Cleavage of p53-Vimentin Complex Enhances Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand-Mediated Apoptosis of Rheumatoid Arthritis Synovial Fibroblasts (original) (raw)
Abstract
Rheumatoid arthritis synovial fibroblasts (RASFs) contribute to arthritic cartilage degradation. Although RASFs are normally resistant to apoptosis, Apo2L/tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-based gene therapy has been successfully used in a mouse model of arthritis. We investigated this further by treating human RASFs with nontoxic doses of the proteasome inhibitor lactacystin. Treatment induced cytosolic accumulation of p53 and enhanced the susceptibility of RASFs to apoptosis mediated by TRAIL-R2 (DR5) but not Fas. A specific role for p53 in TRAIL-R2-mediated apoptosis was indicated by the ability of p53 siRNA to significantly reduce RASF apoptosis and by the reduced apoptosis of RASFs bearing p53 mutations on treatment with anti-DR5 antibody or anti-DR5 antibody plus lactacystin. p53 immunoprecipitation followed by mass spectrometry identified a vimentin-p53 complex, an interaction that was confirmed by reciprocal vimentin-p53 immunoprecipitation and by co-immunofluorescence. Interestingly, human caspase-4 cleaved human vimentin, and blockade of caspase-4 with a chemical inhibitor or with specific siRNA significantly inhibited TRAIL-R2-mediated apoptosis of RASFs. Furthermore, blockade of caspase-4 was paralleled by persistence of a cytosolic pattern of p53 and absence of p53 translocation to the nucleus. Taken together, our findings suggest a unique role for caspase-4 in cleaving vimentin and releasing cytosolic p53 for nuclear translocation, events that may regulate the sensitivity of RASFs to receptor-mediated apoptosis.
Tumor necrosis factor-related apoptosis-inducing ligand (Apo2L/TRAIL)-mediated apoptosis has been studied extensively in tumor cells, which are generally more sensitive to Apo2L/TRAIL-mediated apoptosis than normal cells.1–3 Rheumatoid arthritis synovial fibroblasts (RASFs) proliferate in the joint space, and hyperplasia of the synovium contributes to the degradation of cartilage characteristic of this disease.4–6 Although the sensitivity of RASFs to Apo2L/TRAIL-mediated apoptosis has not been studied fully, the therapeutic potential of Apo2L/TRAIL-based gene therapy has been demonstrated in the collagen-induced mouse model of arthritis.7,8
Apo2L/TRAIL binds to the functional receptors, TRAIL-R1 (DR4) and TRAIL-R2 (DR5) and induces apoptosis by activating caspase-8-associated proapoptotic pathways.2,9 On activation, functional TRAIL receptors bind to the cytoplasmic adapter molecule, Fas-associated death domain (FADD), which, in turn, recruits the cysteine protease, caspase-8. The binding of Apo2L/TRAIL to TRAIL-R1 (DR4) and TRAIL-R2 (DR5) also activates a nuclear factor (NF)-κB-associated anti-apoptotic pathway.10–12 The analysis of the preferential induction of apoptosis of transformed cells by Apo2L/TRAIL may provide clues as to the mechanisms that regulate the sensitivity of cells to Apo2L/TRAIL-mediated apoptosis. Such sensitivity may be regulated at several points during the process of apoptosis; modulation of TRAIL receptor signaling as well as transcriptional and posttranscriptional regulation of the expression of the molecules involved in the apoptosis cascade, such as p53,13–16 are potential mechanisms.
The involvement of p53 in Apo2L/TRAIL-mediated apoptosis has been proposed, but the mechanisms that regulate its functional availability are less clear.16–19 Control of intracellular expression of p53 is achieved primarily through degradation shortly after its synthesis. p53 stability is regulated predominantly by the oncoprotein, Mdm2, which mediates the ubiquitinylation of p53 and subsequent rapid degradation by the 26S proteasome.20–24 In addition to targeting p53 for rapid degradation, Mdm2 decreases p53 function by concealing the transcriptional activation domain of p53.25,26 p53 can also be inactivated by sequestration, and the interaction of p53 with the intermediate filament protein, vimentin, in the cytosol contributes to the resistance of tumor cells to apoptosis.27
Intermediate filaments support cellular integrity and provide resistance against mechanical stresses. In recent years, however, live cell imaging has made it clear that these filaments are highly dynamic in their assembly and disassembly.28–30 For example, vimentin is cleaved by a caspase-3/7-like protease during apoptosis and subsequently by caspase-6 at additional sites, including Asp259 (IDVD259-V).31,32 Thus, orchestrated cleavage of vimentin precedes the dramatic reorganization of the cytoskeleton that typifies apoptotic cell death, and through this mechanism, vimentin could play a role in apoptosis and its regulation.31–37
Caspase-4 is a member of the interleukin-1-converting enzyme family of proteases that promotes a proinflammatory response primarily through its action in cleavage and activation of the precursors of the proinflammatory cytokines.38,39 All of the proteases in the interleukin-1-converting enzyme family act together in a cascade that results in the rapid degradation of basic components of the cell, including the laminins (A and B), fodrin, retinoblastoma protein, and poly(ADP-ribose) polymerase.40–42 The role of caspase-4 during the execution phase of Apo2L/TRAIL-mediated apoptosis remains obscure.
Here, we report that RASFs become highly susceptible to TRAIL-R2-mediated apoptosis when treated with nontoxic doses of the proteasome inhibitor, lactacystin. Although the prevention of ubiquitin-mediated protein degradation resulted in the accumulation of cytosolic p53, this was not in itself sufficient to induce apoptosis of RASFs. However, cross-linking TRAIL-R2 (DR5) with anti-DR5 antibody in the context of lactacystin was able to induce RASF apoptosis. Apo2L/TRAIL-mediated activation of caspase-4 led to the release of p53 sequestered in the cytoplasm by vimentin and to the subsequent enhancement of Apo2L/TRAIL-mediated apoptosis of RASFs.
Materials and Methods
Patient Selection
Tissues from patients undergoing total knee replacement at the University of Alabama at Birmingham Clinics were obtained through the Tissue Procurement Facility. All patients met the American College of Rheumatology 1987 criteria for rheumatoid arthritis, and the protocol for use of synovium was approved by the institutional review board. Rheumatoid synovial fibroblasts were isolated from the synovia of eight females ranging in age from 60 to 73 years, with a mean age of 65 years. The individual RASF cell line, obtained from all eight patients, was used throughout this study. In addition, six individual RASF cell lines from six RA patients were obtained from Dr. Firestein’s laboratory (University of California at San Diego).43,44 Of the six RASFs, three exhibited p53 mutations and three had wild-type p53. All were used between passage 4 and 8.
Cell Culture
RASFs were isolated from synovial tissues as described previously.45 In brief, the cell suspensions were cultured in Dulbecco’s-modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin (Invitrogen, Carlsbad, CA) at 37°C in a humidified 5% CO2 incubator. After overnight culture, the nonadherent cells were washed, and only adherent synoviocytes were cultured. Cells between passage 4 and 8 were used for experiments. WI38 cells were obtained from Dr. Trygve Tollefsbol (University of Alabama at Birmingham, Birmingham, AL)46 and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum.
Dose Titration of Cytotoxicity of the Anti-DR5 Antibody and Lactacystin
Noncytototoxic concentrations of anti-DR5 antibody (Alexis Biochemicals, San Diego, CA), anti-Fas antibody (CH11; Upstate Biotechnology, Lake Placid, NY), and the proteasome inhibitors, lactacystin and MG132 (Boston Biotech, Boston, MA) were determined directly by titration of each reagent in the ATPLite assay. In brief, 1 × 103 primary RASFs were plated in 96-well plates and cultured as described above. The cells were then treated with different amounts of the anti-DR5 antibody, anti-Fas CH11 antibody, lactacystin, or MG132 for 24 hours. The cytotoxicity was assessed using the ATPLite assay as described previously.47 In this assay, ATP was determined as light units measured as counts per second (cps) in a luminescence counter (Packard Instruments, Meriden, CT). The percentage of cytotoxicity is therefore expressed as (1 − cps sample/cps control) × 100.
Induction of Apoptosis of RASFs
To determine whether blockade of the ubiquitin-proteasome pathway can enhance the apoptosis of RASFs treated with anti-DR5 antibody or anti-Fas (CH11) antibody, the RASFs were grown to 80% confluence in 96-well plates and treated with anti-DR5 antibody or anti-Fas antibody (100 ng/ml, a dose determined by titration as described above) in the presence, or absence, of different concentrations of lactacystin or MG132 for 15 hours. Hoechst staining was then performed to determine the percentage of apoptotic cells as described previously.47
Transfection of RASFs with siRNA and Plasmid DNA Vimentin
siRNAs including p53 and scrambled siRNA (Dharmacon Research, Inc., Lafayette, CO) were purchased as single strands, deprotected, and annealed according to the manufacturer’s instructions. A mixture of four human casapase-4 siRNAs (Dharmacon Research, Inc.), which bind to different regions of caspase-4 mRNA was supplied as duplexes, and resuspended in siRNA buffer according to the manufacturer’s instructions before use. The selection of siRNA sequences is based on 1) locating the first AA dimer downstream of 50 to 100 bases from the start codon; 2) recording the next 19 nucleotides following the AA dimer; 3) ascertaining that the G/C content of the AA-N19 sequence was between 30 to 70%; and 4) running a BLAST search to ensure that no other genes were targeted by the selected sequence. RASF cells were plated into 24-well plates at a density of 3 × 104 cells/well in antibiotic-free media. The next day, the cells were transfected with either 25 nmol/L p53 siRNA, or caspase-4 siRNA, or scrambled siRNA as a control, using LipofectAmine2000 (Invitrogen) for 36 hours. Then, the cells were treated with anti-DR5 antibody in the presence of lactacystin (10 μmol/L) for an additional 15 hours. The induction of apoptosis in siRNA-transfected RASFs was analyzed using Hoechst staining. To determine the transfection efficiency, RASFs were also transfected with a fluorescent-labeled si_GLO_ siRNA control (Dharmacon Research, Inc.). The transfection efficiency was determined by calculation of the percentage of fluorescent-positive cells among the total RASFs counted (80 to 90% transfection efficiency was achieved reproducibly).
Caspase Inhibition Assays
RASFs were cultured in 96-well plates containing Dul-becco’s modified Eagle’s medium supplemented with 10% FCS. Caspase inhibitors, including inhibitors of caspase-1 through caspase-10 and caspase-13, which were added according to the manufacturer’s instructions (R&D Systems, Indianapolis, IN). In brief, each fluoro-methyl ketone-derivatized caspase inhibitor, at a final concentration of 10 μmol/L, was added to triplicate wells, and the RASFs were then incubated at 37°C for 16 hours. Apoptosis was quantified using the Hoechst staining method as described previously.47
Caspase Activation Assay
Caspase-8, -4, and -3 activity was measured using colorimetric assay kits purchased from Chemicon Int., Inc. (Temecula, CA). Anti-DR5 antibody plus lactacystin (10 μmol/L)-treated RASF cells were harvested at the appropriate times, washed twice with phosphate-buffered saline (PBS), and subsequently treated with lysis buffer for 10 minutes on ice. The lysates were then frozen at −70°C. On the day of the experiment, the lysates were thawed in an ice bath and then spun at 1500 rpm for 10 minutes to remove cellular debris. Protein determination was performed using the Bio-Rad Bradford assay kit (Bio-Rad, Hercules, CA) and 30 μg of protein was used in each assay. The assay was based on the addition of a caspase-specific peptide conjugated to a color reporter molecule, _p_-nitroanalide (pNA). The cleavage of the peptide by the caspases was determined by the absorbance of the chromophore pNA at 405 nmol/L using a Dynex plate reader (Molecular Devices, Sunnyvale, CA). The data are expressed as the ratio of the absorbance of pNA of a treated sample to the absorbance of pNA of an untreated control.
