The Requirement of Sterol Glucoside for Pexophagy in Yeast Is Dependent on the Species and Nature of Peroxisome Inducers (original) (raw)
Abstract
Sterol glucosyltransferase, Ugt51/Atg26, is essential for both micropexophagy and macropexophagy of methanol-induced peroxisomes in Pichia pastoris. However, the role of this protein in pexophagy in other yeast remained unclear. We show that oleate- and amine-induced peroxisomes in Yarrowia lipolytica are degraded by Atg26-independent macropexophagy. Surprisingly, Atg26 was also not essential for macropexophagy of oleate- and amine-induced peroxisomes in P. pastoris, suggesting that the function of sterol glucoside (SG) in pexophagy is both species and peroxisome inducer specific. However, the rates of degradation of oleate- and amine-induced peroxisomes in P. pastoris were reduced in the absence of SG, indicating that P. pastoris specifically uses sterol conversion by Atg26 to enhance selective degradation of peroxisomes. However, methanol-induced peroxisomes apparently have lost the redundant ability to be degraded without SG. We also show that the P. pastoris Vac8 armadillo repeat protein is not essential for macropexophagy of methanol-, oleate-, or amine-induced peroxisomes, which makes PpVac8 the first known protein required for the micropexophagy, but not for the macropexophagy, machinery. The uniqueness of Atg26 and Vac8 functions under different pexophagy conditions demonstrates that not only pexophagy inducers, such as glucose or ethanol, but also the inducers of peroxisomes, such as methanol, oleate, or primary amines, determine the requirements for subsequent pexophagy in yeast.
INTRODUCTION
Growth of yeast on certain carbon sources, such as methanol or oleate, requires proliferation of peroxisomes housing the enzymes for use of these compounds (Yan et al., 2005). However, when a carbon source that does not require peroxisomes for its oxidation (e.g., glucose or ethanol) becomes available, the selective degradation of superfluous peroxisomes occurs in the yeast vacuole (Bellu and Kiel, 2003). Selective degradation of peroxisomes greatly relies on the molecular machinery of bulk autophagy and is called pexophagy (Dunn et al., 2005). There are 29 known _A_u_T_opha_G_y-related (ATG) genes (Klionsky et al., 2003; Kawamata et al., 2005; Stasyk et al., 2006), but until recently the molecular mechanisms of pexophagy were studied exclusively in yeast. However, recent studies on the ATG7 conditional knockout mice indicate that the autophagic machinery is also essential for pexophagy in mammals (Iwata et al., 2006). Furthermore, peroxisomes are present in murine spermatogonia and disappear during the course of spermatogenesis in a selective and synchronized manner (Luers et al., 2006).
Yeast have developed two morphologically different mechanisms to sequester the superfluous peroxisomes for vacuolar degradation: 1) macropexophagy, induced after the transfer of i) P. pastoris cells from methanol/ammonia to ethanol/ammonia medium, ii) Y. lipolytica cells from oleate/ethylamine to glucose/ammonia medium, and iii) Hansenula polymorpha cells from methanol/ammonia to either ethanol/ammonia or glucose/ammonia medium; and 2) micropexophagy, induced after the transfer of P. pastoris cells from methanol/ammonia to glucose/ammonia medium (Kiel et al., 2003; Farré and Subramani, 2004; Dunn et al., 2005). P. pastoris has two unique properties that make it one of the most attractive peroxisome homeostasis models: 1) peroxisome proliferation can be induced by both methanol and oleate (Liu et al., 1992) and 2) methanol-induced peroxisomes are degraded by both macro- and micropexophagy (Tuttle and Dunn, 1995). The switch between macropexophagy and micropexophagy depends in P. pastoris on the intracellular ATP level with macropexophagy being the default pexophagy mode and micropexophagy operating when a higher level of ATP is available (Ano et al., 2005). However, the mode and molecular mechanisms of degradation of oleate-induced peroxisomes in this versatile model system were unknown until this study was undertaken.
Both macropexophagy and micropexophagy require the formation of additional membrane structures for sequestration of peroxisomes from the cytosol. During macropexophagy, the individual peroxisomes are enclosed within double-membrane pexophagosomes before fusion with the vacuole. In contrast, during micropexophagy whole clusters of peroxisomes are engulfed by the vacuolar sequestering membranes (VSMs). Fusion of the VSMs requires the assistance of micropexophagic membrane apparatus (MIPA). Both pexophagosome and MIPA membranes may originate from the preautophagosomal structure (PAS), which is considered to be the organizing center for vesicles in all autophagy-related pathways (Noda et al., 2002; Kiel et al., 2003; Farré and Subramani, 2004; Dunn et al., 2005). Because 1) most P. pastoris proteins required for micropexophagy are also essential for macropexophagy (Dunn et al., 2005) and 2) the MIPA and pexophagosome both contain PpAtg8 (Mukaiyama et al., 2004), it is not clear what the mechanistic and membrane differences are between these two pathways of peroxisome sequestration. Proteins specifically required for one pexophagy mode but not for the other would shed light on this long-standing question. The only candidate known to confer such specificity is Vac8, which is required for micropexophagy but not for starvation-induced autophagy in P. pastoris (Chang et al., 2005). However, the role of this protein in macropexophagy was not studied until now.
