AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals (original) (raw)
Abstract
Introduction of transgene DNA may lead to specific degradation of RNAs that are homologous to the transgene transcribed sequence through phenomena named post-transcriptional gene silencing (PTGS) in plants, quelling in fungi, and RNA interference (RNAi) in animals. It was shown previously that PTGS, quelling, and RNAi require a set of related proteins (SGS2, QDE-1, and EGO-1, respectively). Here we report the isolation of Arabidopsis mutants impaired in PTGS which are affected at the Argonaute1 (AGO1) locus. AGO1 is similar to QDE-2 required for quelling and RDE-1 required for RNAi. Sequencing of ago1 mutants revealed one amino acid essential for PTGS that is also present in QDE-2 and RDE-1 in a highly conserved motif. Taken together, these results confirm the hypothesis that these processes derive from a common ancestral mechanism that controls expression of invading nucleic acid molecules at the post-transcriptional level. As opposed to_rde-1_ and qde-2 mutants, which are viable, ago1 mutants display several developmental abnormalities, including sterility. These results raise the possibility that PTGS, or at least some of its elements, could participate in the regulation of gene expression during development in plants.
In most eukaryotes, transgenes are not always expressed as expected. In plants, silencing can occur through a block of transgene transcription (transcriptional gene silencing) or through the specific degradation of transgene RNA (post-transcriptional gene silencing: PTGS) (1–5). When transgene RNAs are homologous to RNAs encoded by endogenous genes, both types of RNA are degraded, a phenomenon referred to as cosuppression (6). In the fungus Neurospora crassa, a phenomenon similar to cosuppression has been described and called quelling (7–9). Subsequently, PTGS of endogenous genes mediated by introduction of double-stranded RNA (dsRNA) was demonstrated in many organisms, including worms, flies, and mammals, and called RNA interference (RNAi) (10–13). To trigger RNAi, dsRNA can be supplied exogenously (14) or transcribed from transgenes carrying an internal inverted repeat (15). In plants, PTGS mediated by internal inverted repeat transgenes or by co-expression of sense and antisense transgenes was also reported (16–18), suggesting that PTGS, quelling, and RNAi could be related phenomena deriving from an ancestral mechanism directed against invading nucleic acids.
Models that explain PTGS/quelling/RNAi involve dsRNA molecules as a key intermediate leading to specific RNA degradation (5, 13). When dsRNAs are not directly produced by an internal inverted repeat transgene or exogenously supplied, the unintended transcription of antisense RNA from a neighboring promoter or read-through from adjacent transgenes arranged as an inverted repeat (5) could explain the production of dsRNA. In addition, the synthesis of complementary RNA (cRNA) by an enzyme called RNA-dependent RNA polymerase (RdRP) (19) could also participate in the production of dsRNA. This latter hypothesis was confirmed first by the identification of small cRNA, 21–25 nucleotides long, in post-transcriptionally-silenced (PTG-silenced) plants (20), second by the demonstration that proteins similar to tomato RdRP are required for quelling in_Neurospora_ (QDE-1) (21) and PTGS in Arabidopsis(SGS2) (22). Surprisingly, a protein similar to tomato RdRP was also shown to be required for RNAi in Caenorhabditis elegans (EGO-1) (23), thus indicating that RdRP could participate not only in the synthesis of cRNA (and consequently to the formation of dsRNA), but also in the amplification of the dsRNA signal, allowing silencing to spread throughout the organism (8, 10, 24, 25). How dsRNA leads to the degradation of homologous RNA remains unclear. It has been proposed that dsRNA is targeted by a dsRNA endonuclease to generate short dsRNA pieces, 21–25 nucleotides long (13). Then, annealing of these short dsRNA pieces with mRNA, followed by strand exchange, would generate new substrates cleaved at the same sites by the ribonuclease.