Preparation of Nuclear Proteins
Nuclear extracts from RASF cells were isolated using a nuclear extract buffer kit according to the manufacturer’s instructions (Panomics, Redwood City, CA). In brief, RASFs were treated as described above for 3 hours and then washed with cold PBS once. The cellular membrane was then lysed by adding buffer A and incubating on ice for 5 minutes. After centrifugation at 15,000 × g for 5 minutes, the pellets were resuspended in buffer B and shaken for 2 hours at 4°C to release the nuclear proteins. After centrifugation at 15,000 × g for 5 minutes, the supernatant was aliquoted and stored at −80°C until use.
Western Blot Analysis
RASFs were lysed and the total protein was quantified using the Bradford assay method (Bio-Rad). The proteins (50 μg of total proteins per lane) were then resolved using a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane (Millipore, Danvers, MA). The membrane was first incubated overnight at 4°C with a primary antibody, followed by incubation for 1 hour at room temperature with the peroxidase-conjugated secondary antibody. Immunoreactive proteins were detected by enhanced chemiluminescence (Amersham, Piscataway, NJ). The films were scanned using a Scan-Jet 3c/ADF (Hewlett Packard) and the signals were quantified using Light SPOT imaging. The following antibodies were used for the Western blots: vimentin mAb V9 (1:1000; Sigma, St. Louis, MO.), β-actin (1:5000, Sigma, St. Louis, MO), caspase-3, and caspase-8 (1:1000; BD Pharmingen, San Diego, CA), caspase-4 (1:500; Biomol Research Laboratories Inc., Plymouth Meeting, PA), α-tubulin (1:2000, Sigma), lamin-A/C, and p53 (clone DO1, 1:1000; Santa Cruz Biotechnology, San Diego, CA).
Immunoprecipitation
Antibodies were first crosslinked to protein G-agarose using a Seize X immunoprecipitation kit according to the manufacturer’s instructions (Pierce, Rockford, IL). In brief, protein G-agarose was dispensed into a spin column and washed two to three times with modified Dulbecco’s phosphate-buffered saline (PBS; 8 mmol/L Na2PO3, 2 mmol/L K2PO3, 140 mmol/L NaCl, and 10 mmol/L KCl, pH 7.4). A 100-μg aliquot of monoclonal antibody was incubated with 400 μl of protein G-agarose (50% slurry) for 1 hour at room temperature. After the unbound antibodies were washed away with modified Dulbecco’s PBS, antibody-bound protein G-agarose was resuspended in 400 μl of modified Dulbecco’s PBS. Bound antibodies were covalently attached to the protein G-agarose by adding 0.1 ml of 13 mg/ml DSS cross-linker that was freshly prepared in dimethyl sulfoxide and incubated at room temperature for 1 hour. The excess DSS was removed by washing the resin four times with 400 μl of Tris-buffered saline (25 mmol/L Tris, and 150 mmol/L NaCl, pH 7.2), four times with 0.1 mol/L glycine (pH 2.8) to remove free antibody, and finally three times with Tris-buffered saline. The cross-linking efficiency was evaluated by measuring the optical density at 280 nm. The antibody-protein G-agarose was stored as 50% slurry at 4°C with 0.05% sodium azide.
An aliquot of RASF cell lysate (2.5 mg protein) was diluted in 1 ml of immunoprecipitation buffer (50 mmol/L Tris-HCl, pH 7.4, 150 mmol/L NaCl, 2 mmol/L NaF, 1 mmol/L EDTA, 1 mmol/L EGTA, 1 mmol/L NaVO4, 1 mmol/L phenylmethyl sulfonyl fluoride, 1% Triton X-100) and incubated overnight at 4°C with either 100 μl of primary antibody or a control nonimmune mouse IgG coupled to protein G-agarose beads. After 4× washes with washing buffer (Pierce), the protein complexes were eluted with 100 μl of 0.1 mol/L glycine (pH 2.8), mixed with a sample buffer with 2% β-mercaptoethanol, heated to 95°C, centrifuged, and the supernatant subjected to 4 to 15% SDS-polyacrylamide gel electrophoresis (PAGE) gradient gel followed by colloidal blue staining (Invitrogen) or transferred to a nitrocellulose membrane (Bio-Rad) followed by Western blot analysis. The following antibodies were used for immunoprecipitation and Western blots: vimentin mAb V9, keratin10 mAb, clone 8.6 (Sigma) and p53 (clone DO1, Santa Cruz Biotechnology).
Mass Spectrometry Identification of Proteins
The protein bands of interest from the colloidal blue-stained SDS-PAGE gel were excised, destained, dried, and then rehydrated and digested in-gel with trypsin overnight at 37°C. Aliquots (1 μl) of the trypsin digests were mixed with an equal volume of a saturated solution of α-cyano-4-hydroxy-cinnamic acid in 50% aqueous acetonitrile and 1 μl was spotted onto a stainless steel sample plate followed by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS). The parent polypeptides were identified by comparing the profile of tryptic peptide masses generated by the mass spectrometer with predicted tryptic peptides from all known polypeptides using MASCOT (http://www.matrixscience.com).
Confocal Analysis of p53 and Vimentin
RASFs were fixed with 3% formaldehyde (Tousimis Corp., Rockville, MD) in PBS for 45 minutes at room temperature, permeabilized with 0.5% Triton X-100 (Sigma) in PBS for 3 minutes at room temperature, and further incubated for 1 hour in 1% bovine serum albumin in PBS to prevent nonspecific staining. The cells were then incubated overnight at 4°C with either the primary mouse monoclonal, anti-p53 antibody (1:400, Santa Cruz Biotechnology) labeled with Alexa Fluor 488 (Molecular Probes, Inc., Eugene, OR) or mouse monoclonal anti-vimentin (1:400; NeoMarkers, Fremont, CA) labeled with Alexa Fluor 594 (Molecular Probes). The stained cells were then washed and the staining was visualized and assessed using a Zeiss LSM 510 confocal microscope with a digital image analysis system (Pixera, San Diego, CA).
Recombinant Vimentin Cleavage Assay
The cleavage of vimentin by caspases was assayed using recombinant vimentin. Recombinant vimentin (1 μg) (Research Diagnostics, Flanders NJ) was incubated with different concentrations of caspase-4 (Biomol) in 20 μl of reaction buffer (50 mmol/L Hepes, pH 7.4, 100 mmol/L NaCl, 0.1% CHAPS, 1 mmol/L EDTA, 10 mmol/L dithiothreitol) for 1 hour at 30°C. To determine the specificity of the caspase-4 cleavage of vimentin, recombinant vimentin (1 μg) was incubated with 10 U of caspase-4 in the presence of a caspase-4-specific inhibitor (10 μmol/L, R&D Systems) or different concentrations of caspase-8 (BD Pharmingen) in 20 μl of reaction buffer (50 mmol/L Hepes, pH 7.2, 50 mmol/L NaCl, 0.1% CHAPS, 10 mmol/L EDTA, 10 mmol/L dithiothreitol) for 1 hour at 37°C. In some experiments, a recombinant pro-caspase-3 (BD Pharmingen) instead of vimentin was also used as a positive control for caspase-8 cleavage. The reactions were terminated by the addition of the protein sample loading buffer and the cleaved products resolved using 15% SDS-PAGE. To enhance the visualization of the cleaved products, the gel was Western blotted and the polyvinylidene difluoride membrane probed with anti-vimentin.
Identification of Vimentin Sequence Cleaved by Caspase-4
Based on the fact that human recombinant caspase-4 does not cleave bovine vimentin (data not shown), and that the amino acid sequences of bovine and human vimentins are highly homologous (more than 90%), we predicted that the sequence at position 148 is most probably recognized by human caspase-4. Therefore, the nucleic acid sequences corresponding to this were specifically mutated using pcDNA3-vimentin as a template (a kind gift from Dr. Vincent L. Cryns, Northwestern University, Chicago, IL).32 A mutant human vimentin construct specifically altered at the potential caspase-4 cleavage site, D148Q, was made using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer’s instructions with the following oligonucleotide primers: 5′-cgcgcctggggCAGctctacgaggaggaga-3′ and 5′-tctcctcctcgtagagCTGccccaggcgcg-3′ (D148Q). Mutations were verified by automated DNA sequencing of both strands. Radioactive vimentin products were then made from plasmid pcDNA3-vimentin with and without mutation using in vitro transcription/translation (the TnT T7 Quick Coupled transcription/translation system; Promega, Madison, WI) according to the manufacturer’s instructions. Caspase cleavages were then performed in vitro using purified recombinant caspase-4 as described above. Briefly, 10 μl of TnT reaction product were incubated (37°C) with caspase-4 (20 U). Caspase reactions were performed in 20 mmol/L PIPES (pH 7.2), 100 mmol/L NaCl, 1% CHAPS, 10% sucrose, 10 mmol/L dithiothreitol, and 0.1 mmol/L EDTA. Cleavage reaction products were separated by SDS-PAGE, dried onto filter paper, and placed on X-ray film overnight.
To determine whether overexpression of vimentin with caspase-4 cleavage site mutation (VimentinM) can attenuate apoptosis of RASFs induced by anti-DR5 treatment in the presence of lactacystin, 1 × 103 cells/well of RASF cells in 96-well plates were transfected with either 1 μg of pcDNA3-vimentinM, or 1 μg of pcDNA3 using LipofectAmine 2000 (Invitrogen) for 15 hours. The cells were then treated with anti-DR5 antibody in the presence of lactacystin (10 μmol/L) for an additional 8 hours. The induction of apoptosis in transfected RASFs was analyzed using Hoechst staining.
TaqMan Reverse Transcriptase-Polymerase Chain Reaction (PCR)
To determine whether blocking p53 degradation results in the up-regulation of p53 response genes including Bax, p21, and DR5, the RASFs were grown to 80% confluence in 10-cm cell culture dishes and treated with anti-DR5 antibody (100 ng/ml) in both the presence and absence of lactacystin (10 μmol/L) for 15 hours. Total RNA was then isolated from treated RASFs using the TRIzol extraction method as described previously.48 RNA was quantified according to optical density at 260 nm and 280 nm and adjusted to 100 μg/ml. All probes and primers were provided by Applied Biosystems (Foster City, CA). All probes were labeled with the reporter dye, 6-carboxyfluorescein, at the 5′ end and with a conjugated minor groove-binder quencher dye at the 3′ end. All probes were designed to span an intron to prevent amplification of DNA potentially present in the sample.
An ABI Prism 7700 sequence detection system (Applied Biosystems) was used for amplification and detection of the genes of interest. Replicate dilutions of the unknown sample cDNA from 40 ng of total RNA were combined with a mixture of primers/TaqMan probes, Taq polymerase, and nucleotides, as supplied by Applied Biosystems. Each sample (25-μl total volume) was assayed in an optical tube designed for the 96-well format of the ABI Prism 7000 sequencer (Applied Biosystems). Each PCR amplification was performed in duplicate using the following conditions: 2 minutes at 50°C and 10 minutes at 94°C, followed by a total of 40 cycles of 15 seconds at 94°C and 1 minute at 60°C.