The selectivity of macropexophagy and micropexophagy toward peroxisomes might be provided by Atg11 (Kim et al., 2001; Mukaiyama et al., 2002; Komduur, 2004), Atg26 (Oku et al., 2003; Stasyk et al., 2003), and Atg28 (Stasyk et al., 2006), the proteins required for selective transport of peroxisomes to the vacuole but not for autophagy of bulk cytosol in P. pastoris, H. polymorpha, and S. cerevisiae. Atg11 and Atg28 are coiled-coil proteins that localize to the PAS and VSM (Kim et al., 2001; Monastyrska et al., 2005; Stasyk et al., 2006). Atg11 and Atg26 are required for pexophagosome and MIPA formation (Komduur, 2004; Dunn et al., 2005; Yamashita et al., 2006). Furthermore, the localization of Atg26 to the MIPA, mediated by its GRAM domains that bind phosphatidylinositol 4′-monophosphate, is essential for micropexophagy (Oku et al., 2003; Yamashita et al., 2006). Atg26 is a UDP-glucose:sterol glucosyltransferase that synthesizes a minor membrane lipid, sterol glucoside (SG) (Warnecke et al., 1999; Sakaki et al., 2001). SG synthesis on the MIPA is essential for micropexophagy and is needed for elongation of this micropexophagy-specific membrane structure (Oku et al., 2003; Yamashita et al., 2006). Although Atg26 is required for macro- and micropexophagy of methanol-induced peroxisomes in P. pastoris, this protein is dispensable for the cytoplasm-to-vacuole targeting (Cvt) pathway in Saccharomyces cerevisiae and glucose/ammonia catabolite inactivation of ethanol/ethylamine- or oleate/ethylamine-induced peroxisomal enzymes in Y. lipolytica (Oku et al., 2003; Stasyk et al., 2003). Whether Atg26 is required for all modes of pexophagy in other yeasts was unknown. The answer to this particularly important question is relevant to the issue of whether pexophagy mechanisms are identical or partly different in different model systems.
Here, we have developed biochemical assays and used microscopy to follow the selective degradation of oleate- and amine-induced peroxisomes in Y. lipolytica and P. pastoris. Our studies show that peroxisomes induced by oleate or primary amines are degraded by macropexophagy in both species. However, YlAtg26 is not required for macropexophagy of oleate- or amine-induced peroxisomes. Surprisingly, PpAtg26, which is essential for macropexophagy of methanol-induced peroxisomes, is nonessential for macropexophagy of oleate- and amine-induced peroxisomes, suggesting that the function of SG in pexophagy depends on both species and the nature of peroxisome inducers. Moreover, our data show that PpVac8 is nonessential for macropexophagy of methanol-, oleate- and amine-induced peroxisomes. Therefore, the function of Vac8 in peroxisome degradation is micropexophagy specific.
MATERIALS AND METHODS
Strains, Plasmids, and Transformation
The Y. lipolytica and P. pastoris strains used in this study are listed in Table 1. The Y. lipolytica strains were transformed with 0.3 μg of NotI-digested pYEG1hyg-POX3-EYFP plasmid (Nazarko et al., 2005b) by the LiAc/LiCl method (Barth and Gaillardin, 1996). The Y. lipolytica HygBR-transformants expressing the Aox3-EYFP fusion were selected on YPD (1% wt/vol yeast extract, 1% wt/vol peptone, 1% wt/vol glucose, and 2% wt/vol agar) plates with 200 μg/ml hygromycin B for H222 and N155 strains and 50 μg/ml hygromycin B for Ain16 strain. Two independent HygBR-transformants of each strain were examined by fluorescence microscopy for degradation of peroxisomes under all pexophagy conditions.
Table 1.
Y. lipolytica and P. pastoris strains used in this study
| Name | Genotype | Background | Reference |
|---|---|---|---|
| Y. lipolytica | |||
| H222 | MATA ylT1-free wild type | WT | Barth and Gaillardin (1996) |
| STN001 | H222::PFC2 (_zeta_-P_hp4d_-_HygBR_-P_POX2_-_POX3_-EYFP) #3 | WT | This study |
| STN002 | H222::PFC2 (_zeta_-P_hp4d_-_HygBR_-P_POX2_-_POX3_-EYFP) #4 | WT | This study |
| N155 | H222–41 atg26::zeta-URA3 | atg26 | Stasyk et al. (2003) |
| STN003 | N155::PFC2 (_zeta_-P_hp4d_-_HygBR_-P_POX2_-_POX3_-EYFP) #1 | atg26 | This study |
| STN006 | N155::PFC2 (_zeta_-P_hp4d_-_HygBR_-P_POX2_-_POX3_-EYFP) #4 | atg26 | This study |
| Ain16 | H222-S4 trs85-1::zeta-URA3 | trs85-1 | Nazarko et al. (2005a) |
| STN011 | Ain16::PFC2 (_zeta_-P_hp4d_-_HygBR_-P_POX2_-_POX3_-EYFP) #1 | trs85-1 | This study |
| STN012 | Ain16::PFC2 (_zeta_-P_hp4d_-_HygBR_-P_POX2_-_POX3_-EYFP) #3 | trs85-1 | This study |
| Ain19 | H222-S4 trs85-2::zeta-URA3 | trs85-2 | Nazarko et al. (2005a) |
| P. pastoris | |||
| GS115 | his4 | WT | Cregg et al. (1985) |
| STN017 | GS115 his4::pTW74 (P_GAPDH_-_GFP_-SKL, HIS4) #1 | WT | This study |
| STN018 | GS115 his4::pTW74 (P_GAPDH_-_GFP_-SKL, HIS4) #2 | WT | This study |
| STN025 | GS115 his4::pTW51 (P_AOX1_-_GFP_-SKL, HIS4) #2 | WT | This study |
| STN026 | GS115 his4::pTW51 (P_AOX1_-_GFP_-SKL, HIS4) #3 | WT | This study |
| GS200 | arg4 his4 | WT | Waterham et al. (1996) |
| Pdg3D | GS200 Δ_atg26_::ScARG4 | Δ_atg26_ | Stasyk et al. (2003) |
| STN019 | Pdg3D his4::pTW74 (P_GAPDH_-_GFP_-SKL, HIS4) #1 | Δ_atg26_ | This study |
| STN020 | Pdg3D his4::pTW74 (P_GAPDH_-_GFP_-SKL, HIS4) #3 | Δ_atg26_ | This study |
| STN027 | Pdg3D his4::pTW51 (P_AOX1_-_GFP_-SKL, HIS4) #2 | Δ_atg26_ | This study |