In addition to EGO-1, two other proteins (RDE-1 and MUT-7) required for RNAi in C. elegans have been identified (26, 27). RDE-1 is similar to rabbit eIF2C, which is assumed to participate in the control of translation initiation (28). RDE-1 may be brought to the target mRNA (via the interaction with the interfering dsRNA) and displace or perturb positioning of eIF2C (to which RDE-1 is most similar) in the translation machinery complex, thus preventing translation of the target mRNA (26). MUT-7 has homology with the catalytic domain of_Escherichia coli_ RNase D. MUT-7, guided by dsRNA, could degrade specific mRNA targets (27). In Neurospora, quelling requires QDE-2, which is similar to C. elegans RDE-1, thus indicating that quelling and RNAi share this step as well as the RdRP-controlled step (29). Although eight homologs of RDE-1/QDE-2 exist in Arabidopsis (refs. 26, 29–32, and this work), there was so far no evidence for their role in PTGS. Here, we report the isolation of new PTGS mutants of Arabidopsis that are impaired in one of these homologs encoded by the AGO1 gene (30). Similarities between AGO1, QDE-2, and RDE-1 therefore reinforce the idea that PTGS is mechanistically linked to quelling and RNAi.
Materials and Methods
Plant Material and Growth Conditions.
After seed sterilization, plants of _Arabidopsis thaliana_Heyn, ecotype Columbia, from which were derived the PTG-silenced 35S-β-glucuronidase (35S-GUS) transgenic lines L1 and L2 (33), the PTG-silenced 35S-NIA2 line 2a3 (33), and the_ago1_-3 mutant (30), were grown in sterile medium under a 16 h light/8 h dark regime at 100 μmol⋅m−2⋅s−1. Plants were transferred to soil and grown under the same light regime.
Mutant Selection.
Ten thousand plants derived from the self-progeny of 500 ethyl methanesulfonate-mutagenized seeds of the PTG-silenced 35S-GUS transgenic line L1 were grown in sterile medium. Because this ethyl methanesulfonate library had been screened previously for PTGS_sgs_ mutants that develop as wild type in the greenhouse, plants with a wild-type phenotype having developed a flowering stem after 4 weeks of growth in sterile medium were eliminated. The remaining plants were allowed to grow for another 2 weeks. GUS activity [in nmol of 4-methylumbelliferone (MU) per min per μg of total protein] was then measured in the leaves as described before (33).
Genetic Analysis.
Because ago1 mutants are infertile, allelism tests were performed by crossing parents heterozygous for each allele. Introgression of L1, L2, and 2a3 loci into the _ago1_background was performed by crossing homozygous transgenic lines with parents heterozygous for the ago1_-3 allele. F1 hybrids were allowed to self-fertilize, and those segregating 25% ago1 mutants after self-fertilization were retained. F2 plants carrying the L1 and L2 loci were selected by germinating seeds on medium supplemented with kanamycin. GUS activity was measured in ago1 mutants and wild-type siblings after 6 weeks of growth. F2plants derived from the crosses between line 2a3 and the_ago1 mutants were germinated without selection. The number of ago1 mutants and wild-type siblings dying because of PTGS of endogenous NIA genes and 35S-_NIA2_transgene was scored after 6 weeks of growth.
Molecular Characterization.
Genomic DNA was extracted by the standard cetyltrimethylammonium bromide extraction method. Sequencing of the_ago1_-24 allele was performed on PCR products. Methylation analysis by Southern blot was performed by using the methylation-sensitive _Hpa_II restriction enzyme as described previously (33).
Results
Identification of PTGS Mutants Resembling_ago1_ Mutants.
We previously reported the isolation of PTGS _Arabidopsis_mutants that define three genetic loci called sgs1,sgs2, and sgs3 (for_s_uppressor of _g_ene_s_ilencing) (22, 33). These mutants were isolated after ethyl methanesulfonate mutagenesis of the _Arabidopsis_transgenic L1 line exhibiting PTGS of a reporter 35S-GUS transgene. Only mutants that developed as wild-type plants in the greenhouse were selected during this initial screen. A second screen was therefore performed in sterile medium to identify additional PTGS mutants that would show a delay in germination or development in soil. In the progeny of three independent mutagenized batches (named 23-1, 46-3, and 60-1) we identified 12, 17, and 9 plants, respectively, that displayed the phenotype of ago1 mutants—i.e., they formed unexpanded cotyledons, narrow leaves, and a unique stem with abnormal inflorescences bearing infertile flowers (30). All of the 38_ago1_-like plants identified exhibited GUS activity in leaves similar to that of sgs mutants (ca. 5,000 nmol of MU per min per μg of protein) whereas GUS activity in sibling wild-type plants was less than 5 nmol of MU per min per μg of protein), as in L1 plants (Table 1 and data not shown). GUS staining of cross sections of leaves, hypocotyls and roots revealed uniform expression of the 35S-GUS transgene (data not shown), suggesting complete impairment of PTGS in these organs. Like ago1 mutants (30), the 38 _ago1_-like [GUS+] plants were self-sterile.