In each TaqMan run, serial fivefold dilutions of a single-stranded cDNA derived from a commercially available human positive control RNA (Applied Biosystems) were amplified to create a standard curve, and values of unknown samples were estimated relative to this standard curve. PCR reactions for each sample were run in duplicate in three fivefold serial dilutions. The mean value of the two reactions was defined as being representative of the sample. The volumes described above yielded standard curves with a linear relationship between the copy number (defined as 1 ng of 1000-bp DNA = 9.1 × 1011 molecules) of the original internal standard that was added and the number of PCR cycles that were required to exceed a preset threshold, according to the method described by Dreskin and colleagues.49 From these standard curves, the relative amount of cDNA for the 18S ribosomal RNA (rRNA), GAPDH, and each gene was determined for each sample and expressed as a ratio of the amount of this material in the anti-DR5 antibody, or lactacystin, or anti-DR5 antibody plus lactacystin versus untreated cells after samples were standardized to the relative expression of the 18S rRNA and GAPDH.
Expression of Data and Statistical Analyses
The results are expressed as the mean ± SEM of three to four separate experiments performed in triplicate. Differences between the experimental groups were evaluated using one-way analysis of variance.
Results
Apo2L/TRAIL-, but not Fas-Mediated Apoptosis of Human RASFs Is Enhanced by Blockade of the Ubiquitin-Proteasome Pathway
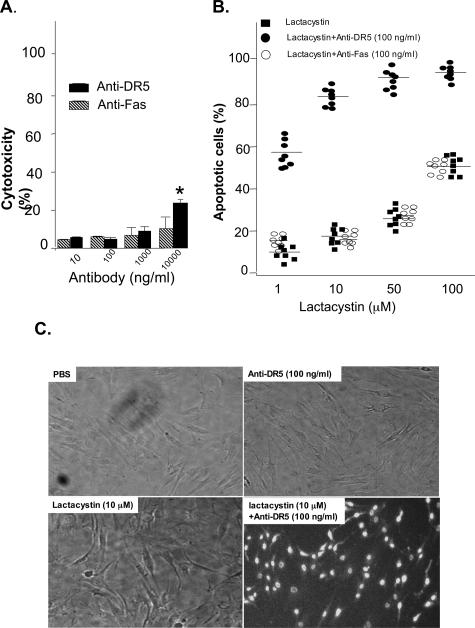
Primary RASFs were isolated, cultured for 24 hours to 80% confluence, and then incubated with anti-DR5 antibody or anti-Fas (CH11) antibody for an additional 15 hours. Induction of cytotoxicity required at least 10,000 ng/ml of anti-DR5 antibody or anti-Fas antibody (Figure 1A). Because blockade of the proteasome degradation pathway may affect apoptosis, we determined if the apoptosis sensitivity of RASFs to a Apo2L/TRAIL- or Fas-mediated signal could be enhanced in the presence of the proteasome inhibitor, lactacystin. Titration of lactacystin showed that ≤50 μmol/L lactacystin alone did not induce apoptosis of RASFs (data not shown). Treatment of the RASFs with a combination of anti-Fas and lactacystin did not result in cell death. However, treatment with a combination of anti-DR5 and lactacystin resulted in the cell death of a significant percentage of RASFs derived from each of eight individual donors (Figure 1B), and the majority of cell death could be attributed to apoptosis (Figure 1C). Similar effects were observed (data not shown) when another inhibitor of the protease pathway, MG132, was used instead of lactacystin, suggesting that inhibition of protein degradation results in the enhancement of Apo2L/TRAIL-mediated apoptosis of RASFs.
Figure 1.
The proteasome inhibitor lactacystin enhances Apo2L/TRAIL receptor DR5-mediated apoptosis of RASFs. A: Primary RASFs (1 × 103 cells per well) at passage number between 4 to 8 in 96-well plates were treated with different concentrations of anti-DR5 antibody or anti-Fas (CH11) for 15 hours and cytotoxicity was evaluated using the ATPLite assay. The results presented are the mean of three experiments. B: Anti-DR5 antibody or anti-Fas antibody at the noncytotoxic doses (100 ng/ml) or PBS were then used in treatment of primary RASF cells (1 × 103 per well) in the presence of different concentration of lactacystin. After incubation for 15 hours at 37°C, the cells were fixed with 10% buffered formalin and stained with Hoechst dye, which is taken up by apoptotic cells. Quantification of the percentage of apoptotic cells by counting the total number of cells and the number of Hoechst-stained cells in at least five randomly selected fields of view indicated a higher percentage of apoptotic RASFs after treatment with anti-DR5 and lactacystin. Each filled square, open circle, or solid circle represents the mean from triplicate samples of RASFs derived from each of eight individuals at passage number between 4 and 8 with different treatments. C: Hoechst-stained apoptotic cells with typical nuclear condensation and extensive blue nuclear staining are apparent on treatment of the RASFs with anti-DR5 (100 ng/ml) plus lactacystin (10 μmol/L), but not in the other treatment groups.
The Threshold of Susceptibility to Induction of Apoptosis after Anti-DR5 Antibody Plus Lactacystin Treatment Is Lower in RASFs than Normal Fibroblasts
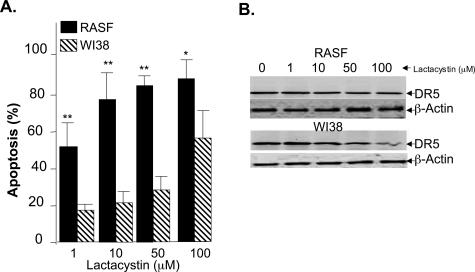
Unlike normal fibroblasts, RASFs are highly proliferative and are generally considered to be semitransformed cells. To determine whether blocking proteasome activity also results in the enhancement of TRAIL-mediated apoptosis of normal fibroblasts, the sensitivity to induction of apoptosis of a normal fibroblast cell line (WI38) was determined by incubating the cells with anti-DR5 plus lactacystin for 15 hours. The results indicated that WI38 cells are significantly less sensitive than RASFs in terms of the percentages of cells undergoing apoptosis after treatment (Figure 2A). The relative insensitivity of WI38 to the treatment could not be attributed to a lack of DR5 receptor expression because the intensity of the signals for DR5 in Western blots of the cells were similar (Figure 2B, bottom versus top panels). Also, enhancement of the TRAIL-mediated apoptosis of RASFs is unlikely to be due to lactacystin-induced changes in DR5 expression because treatment with anti-DR5 antibody plus lactacystin did not alter the intensity of the signal as compared to the signal observed on treatment with anti-DR5 antibody or lactacystin alone (Figure 2B, top).
Figure 2.
Normal fibroblasts are less sensitive than RASFs to lactacystin-enhanced DR5-mediated apoptosis. The normal fibroblast WI38 cell line or RASFs (1 × 103 cells per well of 96-well plates) were cultured in Dulbecco’s modified Eagle’s medium media supplemented with 10% fetal bovine serum for 6 hours, and then treated with anti-DR5 antibody (100 ng/ml) in the presence of different concentration of lactacystin for an additional 15 hours at 37°C. The cells were then fixed with 10% buffered formalin and stained with Hoechst dye. The percentage of apoptotic cells was determined using the method as described in the legend of Figure 1. Each column represents the mean from five replicas of each treatment of either the WI38 cell line or RASFs derived from one individual as described in the legend of Figure 1A. **P < 0.01. B: To determine the expression of DR5 receptor on the WI38 and RASFs, 50 μg of the total protein lysates of RASF (top) or WI38 (bottom) cells treated as indicated in A was separated on the 10% SDS-PAGE gel, and DR5 and β-actin Western blot analyses were performed. A representative gel is shown.
p53 Plays a Role in the Enhancement of DR5-Mediated Apoptosis of RASFs by Lactacystin
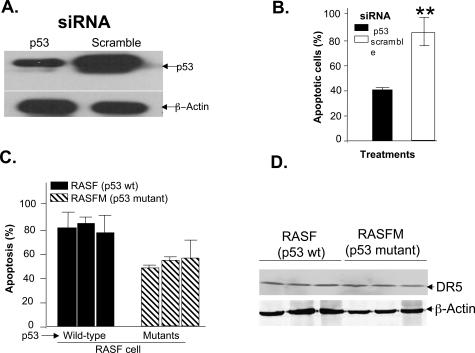
p53 plays a key role in cell proliferation and apoptosis, and proteasome-mediated degradation of p53 is known to contribute to the relative resistance of cells to apoptosis. Therefore, we determined whether p53 is required for the lactacystin enhancement of DR5-mediated apoptosis of RASFs. p53 siRNA, but not scrambled siRNA, decreased p53 expression (Figure 3A, lane 1). No change was observed in β-actin protein expression, indicating that the reduction in p53 was specific (Figure 3A, bottom). DR5-mediated apoptosis of RASFs, measured by Hoechst staining, was attenuated in the RASFs treated with 25 nmol/L p53 siRNA, but not in those treated with scrambled siRNA (Figure 3B). Although RASFs in which the endogenous p53 is mutated do exhibit enhanced apoptosis on treatment with anti-DR5 antibody treatment in the presence of lactacystin, the magnitude of this effect is significantly lower than that induced in the RASFs with wild-type p53 (P < 0.042, Figure 3C). This difference is unlikely to be due to defective expression of DR5 in the RASFs as the results of Western blot analysis of DR5 indicate that the expression of the DR5 receptor is similar in RASFs with mutated p53 and RASFs with wild-type p53 (Figure 3D).
Figure 3.
p53 enhances DR5-signaling-mediated apoptosis of RASFs. RASFs were transfected with p53 siRNAs or scrambled siRNAs using LipofectAmine 2000 for 15 hours, and then treated with anti-DR5 antibody in the presence of lactacystin for an additional 15 hours. Fifty μg of total cell lysates were used for Western blot analysis of p53 expression (A) and apoptotic cells were assessed by Hoechst 33342 staining (B). RASFs with or without endogenous p53 mutations were incubated with anti-DR5 antibody (100 ng/ml) plus lactacystin (10 μmol/L) for 15 hours. RASF cells from three patients with RA exhibited p53 mutations as determined by Dr. Firestein and colleagues,43 whereas the RASF cells from the other three have wild-type p53. C: The induction of apoptosis was determined by the Hoechst staining method. Each histogram represents the mean ± SEM for five samples. The data presented represent the mean ± SEM from a minimum of three different samples. **P < 0.01. D: The expression of DR5 in these cells was determined by DR5 Western blot analysis as described in Materials and Methods.