| STN028 | Pdg3D his4::pTW51 (P_AOX1_-_GFP_-SKL, HIS4) #8 | Δ_atg26_ | This study |
| SMD1163 | his4 pep4 prb1 | pep4 prb1 | Tuttle and Dunn (1995) |
| R12 | GS115 atg1-1::zeoR | atg1-1 | Stromhaug et al. (2001) |
| STN021 | R12 his4::pTW74 (P_GAPDH_-_GFP_-SKL, HIS4) #1 | atg1-1 | This study |
| STN022 | R12 his4::pTW74 (P_GAPDH_-_GFP_-SKL, HIS4) #2 | atg1-1 | This study |
| STN029 | R12 his4::pTW51 (P_AOX1_-_GFP_-SKL, HIS4) #1 | atg1-1 | This study |
| STN030 | R12 his4::pTW51 (P_AOX1_-_GFP_-SKL, HIS4) #3 | atg1-1 | This study |
| WDY53 | GS200 Δ_vac8_::zeoR | Δ_vac8_ | Chang et al. (2005) |
| STN023 | WDY53 his4::pTW74 (P_GAPDH_-_GFP_-SKL, HIS4) #1 | Δ_vac8_ | This study |
| STN024 | WDY53 his4::pTW74 (P_GAPDH_-_GFP_-SKL, HIS4) #2 | Δ_vac8_ | This study |
| STN031 | WDY53 his4::pTW51 (P_AOX1_-_GFP_-SKL, HIS4) #2 | Δ_vac8_ | This study |
| STN032 | WDY53 his4::pTW51 (P_AOX1_-_GFP_-SKL, HIS4) #3 | Δ_vac8_ | This study |
The P. pastoris strains were transformed with 0.3 μg of StuI-digested pTW74 plasmid (Luers et al., 1998) and 0.3 μg of SalI-digested pTW51 plasmid (Wiemer et al., 1996) by electroporation (Cregg and Russell, 1998). The P. pastoris His+-transformants obtained with both pTW74 and pTW51 were selected on SD (1.7 g/l YNB without amino acids and ammonium sulfate, 2% wt/vol glucose, 0.5% wt/vol ammonium sulfate, and 2% wt/vol agar) plates without histidine. Arginine (50 mg/l) was added, when needed. Two independent His+-transformants of each strain with each plasmid were examined for degradation of peroxisomes by fluorescence microscopy under all pexophagy conditions.
Peroxisome and Pexophagy Induction Conditions
Both Y. lipolytica and P. pastoris strains were pregrown overnight in liquid YPD medium, diluted 20-fold in the morning with fresh YPD and grown for an additional 6–8 h. The strains were then washed with water and inoculated into peroxisome-induction medium at an OD600 of 0.1–0.5 (for media with both carbon and nitrogen sources) or 2.0 (for media without any carbon source) for 12 (Y. lipolytica oleate media) or 15 h. The next morning, the yeast cells were washed with water (P. pastoris) or 50 mM phosphate buffer, pH 6.8 (PiB) (Y. lipolytica) and inoculated into pexophagy induction medium at an OD600 of 0.5–1 for 3–24 h. The incubation times in pexophagy induction media varied depending on the organism and experiment (see below and figures for details). All peroxisome and pexophagy induction media contained 1.7 g/l YNB without amino acids and ammonium sulfate. The Y. lipolytica media were prepared in PiB to maintain a stable pH. Histidine (40 mg/l), arginine (40 mg/l), or both were added to P. pastoris media, when needed. All peroxisome but not pexophagy induction media for both yeast species contained 0.05% wt/vol yeast extract. Peroxisome and pexophagy induction media had the following carbon sources: 1% wt/vol glucose for both yeast species, oleate (1% vol/vol for Y. lipolytica and 0.5% vol/vol for P. pastoris), 1% vol/vol methanol, or 0.5% vol/vol ethanol for P. pastoris. The nitrogen sources were as follows: 0.134% wt/vol ammonium chloride for Y. lipolytica, 0.25% wt/vol ammonium sulfate for P. pastoris, and methylamine or ethylamine hydrochloride (0.2% wt/vol for Y. lipolytica and 0.1% wt/vol for P. pastoris). The oleate stock emulsion contained 20% vol/vol oleate and 0.5% vol/vol Tween 80. The methylamine and ethylamine peroxisome-induction media did not contain any other carbon source, and the glucose and ethanol pexophagy induction media did not contain any source of nitrogen for both yeast species.
Biochemical Studies of Pexophagy
For biochemical studies of pexophagy, yeast cells were washed twice with water (P. pastoris) or PiB (Y. lipolytica) after growth on peroxisome induction medium and transferred to fresh glucose/(−N) or ethanol/(−N) medium at an OD600 of 0.5 to induce pexophagy. The cells from 0.5-ml culture samples were collected by centrifugation after 0, 6, 9, 12, and 24 h (when peroxisomes were induced in the medium with both carbon and nitrogen sources) or 0, 3, 6, 9, and 12 h (when peroxisomes were induced in the medium without any carbon source) of glucose or ethanol adaptation. Crude extracts were prepared in the presence of trichloroacetic acid (Baerends et al., 2000). SDS-PAGE and immunoblotting were performed as described previously (Laemmli, 1970; Kyhse-Andersen, 1984). For Y. lipolytica studies, we used antibodies against 1) the Y. lipolytica thiolase (THI), kindly provided by R. A. Rachubinski (University of Alberta, Edmonton, Alberta, Canada) and 2) the GroEL protein of Escherichia coli with cross-reactivity against the mitochondrial Hsp60 protein of Y. lipolytica (Gunkel et al., 1999), kindly provided by M. Veenhuis (University of Groningen, Haren, The Netherlands). For P. pastoris studies, we used antibodies against 1) the S. cerevisiae THI with cross-reactivity against THI of P. pastoris (Liu et al., 1995) and 2) the alcohol oxidase (AOX) of H. polymorpha with cross-reactivity against AOX of P. pastoris (Liu et al., 1995), kindly provided by M. Veenhuis. Antigen–antibody complexes were detected by enhanced chemiluminescence.