Table 1.
GUS activity in L1 plants, sgs2-1 mutants, and_ago1-22_, ago1-23, and _ago1-24_mutants (isolated from batches 23-1, 46-3, and 60-1, respectively, of the EMS-mutagenized L1 library
| GUS activity, nmol per min per μg | ||||
|---|---|---|---|---|
| L1 | sgs2-1 | ago1-22 | ago1-23 | ago1-24 |
| 2.1 | 5,897 | 4,867 | 7,895 | 5,723 |
| 3.5 | 6,334 | 7,133 | 6,945 | 5,332 |
| 1.7 | 5,923 | 5,862 | 7,338 | 6,336 |
| 8.3 | 5,287 | 6,335 | 7,251 | 5,946 |
| 4.7 | 7,429 | 5,384 | 6,586 | 5,963 |
ago1_-Like PTGS Mutants Are Affected at the_AGO1 Locus.
Genetic analysis was performed to determine whether these new PTGS mutants were actually affected at the AGO1 locus. Because they are self-sterile like ago1 mutants, allelism tests were performed by crossing parents heterozygous for each allele. Parents segregating 25% [GUS+] _ago1_-like mutants and 75% [GUS−] wild-type plants after self-fertilization were identified in each batch. These plants were crossed together or with parents heterozygous for the ago1_-3 null allele (30). Each cross generated 25% of plants with the phenotype of_ago1 mutants. Kanamycin-resistant plants exhibited GUS activity comparable to that of sgs mutants in leaves of 8 weeks old plants (data not shown), thus strongly suggesting that these three PTGS mutants were indeed affected at the AGO1 locus. Following the published literature (30, 32, 34), they were designated as _ago1_-22, _ago1_-23, and_ago1_-24 (derived from batches 23-1, 46-3, and 60-1, respectively).
An Essential Amino Acid of AGO1 Is also Present in RDE-1 and QDE-2 in a Highly Conserved Motif.
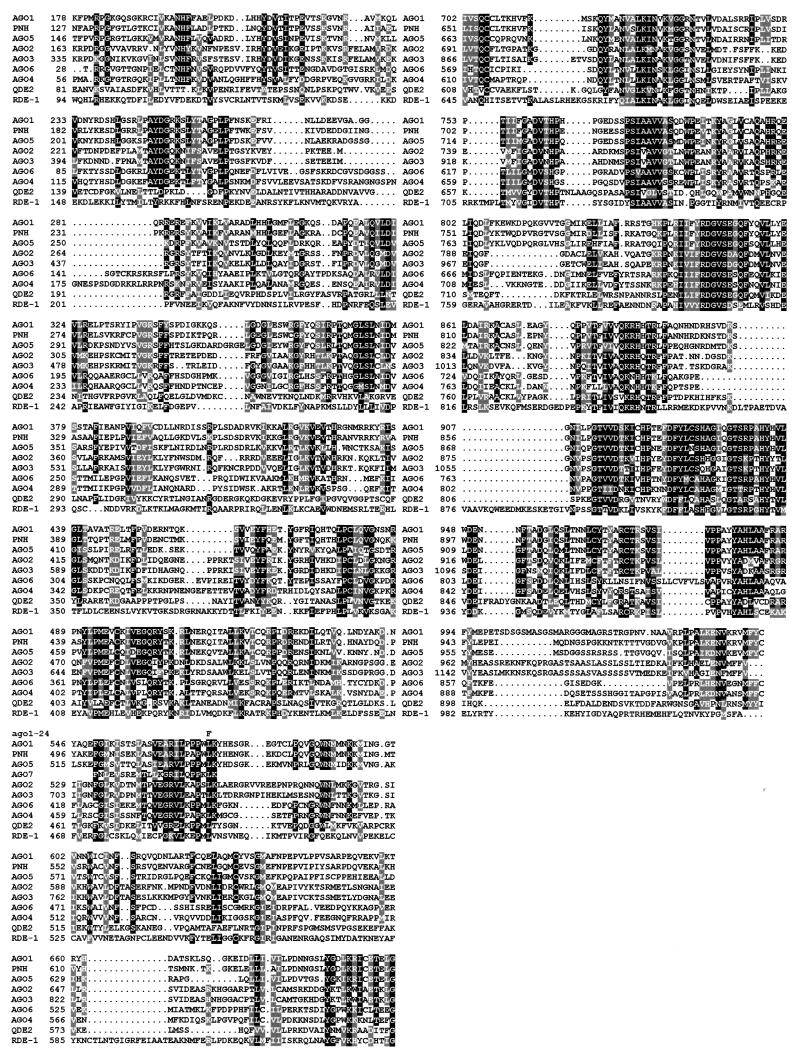
To confirm allelism tests, sequencing of the entire _AGO1_gene was performed in _ago1_-24. It revealed a single CG → TA transition at the 5′ end of exon 10, leading to a Leu → Phe substitution at position 571 of the AGO1 protein (Fig.1). Alignment of AGO1 with RDE-1 and QDE-2 revealed that this essential Leu is also present in RDE-1 and QDE-2 within one of the highly conserved motifs of these related proteins (Fig. 1), suggesting that it could also participate in the silencing function of these two proteins.
Figure 1.
