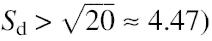Identification, characterization and molecular phylogeny of U12-dependent introns in the Arabidopsis thaliana genome (original) (raw)
Abstract
U12-dependent introns are spliced by the minor U12-type spliceosome and occur in a variety of eukaryotic organisms, including Arabidopsis. In this study, a set of putative U12-dependent introns was compiled from a large collection of cDNA/EST- confirmed introns in the Arabidopsis thaliana genome by means of high-throughput bioinformatic analysis combined with manual scrutiny. A total of 165 U12-type introns were identified based upon stringent criteria. This number of sequences well exceeds the total number of U12-type introns previously reported for plants and allows a more thorough statistical analysis of U12-type signals. Of particular note is the discovery that the distance between the branch site adenosine and the acceptor site ranges from 10 to 39 nt, significantly longer than the previously postulated limit of 21 bp. Further analysis indicates that, in addition to the spacing constraint, the sequence context of the potential acceptor site may have an important role in 3′ splice site selection. Several alternative splicing events involving U12-type introns were also captured in this study, providing evidence that U12-dependent acceptor sites can also be recognized by the U2-type spliceosome. Furthermore, phylogenetic analysis suggests that both U12-type AT-AC and U12-type GT-AG introns occurred in Na+/H+ antiporters in a progenitor of animals and plants.
INTRODUCTION
U12-dependent introns, initially discovered by Jackson (1) and Hall and Padgett (2), are a class of low-abundance introns which are spliced by a minor class (U12-dependent) spliceosome and are distributed in vertebrates, insects and plants (3). This rare class of introns is characterized by highly conserved consensus sequences /[AG]TATCCTT (where / denotes the exon end and [AG] indicates A or G) and TCCTTAAC at the donor and branch sites, respectively (2), in contrast to the much more degenerate splice signals in the major class (U2 type) introns that are spliced by the U2-type spliceosome (reviewed in 4,5). Correspondingly, the U12-type spliceosome consists of specific U11, U12, U4atac and U6atac snRNAs that recognize the U12-type splice signals (6–8). In addition, U12-type introns lack a polypyrimidine tract between the branch site sequence (BSS) and the 3′ splice site (3′ss). Despite these differences, the U12-type spliceosome resembles the conventional spliceosome in many ways (4). For instance, irrespective of the lack of sequence similarity, U11, U12, U4atac and U6atac snRNAs are likely to have roles in the U12-dependent spliceosome that are analogous to the roles of U1, U2, U4 and U6 snRNAs in the U2-dependent spliceosome, respectively. Recent experimental data proved that the stem–loop structure within the U6 snRNA can functionally substitute the U6atac snRNA stem–loop (9). Moreover, not only is U5 snRNA common in each of the two spliceosomes, but a growing number of auxiliary proteins have been confirmed to be shared by both spliceosomes (10–13). U12-type introns typically coexist with U2-type introns in alternate patterns in the same gene (3,14), and the splicing efficiency of U12-type introns can be promoted by splicing the flanking U2-type introns via the exon definition mechanism (15–17). U11/U12 di-snRNAs were found to bridge the 5′ splice site (5′ss) and the BSS in the initial recognition of U12-type introns, suggesting that the mechanism of intron definition also functions in the splicing of minor introns (18).
The U12-dependent spliceosome may have coexisted with the conventional spliceosome in the common ancestor of higher eukaryotes (19). The fact that vertebrates and higher plants share conserved features in the functional regions of U6atac and U12 snRNAs also provides evidence indicating an early origin of the U12-dependent splicing system (20). The differences between the two splicing machineries imply that the two spliceosomes evolved parallel to each other in separate lineages and then merged prior to the divergence of the animal and plant kingdoms (3).
Another distinguishing feature of U12-type introns is that the distance between the branch site adenosine and the acceptor site (DistBA) is unusually short, between 10 and 20 bp (21), while the DistBA of the U2-type introns can be over 100 bp (22). It has also been experimentally confirmed that spacing mutations that generate a predicted DistBA <10 nt or >20 nt strongly activate cryptic 3′ss (23). As a result, Dietrich et al. (23) proposed a local diffusion model to explain acceptor site selection in U12-type introns.
The initial recognition of the U12-type introns arose from its non-canonical dinucleotide termini AT-AC (1), distinct from the conventional GT-AG intron borders. Further research indicated that GT-AG introns can be spliced by U12-type spliceosomes, and, conversely, AT-AC introns can be spliced by U2-type spliceosomes (24,25). Therefore, intron type cannot be simply determined by the dinucleotide termini. This raises the question of how to distinguish U12-type introns from U2-type introns. Based on conserved motifs of the donor site and the branch site in the U12-type introns, Burge et al. (3) designed a computer program, named U12Scan, to address the issue of the identification of U12-type introns and conducted a survey in a variety of species based on the GenBank gene structure annotation. Later, Levine and Durbin (14) adopted a slightly different strategy to recognize human U12-type introns. They predicted U12-type introns in the human genome first, and confirmed the hypothetical introns by expressed sequence data, requiring a 64 bp perfect match between a transcript sequence fragment and the 32 bp flanking sequences of a predicted U12-type intron in both directions. The latter approach did not suffer from the incompleteness or likely errors in the GenBank annotation, but has its own problems. For example, any U12-type intron flanked by exons shorter than 32 bp would not be located. In addition, both analyses restricted their search of DistBA within a short region of the introns consistent with the assumption that no U12-type introns have DistBAs shorter than 8 nt or longer than 21 nt.
In a recent study, we mapped 176 915 Arabidopsis ESTs on the Arabidopsis genome, and 45 U12-type introns were identified in the set of EST-confirmed introns (26). Here, we have undertaken a more sophisticated analysis including 26 961 Arabidopsis full-length cDNAs in addition to the EST set used in the previous study. A total of 165 distinct U12-type introns were identified, including 50 AT-AC introns, one AT-AA intron, one GT-AT intron and 113 GT-AG introns, comprising many more than the overall number of U12-type introns previously reported in plants. Our analysis indicates that Arabidopsis U12-type introns not only share similar features with Arabidopsis U2-type introns in intron length distribution and low GC content relative to the flanking exons, but they also share almost identical splice signals with U12-type introns from other species. One significant discovery is that five U12-type AT-AC introns and seven U12-type GT-AG introns have DistBAs longer than 21 nt, the longest observed distance being 35 nt. When further extending the BSS search region, another novel U12-type GT-AG intron was identified with a DistBA of 39 bp. The presumed 21 bp maximum limit appears incorrect, even though the distribution of DistBAs of the U12-type introns shows a peak at 12 nt. Several alternative splicing events involving U12-type introns were also found in this study and provide evidence that U12-dependent 3′ss could be recognized by the U2-type spliceosome. Analysis of the cases of alternative splicing combined with dinucleotide preference analysis also demonstrates that the sequence context of the potential acceptor site may also have an important role in 3′ss selection, in addition to the spacing constraint. Furthermore, phylogenetic analysis provided an example of conservation of U12-type introns (placement and signals) throughout the plant and animal kingdoms in a family of Na+/H+ antiporters, suggesting that both U12-type AT-AC and U12-type GT-AG introns occurred in the same gene in a progenitor of animals and plants.