Treatment with Lactacystin Results in Sequestration of p53 in the Cytoplasmic Compartment
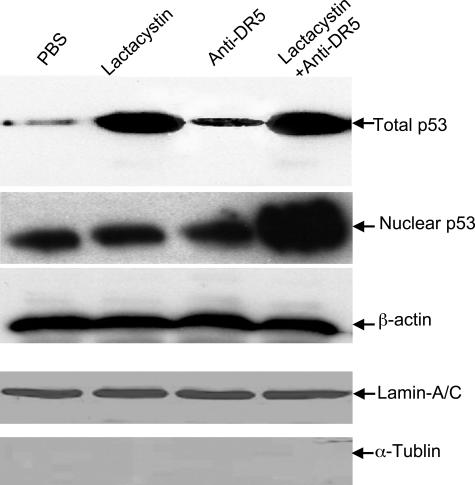
To determine the fate of p53 rescued from degradation by the proteasome inhibitor, the RASFs were treated with lactacystin, in the presence or absence of anti-DR5 antibody, and then either total protein or nuclear extracts were separated on a 10% SDS-PAGE gel, and p53 Western blot analysis was performed. Although p53 accumulated in RASFs treated with lactacystin (Figure 4, top), Western blot results indicated that p53 accumulated in the nucleus only in RASFs treated with lactacystin plus anti-DR5 (Figure 4, the second panel from the top). These apparent differences in the distribution of p53 were not due to unequal protein loading because no differences in β-actin (Figure 4, the third panel from the top) or in nuclear protein marker, lamin-A/C (Figure 4, the fourth panel from the top) were observed. The low amounts of α-tubulin in the nuclear extracts indicate very little cytoplasmic protein contamination of the nuclear extracts (Figure 4, bottom). Nuclear translocation of p53 can lead to up-regulation DR5 as well as a number of other genes, including bax and p21. Therefore, the expression of these genes was quantified using a real-time PCR method. The results indicate that the expression of both bax and p21 is up-regulated by treatment of RASFs with anti-DR5 antibody plus lactacystin for 15 hours (Table 1), but not by treatment with either anti-DR5 antibody or lactacystin alone (Table 1). As suggested by the Western blot analyses, the expression of DR5 was not enhanced either (data not shown). Taken together, these results indicate that p53 accumulates in RASFs treated with lactacystin alone, and is translocated to the nucleus only on treatment of RASFs with anti-DR5 antibody plus lactacystin.
Figure 4.
p53 accumulation in the cytosol of RASFs after lactacystin treatment. RASF cells were treated with PBS or anti-human DR5 antibody (100 ng/ml), or lactacystin (10 μmol/L), or anti-human DR5 antibody (100 ng/ml) plus lactacystin (10 μmol/L) for 3 hours. Protein lysates or nuclear extracts of RASFs were treated as indicated and a 50-μg aliquot of each was separated by 10% SDS-PAGE, and p53, lamin-A/C, α-tubulin, and β-actin Western blot analyses were performed. Representative gels are shown.
Table 1.
TaqMan Real-Time RT-PCR Analysis
| Gene name | RASF treatment/untreated* | Anti-DR5/lactacystin/lactacystin + anti-DR5 | |||
|---|---|---|---|---|---|
| Anti-DR5 | Lactacystin | Lactacystin + anti-DR5 | Fold increase | P | |
| Bax | 58.4/55.1 | 62.8/58.1 | 371.8/65.4 | 1.0/1.0/5.7 | 0.99/0.94/0.023 |
| p21 | 33.4/30.8 | 44.2/38.9 | 204.5/60.2 | 1.1/1.0/3.3 | 0.90/0.92/0.029 |
| DR5 | 29.8/27.5 | 36.6/33.7 | 52.4/44.9 | 1.0/1.1/1.1 | 0.91/0.92/0.98 |
In the Absence of DR5 Signaling, p53 Co-Localizes with Vimentin in the Cytosol
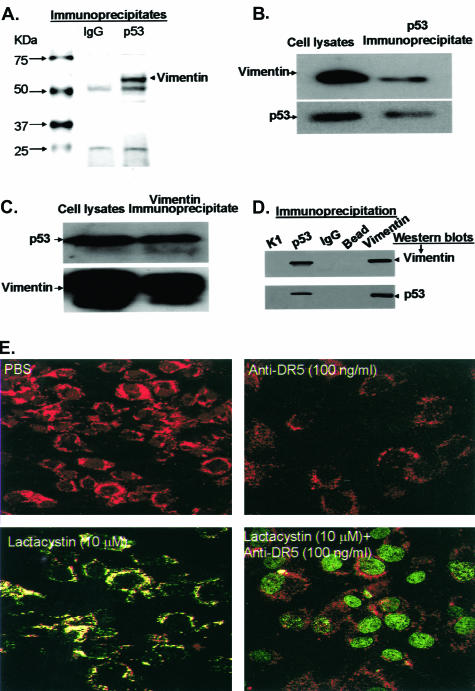
We considered the possibility that, in the absence DR5 signaling, p53 might be retained in the cytosol of the RASFs through interactions with other protein partners. This possibility was tested by immunoprecipitation of p53 with protein G-agarose-conjugated anti-p53 and analysis of the proteins resolved using a 4 to 15% gradient SDS-PAGE gel. Colloidal blue staining revealed a protein with an estimated molecular weight of 58 kd that is associated with the p53-immune complex (Figure 5A, lane 2), but absent in the control mouse IgG-immune complex (Figure 5A, lane 1). This 58-kd band was excised, trypsinized, analyzed by MALDI-TOF C-MS, and identified as vimentin.
Figure 5.
Vimentin interacts with p53. RASF cells were treated with lactacystin (10 μmol/L) for 3 hours. The cells were lysed using an immunoprecipitation lysate buffer and 2.5-mg aliquots of the total protein lysates were immunoprecipitated with either mouse IgG-conjugated (A, lane 2) or mouse anti-p53 (clone DO1)-conjugated (A, lane 3) agarose beads, and the precipitates were separated by 4 to 15% SDS-PAGE, and stained with colloidal blue. Total protein lysates (50 μg) (B and C, lane 1) or the immunoprecipitated complexes (B and C, lane 2) were separated by 10% SDS-PAGE, and Western blotted with anti-vimentin or anti-p53 as indicated in A and B. To confirm the specificity of the interaction, 2.5 mg of total protein lysates from the same samples used in A, B, and C were immunoprecipitated with anti-p53 (D, lane 2), anti-vimentin (D, lane 5), anti-keratin 10 (D, lane 1), anti-mouse IgG (D, lane 3), or beads alone (lane 4) and Western blotted with anti-vimentin (D, top) or anti-p53 (D, bottom). For confocal microscopic analysis, RASF cells were cultured to 80% confluency, and treated as indicated for an additional 3 hours. E: The treated RASF cells were then fixed with 3% formaldehyde, immunostained with anti-vimentin (red fluorescence) and anti-p53 (green fluorescence), and analyzed by confocal microscopy to determine the intracellular localization of p53. Each photograph is representative of five different independent experiments.
To confirm the interaction of p53 with vimentin, the RASFs were treated with lactacystin (10 μmol/L) for 6 hours and then reciprocal immunoprecipitation with p53 or vimentin was performed. Western blot analysis of immunoprecipitates of p53 indicated that p53 co-immunoprecipitates with vimentin (Figure 5B, lane 2). To confirm the interaction of vimentin with p53, reciprocal immunoprecipitation and Western blots indicated that vimentin also co-immunoprecipitates with p53 (Figure 5C, lane 2). The co-precipitation would appear to be specific because neither p53 nor vimentin was detected in the immune complex precipitated with an irrelevant anti-keratin 10 antibody (Figure 5D, lane 1), mouse IgG (Figure 5D, lane 3), or beads alone (Figure 5D, lane 4). The p53/vimentin interaction also was demonstrated by confocal analysis. In the absence of lactacystin, co-localization of the sparse p53 and abundant vimentin was not detected in the cytosol of either the PBS- or anti-DR5-treated cells (Figure 5E, top). On treatment of the RASF cells with lactacystin, p53 accumulation was observed in the cytosol and this p53 was co-localized with the vimentin (Figure 5E, bottom left), together with enhanced localization of p53 in the peripheral nuclear membranes. Treatment of these cells with the combination of anti-DR5 antibody and lactacystin led to remarkable translocation of p53 into the nucleus and an absence of co-localization with vimentin in the cytosol (Figure 5E, bottom right).
Activation of Caspase-8, Caspase-4, and Caspase-3 Plays a Role in the Apo2L/TRAIL-Mediated Apoptosis of Human RASFs
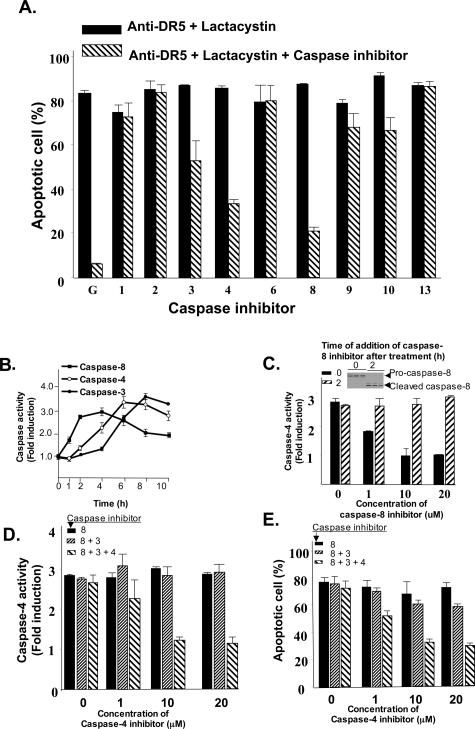
To determine whether the apoptosis induced in RASFs by treatment with anti-DR5 antibody plus lactacystin is dependent on the activation of caspases, RASFs were cultured in 96-well plates for 15 hours and then treated with anti-DR5 antibody plus lactacystin in the presence or absence of highly selective caspase inhibitors for 15 hours. The induction of apoptosis in RASFs was analyzed using the Hoechst staining method as previously described.47 Blocking of caspase activity by ∼70% for caspase-8, 40% for caspase-4, and 25% for capase-3 was observed, and was found to decrease apoptosis, suggesting that the Apo2L/TRAIL-mediated apoptosis of RASFs (Figure 6A) is caspase-dependent.
Figure 6.
Activation of caspase-8 and caspase-4 is required for enhancement of anti-DR5-mediated apoptosis of RASFs in the presence of the proteasome inhibitor, lactacystin. Primary RASF cells (1 × 103) per well in 96-well plates were cultured for 15 hours and treated with lactacystin (10 μmol/L) followed by treatment with an anti-DR5 antibody (100 ng/ml) in the presence or absence of different caspase inhibitors as indicated and incubated at 37°C for an additional 15 hours. A: The percentage of apoptotic cells was quantified by counting the total number of cells and the number of Hoechst-stained cells in at least five randomly selected fields of view. The data represent mean values (mean ± SEM) from three different experiments. The activities of caspase-3, -4, and -8, that were found to participate in the enhancement of anti-DR5-mediated apoptosis of RASFs in the presence of lactacystin was further analyzed kinetically in RASF cells treated with anti-DR5 antibody (100 ng/ml) plus lactacystin (10 μmol/L). RASF cells were lysed at the indicated times after the treatments and lysates (30 μg of protein) used for colorimetric caspase assays as described in Materials and Methods using IETD-pNA, LEVD-pNA, and DEVD-pNA substrates. B: IETD, LEVD, and DEVD are the preferred peptide substrates for caspase-8, -4, and -3, respectively. The changes of activity after the treatment were compared with the activity before treatment and are expressed as the fold increase. The data represent mean value (mean + SEM) from five individual RASF samples. RASF cells were cultured as described in B for 15 hours, and then treated with anti-DR5 antibody (100 ng/ml) plus lactacystin (10 μmol/L) and caspase-8 inhibitor (10 μmol/L) was added at 0 or 2 hours after treatment. C: Ten hours later caspase-4 activity was measured using LEVD-pNA as the substrate. The activation of caspase-8 in the RASFs before addition of caspase-8 inhibitor was determined by Western blot analysis as shown in the inset of C. The data are presented as described for B. At hour 2 after treatment of RASF cells with anti-DR5 antibody (100 ng/ml) plus lactacystin (10 μmol/L), the cells were treated with caspase-8 inhibitor (10 μmol/L) with or without caspase-3 inhibitor (10 μmol/L) and various concentrations of a caspase-4 inhibitor as indicated in the figure legend, for 8 hours. D: The activity of caspase-4 was then assessed by a colorimetric assay using the caspase-4 substrate LEVD-pNA. The changes of caspase-4 activity were compared between samples treated with or without a caspase-4 inhibitor. Experimental values are reported as means ± SEM of three individual replica samples. E: Induction of apoptosis was estimated by counting Hoechst-stained apoptotic cells. The percentage of apoptotic cells was quantified by counting the total number of cells and the number of Hoechst-stained cells in at least five randomly selected fields of view.