Alignment of AGO1 with RDE-1, QDE-2, and the seven AGO1-related putative proteins encoded by the Arabidopsis genome. The predicted amino acid sequence of AGO1 protein (GenBank accession numberU91995, protein number AAC18440.1) (16) is aligned with that of RDE-1 (AF180730, AAF06159.1), QDE-2 (AF217760, AAF43641.1), ZLL/PNH (AJ223508, CAA11429.1/AF154272, AAD40098.1) (31, 32), AGO2 (AC007654,AAF24585.1), AGO3 (AC007654, AAF24586.1), AGO4 (AC005623, AAC77862.1), AGO5 (AC006929, AAD21514.1), AGO6 (AC003033, AAB91987.1), and with partial predicted amino acid sequence of AGO7 (AC073178) the sequencing of which is in progress. Sequence identities are indicated by filled boxes and conservative changes are shaded. Amino acid coordinates are indicated on the left. Dashes indicated gaps introduced by the MultiAlin (35) algorithm to maximize alignment. The alignment was processed by boxshade Version 3.21. The replacing amino acid in the sequenced _ago1_-24 allele is indicated above the AGO1 sequence.
Computer analysis revealed the presence of seven_AGO1_-related putative genes in the Arabidopsis_genome (92.5% sequenced at this date). One of these genes was previously identified twice as ZWILLE (ZLL) and_PINHEAD (PNH), and has overlapping function with_AGO1_ in the meristem, in the embryo, and in the vascular tissues (31, 32). The six other genes are putative. Arbitrarily, we named them AGO2 (GenBank accession number AC007654, protein number AAF24585.1), AGO3 (AC007654, AAF24586.1),AGO4 (AC005623, AAC77862.1), AGO5 (AC006929,AAD21514.1), AGO6 (AC003033, AAB91987.1), and_AGO7_ (AC073178). Another related gene was also identified (AC069325) but was not considered as an _AGO1_-like gene because it encodes a protein lacking 200 amino acids at the highly conserved C terminus of the AGO1 protein family. Only expressed sequence tags corresponding to AGO1,ZLL/PNH, AGO4, and AGO7_genes were found in the database, suggesting that the other members could be pseudogenes or could be transcribed at very low levels. Alignment of AGO1 with the complete translation products of_ZLL/PNH and five putative genes (AGO2–AGO6) and partial translation of one gene of which sequencing is in progress (AGO7) revealed that the essential Leu-571 of AGO1, which is conserved in RDE-1 and QDE-2, is also present in all known members of the AGO1 family (Fig. 1). However, the impairment of PTGS in ago1 mutants suggests that ZLL/PNH and the six other putative proteins cannot (fully) substitute for AGO1 silencing functions in ago1 mutants.
ago1 Mutants Are Impaired in Both PTGS and Cosuppression.