Fluorescence Microscopy to Monitor Pexophagy
For fluorescence microscopy of pexophagy, yeast cells were incubated in peroxisome-induction medium with or without (Y. lipolytica oleate media) 5 μg/ml _N_-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl) pyridinium dibromide (FM 4-64), diluted from 1 mg/ml stock solution in dimethyl sulfoxide. Then, cells were washed once with water (P. pastoris) or PiB (Y. lipolytica) and transferred to fresh glucose/(−N) or ethanol/(−N) medium at an OD600 of 1.0 to induce pexophagy. The glucose/(−N) medium for oleate-induced cells of Y. lipolytica contained 5 μg/ml FM 4-64. After 0 and 9 h (for oleate-induced cells of both yeast species and methylamine-induced cells of Y. lipolytica), or 0 and 3 h (for methylamine- and methanol-induced cells of P. pastoris) under pexophagy conditions, cells were collected by centrifugation and placed on ice until observation. The progress of pexophagy was determined immediately after each time point. The optimal exposition time for each fluorophore was determined at time point “0 h” and applied for all subsequent time points. Images were captured on a motorized fluorescence microscope (Axioskop 2 MOT; Carl Zeiss MicroImaging, Jena, Germany) coupled to a monochrome digital camera (AxioCam MRm; Carl Zeiss MicroImaging) and processed using the AxioVision 4.5 (Carl Zeiss MicroImaging) and Adobe Photoshop 7.0 software (Adobe Systems, Mountain View, CA).
RESULTS
The Y. lipolytica ATG26 Gene Is Not Required for Macropexophagy of Oleate-induced Peroxisomes
Recently, it was proposed that sterol glucosyltransferase, Ugt51/Atg26, may play different functional roles in P. pastoris and Y. lipolytica based on the observation that inactivation of two peroxisomal enzymes, amine oxidase and isocitrate lyase, was not affected in the Y. lipolytica atg26 mutant after the transfer of cells from peroxisome induction medium (ethanol/ethylamine or oleate/ethylamine) to pexophagy conditions (glucose/ammonia medium) (Stasyk et al., 2003).
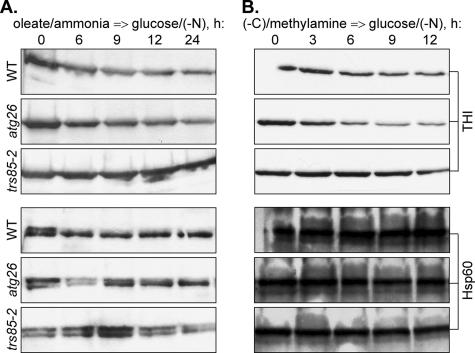
To understand the basis of this observation, we studied the levels of THI after the transfer of Y. lipolytica cells from oleate/ammonia- and oleate/ethylamine-containing media to glucose medium without nitrogen [glucose/(−N)] (Figure 1A and Supplemental Figure 1A). Pexophagy in Y. lipolytica is a rather slow process (Nazarko et al., 2005b) requiring that it be monitored over a long time (24 h). However, under the glucose growth conditions, the rates of disappearance of peroxisomes due to cell growth and division are much faster than the rates of disappearance of peroxisomes due to pexophagy. Therefore, as in S. cerevisiae (Hutchins et al., 1999), it was not possible to study degradation of oleate-induced peroxisomes in Y. lipolytica in glucose/ammonia-containing medium over a prolonged period of time. In contrast, the exclusion of nitrogen from glucose medium allowed us to follow the Y. lipolytica THI levels over a 24-h time course. The decrease of THI levels in the Y. lipolytica wild-type (WT) strain was caused mostly by selective degradation of peroxisomes, because the level of a mitochondrial protein, heat-shock protein of 60 kDa (Hsp60), was stable during the whole time course. In contrast, the Y. lipolytica mutant affected in the TRS85 gene that is required for all known autophagy-related pathways in both Y. lipolytica and S. cerevisiae (Nazarko et al., 2005a; Meiling-Wesse et al., 2005) was completely blocked in the degradation of THI. Importantly, the trs85-2 mutant had the same stable level of Hsp60 protein as the WT strain, supporting the negligible role of autophagy under our experimental setup. Degradation of oleate-induced peroxisomes seemed to be normal in the Y. lipolytica atg26 mutant. The peroxisome degradation rates in both WT and atg26 strains did not depend on the nature of nitrogen source present in oleate medium (compare Figure 1A and Supplemental Figure 1A). Therefore, selective degradation of oleate-induced peroxisomes in Y. lipolytica does not require the function of Atg26.
Figure 1.
The Y. lipolytica ATG26 gene is not required for selective degradation of oleate- and amine-induced peroxisomes. Y. lipolytica WT (H222), atg26 (N155), and trs85-2 (Ain19) cells were induced in (A) oleate/ammonia or (B) (−C)/methylamine medium and transferred to glucose/(−N) medium to induce pexophagy. At the indicated time points, the culture samples were collected and processed for immunoblotting with antibodies against peroxisomal THI and mitochondrial Hsp60 proteins as described in Materials and Methods.