To further determine the sequence of activation of the caspases, a colorimetric assay based on substrate cleavage was used. The results showed a time-dependent increase in caspase-8 activity, indicated by cleavage of its substrate IETD-pNA, starting at 1 hour and reaching peak levels between 2 and 4 hours. The kinetics of caspase-4 activity, indicated by cleavage of its substrate-LEVD-pNA, showed a linear, time-dependent increase between 2 and 6 hours. Similarly, caspase-3 activity indicated by cleavage of its substrate DEVD-pNA, was observed starting at 2 hours, and reached peak levels at 8 hours (Figure 6B).
To determine the time required for caspase-8 to activate caspase-4, caspase-8 activation was blocked either immediately or 2 hours after treatment of RASFs with anti-DR5 in the presence of lactacystin, with capase-4 activity being measured 8 hours later. The results of Western blot analysis of caspase-8 confirmed that addition of caspase-8 inhibitor simultaneously with the addition of anti-DR5 and lactacystin blocked the activation of caspase-8, but that addition of the caspase inhibitor 2 hours after treatment with anti-DR5 and lactacystin did not (Figure 6C, inset). Blockade of caspase-8 activity by simultaneous addition of the inhibitor resulted in a decrease in caspase-4 activity in a dose-dependent manner (Figure 6C). Caspase-4 activity was not reduced when the caspase-8 inhibitor was applied 2 hours after addition of the anti-DR5 and lactacystin (Figure 6C). Because blocking caspase-8 activity at 2 hours after treatment with anti-DR5 and lactacystin did not significantly affect the caspase-4 activity, the contribution of activated caspase-4 to the apoptosis of RASFs treated with anti-DR5 antibody plus lactacystin (10 μmol/L) was determined. Neither blocking the activity of caspase-8, nor blocking the activity of caspase-8 and caspase-3, at 2 hours after addition of anti-DR5 and lactacystin had any effect on the activity of caspase-4 regardless of the concentrations of the inhibitors. Addition of a caspase-4 inhibitor at this time point did decrease caspase-4 activity in a dose-dependent manner (Figure 6D) and the caspase-4 activity was correlated with the induction of apoptosis of RASFs at 8 hours after treatment (Figure 6E). Collectively, these data suggest that preactivation of caspase-8 is required for activation of casapse-4, and activity of caspase-4 plays a role in the enhancement of Apo2L/TRAIL-mediated apoptosis of RASFs in the presence of a proteasome inhibitor. We also analyzed the sequential activation of caspase-8, caspase-4, and caspase-3 in the three RASFs with p53 mutations. Similar results were observed (data not shown), suggesting that the DR5-mediated activation of caspase-4 is independent of p53 status or signals upstream of p53.
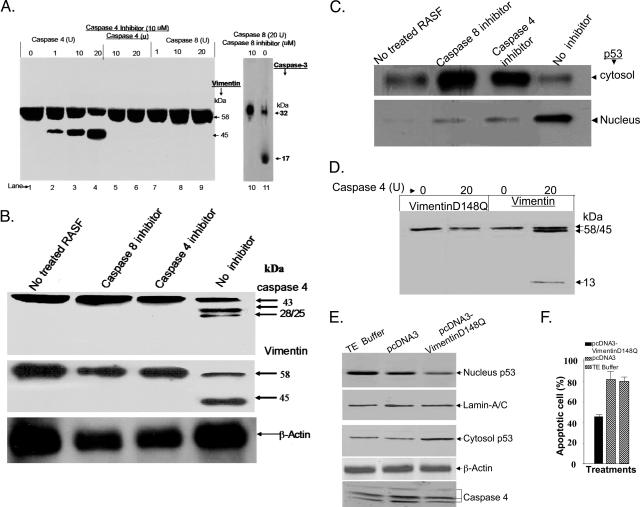
Activated Caspase-4 Can Cleave Vimentin
Because vimentin is co-immunoprecipitated with p53 and p53 is co-localized with vimentin, the ability of p53 to undergo translocation from the cytosol to the nucleus in RASFs treated with lactacystin plus anti-DR5 might result from the caspase-mediated cleavage of vimentin. To explore this possibility, HPLC-purified recombinant human vimentin protein was incubated with different concentrations of caspase-8 or caspase-4 for 1 hour at 37°C. The cleaved products were then visualized by Western blot analysis with anti-vimentin. Caspase-8 cleaved caspase-3 (Figure 7A, lane 11) but did not cleave vimentin at the concentrations tested (Figure 7A, lanes 7 to 9). Caspase-8 cleavage activity was specific and completely blocked by the caspase-8-specific inhibitor (Figure 7A, lane 10). Interestingly, caspase-4 led to a dose-dependent cleavage of human vimentin (Figure 7A) that was blocked by a specific caspase-4 inhibitor (Figure 7A, lanes 5 and 6). This role of caspase-4 in vimentin cleavage was also supported by in vivo caspase-4 inhibition assays. The primary RASF cells were treated with either PBS or anti-DR5 plus lactacystin in the presence of the indicated caspase inhibitors for 3 hours. Caspase-4 was activated in the RASFs treated with anti-DR5 plus lactacystin (Figure 7B, lane 4), but not in RASFs treated with PBS (Figure 7B, lane 1). Addition of a caspase-8 inhibitor (Figure 7B, lane 2) completely blocked the cleavage of caspase-4, indicating that the activation of caspase-8 is required for the activation of caspase-4. Therefore, the activation of caspase-4 may be responsible for the cleavage of vimentin directly or indirectly (Figure 7B, middle). This possibility was further supported by the observation that the blockade of caspase-4 activation, as well as of caspase-8, which is the precursor to caspase-4 activation, led to the retention of p53 in the cytosol and attenuation of nuclear translocation of p53 (Figure 7C). Because in vitro analysis had indicated that human caspase-4 cleaved human vimentin, but not bovine vimentin (data not shown), we determined if the unique sequence (amino acid residue 148) present in human vimentin is recognized and cleaved by human caspase-4. An in vitro translated and [35S]-methionine-labeled recombinant wild-type protein or mutated human vimentin protein was incubated with caspase-4 (20 U) for 1 hour at 37°C and the cleaved products visualized by autoradiography. The results confirmed that wild-type human vimentin (Figure 7D, lane 4) but not human vimentin with mutation at position of D148 (D148Q) (Figure 7, lane 2) is cleaved by human casapse-4.
Figure 7.
Caspase-4-mediated cleavage of vimentin promotes p53 nuclear translocation. A: Different amounts of caspase-4 were added into a 20-μl volume of reaction buffer (50 mmol/L Hepes, pH 7.4, 100 mmol/L NaCl, 0.1% CHAPS, 1 mmol/L EDTA, 10 mmol/L dithiothreitol) containing 1 μg of vimentin in the absence of (lanes 2 to 4) or in the presence of (lanes 5 and 6) caspase-4 inhibitor (10 μmol/L), and incubated for 1 hour at 30°C, followed by gel electrophoresis and Western blotting of the cleavage products of vimentin. Analysis of caspase-8 cleavage of vimentin also was performed. A recombinant caspase-3 (1 μg) was also co-incubated with caspase-8 in the presence (10 μmol/L, lane 10) or in the absence of caspase-8 inhibitor as a positive control. The products were then separated on 10% SDS-PAGE gel, followed by vimentin (lanes 1 to 9) or by caspase-3 (lanes 10 and 11) Western blot analyses. B: RASF cells were treated as labeled for 3 hours. The protein lysates were separated on the 10% SDS-PAGE gel and transferred onto a polyvinylidene difluoride membrane. The membrane was then immunoblotted with caspase-4 antibody (top), or vimentin antibody (middle), or β-actin antibody (bottom). C: Three hours after RASF cells were treated, both total protein (top) and nuclear extracts (bottom) were separated on a 10% SDS-PAGE gel, and p53 Western blot analysis was performed. D: 35S-methionine-labeled human vimentin and vimentin mutant D148Q was in vitro translated and 10 μl of the translated proteins were incubated with control buffer, or 20 U of caspase-4 for 1 hour at 37°C, and the cleavage reactions were separated by 10 to 15% SDS-PAGE and visualized by autoradiography as detailed in Materials and Methods. RASF cells (5 × 106) were transfected with 10 μg of plasmid DNAs including pcDNA3-vimentin and pcDNA3-vimentinD148Q or DNA diluent T.E. buffer as labeled for 15 hours and then treated with anti-DR5 antibody (100 ng/ml) plus lactacystin (10 μmol/L) for 8 hours. The cells were then either lysed for Western blot analyses of nuclear and cytosol p53 or caspase-4 cleavage using a method as described in the legend to Figure 2 (E) or quantified for percentages of apoptotic cells using a Hoechst staining method as described in the Materials and Methods (F). The data presented represent the mean ± SEM from five different RASF samples. **P < 0.01, *P < 0.05.
We therefore overexpressed mutant vimentin and determined its effect on p53 nuclear translocation and Apo2L/TRAIL-mediated apoptosis. RASF cells were transiently transfected with a vimentin mutant expression vector pcDNA3vimentin-D148Q. The RASF cells were then treated with anti-DR5 antibody plus lactacystin for 8 hours. Western blot analysis of p53 indicated that overexpression of mutant vimentin led to the prevention of p53 nuclear translocation (Figure 7E, top), and accumulation of p53 in the cytosol (Figure 7E, the third panel from the top), which was accompanied by a reduction in apoptosis of RASF cells (Figure 7F). The different amounts of p53 detected in the cell lysates were not due to differences in the protein loading because levels of lamin A/C (Figure 7E, the second panel from the top) in the nuclear extracts and β-actin (Figure 7E, bottom) in the cytosol were equivalent.
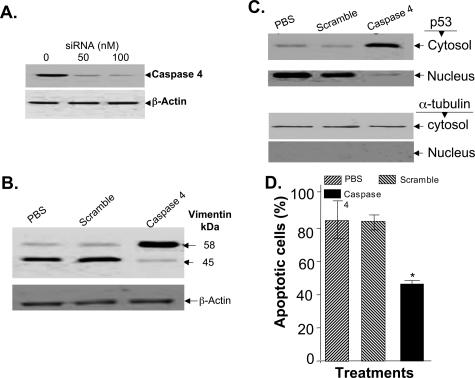
Forced Reduction of the Levels of Caspase-4 Attenuates Vimentin Cleavage, p53 Nuclear Translocation, and DR5-Mediated Apoptosis of RASFs
To further establish the role of caspase-4 in DR5-mediated apoptosis of RASFs, we blocked expression of caspase-4 using siRNA. The ability of 50 nmol/L siRNA specific for caspase-4, to reduce caspase-4 protein levels was demonstrated by Western blot analysis (Figure 8A). RASFs at 80% confluence were transfected with 50 nmol/L siRNA caspase-4 and subsequently incubated with anti-DR5 antibody (100 ng/ml) plus lactacystin (10 μmol/L) for an additional 15 hours. The effectiveness of capase-4 siRNA in blocking the cleavage of vimentin and preventing p53 nuclear translocation were analyzed by Western blot analysis and its effectiveness in reducing apoptosis of treated RASFs was measured using Hoechst staining. Caspase-4 siRNA treatment reduced the cleavage of vimentin (Figure 8B), led to the accumulation of p53 in the cytosol (Figure 8C, lane 3, top panel), and reduced apoptosis mediated by DR5 plus lactacystin (10 μmol/L) (Figure 8D, P < 0.05). Caspase-4 siRNA treatment was specific because scrambled siRNA had no effects on the cleavage of vimentin (Figure 8B, lane 2), p53 nuclear translocation (Figure 8C, lane 2), or the induction of apoptosis (Figure 8D, column 2) compared with RASFs treated with PBS control [Figure 8, B (lane 1) and D (column 1)]. The observed nuclear translocation of p53 was unlikely to be due to contamination with cytosolic p53 because a cytoplasmic marker, α-tubulin, was not detected in the nucleus (Figure 8C, bottom).