To definitively prove that the AGO1 gene is required for PTGS and not only for silencing of the 35S-GUS transgene of line L1, we crossed parents heterozygous for the previously identified_ago1_-3 null allele with three different PTG-silenced lines. Lines L1 and L2 carry the same 35S-GUS reporter transgene but inserted at two different positions within the genome (33). Line 2a3 carries a 35S-NIA2 transgene that triggers cosuppression of homologous NIA endogenous genes and transgenes (33). These three loci were chosen because PTGS mediated by L1, L2, and 2a3 is released in sgs1, sgs2, and_sgs3_ mutants (22, 33). GUS activity was measured in_ago1_ mutants and wild-type siblings of the F2 progeny of the cross with line L1 grown on medium supplemented with kanamycin to select for the presence of the L1 locus. All _ago1_-3 mutants (45 plants tested) were [GUS+], whereas all wild-type siblings (30 tested) were [GUS−], indicating that GUS PTGS mediated by the L1 locus is released in_ago1_-3 as it is in _ago1_-22,_ago1_-23, and _ago1_-24_mutants. Similarly, GUS PTGS mediated by the L2 locus (52_ago1 plants tested) and NIA cosuppression mediated by the 2a3 locus (92 ago1 plants tested) were released in all _ago1_-3 mutants tested but not in wild-type siblings, thus confirming that the AGO1 gene is required for both PTGS of a foreign transgene and cosuppression of homologous endogenous genes and transgenes. Moreover, these results suggest that the action of AGO1 on PTGS does not depend on transgene position within the genome.
PTGS Deficiency Correlates with a Decrease of Transgene Methylation in ago1 Mutants.
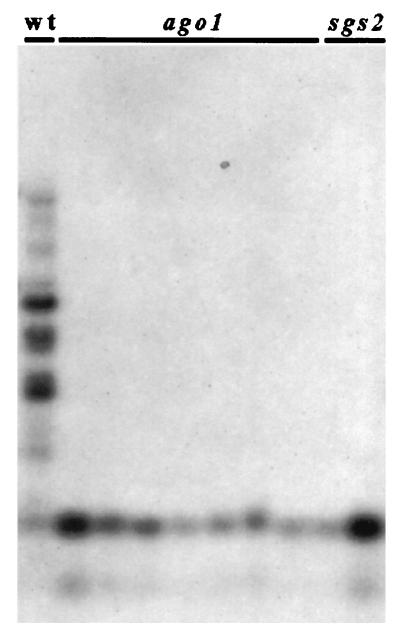
PTGS correlates with methylation in the transgene transcribed sequence (33, 36–38). Although the exact role played by methylation in PTGS is still not known [methylation is dispensable for quelling in_Neurospora_ (8) and for RNAi because methylation is absent in_C. elegans_], it is assumed that this type of imprint of transgene DNA is involved in the maintenance of the production of aberrant RNA (and subsequently of dsRNA) that trigger sequence-specific RNA degradation (4, 39). Methylation is (at least partially) reduced in_sgs_ mutants (22, 33), whereas it is maintained in plants infected by viruses that counteract PTGS (40–42), suggesting that_sgs_ mutants are impaired in PTGS steps that control both RNA degradation and DNA methylation, whereas viruses impede only step(s) that control RNA degradation. To determine the step at which AGO1 protein acts in PTGS, we compared methylation of the GUS reporter transgene in ago1 and sgs mutants with that of PTG-silenced L1 plants. As in sgs2 and _sgs3_mutants (22), methylation is strongly reduced in _ago1_mutants (Fig. 2), indicating that AGO1 also controls a step that is necessary for both RNA degradation and DNA methylation.
Figure 2.
Methylation analysis of sgs2 and _ago1_mutants compared with L1 plants. Methylation was evaluated by Southern blots of DNA extracted from leaves of adult L1 plants,ago1 mutants, and sgs2 mutants, digested with _Hpa_II and hybridized with the GUS3 probe corresponding to the 3′ part of the GUS ORF (33).