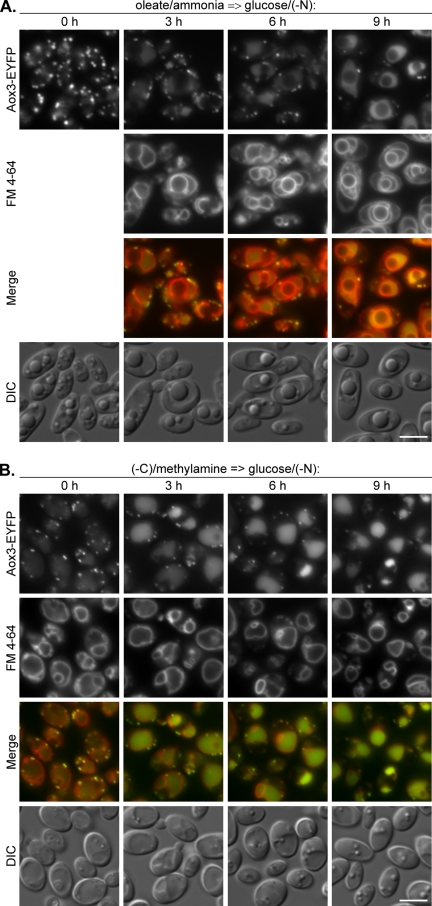
Degradation of Y. lipolytica oleate/ethylamine-induced peroxisomes in glucose/ammonia-containing medium was suggested to proceed via macropexophagy (Gunkel et al., 1999; Nazarko et al., 2005b). Using a fusion of the peroxisomal acyl-CoA oxidase 3 and the enhanced yellow fluorescent protein (Aox3-EYFP), in combination with the vacuolar membrane dye FM 4-64, we followed macropexophagy in Y. lipolytica by fluorescence microscopy (Nazarko et al., 2005b). We used the Aox3-EYFP/FM 4-64 double labeling to study the rates of delivery of oleate/ammonia-induced peroxisomes to the vacuole after the transfer of WT cells to glucose/(−N) (Figure 2A). Because the Y. lipolytica cells do not internalize FM 4-64 in oleate media (Nazarko et al., 2005b), FM 4-64 was added at time point 0 h of glucose adaptation and delineated the vacuolar membrane at 3–9 h. After oleate induction, the Y. lipolytica cells contained 10–15 peroxisomes per cell. The number of peroxisomes decreased drastically to two to five per cell after 9 h of glucose adaptation. Furthermore, the transient accumulation of peroxisomal Aox3-EYFP was observed inside the vacuoles during the 3–9 h of pexophagy. Accumulation of Aox3-EYFP in the vacuoles was also consistent with the low rates of THI degradation in our biochemical experiments under the same conditions (Figure 1A). Therefore, we conclude that the rates of THI degradation in Y. lipolytica do not reflect the rates of peroxisome delivery to the vacuole. We also did not notice any invagination of the vacuolar membrane at the sites of peroxisome uptake (Figure 2A) as we did after shift from oleate/ethylamine to glucose/ammonia (Nazarko et al., 2005b). This observation is in line with an earlier conclusion that in Y. lipolytica, oleate-induced peroxisomes are delivered to the vacuoles by macropexophagy during adaptation of cells to glucose (Nazarko et al., 2005b).
Figure 2.
Oleate- and amine-induced peroxisomes in Y. lipolytica are delivered to the vacuoles by macropexophagy. (A) Y. lipolytica WT (STN001) cells were induced in oleate/ammonia medium and transferred to glucose/(−N) medium with FM 4-64. (B) Y. lipolytica WT (STN002) cells were induced in (−C)/methylamine medium with FM 4-64 and transferred to glucose/(−N) medium. At the indicated time points, the progress of pexophagy was determined by fluorescence microscopy as described in Materials and Methods. Peroxisomes were labeled with Aox3-EYFP and vacuolar membranes with FM 4-64. Bar, 5 μm.
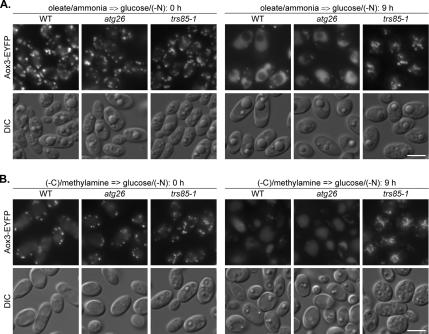
Because the rates of peroxisomal THI degradation in Y. lipolytica do not reflect the rates of peroxisome delivery to the vacuole, the possibility existed that the YlATG26 gene might be required for peroxisome delivery to the vacuole. Therefore, we compared the peroxisome-vacuole dynamics in the Y. lipolytica WT, atg26, and trs85-1 strains 0 and 9 h after shift from oleate/ammonia to glucose/(−N) (Figure 3A). Both Y. lipolytica WT and atg26 strains were characterized by a pronounced decrease in peroxisome number and accumulation of Aox3-EYFP in the vacuoles, indicating that peroxisomes are delivered normally to the vacuoles in the absence of YlAtg26. In contrast, peroxisomes remained outside the vacuoles in the trs85-1 mutant suggested to be blocked in pexophagy before the pexophagosome docking and fusion stage (Nazarko et al., 2005a). Altogether, these results demonstrate that oleate-induced peroxisomes in Y. lipolytica are degraded by Atg26-independent macropexophagy.
Figure 3.
Macropexophagy of oleate- and amine-induced peroxisomes in Y. lipolytica does not depend on Atg26. (A) Y. lipolytica WT (STN001), atg26 (STN003), and trs85-1 (STN012) cells were induced in oleate/ammonia medium and transferred to glucose/(−N) medium. (B) Y. lipolytica WT (STN001), atg26 (STN003), and trs85-1 (STN011) cells were induced in (−C)/methylamine medium and transferred to glucose/(−N) medium. After 9 h, the progress of pexophagy was determined by fluorescence microscopy. Peroxisomes were labeled with Aox3-EYFP. Bar, 5 μm.