MATERIALS AND METHODS
cDNA/EST-confirmed introns in the Arabidopsis thaliana genome
The A.thaliana genome sequence (release of August 20, 2002) was retrieved from GenBank (http://www.ncbi.nih.gov/GenBank/), with accession nos NC_003070, NC_003071, NC_003074, NC_003075 and NC_003076 for the five chromosomes, respectively. Arabidopsis full-length cDNA sequences were also downloaded from GenBank (dated April 11, 2002), and Arabidopsis ESTs were downloaded from NCBI dbEST (http://www.ncbi.nlm.nih.gov/dbEST/) in December 2002. All 27 288 putative Arabidopsis proteins (data label: ATpep, version: July 25, 2002) were downloaded from The Institute of Genome Research (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/SEQUENCES/ATH1.pep), corresponding to the annotation of the same Arabidopsis genome release as the one used in this study.
A total of 26 961 full-length cDNAs and 176 915 ESTs were aligned with the Arabidopsis genome sequence using the GeneSeqer spliced alignment program (27) at high stringency in order to generate a reliable data set of Arabidopsis introns. The cDNA/EST-confirmed introns originated from the putative cognate spliced alignments with local similarity scores higher than 0.9 (26), and qualified introns were merged into a non-redundant intron set for subsequent analysis.
Identification of Arabidopsis U12-type introns
The identification procedure used follows the procedure established by Burge et al. (3). A brief description is given in the following. First, weight matrices for the splice sites of U12- and U2-type introns were derived from subsets of the transcript-confirmed Arabidopsis introns. The U12-type intron subset consisted of 47 introns with AT-AC termini and two introns with AT-AA termini that display strong U12-type splice signals as previously identified in a variety of species (3,14). The weight matrices for the recognition of U12-type introns in the subsequent analysis were generated from this subset with the MEME program (28). In addition, 70 189 cDNA/EST-confirmed GT-AG introns which lack the U12-type consensus sequence ATCC in positions +3 to +6 relative to the 5′ss were utilized as a training set to construct the corresponding weight matrixes for U2-type introns. The probabilities of the intron-type signals were then computed as the products of the corresponding position-specific probabilities, based on the observed residue frequencies derived from the transcript-confirmed introns. The log-odds ratio of the score derived from the U12-type splice signals versus that from the U2-type splice signals was computed for the 5′ss and the BSS of all the transcript-confirmed introns included in the training sets. The log-odds ratios were further normalized by subtracting the sample mean and dividing by the corresponding standard deviation. The normalized scores are referred to as S x, where x is d or b, denoting the donor site and the BSS, respectively. Then, introns with large values for both _S_d and _S_b were selected as U12-type introns [see Fig.1; also see fig. 2 in Burge _et al_. (3)]. Because one of the U12-type likely AT-AC introns in the training set has DistBA as long as 35 nt, we set the search region for the branch-site motif within the intron relative to the confirmed 3′ss corresponding to DistBA values in the range 6–35. The region [–5, +5] relative to the 5′ss was also scanned for possible ambiguities in assignment of the precise exon–intron junctions in cases where sequence repeats yield alternative spliced alignments. If necessary, such ambiguous cases were corrected manually to report the correct exon–intron borders.
Gene duplications
3044 gene pairs were identified in a recent study of large-scale gene duplications in the Arabidopsis genome (29). The gene pairs and their related information were downloaded from http://wolfe.gen.tcd.ie/athal/dup and used to study the fate of the U12-type introns after gene duplication.
Sequence alignment and phylogenetic tree construction
Homologous protein sequences were aligned by Clustal X (30) with default settings. The neighbor-joining trees were constructed based on the multiple alignments using MEGA2.1 (31), after removal of all columns in the alignment containing one or more gaps. The trees were based on the _p_-distance, which is the proportion of amino acid sites at which the two sequences compared are different. Confidence of the tree topology was assessed by a bootstrap test with 500 replicates.
Dinucleotide relative abundance in the proximity of the acceptor site of U12-type introns
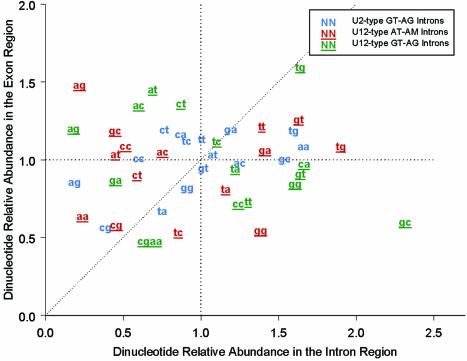
Let f x and f xy represent the frequency of the nucleotide x and the frequency of the dinucleotide xy, respectively. The dinucleotide relative abundance is defined as ρ_xy_ = f xy / f x f y, as a common assessment of dinucleotide bias (32). Dinucleotide relative abundances were calculated for the region between 10 bp downstream of the branch site adenosine and 1 bp upstream of the 3′ss and also in the equally sized region immediately downstream of the 3′ss within the exon. As a control, dinucleotide relative abundances were also derived for the U2-type GT-AG intron sequences in the 10 bp regions immediately preceding and succeeding the 3′ terminal dinucleotide. Hence, if the 5′-most AC downstream of the BSS is almost always selected as the 3′ss in U12-type AT-AC introns, the dinucleotide AC should be under-represented between the BSS and the 3′ss, that is, ρ_AC_ should be significantly smaller than 1.