Figure 8.
Caspase-4 siRNA treatment attenuates DR5-mediated apoptosis of RASFs in the presence of lactacystin. RASFs were transfected with different concentrations of caspase-4 siRNA using LipofectAmine 2000 for 48 hours, and then lysed for Western blot analysis of caspase-4. Total cell lysates (50 μg) were used for Western blot analysis of caspase-4 expression (A, top) or β-actin (A, bottom). RASFs were cultured and treated as described in A with caspase-4 siRNA or scrambled siRNA at a concentration of 50 nmol/L or PBS for 36 hours, and then treated with anti-DR5 antibody in the presence of lactacystin for an additional 15 hours. Cleavage of vimentin (B) or translocation of p53 (C) was assessed by Western blot analysis, percentage of Hoechst 33342-stained apoptotic cells was counted, and the data presented represent the mean ± SEM from a minimum of three different samples (D). **P < 0.01, *P < 0.05.
Discussion
RASFs are normally resistant to apoptosis; here, we show that treatment of RASFs with lactacystin, a blocker of proteasome-mediated degradation, enhances the sensitivity of RASFs to Apo2L/TRAIL-mediated apoptosis. This effect appears to be regulated through p53, which accumulates in the cytosol in a vimentin-sequestered manner in response to the lactacystin signaling alone, and is released from the vimentin and translocated to the nucleus on additional signaling by anti-DR5. These studies, therefore, indicate that two actions, prevention of proteasome-mediated degradation of p53 and caspase-mediated release of p53 sequestered as a vimentin-p53 complex, are required for the net effect of lactacystin enhancement of DR5-mediated apoptosis.
The importance of p53 in TRAIL-mediated apoptosis is indicated by the effects of reduction in p53 using small interference RNA technology on Apo2L/TRAIL-mediated apoptosis induced by its receptor, DR5. In both the presence and absence of lactacystin, the knockout of p53 resulted in a reduction in apoptosis of RASFs. Neither blockade of the expression of p53 by p53 siRNA nor the presence of endogenous p53 mutations could completely attenuate the apoptosis of RASFs, indicating that additional ubiquitin-regulated proteins may also participate in the enhancement of DR5-mediated apoptosis signaling.
Our current data are consistent with the concept that the activity of p53 in RASFs is regulated by proteasome-mediated degradation. Proteasome-mediated degradation of p53 is well documented. In nonstressed cells, p53 is maintained in a biologically latent state, primarily due to the action of the proto-oncogene, mdm2. The Mdm2 protein binds to p53 and acts as its major cellular antagonist. This is achieved not only through direct interference of the bound Mdm2 with the transcriptional activities of p53, but also through the ability of Mdm2 to target p53 for rapid proteolysis.16,20,21,24,25 This latter capacity of Mdm2 functions as a p53-specific E3 ubiquitin ligase, which attaches ubiquitin residues onto p53 and targets it for degradation by the 26S proteasom_e_.
Nuclear translocation of p53 leading to the up-regulation of proapoptosis genes, including bax, is considered to be one of the mechanisms underlying p53-mediated apoptosis induction and we found that accumulation of p53 in the nucleus after anti-DR5 antibody plus lactacystin treatment does indeed lead to the up-regulation of bax and p21 mRNA expression. Other investigators have reported that the gene encoding the DR5 receptor also is regulated by p53 in various cell types.50–52 Our real-time PCR and Western blots results failed to show increased expression of the DR5 receptor, suggesting that the expression of the DR5 receptor in RASFs is regulated in a p53-independent manner. Our previous work has shown that the regulation of expression of the DR5 receptor in RASFs differs from its regulation in nonproliferative synovial fibroblasts derived from patients with osteoarthritis (OASF).53 This differential regulation of the DR5 receptor in response to p53 in RASFs and OASF is of interest and needs further clarification.
Our studies are the first to indicate that the activation of caspase-4, which leads to cleavage of vimentin and subsequent release of the p53 from the p53-vimentin complex, is also required for the enhancement of Apo2L/TRAIL-mediated apoptosis of RASFs. Blockade of vimentin cleavage through the use of a construct with a mutated caspase cleavage site, D148Q, conferred protection against apoptotic cell death in the RASFs. Cleavage of vimentin by caspases has been demonstrated by other investigators. After stimulation of HeLa cells with tumor necrosis factor-α, activated caspase-3 leads to vimentin cleavage.32 Morishima54 has shown that after stimulation with anti-Fas antibody in the Jurkat T cell line, vimentin cleavage is associated with activated caspases-8, -3, -6, and -7. The present study, however, demonstrates that DR5-mediated apoptosis signaling can trigger the cleavage of vimentin through activation of caspase-4, and that caspase-4 activation enhances DR5-mediated apoptosis of RASFs. Caspase-4 belongs to the interleukin-1-converting enzyme family of proteins that promote a proinflammatory response primarily through their action in cleaving proinflammatory cytokines. Caspase-4 triggers apoptosis in the presence of Apo2L/TRAIL signaling, but enhances inflammation in the absence of Apo2L/TRAIL signaling. The effect of the inflammatory response on caspase-4 activation has been observed after stimulation of the human cholangiocarcinoma cell line, Sk-ChA-1, with interferon-γ,55 and after stressing the endoplasmic reticulum (ER) of human neuroblastoma SK-N-SH cells.56 Recent data indicate that Apo2L/TRAIL-knockout mice develop arthritis spontaneously and that local injection of a vector expressing Apo2L/TRAIL can attenuate collagen II-induced arthritis.7,8,57 Thus, it would be of interest to determine whether the activity of caspase-4 is elevated in the synovial tissues of patients with RA during the acute and chronic phases, and whether Apo2L/TRAIL gene therapy leads to the attenuation of inflammation in these tissues.
Caspase-8 also plays a role in lactacystin enhancement of anti-DR5-mediated apoptosis of RASFs. Caspase-8 is the key initiator caspase in the death-receptor pathway. On ligand binding, death receptors such as DR5 aggregate and form membrane-bound signaling complexes. These complexes then recruit, through adapter proteins, several molecules of procaspase-8, resulting in a high local concentration of zymogen and allowing the various proenzyme molecules to mutually cleave and activate each other. Whether caspase-4 is recruited to the caspase-8 complexes and caspase-8 is required to activate caspase-4 has not been established, and additional levels of regulation may exist in vivo to modulate the process. Although our results support the concept that the activation of caspase-8 is required for caspase-4-mediated cleavage of vimentin, an in vitro cleavage reaction (data not shown) failed to show that caspase-8 cleaves caspase-4 directly. This would suggest that a factor, or factors, downstream of caspase-8 also participate in caspase-8-mediated activation of caspase-4. Similarly, it remains unclear whether caspase-8 can cleave vimentin directly. Nakanishi and colleagues31 has reported that, during apoptosis, vimentin is first cleaved at Asp259 by a caspase-8-like protease and subsequently at Asp85 by a caspase-3-like protease. Byun and colleagues,32 have reported that vimentin is cleaved by caspase-6 in vitro, but not by caspase-8. Our data show that after caspase-4 is activated by caspase-8, inhibition of caspase-8 activity does not lead to the reduction of apoptosis of RASFs stimulated with anti-DR5 antibody in the presence of a proteasome inhibitor, suggesting that caspase-8 does not cleave vimentin directly, but that activation of caspase-8 is required for the initiation of activation of caspase-4 that then cleaves vimentin.
We have shown that vimentin-associated sequestration of p53 leads to the attenuation of anti-DR5-mediated apoptosis of RASFs and that activation of caspase-4 results in the cleavage of vimentin and release of p53 from the vimentin-p53 complex. Furthermore, overexpression of vimentin in RASFs led to the retention of p53 in the cytosol and reduction of Apo2L/TRAIL-mediated apoptosis of RASFs. However, mutation of the caspase-4 cleavage site of vimentin attenuates, but does not reverse, the effects produced by treatment of anti-DR5 plus lactacystin. This suggests that additional mechanisms of p53 regulation, such as conversion of monomeric p53 into a tetrameric form58 or Nedd-8 inactivation of p53,59,60 may also play a role in Apo2L/TRAIL-mediated apoptosis. In addition, it is likely that vimentin cleavage also regulates apoptosis of RASFs in a p53-independent manner because vimentin regulates organization of cytoplasmic structure,61 cell migration and response to mechanical stress,62 membrane trafficking, granular secretion, protein kinase activation, and regulation of stress response proteins.63 The present results suggest, however, that the major affect of vimentin in regulation of apoptosis after DR5 signaling is the release of p53, which can be significantly inhibited by expression of mutant vimentin, blocking of caspase-4 activation, or blocking of p53 function.
Blocking of the proteasome degradation pathway preferentially enhanced DR5-mediated apoptosis of RASFs without enhancing Fas-mediated apoptosis, suggesting that Fas-mediated apoptosis of RASFs is less dependent on the proteins regulated by the proteasome degradation machinery. Ichikawa and colleagues53 have reported that RASFs are more sensitive to apoptosis induced by DR5 than by Fas ligand, and a molecular basis for this preferential effect has been proposed64,65 but needs further exploration.
A normal fibroblast cell line, WI38, exhibited much less sensitivity to DR5-mediated apoptosis in the presence of lactacystin than RASFs did. Numerous studies indicate that TRAIL preferentially induces the apoptosis of transformed cells and cancer cells, but not normal cells.66–69 The expression profiles of many genes that regulate apoptosis signaling in RASFs have been altered, and RASF is considered to be a semitransformed cell.44,70–74 Therefore, it is not surprising that the sensitivity of RASFs to DR5-mediated apoptosis is different from that of normal fibroblasts. It is of interest in terms of the development of potential therapies for rheumatoid arthritis, that the inhibition of the proteasome degradation pathway enhanced the TRAIL apoptosis of the RASFs without inducing the susceptibility of normal cells.
The potential development of therapeutic strategies is complicated however, by the in vitro and in vivo studies that indicate that TRAIL can either function as a pro- or anti-inflammatory cytokine by participating in either the initiation or resolution of an inflammatory response.75–77 In an inflammatory environment such as the joints of a patient with rheumatoid arthritis, Apo2L/TRAIL apoptosis signaling may be influenced by tumor necrosis factor-α or Fas ligand during inflammation, and Apo2L/TRAIL signaling may, in turn, enhance this inflammatory response. Therefore, it will be of importance to determine the optimal approach for exploitation of Apo2L/TRAIL-mediated apoptosis in the inhibition of synovitis and inflammation in rheumatoid arthritis.