Amine-induced Peroxisomes in Y. lipolytica Are Also Degraded by Atg26-independent Macropexophagy
Two possible explanations exist for the function of Atg26 in pexophagy: either it is P. pastoris specific or it is specific to methanol, the C1 carbon source that was used to induce peroxisomes in P. pastoris (Oku et al., 2003; Stasyk et al., 2003). We could not check the second possibility directly with Y. lipolytica, because it does not grow on methanol. However, it was possible to examine the degradation of Y. lipolytica peroxisomes induced by methylamine. This C1 primary amine is oxidized in peroxisomes and, in addition to ammonia, produces formaldehyde and hydrogen peroxide, two metabolites also produced by the oxidation of methanol (Zwart et al., 1980). As a control for methylamine, we used ethylamine, a C2 primary amine, which does not influence normal degradation of peroxisomes in the Y. lipolytica atg26 mutant, when used as a nitrogen source with oleate, the C18 carbon source that is also metabolized via C2 compounds (Supplemental Figure 1A). Both methylamine and ethylamine were used as the sole sources of carbon and nitrogen in peroxisome induction media. These carbon limited (−C) conditions were followed by glucose/(−N) adaptation to induce pexophagy. Because both peroxisome biogenesis and degradation were induced under nutrient-limiting conditions, we expected to observe the selective degradation of proliferated peroxisomes. Indeed, the levels of THI, which seemed to be derepressed both in (−C)/methylamine and (−C)/ethylamine media, gradually decreased in the Y. lipolytica WT strain during a 12-h time course of glucose adaptation (Figure 1B and Supplemental Figure 1B). The rates of degradation of both methylamine- and ethylamine-induced peroxisomes were comparable with the rates of degradation of Y. lipolytica peroxisomes induced by oleate. In contrast to the WT, peroxisome degradation was completely blocked in the Y. lipolytica trs85-2 mutant, indicating that in this yeast amine-induced peroxisomes are also degraded by an autophagy-related mechanism. Surprisingly, the Y. lipolytica atg26 mutant exhibited reproducibly higher rates of degradation of both methylamine- and ethylamine-induced peroxisomes relative to the WT strain. At the same time, the levels of mitochondrial Hsp60 protein remained stable in all strains tested (Figure 1B and Supplemental Figure 1B), suggesting that the differences we observed between the stabilities of THI and Hsp60 in WT and atg26 strains were caused by pexophagy. These data demonstrate that the ATG26 gene is not required for selective degradation of amine-induced peroxisomes in Y. lipolytica.
The Aox3-EYFP fusion that was used to label the Y. lipolytica oleate-induced peroxisomes is expressed under the promoter of acyl-CoA oxidase 2 gene. As was the case with THI, the Aox3-EYFP fusion was derepressed under the carbon-limited conditions in (−C)/methylamine medium and labeled the methylamine-induced peroxisomes (Figure 2B, 0 h). However, some Aox3-EYFP was already present in the vacuolar matrix, explaining the faster saturation of vacuoles with Aox3-EYFP during the pexophagy time course relative to oleate/ammonia-grown cells (Figure 2, compare A with B). In contrast to oleate medium that does not allow the Y. lipolytica cells to internalize FM 4-64 (Nazarko et al., 2005b), it was possible to label the Y. lipolytica vacuole membranes with FM 4-64 in methylamine medium and follow the vacuolar membrane dynamics from time point 0 h of glucose adaptation. As for oleate-induced cells transferred to glucose/(−N), two major lines of evidence were observed for the vacuolar degradation of methylamine-induced peroxisomes: 1) the gradual decrease of peroxisome number from 5–10 to 1–3 per cell in a 9-h time course and 2) the drastic increase in vacuolar Aox3-EYFP fluorescence after 3 h of glucose adaptation (Figure 2B). Transient accumulation of peroxisomal Aox3-EYFP inside the vacuoles during the 3–9 h of pexophagy induction was also consistent with the low rates of THI degradation in our biochemical studies under the same experimental setup (Figure 1B). Similar to oleate-induced cells, we also never observed any changes in vacuole membrane curvature around methylamine-induced peroxisomes (Figure 2B). Therefore, our results are more consistent with the vesicular transport of methylamine-induced peroxisomes to the vacuole, which is a characteristic feature of macropexophagy.
Because in Y. lipolytica the rates of peroxisomal THI degradation (Figure 1B and Supplemental Figure 1B) were lower than the peroxisome delivery rates (Figure 2B), the possibility still existed that the YlATG26 gene might be required for the delivery of methylamine-induced peroxisomes to the vacuole. Therefore, we compared the number of methylamine-induced peroxisomes in Y. lipolytica WT, atg26, and trs85-1 strains after 0 and 9 h of glucose adaptation (Figure 3B). The trs85-1 mutant was completely blocked in the delivery of peroxisomes to the vacuoles. Even after 9 h of pexophagy induction all the peroxisomes remained outside the vacuoles, and the vacuole lumen remained nonfluorescent in the trs85-1 strain. In contrast, the atg26 strain demonstrated both hallmarks of normal pexophagy observed in the WT strain, supporting the conclusion that the ATG26 gene is not required for macropexophagy of amine-induced peroxisomes in Y. lipolytica. The combined Y. lipolytica pexophagy data presented in this article demonstrate that Atg26 and the product of its enzymatic reaction, sterol glucoside, have a species-specific function in peroxisome degradation in yeast. Atg26 is nonessential for macropexophagy of both oleate- and amine-induced peroxisomes in Y. lipolytica, in direct contrast to its absolute requirement for degradation of methanol-induced peroxi somes in P. pastoris.