RESULTS
Identification and characteristics of U12-dependent introns
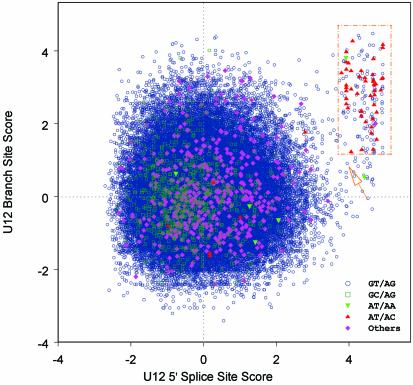
There are 53 introns with AT-AC terminal dinucleotides and six introns with AT-AA termini in the non-redundant transcript-confirmed Arabidopsis intron set that were identified as candidate U12-type introns. Because of the absence of the typical U12-type motifs for both the donor site and the branch site, six AT-AC introns and four AT-AA introns were removed. The remaining 49 introns were utilized to build weight matrices for U12-type 5′ss and BSS. The weight matrices of U2-type introns 5′ss and BSS were also constructed based on transcript-confirmed U2-type GT-AG introns (see Materials and Methods). On the basis of the derived U12- and U2-type weight matrices, the pairs of the standardized scores (_S_d, _S_b) of 75 717 transcript-confirmed introns were computed (see Materials and Methods), projecting these introns into points in the two-dimensional plane (Fig. 1). As expected, the 49 U12-type like AT-AM (M represents A or C) introns from the training set map in the upper-right corner in the plot, accompanied by one GT-AT intron and hundreds of GT-AG introns. Consistent with the manual inspection mentioned above, six AT-AC introns and four AT-AA introns map close to the origin in the plane and are thus predicted to be spliced by the U2-type spliceosome. One AT-AC intron and one AT-AA intron included in the training data set have relatively low values in either _S_d or _S_b when compared with the other 47 U12-type AT-AM introns. We conservatively excluded these two introns from further analysis. The remaining 47 AT-AM introns were selected as authentic U12-type introns for reference. Because there is not an obvious cluster to separate the putative U12-type introns from U2-type introns, the determinant of U12-type introns versus U2-type introns was empirically defined with respect to the standardized scores of the 47 introns in the reference set, such that the U12-type intron should satisfy the condition that _S_d and _S_b are no less than the minimum value of _S_d (= 3.79) and _S_b (= 1.23) from the 47 U12-type AT-AM introns, respectively. The selected introns roughly enclosed by the yellow rectangle in Figure 1 include 110 GT-AG introns, 46 AT-AC introns, one AT-AA intron and one GT-AT intron.
Figure 1.
Identification of U12-type introns. Each transcript-confirmed intron is represented by a point at coordinates (_S_d, _S_b) where _S_d and _S_b are the statistical score for donor site and branch site, respectively. The yellow rectangle identifies introns that were empirically classified as U12 type. In addition, a yellow arrow indicates a U12-type likely GT-AG intron not included in the selection (see text for details).
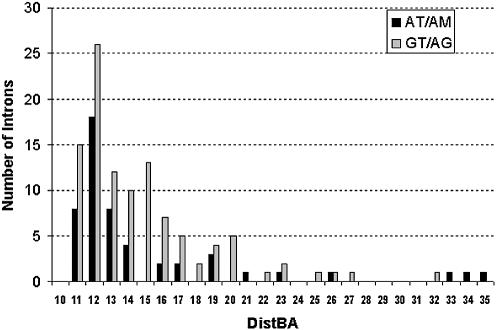
All 158 predicted U12-type introns and related information are listed in a table provided as Supplementary Material, together with another four U12-type AT-AC introns and three U12-type GT-AG introns identified by non-cognate transcripts described in the next section. As shown in Table 1, there are four U12-type GT-AG introns that are alternatively spliced with cryptic acceptor sites in the proximity of the normal 3′ss, which leads to ambiguity in the determination of DistBA in these cases. Thus, the analysis of the DistBA distribution was based on the remaining 157 distinct introns (51 AT-AM introns and 106 GT-AG introns) after excluding the four pairs of introns involved in alternative splicing (including seven GT-AG introns and one GT-AT intron; also see Table 1). As shown in Figure 2, the DistBA distribution of the U12-type AT-AM introns seems similar to that of the U12-type GT-AG introns. In particular, both distributions have the mode at 12 nt, and in both sets the shortest DistBA is 11 nt. Interestingly, 12 U12-type introns (five AT-AC introns and seven GT-AG introns) have distances longer than 21 nt, the maximum distance previously reported (14,23). To be conservative in assessing the authenticity of the U12-type introns in this study, the introns with long DistBA (>21 bp) were left out in the subsequent analysis of the sequence characteristics of U12-type introns.
Table 1. U12-type introns involved in alternative splicing.
| | ID | Chromosome | Location | Termini | DistBA | Evidence | Gene ID | Description | | | | ------------------------- | ---------- | -------- | -------- | -------- | -------- | ------- | ----------- | ---------------------------------------- | ---------------------------------------- | | | | | Start | End | | | | | | | | Alternative acceptor site | 40 | 2 | 11193817 | 11193727 | gt_ag | 27 | 2 | At2g26430 | Putative cyclin | | | 41 | 2 | 11193817 | 11193734 | gt_ag | 20 | 11 | | | | | | 68 | 3 | 4388199 | 4387910 | gt_ag | 26 | 1 | At3g13460 | Unknown protein | | | | 69 | 3 | 4388199 | 4387919 | gt_ag | 17 | 9 | | | | | | 83 | 3 | 19360482 | 19360367 | gt_ag | 14 | 5 | At3g52180 | Putative protein | | | | 84 | 3 | 19360482 | 19360395 | gt_ag | ? | 1 | | | | | | 102 | 4 | 5098265 | 5098368 | gt_at | 10 | 1 | At4g09720 | GTP-binding protein, putative | | | | 103 | 4 | 5098265 | 5098373 | gt_ag | 15 | 2 | | | | | Alternative donor site | 58 | 2 | 18375661 | 18375494 | gt_ag | 13 | 2 | At2g44680 | Putative casein kinase II beta subunit | | | NA | 2 | 18375664 | 18375494 | gc_ag | ? | 6 | | | | | | 90 | 3 | 22278983 | 22278836 | gt_ag | 14 | 2 | At3g60250 | Regulatory subunit of protein kinase CK2 | | | | NA | 3 | 22278986 | 22278836 | gc_ag | ? | 1 | | | | | Exon skipping | 18 | 1 | 17781408 | 17781268 | gt_ag | 22 | 1 | At1g49160 | Putative serine/threonine protein kinase | | | NA | 1 | 17781576 | 17781268 | gt_ag | ? | 2 | | | |
Figure 2.
Histogram of branch site to acceptor site distances (DistBA) of U12-type introns. The distances were compiled from 51 U12-type AT-AC or AT-AA introns (black bars) and 106 U12-type GT-AG introns (gray bars) listed in Supplementary Material. Note that 12 U12-type introns (five AT-AC introns and seven GT-AG introns) have branch site to 3′ss distances that are longer than 21 bp.
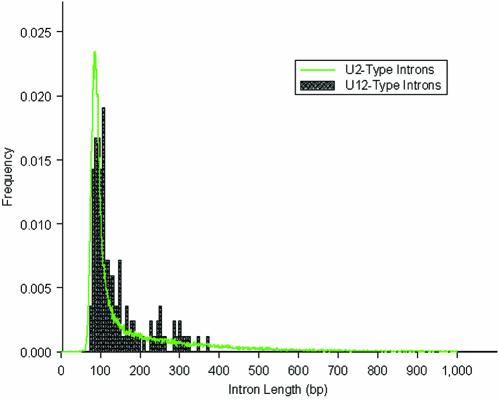
Thus, 46 U12-type AT-AM and 99 U12-type GT-AG introns comprise the set for further analysis. The Arabidopsis U12-dependent splice signals display a similar base composition to that of previously identified U12-type introns from various species (3). There is no significant difference in the length distribution between the U12-type introns and U2-type introns in Arabidopsis (Fig. 3), in contrast to the reported lack of short U12-type introns relative to U2-type introns in human (14). Furthermore, plant introns are characterized by low GC content when compared with the flanking exons (33), and our analysis shows that U12-type introns have this trait in common with the U2-type introns in Arabidopsis (data not shown).