Discussion
By a genetic approach, a number of mutants affected in quelling, RNAi, or PTGS have been isolated, leading to the identification of eight genes controlling these phenomena (21–23, 26, 27, 29, 33, 43,44). QDE-1, QDE-2, and QDE-3 genes, required for quelling in the fungus N. crassa, encode proteins similar to tomato RNA-directed RNA polymerase, rabbit translation initiation factor eIF2C, and E. coli RecQ DNA helicase, respectively (21, 29, 44), whereas EGO-1, RDE-1, and _MUT-7_genes required for RNAi in the nematode C. elegans encode proteins similar to tomato RNA-directed RNA polymerase, rabbit translation initiation factor eIF2C, and E. coli RNaseD, respectively (23, 26, 27). SGS2 and SGS3 genes, required for PTGS in A. thaliana, encode a protein similar to tomato RNA-directed RNA polymerase and a protein of unknown function, respectively (22). The finding of one set of related proteins shared by PTGS, quelling, and RNAi (SGS2/QDE-1/EGO-1), therefore, suggested that these three mechanisms could be mechanistically linked. However, evidence for the role of RdRP in RNAi is restricted to the germ line because ego-1 mutants are still able to undergo RNAi in somatic tissues (23). Whether RdRP is dispensable for RNAi in somatic tissues or whether RNAi in somatic tissues involves one of the other known RdRPs in C. elegans remains to be determined. In addition, PTGS does not occur in the meristems from which derive the germ line in plants (25, 41). Therefore, SGS2 and EGO-1 provide a molecular link between PTGS and RNAi, but they act in different tissues. Here we report the finding of a second set of highly related proteins (AGO1/QDE-2/RDE-1) required for PTGS/quelling/RNAi. They are all necessary for silencing in similar (somatic) tissues and they all carry a Leu in a highly conserved domain (Fig. 1) that was shown here to be essential for the role of AGO1 in PTGS, further establishing that these three phenomena are mechanistically linked.
Developmental defects are associated with mutations in some but not all proteins required for PTGS/quelling/RNAi. Indeed, among the two sets of related proteins shared by PTGS, quelling and RNAi (SGS2/QDE-1/EGO-1 and AGO1/QDE-2/RDE-1), four proteins are dispensable for development in standard conditions of growth (SGS2/QDE-1 and QDE-2/RDE-1). Conversely, mutations in AGO1 have pleiotropic effects on development and fertility that strongly compromise the life and reproduction of Arabidopsis in standard conditions of growth (30, 32). Similarly, ego-1_mutants, impaired in RNAi in the germ line, also show gametogenesis defects and sterility in C. elegans (23). This result could indicate that PTGS and RNAi, but not quelling, regulate some step(s) of gene expression during development. However, other PTGS mutants, like_sgs1, sgs2, and sgs3, are viable (22, 33), as well as other RNAi-deficient mutants like rde-1 and_mut-7_ (26, 27), indicating that PTGS and RNAi, as a whole, are dispensable for development. The participation of _AGO1_and EGO-1 in developmental functions simultaneously with silencing therefore suggests that PTGS/RNAi and development simply share common enzyme(s) or pathway(s).