The P. pastoris ATG26 Gene Is Not Essential for Macropexophagy of Oleate-induced Peroxisomes, but It Increases Its Efficiency
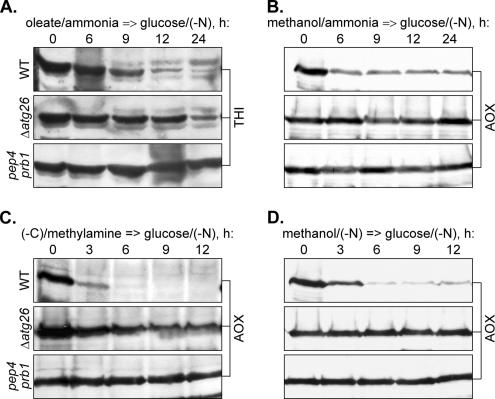
We and others have reported that Atg26 is essential for both micro- and macropexophagy of methanol-induced peroxisomes but not for starvation-induced autophagy in P. pastoris (Oku et al., 2003; Stasyk et al., 2003). However, there are still two possible scenarios for PpAtg26 function: either it is a general factor essential for degradation of P. pastoris peroxisomes induced by any substrate, or its requirement may be specific to methanol-induced peroxisomes. If Atg26 has a species-specific function in P. pastoris pexophagy, one would expect the same requirement of Atg26 for degradation of peroxisomes induced by oleate. To check this possibility, we induced P. pastoris cells by either oleate/ammonia or oleate/ethylamine, transferred them to glucose/(−N), and followed the levels of THI in the 24-h time course (Figure 4A and Supplemental Figure 1C). In the control P. pastoris cells shifted to glucose/(−N) from methanol/ammonia medium (Figure 4B), the level of AOX in the WT strain decreased to the basal level in 6 h of glucose adaptation. However, it remained stable for at least 24 h in both P. pastoris pep4 prb1 (Tuttle and Dunn, 1995) (deficient in vacuolar proteases A and B) and Δ_atg26_ strains. The complete block in the degradation of methanol-induced peroxisomes in the P. pastoris Δ_atg26_ strain in glucose/(−N) medium indicates that peroxisomes in P. pastoris, as in S. cerevisiae (Hutchins et al., 1999) and Y. lipolytica (this study), are degraded by selective pexophagy, rather than by nonselective autophagy. Therefore, we conclude that glucose/(−N) medium is appropriate for the studies of selective degradation of oleate-induced peroxisomes in P. pastoris. However, the P. pastoris oleate-induced cells seemed to degrade THI faster than the Y. lipolytica cells, irrespective of the nitrogen source used in oleate medium (compare Figure 1A with 4A). We could usually observe the complete degradation of oleate-induced peroxisomes in the P. pastoris WT strain after 24 h of glucose adaptation, but peroxisome degradation was completely blocked, as expected, in the pep4 prb1 mutant. Surprisingly, the P. pastoris Δ_atg26_ strain reproducibly exhibited an intermediate phenotype with degradation of both oleate/ammonia- and oleate/ethylamine-induced peroxisomes delayed by at least 9 h relative to the WT strain (Figure 4A and Supplemental Figure 1C). This led us to conclude that the P. pastoris ATG26 gene is not essential for degradation of oleate-induced peroxisomes but that it might increase its efficiency.
Figure 4.
The P. pastoris ATG26 gene is not essential for selective degradation of oleate- and amine-induced peroxisomes, but it increases its efficiency. P. pastoris WT (GS200), Δ_atg26_ (Pdg3D), and pep4 prb1 (SMD1163) cells were induced in oleate/ammonia (A), methanol/ammonia (B), (−C)/methylamine (C), or methanol/(−N) (D) medium and transferred to glucose/(−N) medium to induce pexophagy. At the indicated time points, the culture samples were collected and processed for immunoblotting with antibodies against peroxisomal THI or AOX.
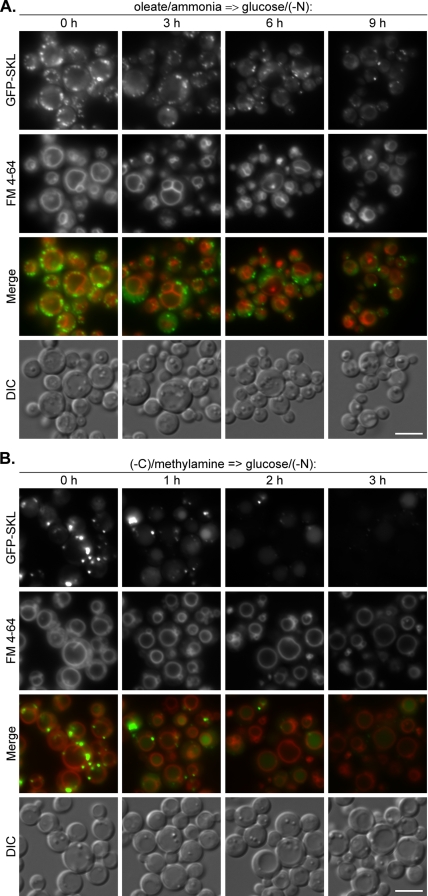
To study the morphological mechanism and kinetics of delivery of oleate-induced peroxisomes to the vacuole in P. pastoris, we used 1) a fusion of a brighter mutant of the green fluorescent protein (GFP), GFP-S65T, to the peroxisomal targeting signal 1, Ser-Lys-Leu (SKL), under the control of the constitutive P. pastoris glyceraldehyde-3-phosphate dehydrogenase gene promoter to label peroxisomes; and 2) FM 4-64 to label the vacuolar membrane (Sakai et al., 1998). Interestingly, unlike Y. lipolytica, the P. pastoris cells internalized FM 4-64 in oleate medium, allowing us to label the vacuole membrane before the onset of pexophagy. P. pastoris WT cells were transferred from oleate/ammonia to glucose/(−N) and peroxisome-vacuole dynamics were studied during 9 h of glucose adaptation (Figure 5A). As in Y. lipolytica, the P. pastoris cells contained 10–15 peroxisomes per cell after oleate induction, and this number decreased to two to five per cell after 9 h in glucose/(−N). However, in contrast to Y. lipolytica, we could not observe any accumulation of GFP-SKL inside the P. pastoris vacuoles at any time point of glucose adaptation (compare Figure 2A with 5A). As in Y. lipolytica, we did not observe any invaginations of vacuolar membrane at the sites of peroxisome uptake in P. pastoris cells. These experiments demonstrate that oleate-induced peroxisomes in P. pastoris are also delivered to the vacuoles by macropexophagy during adaptation of cells to glucose.
Figure 5.