Figure 3.
Length distribution of the U12- and U2-type introns. The histogram for the U2-type introns was derived from 70 189 transcript-confirmed Arabidopsis introns (plotted in green line). The histogram for U12-type introns (filled column) is based on 145 sequences.
Gene duplications and molecular phylogeny analysis
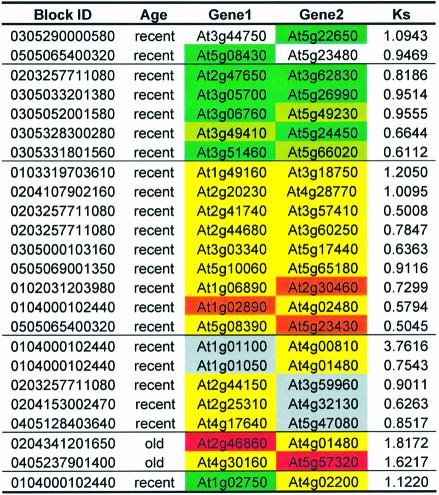
A recent study suggested that the Arabidopsis genome has undergone at least two large-scale duplications (29). The authors identified 3044 gene pairs divided into 91 chromosomal blocks and concluded that one event was a recent polyploidy which occurred 24–40 millions years ago (mya) and that the other event was an older one which happened after the monocot/dicot split. We found that among this set there are 24 gene pairs that have at least one U12-type intron in one or the other gene (Table 2). Based on our stringent criteria, the candidate U12-type introns are highly likely to be authentic U12-type introns. We cannot be sure, however, that the remaining transcript-confirmed introns are necessarily spliced by the major spliceosome, and, moreover, lack of transcript evidence may cause some U12-type introns in the duplicated gene set to remain undetected. In only two of the 24 gene pairs, the U12-type intron cannot be matched up with an intron in the paralog, reflecting an instance of intron loss or gain. Two of the gene pairs conserve a U12-type AT-AC intron, and six gene pairs conserve a U12-type GT-AG intron. Manual inspection led us to clarify three additional introns each as conserved AT-AC and GT-AG U12-type introns, respectively. In the remaining eight gene pairs there are five cases of uncertain GT-AG U12-type conservation (one of the introns in each pair having below-threshold _S_d and _S_b scores), two possible conversions of U12-type GT-AG introns to U2-type GT-AG introns (occurring in the only two gene pairs derived from the ancient large-scale gene duplications), and one case of a GT-AG/AT-AC U12-type pair. Excluding the five ambiguous cases, U12-type introns were stably conserved in 15 out of the remaining 19 pairs since the onset of gene divergence ∼24–40 mya (29). Thus, U12-type introns seem to be very stable in recent gene duplications, but are likely to be converted into U2-type introns in the long run.
Table 2. The fate of U12-type introns after large-scale segmental duplications in the Arabidopsis genome.
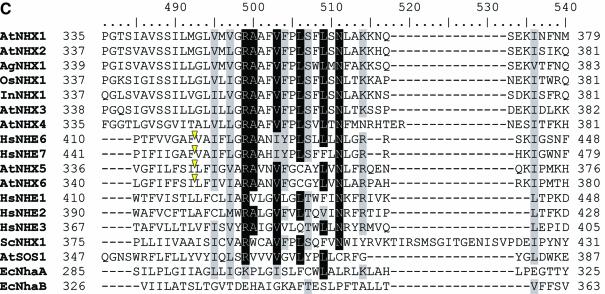
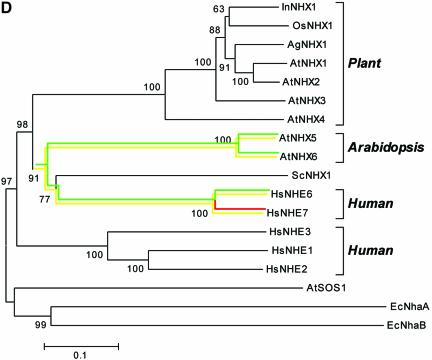
Because occasional (random) gene duplications occur in addition to the large-scale segmental genome duplications, we searched all the genes containing U12-type AT-AM introns against ATpep using BLASTP (34) and identified additional duplicated genes. This resulted in the detection of another novel U12-type AT-AC intron in the gene At1g76170, supported by spliced alignment of the non-cognate EST gi:23303104 from its paralogous gene At2g44270. The BLAST search also revealed more cases of the conservation of U12-type introns. The gene At1g79610, which encodes a low abundance Na+/H+ antiporter (AtNHX5) active in shoots and roots in Arabidopsis (35), contains a total of two U12-type AT-AC introns (intron 3 and 10), one U12-type GT-AG intron (intron 15) and 18 U2-type introns (see Supplementary Material for detailed descriptions of the U12-type introns). AtNHX5 shares its highly conserved sequence and gene structure with another family member, AtNHX6 (from gene At1g54370), which has two matching U12-type AT-AC introns (introns 3 and 10; listed in Supplementary Material). The intron type of the counterpart (intron 15) in AtNHX6 of the U12 GT-AG intron of AtNHX5 is uncertain, because the intron has a strong U12-dependent donor site but a relatively weak U12-dependent branch signal (TTCATGAC; _S_b = 0.89) with an 11 bp DistBA (indicated by the yellow hollow arrow in Fig. 1). However, the intron has a strong U12-dependent branch site signal (TCCTTGAC; _S_b = 3.40) with a 39 bp DistBA, whereas the branch site of the corresponding U12-type GT-AG intron in AtNHX5 has a DistBA of 32 bp. Whether the intron is an authentic U12-type intron and whether the high score branch site is functional in vivo will have to be determined by experimental methods. It would be the longest DistBA for the U12-type introns identified to date, if confirmed. There are four other members of Na+/H+ antiporter in Arabidopsis (AtNHX1–4). AtNHX5 and AtNHX6 have more sequence similarity with the human Na+/H+ exchangers HsNHE6 and HsNHE7 (Fig. 4; also see fig. 2 in Yokoi et al. (35)]. Interestingly, there are two U12-type GT-AG introns and one U12-type AT-AC intron reported in HsNHE6 (14). The Ensembl annotation indicates that the two corresponding U12-type GT-AG introns also exist in HsNHE7 (http://www.ensembl.org/Homo_sapiens/exonview?transcript=ENST00000163256&db=core). The annotation also displays an U2-type GT-AG intron near the location of the corresponding U12-type AT-AC intron in the locus of HsNHE7 (see Fig. 4A). However, this intron has intact U12-type splice site sequences, suggesting that the U12-type AT-AC introns may still be functional in HsNHE7 (Fig. 4B). Additionally, there are no U12-type introns in AtNHX1–4, which actually have almost completely different gene structures compared with AtNHX5–6. As shown in Figure 4A–C, the U12-type AT-AC intron and one of U12-type GT-AG introns have conserved locations between AtNHX5–6 and HsNHE6–7. With respect to the neighbor-joining tree of Na+/H+ antiporters from Arabidopsis, human, rice, Escherichia coli, yeast and other species [Fig. 4D; also see fig. 2 in Yokoi et al. (35)], we may infer that the appearance of the U12-type introns is dated prior to the divergence of AtNHX5–6 from HsNHE6–7, but after the divergence of AtNHX5–6 and AtNHX1–4.