Most of the genes controlling PTGS/RNAi are members of multigene families. EGO-1/SGS2 genes belong to families of at least 4 members in C. elegans (23) and 7 members in_Arabidopsis_ (22), respectively, whereas_RDE-1_/AGO1 genes belong to families of at least 23 members in C. elegans (26) and 8 members in_Arabidopsis_ (refs. 30–32, and this work), respectively. Although mutations in these four genes are sufficient to abolish PTGS/RNAi, the role of most of the other members of the families is not known. They could participate in silencing or control different biological processes, or could be pseudogenes. In the case of the_AGO1_ family, one related gene (ZLL/PNH) was genetically identified as encoding a protein required for development (31, 32), while six other genes encoding putative proteins strongly similar to AGO1 (Fig. 1) were identified by computer analysis of the Arabidopsis genome (92.5% sequenced at this date). ZLL/PNH and_AGO1_ genes are expressed in part in the same organs (including leaves, roots, and stems; refs. 30–32). However,ago1 and zll/pnh mutants have distinct phenotypes (30–32), suggesting that_ZLL_/PNH and AGO1 genes have specialized (although probably synergistic) functions, at least in development. Among the six other known putative _AGO1_-like genes, only two (AGO4, AGO7) have corresponding expressed sequence tags in the databases, suggesting that the others could be pseudogenes or could be expressed at very low levels. Whether such proteins could overlap with AGO1 silencing functions is unknown. Nevertheless, uniform reexpression of the PTG-silenced 35S-GUS reporter transgene was observed in cross sections of leaves, hypocotyls, and roots of ago1 mutants, suggesting a total impairment of PTGS in these organs. This result fits well with the fact that transgene expression and methylation levels in ago1 mutants are similar to those in sgs2 and sgs3 mutants in leaves, stems, and roots (data not shown), indicating that (at least part of) AGO1 silencing functions cannot be performed by the seven other members of the family. If these proteins have silencing functions similar to AGO1, they are most likely expressed at levels that are too low or in very few cells, or they require the simultaneous presence of a functional AGO1 protein to be active. Nevertheless, the requirement for AGO1 in PTGS does not exclude that any of the AGO1-related proteins also play a role in PTGS, alone or in combination with AGO1. Whether PTGS still occurs in mutants affected in each of the AGO1-related proteins needs to be determined to answer this question.
To conclude, the identification of a second set of proteins shared by PTGS, quelling, and RNAi confirms the mechanistic link existing between these silencing phenomena. The identification of a protein (AGO1) controlling PTGS that is absolutely required for the growth of plants in standard conditions points out the limits of a genetic approach to identify all proteins involved in silencing processes. The fact that most of the genes controlling PTGS/RNAi are members of multigene families is also a limitation for a genetic approach in cases of complete redundancy. Therefore, it would be relevant to use a biochemical approach, by using the two-hybrid system or in vitro systems (from partially purified cell extracts), for example (45, 46), to identify other members of complexes involving already known proteins. Because the rabbit homologue of AGO1/QDE-2/RDE-1, eIF2C (26, 30–32), is supposed to be part of a protein complex that stimulates the start of translation (28), such an approach would help to identify other members of this complex and test their effect on PTGS/quelling/RNAi.
Acknowledgments
We thank Aurélie Dewaelle for technical assistance and lab colleagues for fruitful discussions. M.F. received a grant from the European Union Biotech program (Contract B104-CT96–0253).
Abbreviations
cRNA
complementary RNA
dsRNA
double-stranded RNA
GUS
β-glucuronidase
PTGS
post-transcriptional gene silencing
PTG-silenced
post-transcriptional gene-silenced
RNAi
RNA interference
MU
4-methylumbelliferone
RdRP
RNA-dependent RNA polymerase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.200217597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.200217597
References
- 1.Matzke M A, Matzke A J M. Plant Physiol. 1995;107:679–685. doi: 10.1104/pp.107.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baulcombe D C. Plant Mol Biol. 1996;34:125–137. doi: 10.1007/BF00039378. [DOI] [PubMed] [Google Scholar]
- 3.Depicker A, Van Montagu M. Curr Opin Cell Biol. 1997;9:373–382. doi: 10.1016/s0955-0674(97)80010-5. [DOI] [PubMed] [Google Scholar]
- 4.Vaucheret H, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel J-B, Mourrain P, Palauqui J-C, Vernhettes S. Plant J. 1998;16:651–659. doi: 10.1046/j.1365-313x.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 5.Kooter J M, Matzke M A, Meyer P. Trends Plant Sci. 1999;4:340–347. doi: 10.1016/s1360-1385(99)01467-3. [DOI] [PubMed] [Google Scholar]