In summary, our data suggest that the tumor suppressor p53 is critically involved in the regulation of proliferation and apoptosis in rheumatoid synovial fibroblasts. Based on evidence that inhibition of endogenous p53 degradation leads to enhancement of Apo2L/TRAIL-mediated apoptosis, we suggest that abnormalities in the trafficking of p53 may contribute to defects of RASF apoptosis in general and to the development of synovial hyperplasia and tissue invasion in rheumatoid arthritis. Since proteasome inhibitors are now available for clinical use, the inhibition of endogenous p53 degradation may have a potential for treatment of rheumatoid arthritis.
Acknowledgments
We thank Dr. Fiona Hunter for editorial assistance, Dr. Vincent L. Cryns for providing a vimentin expression vector, Dr. Gary S. Firestein for providing RASF cells, Dr. Trygve Tollefsbol for providing the WI38 cell line, Mr. Landon Wilson for the MALDI-TOF-MS mass fingerprinting analysis. The MALDI-TOF mass spectrometer was purchased with funds from a NCRR Shared Instrumentation grant (S10 RR13795) and the UAB Health Services Foundation General Endowment Fund. Operation of the Mass Spectrometry Shared Facility is supported from NCI Grants (P30 CA-13148 and U54-100949) to the UAB Comprehensive Cancer Center and Center for Nutrient-Gene Interaction in Cancer Prevention.
Footnotes
Address reprint requests to Dr. Huang-Ge Zhang, University of Alabama at Birmingham, 701 South 19th St., LHRB 473, Birmingham, AL 35294-0007. E-mail: huang-ge.zhang@ccc.uab.edu.
Supported in part by the Arthritis Foundation (investigator award to H.-G.Z.), the National Institutes of Health (grants R21 AR43321 and P30 AR48311), the Sankyo Co. Ltd., and the Birmingham Veterans Administration Medical Center (merit review grant to H.-G.Z.).
X. Yang and J. Wang contributed equally to this work.
References
- Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, Goodwin RG. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- Gibson SB, Oyer R, Spalding AC, Anderson SM, Johnson GL. Increased expression of death receptors 4 and 5 synergizes the apoptosis response to combined treatment with etoposide and TRAIL. Mol Cell Biol. 2000;20:205–212. doi: 10.1128/mcb.20.1.205-212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin RG, Rauch CT, Schuh JC, Lynch DH. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Aratani S, Hatta M, Araya N, Daitoku H, Kawahara K, Watanabe S, Nakamura H, Yoshino S, Fujii R, Fujita H, Fukamizu A, Nishioka K, Nakajima T. TNFalpha induces acetylation of p53 but attenuates its transcriptional activation in rheumatoid synoviocytes. Int J Mol Med. 2002;10:269–275. [PubMed] [Google Scholar]
- Perlman H, Pagliari LJ, Volin MV. Regulation of apoptosis and cell cycle activity in rheumatoid arthritis. Curr Mol Med. 2001;1:597–608. doi: 10.2174/1566524013363429. [DOI] [PubMed] [Google Scholar]
- Makarov SS. NF-kappa B in rheumatoid arthritis: a pivotal regulator of inflammation, hyperplasia, and tissue destruction. Arthritis Res. 2001;3:200–206. doi: 10.1186/ar300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Wang S, Gambotto A, Glorioso JC, Evans CH, Robbins PD, Ghivizzani SC, Oligino TJ. Intra-articular adenoviral-mediated gene transfer of TRAIL induces apoptosis of arthritic rabbit synovium. Gene Ther. 2003;10:1055–1060. doi: 10.1038/sj.gt.3301881. [DOI] [PubMed] [Google Scholar]
- Song K, Chen Y, Goke R, Wilmen A, Seidel C, Goke A, Hilliard B. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is an inhibitor of autoimmune inflammation and cell cycle progression. J Exp Med. 2000;191:1095–1104. doi: 10.1084/jem.191.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokhlin OW, Guseva NV, Tagiyev AF, Glover RA, Cohen MB. Caspase-8 activation is necessary but not sufficient for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis in the prostatic carcinoma cell line LNCaP. Prostate. 2002;52:1–11. doi: 10.1002/pros.10074. [DOI] [PubMed] [Google Scholar]
- Secchiero P, Gonelli A, Carnevale E, Milani D, Pandolfi A, Zella D, Zauli G. TRAIL promotes the survival and proliferation of primary human vascular endothelial cells by activating the Akt and ERK pathways. Circulation. 2003;107:2250–2256. doi: 10.1161/01.CIR.0000062702.60708.C4. [DOI] [PubMed] [Google Scholar]
- Spalding AC, Jotte RM, Scheinman RI, Geraci MW, Clarke P, Tyler KL, Johnson GL. TRAIL and inhibitors of apoptosis are opposing determinants for NF-kappaB-dependent, genotoxin-induced apoptosis of cancer cells. Oncogene. 2002;21:260–271. doi: 10.1038/sj.onc.1205048. [DOI] [PubMed] [Google Scholar]
- Oya M, Ohtsubo M, Takayanagi A, Tachibana M, Shimizu N, Murai M. Constitutive activation of nuclear factor-kappaB prevents TRAIL-induced apoptosis in renal cancer cells. Oncogene. 2001;20:3888–3896. doi: 10.1038/sj.onc.1204525. [DOI] [PubMed] [Google Scholar]
- Arizono Y, Yoshikawa H, Naganuma H, Hamada Y, Nakajima Y, Tasaka K. A mechanism of resistance to TRAIL/Apo2L-induced apoptosis of newly established glioma cell line and sensitisation to TRAIL by genotoxic agents. Br J Cancer. 2003;88:298–306. doi: 10.1038/sj.bjc.6600666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B, Yue P, Clayman GL, Sun SY. Evidence that the death receptor DR4 is a DNA damage-inducible, p53-regulated gene. J Cell Physiol. 2001;188:98–105. doi: 10.1002/jcp.1101. [DOI] [PubMed] [Google Scholar]
- Kim K, Takimoto R, Dicker DT, Chen Y, Gazitt Y, El-Deiry WS. Enhanced TRAIL sensitivity by p53 overexpression in human cancer but not normal cell lines. Int J Oncol. 2001;18:241–247. doi: 10.3892/ijo.18.2.241. [DOI] [PubMed] [Google Scholar]
- Sheikh MS, Fornace AJ., Jr Death and decoy receptors and p53-mediated apoptosis. Leukemia. 2000;14:1509–1513. doi: 10.1038/sj.leu.2401865. [DOI] [PubMed] [Google Scholar]
- Zeimet AG, Riha K, Berger J, Widschwendter M, Hermann M, Daxenbichler G, Marth C. New insights into p53 regulation and gene therapy for cancer. Biochem Pharmacol. 2000;60:1153–1163. doi: 10.1016/s0006-2952(00)00442-1. [DOI] [PubMed] [Google Scholar]
- Wang S, El-Deiry WS. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene. 2003;22:8628–8633. doi: 10.1038/sj.onc.1207232. [DOI] [PubMed] [Google Scholar]
- Jang SH, Seol JY, Kim CH, Yoo CG, Kim YW, Han SK, Shim YS, Lee CT. Additive effect of TRAIL and p53 gene transfer on apoptosis of human lung cancer cell lines. Int J Mol Med. 2004;13:181–186. [PubMed] [Google Scholar]
- Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer. 2003;3:102–109. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- Inoue T, Geyer RK, Howard D, Yu ZK, Maki CG. MDM2 can promote the ubiquitination, nuclear export, and degradation of p53 in the absence of direct binding. J Biol Chem. 2001;276:45255–45260. doi: 10.1074/jbc.M107477200. [DOI] [PubMed] [Google Scholar]
- Daujat S, Neel H, Piette J. MDM2: life without p53. Trends Genet. 2001;17:459–464. doi: 10.1016/s0168-9525(01)02369-1. [DOI] [PubMed] [Google Scholar]
- Liang SH, Clarke MF. Regulation of p53 localization. Eur J Biochem. 2001;268:2779–2783. doi: 10.1046/j.1432-1327.2001.02227.x. [DOI] [PubMed] [Google Scholar]
- Ostermeyer AG, Runko E, Winkfield B, Ahn B, Moll UM. Cytoplasmically sequestered wild-type p53 protein in neuroblastoma is relocated to the nucleus by a C-terminal peptide. Proc Natl Acad Sci USA. 1996;93:15190–15194. doi: 10.1073/pnas.93.26.15190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Ostermeyer AG, Haladay R, Winkfield B, Frazier M, Zambetti G. Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol Cell Biol. 1996;16:1126–1137. doi: 10.1128/mcb.16.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotzsche O, Etzrodt D, Hohenberg H, Bohn W, Deppert W. Cytoplasmic retention of mutant tsp53 is dependent on an intermediate filament protein (vimentin) scaffold. Oncogene. 1998;16:3423–3434. doi: 10.1038/sj.onc.1202155. [DOI] [PubMed] [Google Scholar]
- Mucke N, Wedig T, Burer A, Marekov LN, Steinert PM, Langowski J, Aebi U, Herrmann H. Molecular and biophysical characterization of assembly-starter units of human vimentin. J Mol Biol. 2004;340:97–114. doi: 10.1016/j.jmb.2004.04.039. [DOI] [PubMed] [Google Scholar]
- Tsuruta D, Jones JC. The vimentin cytoskeleton regulates focal contact size and adhesion of endothelial cells subjected to shear stress. J Cell Sci. 2003;116:4977–4984. doi: 10.1242/jcs.00823. [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Omary MB. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr Opin Cell Biol. 2002;14:110–122. doi: 10.1016/s0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Maruyama M, Shibata T, Morishima N. Identification of a caspase-9 substrate and detection of its cleavage in programmed cell death during mouse development. J Biol Chem. 2001;276:41237–41244. doi: 10.1074/jbc.M105648200. [DOI] [PubMed] [Google Scholar]
- Byun Y, Chen F, Chang R, Trivedi M, Green KJ, Cryns VL. Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ. 2001;8:443–450. doi: 10.1038/sj.cdd.4400840. [DOI] [PubMed] [Google Scholar]
- Chen F, Chang R, Trivedi M, Capetanaki Y, Cryns VL. Caspase proteolysis of desmin produces a dominant-negative inhibitor of intermediate filaments and promotes apoptosis. J Biol Chem. 2003;278:6848–6853. doi: 10.1074/jbc.M212021200. [DOI] [PubMed] [Google Scholar]
- Muller K, Dulku S, Hardwick SJ, Skepper JN, Mitchinson MJ. Changes in vimentin in human macrophages during apoptosis induced by oxidised low density lipoprotein. Atherosclerosis. 2001;156:133–144. doi: 10.1016/s0021-9150(00)00641-9. [DOI] [PubMed] [Google Scholar]
- Slee EA, Adrain C, Martin SJ. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J Biol Chem. 2001;276:7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- Hsieh JK, Kletsas D, Clunn G, Hughes AD, Schachter M, Demoliou-Mason C. p53, p21(WAF1/CIP1), and MDM2 involvement in proliferation and apoptosis in an in vitro model of conditionally immortalized human vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2000;20:636–644. doi: 10.1161/01.atv.20.3.636. [DOI] [PubMed] [Google Scholar]