Figure 4.
(Opposite and above) Analysis of Na+/H+ antiporters. The sources of the Na+/H+ antiporter protein sequences are as follows (GenBank accession nos in parentheses): AtNHX1 (AAD16946), AtNHX2 (AAM08403), AtNHX3, (AAO41905), AtNHX4 (AAM08405), AtNHX5(AAM08406), AtNHX6 (AAM08407) and AtSOS1 (AAF76139) from A.thaliana; InNHX1 (BAB60899) from Ipomoea nil; OsNHX1 (BAA83337) from rice; AgNHX1 (BAB11940) from Atriplex gmelini; ScNHX1 (NP_010744) from yeast; HsNHE1 (P19634), HsNHE2 (AAD41635), HsNHE3 (P48764), HsNHE6 (Q92581) and HsNHE7 (NP_115980) from human; EcNhaA (P13738) and EcNhaB (P27377) from E.coli. (A) The region [361, 420] of the multiple alignment of the Na+/H+ antiporters. Residues in each column of the alignments are shaded in black or gray if >70% of residues in the column are identical or similar. A phase-0 (i.e. between codons) U12-type AT-AC intron is marked by an upside down green triangle for AtNHX5, AtNHX6 and HsNHE6. Correspondingly, a phase-0 U2-type GT-AG intron is marked by a red triangle in HsNHE7. (B) The nucleotide sequences around the termini of the U2-type GT-AG intron marked by the red triangle in HsNHE7 in (A) (where | represents an exon–intron junction) and the sequence of the translation product. The intact U12-type splice signals are marked by shading. The potential U12-dependent splicing would replace the NAN tripeptide in the translation of the transcript resulting from U2-type splicing with the tetrapeptide VTAL, equal to the sequence in HsNHE6. (C) The region [481, 540] of the multiple alignment of the Na+/H+ antiporters. A phase-0 U12-type GT-AG intron is marked by an upside down yellow triangle for AtNHX5, AtNHX6, HsNHE6 and HsNHE7. (D) Neighbor-joining tree derived from the multiple alignment of the Na+/H+ antiporters using MEGA2 (31). The numbers on the tree branches are bootstrap values, and branch lengths are proportional to the pairwise _p_-distances as indicated by the scale bar in the lower left (see Materials and Methods for details). The branches are colored green, yellow and red corresponding to the occurrences of U12-type AT-AC, U12-type GT-AG and U2-type GT-AG introns, respectively.
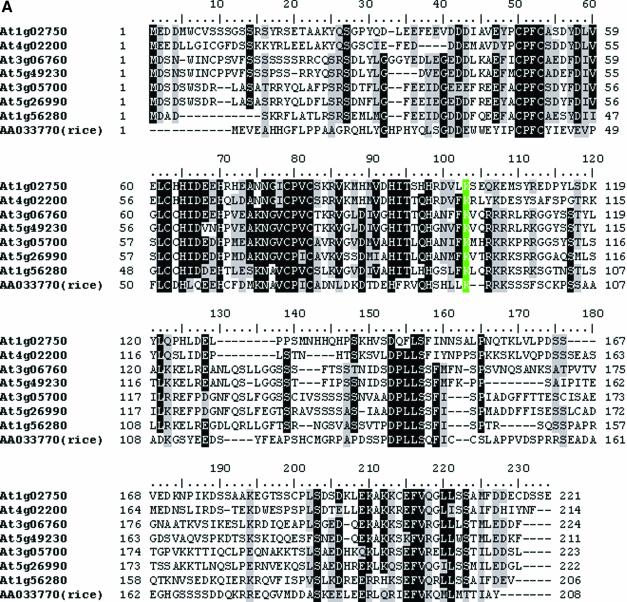
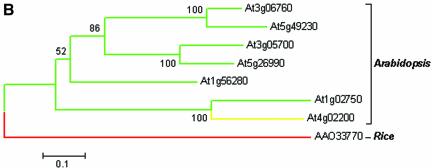
The genes At1g02750, At1g56280, At3g05700, At3g06760, At4g02200, At5g26990 and At5g49230 that encode proteins thought to be drought induced, all have one U12-type AT-AC intron in the same location. The only exception is the gene At4g02200, which has a U12-type GT-AG intron instead. After correcting annotation errors based on the transcript sequence data, the protein sequences of the seven genes and a homologous gene from rice (accession no. AAO33770) were aligned by Clustal X [Fig. 5A; for Clustal X see Jeanmougin et al. (30)] and a neighbor-joining tree was constructed based on the multiple alignment using MEGA2.1 [Fig. 5B; for MEGA2.1 see Kumar et al. (31)]. Detailed analysis indicates that the gene structures are highly conserved among the seven Arabidopsis genes and the rice gene. The identified U12-type introns are all in coding phase 0 (i.e. splitting two neighboring codons) and the same position starting after the conserved lysine (K103, highlighted in green in Fig. 5A), where an U2-type GT-AG intron is located in the rice homolog. We were particularly interested in this example to find out how the U12-type AT-AC intron switches to the U12-type GT-AG intron in the gene At4g02200 since the divergence from the gene At1g02750. The multiple alignment of At4g02200, At3g05700 and At1g02750 suggests that the conversion was probably initiated by the mutation of the 5′ terminal AT to GT, with subsequent activation of an AG downstream of the original acceptor site as the canonical 3′ss (Fig. 5C).
Figure 5.
(Opposite and above) Putative drought-induced proteins. (A) Alignment of the protein sequences from Arabidopsis and rice. There is a phase-0 intron conservatively located immediately after the green colored column K103 in all of the genes. At that location, the rice gene AAO33770 has a U2-type GT-AG intron and the Arabidopsis gene At4g02200 has a U12-type GT-AG intron, whereas the remaining six genes each have a U12-type AT-AC intron. (B) Neighbor-joining tree derived from the alignment in (A). The figure scheme follows Figure 4D. (C) Alignment of the U12-type intron sequences in the genes At4g02200, At1g02750 and At3g05700. Only terminal alignments are displayed and splicing signals are indicated by shading.