- 6.Napoli C, Lemieux C, Jorgensen R. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romano N, Macino G. Mol Microbiol. 1992;6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 8.Cogoni C, Irelan J, Schumacher M, Selker E, Macino G. EMBO J. 1996;15:3153–3163. [PMC free article] [PubMed] [Google Scholar]
- 9.Cogoni C, Macino G. Trends Plant Sci. 1997;2:438–443. [Google Scholar]
- 10.Fire A. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- 11.Sharp P A. Genes Dev. 1999;13:139–141. [PubMed] [Google Scholar]
- 12.Bosher J M, Labouesse M. Nat Cell Biol. 2000;2:E31–E36. doi: 10.1038/35000102. [DOI] [PubMed] [Google Scholar]
- 13.Bass B L. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- 14.Fire A, Xu S, Montgomery M, Kostas S, Driver S, Mello C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 15.Tavernarakis N, Wang S L, Dorovkov M, Ryazanov A, Driscoll M. Nat Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- 16.Waterhouse P M, Graham M W, Wang M-B. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton A, Brown S, Yuanhai H, Ishizuka M, Lowe A, Alpuche Solis A-G, Grierson D. Plant J. 1998;15:737–746. doi: 10.1046/j.1365-313X.1998.00251.x. [DOI] [PubMed] [Google Scholar]
- 18.Chuang C-F, Meyerowitz E M. Proc Natl Acad Sci USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindbo J A, Silva-Rosales L, Proebsting W M, Dougherty W G. Plant Cell. 1993;5:1749–1759. doi: 10.1105/tpc.5.12.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton A, Baulcombe D. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 21.Cogoni C, Macino G. Nature (London) 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 22.Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel J-B, Jouette D, Lacombe A-M, Nikic S, Picault N, et al. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 23.Smardon A, Spoerke J M, Stacey S C, Klein M E, Mackin N, Maine E M. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 24.Palauqui J-C, Elmayan T, Pollien J-M, Vaucheret H. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voinnet O, Vain P, Angell S, Baulcombe D C. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- 26.Tabara H, Sarkissian M, Kelly W, Fleenor J, Grishok A, Timmons L, Fire A, Mello C. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 27.Ketting R F, Haverkamp T H, van Luenen H G, Plasterk R H. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 28.Zou C, Zhang Z, Wu S, Osterman J C. Gene. 1998;211:187–194. doi: 10.1016/s0378-1119(98)00107-3. [DOI] [PubMed] [Google Scholar]
- 29.Catalanotto C, Azzalin G, Macino G, Cogoni C. Nature (London) 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- 30.Bohmert K, Camus I, Bellini C, Caboche M, Benning C. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moussian B, Schoof H, Haecker A, Jürgens G, Laux T. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton M. Development. U.K.: Cambridge; 1999. 126, 469–481. [DOI] [PubMed] [Google Scholar]
- 33.Elmayan T, Balzergue S, Béon F, Bourdon V, Daubremet J, Guénet Y, Mourrain P, Palauqui J-C, Vernhettes S, Vialle T, Wostrikoff K, Vaucheret H. Plant Cell. 1998;10:1447–1457. doi: 10.1105/tpc.10.10.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camus I. Ph.D. thesis. 1999. , Univ. of Paris VI, Paris. [Google Scholar]
- 35.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingelbrecht I, Van Houdt H, Van Montagu M, Depicker A. Proc Natl Acad Sci USA. 1994;91:10502–10506. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.English J J, Mueller E, Baulcombe D C. Plant Cell. 1996;8:179–188. doi: 10.1105/tpc.8.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones A L, Thomas C L, Maule A J. EMBO J. 1998;17:6385–6393. doi: 10.1093/emboj/17.21.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones J, Hamilton A J, Voinnet O, Thomas C L, Maule A J, Baulcombe D C. Plant Cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anandalakshmi R, Pruss G, Ge X, Marathe R, Mallory A, Smith T, Vance V. Proc Natl Acad Sci USA. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Béclin C, Berthome R, Palauqui J-C, Tepfer M, Vaucheret H. Virology. 1998;252:313–317. doi: 10.1006/viro.1998.9457. [DOI] [PubMed] [Google Scholar]
- 42.Voinnet O, Pinto Y M, Baulcombe D. Proc Natl Acad Sci USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cogoni C, Macino G. Proc Natl Acad Sci USA. 1997;94:10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cogoni C, Macino G. Science. 1999;286:2342–2344. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- 45.Tuschl T, Zamore P D, Lehmann R, Bartel D P, Sharp P A. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zamore P D, Tuschl T, Sharp P A, Bartel D P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]