- Prasad S, Soldatenkov VA, Srinivasarao G, Dritschilo A. Intermediate filament proteins during carcinogenesis and apoptosis. Int J Oncol. 1999;14:563–570. doi: 10.3892/ijo.14.3.563. [DOI] [PubMed] [Google Scholar]
- Humke EW, Ni J, Dixit VM. ERICE, a novel FLICE-activatable caspase. J Biol Chem. 1998;273:15702–15707. doi: 10.1074/jbc.273.25.15702. [DOI] [PubMed] [Google Scholar]
- Kamada S, Washida M, Hasegawa J, Kusano H, Funahashi Y, Tsujimoto Y. Involvement of caspase-4(-like) protease in Fas-mediated apoptotic pathway. Oncogene. 1997;15:285–290. doi: 10.1038/sj.onc.1201192. [DOI] [PubMed] [Google Scholar]
- Henkart PA. ICE family proteases: mediators of all apoptotic cell death? Immunity. 1996;4:195–201. doi: 10.1016/s1074-7613(00)80428-8. [DOI] [PubMed] [Google Scholar]
- Wong WW. ICE family proteases in inflammation and apoptosis. Agents Actions Suppl. 1998;49:5–13. doi: 10.1007/978-3-0348-8857-8_2. [DOI] [PubMed] [Google Scholar]
- Concha NO, Abdel-Meguid SS. Controlling apoptosis by inhibition of caspases. Curr Med Chem. 2002;9:713–726. doi: 10.2174/0929867023370761. [DOI] [PubMed] [Google Scholar]
- Yamanishi Y, Boyle DL, Rosengren S, Green DR, Zvaifler NJ, Firestein GS. Regional analysis of p53 mutations in rheumatoid arthritis synovium. Proc Natl Acad Sci USA. 2002;99:10025–10030. doi: 10.1073/pnas.152333199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemayer CA, Kuchen S, Neidhart M, Kuenzler P, Rihoskova V, Neumann E, Pruschy M, Aicher WK, Muller-Ladner U, Gay RE, Michel BA, Firestein GS, Gay S. p53 in rheumatoid arthritis synovial fibroblasts at sites of invasion. Ann Rheum Dis. 2003;62:1139–1144. doi: 10.1136/ard.2003.007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HG, Huang N, Liu D, Bilbao L, Zhang X, Yang P, Zhou T, Curiel DT, Mountz JD. Gene therapy that inhibits nuclear translocation of nuclear factor kappaB results in tumor necrosis factor alpha-induced apoptosis of human synovial fibroblasts. Arthritis Rheum. 2000;43:1094–1105. doi: 10.1002/1529-0131(200005)43:5<1094::AID-ANR20>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Casillas MA, Jr, Lopatina N, Andrews LG, Tollefsbol TO. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. doi: 10.1023/a:1025548623524. [DOI] [PubMed] [Google Scholar]
- Zhang HG, Wang Y, Xie JF, Liang X, Liu D, Yang P, Hsu HC, Ray RB, Mountz JD. Regulation of tumor necrosis factor alpha-mediated apoptosis of rheumatoid arthritis synovial fibroblasts by the protein kinase Akt. Arthritis Rheum. 2001;44:1555–1567. doi: 10.1002/1529-0131(200107)44:7<1555::AID-ART279>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Zhang HG, Huang N, Liu D, Bilbao L, Zhang X, Yang P, Zhou T, Curiel DT, Mountz JD. Gene therapy that inhibits nuclear translocation of nuclear factor kappaB results in tumor necrosis factor alpha-induced apoptosis of human synovial fibroblasts. Arthritis Rheum. 2000;43:1094–1105. doi: 10.1002/1529-0131(200005)43:5<1094::AID-ANR20>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Dreskin SC, Dale SN, Foster SM, Martin D, Buchmeier A, Nelson HS. Measurement of changes in mRNA for IL-5 in noninvasive scrapings of nasal epithelium taken from patients undergoing nasal allergen challenge. J Immunol Methods. 2002;268:189–195. doi: 10.1016/s0022-1759(02)00206-5. [DOI] [PubMed] [Google Scholar]
- Meng RD, El-Deiry WS. p53-independent upregulation of KILLER/DR5 TRAIL receptor expression by glucocorticoids and interferon-gamma. Exp Cell Res. 2001;262:154–169. doi: 10.1006/excr.2000.5073. [DOI] [PubMed] [Google Scholar]
- Sheikh MS, Burns TF, Huang Y, Wu GS, Amundson S, Brooks KS, Fornace AJ, Jr, el-Deiry WS. p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor alpha. Cancer Res. 1998;58:1593–1598. [PubMed] [Google Scholar]
- Fei P, Bernhard EJ, El-Deiry WS. Tissue-specific induction of p53 targets in vivo. Cancer Res. 2002;62:7316–7327. [PubMed] [Google Scholar]
- Ichikawa K, Liu W, Fleck M, Zhang H, Zhao L, Ohtsuka T, Wang Z, Liu D, Mountz JD, Ohtsuki M, Koopman WJ, Kimberly R, Zhou T. TRAIL-R2 (DR5) mediates apoptosis of synovial fibroblasts in rheumatoid arthritis. J Immunol. 2003;171:1061–1069. doi: 10.4049/jimmunol.171.2.1061. [DOI] [PubMed] [Google Scholar]
- Morishima N. Changes in nuclear morphology during apoptosis correlate with vimentin cleavage by different caspases located either upstream or downstream of Bcl-2 action. Genes Cells. 1999;4:401–414. doi: 10.1046/j.1365-2443.1999.00270.x. [DOI] [PubMed] [Google Scholar]
- Ahn EY, Pan G, Vickers SM, McDonald JM. IFN-gamma upregulates apoptosis-related molecules and enhances Fas-mediated apoptosis in human cholangiocarcinoma. Int J Cancer. 2002;100:445–451. doi: 10.1002/ijc.10516. [DOI] [PubMed] [Google Scholar]
- Hitomi J, Katayama T, Eguchi Y, Kudo T, Taniguchi M, Koyama Y, Manabe T, Yamagishi S, Bando Y, Imaizumi K, Tsujimoto Y, Tohyama M. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and A{beta}-induced cell death. J Cell Biol. 2004;165:347–356. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamhamedi-Cherradi SE, Zheng SJ, Maguschak KA, Peschon J, Chen YH. Defective thymocyte apoptosis and accelerated autoimmune diseases in TRAIL−/− mice. Nat Immunol. 2003;4:255–260. doi: 10.1038/ni894. [DOI] [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Harper JW. Neddylating the guardian; Mdm2 catalyzed conjugation of Nedd8 to p53. Cell. 2004;118:2–4. doi: 10.1016/j.cell.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Trevor KT, McGuire JG, Leonova EV. Association of vimentin intermediate filaments with the centrosome. J Cell Sci. 1995;108:343–356. doi: 10.1242/jcs.108.1.343. [DOI] [PubMed] [Google Scholar]
- Wang N, Stamenovic D. Mechanics of vimentin intermediate filaments. J Muscle Res Cell Motil. 2002;23:535–540. doi: 10.1023/a:1023470709071. [DOI] [PubMed] [Google Scholar]
- Benitez-King G. PKC activation by melatonin modulates vimentin intermediate filament organization in N1E-115 cells. J Pineal Res. 2000;29:8–14. doi: 10.1034/j.1600-079x.2000.290102.x. [DOI] [PubMed] [Google Scholar]
- Mariani SM, Krammer PH. Differential regulation of TRAIL and CD95 ligand in transformed cells of the T and B lymphocyte lineage. Eur J Immunol. 1998;28:973–982. doi: 10.1002/(SICI)1521-4141(199803)28:03<973::AID-IMMU973>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Falsig J, Latta M, Leist M. Defined inflammatory states in astrocyte cultures: correlation with susceptibility towards CD95-driven apoptosis. J Neurochem. 2004;88:181–193. doi: 10.1111/j.1471-4159.2004.02144.x. [DOI] [PubMed] [Google Scholar]
- Thorburn J, Moore F, Rao A, Barclay WW, Thomas LR, Grant KW, Cramer SD, Thorburn A. Selective inactivation of a FADD-dependent apoptosis and autophagy pathway in immortal epithelial cells. Mol Biol Cell. 2005;16:1189–1199. doi: 10.1091/mbc.E04-10-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouralexis S, Findlay DM, Evdokiou A. Death to the bad guys: targeting cancer via Apo2L/TRAIL. Apoptosis. 2005;10:35–51. doi: 10.1007/s10495-005-6060-0. [DOI] [PubMed] [Google Scholar]
- Yagita H, Takeda K, Hayakawa Y, Smyth MJ, Okumura K. TRAIL and its receptors as targets for cancer therapy. Cancer Sci. 2004;95:777–783. doi: 10.1111/j.1349-7006.2004.tb02181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S, Debatin KM. Modulation of TRAIL signaling for cancer therapy. Vitam Horm. 2004;67:275–290. doi: 10.1016/S0083-6729(04)67015-4. [DOI] [PubMed] [Google Scholar]
- Masuda K, Masuda R, Neidhart M, Simmen BR, Michel BA, Muller-Ladner U, Gay RE, Gay S. Molecular profile of synovial fibroblasts in rheumatoid arthritis depends on the stage of proliferation. Arthritis Res. 2002;4:R8. doi: 10.1186/ar427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedel J, Gay RE, Kuenzler P, Seemayer C, Simmen B, Michel BA, Gay S. FLICE-inhibitory protein expression in synovial fibroblasts and at sites of cartilage and bone erosion in rheumatoid arthritis. Arthritis Rheum. 2002;46:1512–1518. doi: 10.1002/art.10309. [DOI] [PubMed] [Google Scholar]
- Neumann E, Kullmann F, Judex M, Justen HP, Wessinghage D, Gay S, Scholmerich J, Muller-Ladner U. Identification of differentially expressed genes in rheumatoid arthritis by a combination of complementary DNA array and RNA arbitrarily primed-polymerase chain reaction. Arthritis Rheum. 2002;46:52–63. doi: 10.1002/1529-0131(200201)46:1<52::AID-ART10048>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Muller-Ladner U, Kriegsmann J, Franklin BN, Matsumoto S, Geiler T, Gay RE, Gay S. Synovial fibroblasts of patients with rheumatoid arthritis attach to and invade normal human cartilage when engrafted into SCID mice. Am J Pathol. 1996;149:1607–1615. [PMC free article] [PubMed] [Google Scholar]
- Trabandt A, Gay RE, Gay S. Oncogene activation in rheumatoid synovium. APMIS. 1992;100:861–875. doi: 10.1111/j.1699-0463.1992.tb04012.x. [DOI] [PubMed] [Google Scholar]
- Zheng SJ, Wang P, Tsabary G, Chen YH. Critical roles of TRAIL in hepatic cell death and hepatic inflammation. J Clin Invest. 2004;113:58–64. doi: 10.1172/JCI200419255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi QS, Ly D, Lamhamedi-Cherradi SE, Salojin KV, Zhou L, Grattan M, Meagher C, Zucker P, Chen YH, Nagle J, Taub D, Delovitch TL. Blockade of tumor necrosis factor-related apoptosis-inducing ligand exacerbates type 1 diabetes in NOD mice. Diabetes. 2003;52:1967–1975. doi: 10.2337/diabetes.52.8.1967. [DOI] [PubMed] [Google Scholar]
- Hilliard B, Wilmen A, Seidel C, Liu TS, Goke R, Chen Y. Roles of TNF-related apoptosis-inducing ligand in experimental autoimmune encephalomyelitis. J Immunol. 2001;166:1314–1319. doi: 10.4049/jimmunol.166.2.1314. [DOI] [PubMed] [Google Scholar]