In total, non-cognate transcripts helped to identify four U12-type AT-AC introns and three GT-AG introns in this extended search. High conservation of the U12-type introns among paralogous genes was also observed in previous studies (3,14).
Alternative splicing
Seven cases of alternative splicing events related with U12-type introns were captured in this analysis (listed in Table 1). Four of the cases involve alternatively activated cryptic acceptor sites in the proximity of the normal splice sites. In detail, the GT-AG U12-type introns in the genes At2g26430 and At3g13460 both have minor isoforms utilizing CAG/, 7 and 9 nt downstream of the cognate acceptor site TAG/, respectively. This suggests that the distal AG with the favored sequence motif CAG/ may compete with the first AG downstream of the BSS in 3′ss selection of U12-type GT-AG introns. The ‘leaky’ scan revealed by these two alternative splicing events also implies that the spacing constraint in the 3′ss selection may not be as strong as previously assumed. Differing from the previous two examples, the U12-type GT-AG intron in At3g52180 has a cryptic acceptor site 28 nt upstream of the wild-type acceptor and 14 bp preceding the normal U12-dependent branch site. Motif search for U12-type branch site signals suggests the sequence TCCTTCGC as the most likely alternative branch site signal (_S_b = 1.66; with DistBA 28 bp, but G replacing the usual A) or GTTTTCAC (_S_b _= –_1.31; with DistBA 38 bp, unusually long). An alternative explanation is that the transcript isoform is spliced by the U2-type spliceosome rather than the minor spliceosome. The U12-type intron in the gene At4g09720 has a cryptic and unusual acceptor site AT/ (5 nt upstream of the cognate 3′ss) with a DistBA of 10 nt, which is the shortest DistBA found in this study.
Of the remaining three instances of alternative splicing listed in Table 1, one involves a U12-dependent donor site /GT (supported by two sequenced transcripts) 3 nt downstream of the dominant donor site /GC (supported by six sequenced transcripts) in the gene At2g44680. Further analysis revealed that the U12-type GT-AG intron in the paralogous gene At3g60250 is also alternatively spliced with a similar pattern. Interestingly, based on the few transcripts sampled, the major isoform for gene At3g60250 is the U12-type 5′ss /GT instead of the U2-type 5′ss /GC for gene At2g44680. The third example is an exon skipping event in the gene At1g49160, a putative serine/threonine protein kinase. The U12-type GT-AG intron in At1g49160 is alternatively spliced using the donor site of the upstream U2-type GT-AG intron. In all three examples, the alternative transcripts pair the U12-type 3′ss with an U2-type donor site, suggesting that a U12-type 3′ss can also be recognized by the major spliceosome. These examples also reveal a potential pathway for the conversion from U12-type GT-AG introns to U2-type introns via alternatively activating a cryptic U2-type 5′ss that subsequently becomes fixated. By EST evidence, retention of the U12-type AT-AC intron was observed in the genes At1g73350 and At5g63700. Intron retention may be a step preceding the loss of the U12-type AT-AC introns.
Selection of the acceptor site of U12-type introns
In our large-scale analysis on the Arabidopsis genome, we identified only four combinations of terminal dinucleotides for U12-type introns, with the vast majority GT-AG and AT-AC and only one U12-type AT-AA intron and one U12-type GT-AT intron (the latter belonging to a splicing isoform as mentioned above). It seems that the selection of the 3′ terminal dinucleotides of U12-type introns is highly correlated with the selection of the 5′ terminal dinucleotides of U12-type introns, that is /GT is typically matched with AG/, and /AT is paired with AC/ or AA/ occasionally. In order to probe whether the scanning model is applicable in the selection of U12-type 3′ss, we computed the dinucleotide preferences in the region around the U12-type acceptor site (see Materials and Methods; Fig. 6). The results indicate that all dinucleotides starting with adenosine are under-represented upstream of the U12- dependent acceptor site. Further analysis revealed that there are six AC occurrences between the BSS and 3′ss in the total 51 AT-AM introns. All six AC are located in introns with DistBA < 18 nt, and in each case the AC occurs immediately upstream of the 3′ss AC/. In none of these cases is the first AC preceded by a C, so that these sites conform to the consensus pattern [AGT]ACAC/. Thus, CAC/ seems to be strongly preferred as the acceptor site of U12-type AT-AC introns. This suggests that the selection of the U12-dependent acceptor site does not follow a simple scanning mechanism that selects the first AC following the branch site as the acceptor site. Rather, the sequence surrounding the 3′ss also plays an important role in the selection of U12-dependent acceptor sites. In this context, the five U12-type AT-AC introns with DistBA larger than 21 nt all have CAC/ as the acceptor site, further indicating the importance of the surrounding sequence in the selection of U12-type 3′ss.
Figure 6.
Dinucleotide relative abundances in the proximity of the 3′ss of U12- and U2-type introns. The dinucleotide relative abundances (see Materials and Methods for definition) between the BSS and the acceptor site versus the equivalent size region immediately succeeding to the acceptor site were plotted for U12-type AT-AM introns (red fonts with underline), U12-type GT-AG introns (green fonts with underline) and U2-type GT-AG introns (blue fonts).
AG is strongly avoided in the proximal region prior to the acceptor site in either of the two classes of introns. Excluding the U12-type introns involved in alternative splicing, only one dinucleotide AG was found immediately prior to the cognate 3′ss CAG/ (in gene At4g02200). It seems that the scan model may be more applicable to the U12-type GT-AG introns than to the U12-type AT-AC introns.
DISCUSSION
Identification of U12-type introns
Using a computational approach that largely follows the procedure proposed by Burge et al. (3), we identified a total of 165 U12-type introns in the Arabidopsis genome, including 50 AT-AC introns, one AT-AA intron, 113 GT-AG introns and one GT-AT intron. A slight difference in our approach is the definition of the test criterion for intron-type classification.Burge et al. used as the test statistics t = _S_b2 + _S_d2 > 20, where _S_b and _S_d are normalized log-odds ratios discriminating U12-type introns against U2-type introns by branch and donor site characteristics, respectively. This criterion implies that an intron with a strong U12-dependent donor site signal (i.e.
should still be spliced by the U12-type spliceosome, in spite of a possibly weak branch site signal (and vice versa). Experimental evidence indicated that the donor and branch sites interact simultaneously in a U11/U12 di-snRNA complex at the step of initial recognition of the U12-type intron by the minor spliceosome (18). Other studies showed that normal U12-dependent intron splicing can be abolished by mutations in the BSS (16). Thus, both donor and branch site features appear necessary for proper splicing. Therefore, we adopted a more conservative criterion requiring an intron to exceed minimal values of both _S_d and _S_b for it to be classified as U12 type. Compared with Arabidopsis U12-type introns reported in the earlier study (3), 10 out of 11 were accurately recovered in this study, with the exception of one AT-AA intron in gene At3g51830 encoding the transmembrane protein G5p (represented by the green upside down triangle below the yellow rectangle in Fig. 1).
