex schizophrenia families and follow-up of selected regions in all families provides evidence for multiple susceptibility genes. Mol Psychiatry 7:542–559 10.1038/sj.mp.4001051 [DOI] [PubMed] [Google Scholar]
- 5.Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O’Neill FA, Walsh D, Kendler KS (2002) Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet 71:337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Oord EJ, Sullivan PF, Jiang Y, Walsh D, O’Neill FA, Kendler KS, Riley BP (2003) Identification of a high-risk haplotype for the dystrobrevin binding protein 1 (DTNBP1) gene in the Irish study of high-density schizophrenia families. Mol Psychiatry 8:499–510 10.1038/sj.mp.4001263 [DOI] [PubMed] [Google Scholar]
- 7.Schwab SG, Knapp M, Mondabon S, Hallmayer J, Borrmann-Hassenbach M, Albus M, Lerer B, Rietschel M, Trixler M, Maier W, Wildenauer DB (2003) Support for association of schizophrenia with genetic variation in the 6p22.3 gene, dysbindin, in sib-pair families with linkage and in an additional sample of triad families. Am J Hum Genet 72:185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang JX, Zhou J, Fan JB, Li XW, Shi YY, Gu NF, Feng GY, Xing YL, Shi JG, He L (2003) Family-based association study of DTNBP1 in 6p22.3 and schizophrenia. Mol Psychiatry 8:717–718 10.1038/sj.mp.4001287 [DOI] [PubMed] [Google Scholar]
- 9.Kirov G, Ivanov D, Williams NM, Preece A, Nikolov I, Milev R, Koleva S, Dimitrova A, Toncheva D, O’Donovan MC, Owen MJ (2004) Strong evidence for association between the dystrobrevin binding protein 1 gene (DTNBP1) and schizophrenia in 488 parent-offspring trios from Bulgaria. Biol Psychiatry 55:971–975 10.1016/j.biopsych.2004.01.025 [DOI] [PubMed] [Google Scholar]
- 10.Van Den Bogaert A, Schumacher J, Schulze TG, Otte AC, Ohlraun S, Kovalenko S, Becker T, Freudenberg J, Jönsson EG, Mattila-Evenden M, Sedvall GC, Czerski PM, Kapelski P, Hauser J, Maier W, Rietschel M, Propping P, Nöthen MM, Cichon S (2003) The DTNBP1 (dysbindin) gene contributes to schizophrenia, depending on family history of the disease. Am J Hum Genet 73:1438–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams CC, Allison JG, Vidal GA, Burow ME, Beckman BS, Marrero L, Jones FE (2004) The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J Cell Biol 167:469–478 10.1083/jcb.200403155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funke B, Finn CT, Plocik AM, Lake S, DeRosse P, Kane JM, Kucherlapati R, Malhotra AK (2004) Association of the DTNBP1 locus with schizophrenia in a US population. Am J Hum Genet 75:891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Numakawa T, Yagasaki Y, Ishimoto T, Okada T, Suzuki T, Iwata N, Ozaki N, Taguchi T, Tatsumi M, Kamijima K, Straub RE, Weinberger DR, Kunugi H, Hashimoto R (2004) Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Hum Mol Genet 13:2699–2708 10.1093/hmg/ddh280 [DOI] [PubMed] [Google Scholar]
- 14.Li T, Stefansson H, Gudfinnsson E, Cai G, Liu X, Murray RM, Steinthorsdottir V, Januel D, Gudnadottir VG, Petursson H, Ingason A, Gulcher JR, Stefansson K, Collier DA (2004) Identification of a novel neuregulin 1 at-risk haplotype in Han schizophrenia Chinese patients, but no association with the Icelandic/Scottish risk haplotype. Mol Psychiatry 9:698–704 [DOI] [PubMed] [Google Scholar]
- 15.Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, Smith RJ, Hahn CG, Siegel SJ, Trojanowski JQ, Gur RE, Blake DJ, Arnold SE (2004) Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest 113:1353–1363 10.1172/JCI200420425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weickert CS, Straub RE, McClintock BW, Matsumoto M, Hashimoto R, Hyde TM, Herman MM, Weinberger DR, Kleinman JE (2004) Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch Gen Psychiatry 61:544–555 10.1001/archpsyc.61.6.544 [DOI] [PubMed] [Google Scholar]
- 17.The International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796 10.1038/nature02168 [DOI] [PubMed] [Google Scholar]
- 18.Williams NM, Preece A, Morris DW, Spurlock G, Bray NJ, Stephens M, Norton N, Williams H, Clement M, Dwyer S, Curran C, Wilkinson J, Moskvina V, Waddington JL, Gill M, Corvin AP, Zammit S, Kirov G, Owen MJ, O’Donovan MC (2004) Identification in 2 independent samples of a novel schizophrenia risk haplotype of the dystrobrevin binding protein gene (DTNBP1). Arch Gen Psychiatry 61:336–344 10.1001/archpsyc.61.4.336 [DOI] [PubMed] [Google Scholar]
- 19.Sklar P, Gabriel SB, McInnis MG, Bennett P, Lim YM, Tsan G, Schaffner S, Kirov G, Jones I, Owen M, Craddock N, DePaulo JR, Lander ES (2002) Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. Mol Psychiatry 7:579–593 10.1038/sj.mp.4001058 [DOI] [PubMed] [Google Scholar]
- 20.Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 24.Bray NJ, Preece A, Williams NM, Moskvina V, Buckland PR, Owen MJ, O’Donovan MC (2005) Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Hum Mol Genet 14:1947–1954 10.1093/hmg/ddi199 [DOI] [PubMed] [Google Scholar]
- 21.Bray NJ, Buckland PR, Owen MJ, O’Donovan MC (2003) Cis-acting variation in the expression of a high proportion of genes in human brain. Hum Genet 113:149–153 [DOI] [PubMed] [Google Scholar]
- 22.Clopper C, Pearson S (1934) The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26:404–413 10.2307/2331986 [DOI] [Google Scholar]
- 23.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D (2005) Efficiency and power in genetic association studies. Nat Genet 37:1217–1223 10.1038/ng1669 [DOI] [PubMed] [Google Scholar]
- 25.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- 26.Crawford DC, Nickerson DA (2005) Definition and clinical importance of haplotypes. Annu Rev Med 56:303–320 10.1146/annurev.med.56.082103.104540 [DOI] [PubMed] [Google Scholar]