Characteristics of Arabidopsis U12-dependent introns
The identified Arabidopsis U12-dependent introns display patterns almost identical to the motifs of the U12-type introns from various other species, which is in accordance with the postulated early common origin of U12-type introns predating the divergence of animals and plants (3,19). Different from the characteristics of the U12-type introns recently identified from the human genome (14), however, our results illustrate that there is neither an appreciable difference in the distribution of intron length between U2-type introns and U12-type introns in Arabidopsis, nor is there a significant difference in the distribution of DistBAs between U12-type AT-AM introns and U12-type GT-AG introns (see Figs 2 and 3).
The distance between the branch site and the 3′ splice site and the selection of the acceptor site of U12-type introns
Both of the two previous large-scale computational scans for U12-type introns were restricted to DistBA between 8 and 21 bp (3,14), based on the distribution of the DistBAs of naturally occurring U12-type introns (21). Experimental evidence for this range came mainly from the recent study of spacing mutants in the human P120 gene, in which the unfavorable dinucleotide UU with a DistBA of 12 nt was selected as the 3′ss rather than a downstream AC with a DistBA of 27 nt (23; construct +27 AC). However, the authors’ conclusion that the DistBA constraint is extremely strong in U12-type intron splicing seems problematic, because the uncommon guanosine immediately prior to the +27 AC might have precluded this dinucleotide as a functional acceptor site in that construct. Here, we have made three observations that further question that conclusion, at least as a model for Arabidopsis. First, at least 12 U12-type introns have DistBAs larger than 21 bp, even though the mode of the DistBA distribution is 12 nt (see Supplementary Material and Fig. 2). Secondly, only one AT-AA intron and one GT-AT intron were found in addition to the U12-dependent GT-AG and AT-AC introns, suggesting that the combination of GT-AG or AT-AC intron termini is strongly preferred in naturally occurring U12-type introns. Thirdly, there are six cases in which the dinucleotide AC is located immediately prior to the confirmed 3′ss in the U12-type AT-AC introns, indicating that the 3′ss surrounding sequence also plays an important role in the selection of the acceptor site of U12-dependent introns. Similar results were also observed in the U12-type GT-AG introns. Furthermore, the alternative 3′ss events in the U12-type GT-AG introns (Table 1) also confirm that distal acceptor sites with favorable sequence context can compete with the proximal wild-type 3′ss in U12-type intron splicing.
A caveat concerning the existence of long DistBAs is that there might be a weak BSS actually functioning downstream of the predicted BSS. Another possibility is that the introns with long DistBA (>21 bp) may actually be spliced by the U2-type spliceosome. However, there is no experimental support for either of these scenarios, and therefore it seems more reasonable to assume that a small number of U12-type introns have longer DistBAs than previously thought. To further test the allowable DistBA range, we searched for BSS also in an extended window allowing distances up to 45 nt. Only one instance was discovered with a possible longer distance (39 nt in AtNHX6), suggesting that the adopted range is reasonable.
The functional regions of U6atac and U12 snRNAs are highly conserved between human and Arabidopsis (20). Therefore, the longer range DistBA values observed in the Arabidopsis U12-type introns in this study are likely to be also applicable to humans and other species. Hence, rescanning the genomes of humans and other organisms with an extended search region may reveal more novel U12-type introns.
Evolutionary origins and fates of U12-type introns
To date, the fission/fusion model proposed by Burge et al. (3) is well accepted as an explanation of the origin of the U12-dependent spliceosome. According to this model, two splicing systems diverged in two separate lineages, but fused (probably via endosymbiosis) in a progenitor of higher eukaryotes (3). Compared with the parasitic invasion and codivergence model, the fission/fusion model provides a better explanation for the observation of genes with multiple U12-type introns (3). The analysis of Na+/H+ antiporters in this study gives additional support to the fission/fusion model. There are three U12-type introns in AtNHX5–6 and HsNHE6 (and probably also in HsNHE7, if the predicted U12-type AT-AC intron is actually also spliced in vivo; see Fig. 4B). Two out of the three U12-type introns are conserved across human and Arabidopsis, and the remaining one is unrelated between each other. This implies that at least two U12-type introns in AtNHX5–6 and HsNHE6-7 have orthologous origins, and the common ancestor of AtNHX5–6 and HsNHE6–7 may have contained at least four U12-type introns. Phylogenetic analysis of the Na+/H+ antiporter family dated the likely fusion event as being prior to the divergence of the plant kingdom and the animal kingdom and subsequent to the divergence of AtNHX1–4 and AtNHX5–6 [Fig. 4D; also see fig. 2 in Yokoi et al. (35)]. This analysis also gives the evidence that the U12-type introns are evolutionarily stable over one billion years. This amazing stability probably results from the unusual conserved U12-dependent 5′ss and BSS, so that any mutation in the splice signal sequences may easily disrupt the normal splicing of the U12-type AT-AC introns and thereby is strongly selected against. The alternative splicing examples, however, also demonstrate that U12-type AT-AC introns can be lost by intron retention, which is probably caused by mutations in the U12-dependent splice signals. In addition to the experimental evidence indicating that the U12-dependent 5′ss can be exploited by the major spliceosome (24), the alternative splicing events captured in this study also indicate that the 3′ss (probably as well as the BSS) of the U12-type GT-AG introns can also be exploited by the major spliceosome. Therefore, mutations that corrupt the conserved U12- dependent splice signals may easily trigger the conversion from the U12-type GT-AG introns to the U2-type GT-AG intron, while the reverse process is highly improbable.
Besides stability or loss, another fate of U12-type AT-AC introns may involve switching to U12-type GT-AG introns. A plausible mechanism of the switch is as follows. The 5′ terminal dinucleotide /AT mutates to /GT and then the first AG or the distal AG with the favored surrounding sequence in the downstream of the BSS is selected in the U12-type intron splicing. Under this model, the mutation is likely to cause the downstream exon to be truncated or extended with an alternate reading frame. Therefore, the chance of conversion from U12-type AT-AC introns to U12-type GT-AG introns is very low. Another mechanism of the switch proposed by Burge et al. (3) is from AT-AC to AT-AG and then to GT-AG. However, because it requires two mutation events and the occurrence of natural AT-AG introns is extremely low, the chance of successful conversion under this pathway should be also extremely low. At any rate, the switch between the U12-type AT-AC intron and the U12-type GT-AG intron should be much rarer than the conversion from the U12-type GT-AG intron to the U2-type GT-AG intron. However, it is difficult to explain why the U12-type GT-AG introns outnumber the U12-type AT-AC introns. We are forced to infer that U12-type GT-AG introns did not originate from U12-type AT-AC introns but appeared together with the latter one billion years ago. The analysis of the Na+/H+ antiporters supports this conjecture (see Fig. 4D). However, another plausible explanation is that there is some kind of selection against the conversion from the U12-type GT-AG intron to the U2-type GT-AG intron. It was noted that most of the genes containing U12-type introns function in information processing (3). In this study, some of the genes containing U12-type introns were found to be stress reaction related, such as the putative drought-induced proteins (Fig. 5) and AtNHX5, whose expression level increases in response to salt treatment (35). It is possible that the U12-dependent spliceosome system might be activated and therefore regulates the expression level of target genes that contain U12-type introns via changing the speed of U12-type intron splicing (36) in response to stresses in plants or the analogous situations in vertebrates or insects. In this scheme, the potential role of the U12-dependent spliceosome system may result in selective pressure against the conversion from U12-type GT-AG introns to U2-type GT-AG introns.
In spite of the high stability and possible selective advantage, it is likely that the number of U12-type introns has been slowly but continuously reduced by accumulating mutations. Gene duplication, however, may help the U12-type introns propagate within the gene families, for example, the putative drought-induced protein family, Na+/H+ antiporters and dTDP-glucose 4-6-dehydratease like proteins (26).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
[Supplementary Material]
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Jacqueline E. Townsend for help with preparation of the manuscript, Richard A. Padgett for kindly providing reprints, and Shannon D. Schlueter for critical discussions. This work was supported in part by NSF grant DBI-0110254 to V.B.
REFERENCES
- 1.Jackson I.J. (1991) A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res., 19, 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall S.L. and Padgett,R.A. (1994) Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J. Mol. Biol., 239, 357–365. [DOI] [PubMed] [Google Scholar]
- 3.Burge C.B., Padgett,R.A. and Sharp,P.A. (1998) Evolutionary fates and origins of U12-type introns. Mol. Cell, 2, 773–785. [DOI] [PubMed] [Google Scholar]
- 4.Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F., Cech,T. and Atkins,J.F. (eds), The RNA World II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560. [Google Scholar]
- 5.Wu Q. and Krainer,A.R. (1999) AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol. Cell. Biol., 19, 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall S.L. and Padgett,R.A. (1996) Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science, 271, 1716–1718. [DOI] [PubMed] [Google Scholar]
- 7.Tarn W.Y. and Steitz,J.A. (1996) Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science, 273, 1824–1832. [DOI] [PubMed] [Google Scholar]
- 8.Tarn W.Y. and Steitz,J.A. (1996) A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell, 84, 801–811. [DOI] [PubMed] [Google Scholar]
- 9.Shukla G.C. and Padgett,R.A. (2001) The intramolecular stem–loop structure of U6 snRNA can functionally replace the U6atac snRNA stem–loop. RNA, 7, 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo H.R., Moreau,G.A., Levin,N. and Moore,M.J. (1999) The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA, 5, 893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Will C.L., Schneider,C., Reed,R. and Lührmann,R. (1999) Identification of both shared and distinct proteins in the major and minor spliceosomes. Science, 284, 2003–2005. [DOI] [PubMed] [Google Scholar]
- 12.Will C.L., Schneider,C., MacMillan,A.M., Katopodis,N.F., Neubauer,G., Wilm,M., Lührmann,R. and Query,C.C. (2001) A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J., 20, 4536–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider C., Will,C.L., Makarova,O.V., Makarov,E.M. and Lührmann,R. (2002) Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions. Mol. Cell. Biol., 22, 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine A. and Durbin,R. (2001) A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res., 29, 4006–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Q. and Krainer,A.R. (1996) U1-mediated exon definition interactions between AT-AC and GT-AG introns. Science, 274, 1005–1008. [DOI] [PubMed] [Google Scholar]
- 16.Dietrich R.C., Shukla,G.C., Fuller,J.D. and Padgett,R.A. (2001) Alternative splicing of U12-dependent introns in vivo responds to purine-rich enhancers. RNA, 7, 1378–1388. [PMC free article] [PubMed] [Google Scholar]
- 17.Hastings M.L. and Krainer,A.R. (2001) Functions of SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway. RNA, 7, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frilander M.J. and Steitz,J.A. (1999) Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev., 13, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H.J., Gaubier-Comella,P., Delseny,M., Grellet,F., Van Montagu,M. and Rouzé,P. (1996) Non-canonical introns are at least 109 years old. Nature Genet., 14, 383–384. [DOI] [PubMed] [Google Scholar]
- 20.Shukla G.C. and Padgett,R.A. (1999) Conservation of functional features of U6atac and U12 snRNAs between vertebrates and higher plants. RNA, 5, 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp P.A. and Burge,C.B. (1997) Classification of introns: U2-type or U12-type. Cell, 91, 875–879. [DOI] [PubMed] [Google Scholar]
- 22.Smith C.W. and Nadal-Ginard,B. (1989) Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell, 56, 749–758. [DOI] [PubMed] [Google Scholar]
- 23.Dietrich R.C., Peris,M.J., Seyboldt,A.S. and Padgett,R.A. (2001) Role of the 3′ splice site in U12-dependent intron splicing. Mol. Cell. Biol., 21, 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dietrich R.C., Incorvaia,R. and Padgett,R.A. (1997) Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell, 1, 151–160. [DOI] [PubMed] [Google Scholar]
- 25.Wu Q. and Krainer,A.R. (1997) Splicing of a divergent subclass of AT-AC introns requires the major spliceosomal snRNAs. RNA, 3, 586–601. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu W., Schlueter,S.H. and Brendel,V. (2003) Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol., 132, 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Usuka J., Zhu,W. and Brendel,V. (2000) Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics, 16, 203–211. [DOI] [PubMed] [Google Scholar]
- 28.Bailey T.L. and Elkan,C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol., 2, 28–36. [PubMed] [Google Scholar]
- 29.Blanc G., Hokamp,K. and Wolfe,K.H. (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res., 13, 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeanmougin F., Thompson,J.D., Gouy,M., Higgins,D.G. and Gibson,T.J. (1998) Multiple sequence alignment with Clustal X. Trends Biochem. Sci., 23, 403–405. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S., Tamura,K., Jakobsen,I.B. and Nei,M. (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics, 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 32.Burge C., Campbell,A.M. and Karlin,S. (1992) Over- and under-representation of short oligonucleotides in DNA sequences. Proc. Natl Acad. Sci. USA, 89, 1358–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brendel V., Carle-Urioste,J.C. and Walbot,V. (1998) Intron recognition in plants. In Bailey-Serres,J. and Gallie,D.R. (eds), A Look Beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. American Society of Plant Physiology, Rockville, MD, USA, pp. 20–28. [Google Scholar]
- 34.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokoi S., Quintero,F.J., Cubero,B., Ruiz,M.T., Bressan,R.A., Hasegawa,P.M. and Pardo,J.M. (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J., 30, 529–539. [DOI] [PubMed] [Google Scholar]
- 36.Patel A.A., McCarthy,M. and Steitz,J.A. (2002) The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J., 21, 3804–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]