Oleate- and amine-induced peroxisomes in P. pastoris are delivered to the vacuoles by macropexophagy. (A) P. pastoris WT (STN018) cells were induced in oleate/ammonia medium with FM 4-64 and transferred to glucose/(−N) medium. (B) P. pastoris WT (STN026) cells were induced in (−C)/methylamine medium with FM 4-64 and transferred to glucose/(−N) medium. The progress of pexophagy was determined by fluorescence microscopy. Peroxisomes were labeled with GFP-SKL and vacuolar membranes with FM 4-64. Bar, 5 μm.
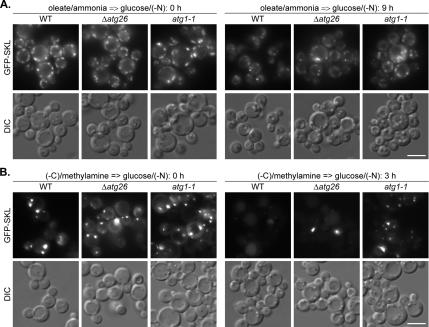
To confirm that the PpATG26 gene is not essential for delivery of oleate-induced peroxisomes to the vacuole, we compared the peroxisome dynamics in the P. pastoris WT, Δ_atg26_ and atg1-1 strains 0 and 9 h after the shift from oleate/ammonia to glucose/(−N) (Figure 6A). After 9 h of glucose adaptation, the majority of oleate-induced peroxisomes disappeared in the P. pastoris WT strain but not in the atg1-1 mutant, disrupted in the gene encoding the serine/threonine protein kinase Atg1, which is required for the pexophagy, autophagy, and Cvt pathways in S. cerevisiae, P. pastoris, and H. polymorpha (Harding et al., 1996; Matsuura et al., 1997; Straub et al., 1997; Stromhaug et al., 2001; Mukaiyama et al., 2002; Komduur et al., 2003). The P. pastoris Δ_atg26_ strain clearly exhibited an intermediate phenotype with a greater number of cells in the population resembling the WT strain but still with a significant number of cells exhibiting a high number of peroxisomes as in the atg1-1 mutant (Figure 6A). Together, our biochemical and microscopy data demonstrate that PpAtg26 protein is not essential for macropexophagy of oleate-induced peroxisomes in P. pastoris but that it enhances its efficiency. Therefore, the function of Atg26 in pexophagy might be also peroxisome inducer specific.
Figure 6.
Macropexophagy of oleate- and amine-induced peroxisomes in P. pastoris can proceed without the ATG26 gene but at slower rates. (A) P. pastoris WT (STN018), Δ_atg26_ (STN020), and atg1-1 (STN022) cells were induced in oleate/ammonia medium and transferred to glucose/(−N) medium for 9 h. (B) P. pastoris WT (STN026), Δ_atg26_ (STN028), and atg1-1 (STN029) cells were induced in (−C)/methylamine medium and transferred to glucose/(−N) medium for 3 h. The progress of pexophagy was determined by fluorescence microscopy. Peroxisomes were labeled with GFP-SKL. Bar, 5 μm.
Macropexophagy of Amine-induced Peroxisomes in P. pastoris Also Proceeds without the ATG26 Gene but at the Slower Rates
If the function of PpAtg26 in pexophagy is dependent on the peroxisome inducer, is the PpAtg26 specifically essential for degradation of methanol-induced peroxisomes or alternatively, are oleate-induced peroxisomes the specific exception? To address this question, we examined the degradation of P. pastoris peroxisomes induced by methylamine and ethylamine. As in the studies with Y. lipolytica, these primary amines were used as the sole sources of carbon and nitrogen in peroxisome-induction media and were followed by glucose/(−N) adaptation for pexophagy induction (Figure 4C and Supplemental Figure 1D). In the control, when we transferred P. pastoris cells from methanol/(−N) to glucose/(−N), the level of AOX in the WT strain decreased to the basal level in 6 h of glucose adaptation, essentially the same as after the shift of cells from methanol/ammonia to glucose/(−N) (compare Figure 4B with D). However, it remained stable for at least 12 h in both P. pastoris pep4 prb1 and Δ_atg26_ strains. The complete block of degradation of methanol-induced peroxisomes in the P. pastoris Δ_atg26_ strain under nitrogen starvation in both methanol and glucose media indicates that peroxisomes in P. pastoris are degraded exclusively by pexophagy, even under starvation conditions. This allowed us to study the selective degradation of P. pastoris peroxisomes after the shift of cells from (−C)/amine to glucose/(−N). As expected, the P. pastoris AOX was derepressed in both (−C)/methylamine and (−C)/ethylamine media, allowing its use as a peroxisomal marker to study the degradation of amine-induced peroxisomes. The rates of degradation of both methylamine- and ethylamine-induced peroxisomes in the P. pastoris WT strain were much faster than the rates of degradation of peroxisomes induced by oleate (compare Figure 4A with C). Interestingly, P. pastoris peroxisomes induced by oleate/ethylamine were degraded like oleate-, rather than like ethylamine-induced peroxisomes (compare Supplemental Figure 1C with D), suggesting a dominant effect of oleate on subsequent pexophagy rates. In contrast to the WT, peroxisome degradation was completely blocked in the P. pastoris pep4 prb1 mutant, suggesting that the P. pastoris amine-induced peroxisomes are degraded in the vacuoles. However, the levels of peroxisomal AOX decreased gradually in the P. pastoris Δ_atg26_ strain during the 12-h time course of glucose adaptation of amine-induced cells (Figure 4C and Supplemental Figure 1D). Therefore, the P. pastoris ATG26 gene is also not essential for selective degradation of peroxisomes induced by primary amines, but it increases the efficiency of the process.
To study the mode of degradation of P. pastoris amine-induced peroxisomes, we labeled peroxisomes with the GFP–SKL fusion protein expressed under control of the promoter of the P. pastoris AOX1 gene and the vacuolar membrane was labeled with FM 4-64 (Sakai et al., 1998). The P. pastoris WT cells were transferred from (−C)/methylamine to glucose/(−N), and peroxisome-vacuole dynamics were studied during the first 3 h of glucose adaptation (Figure 5B). In contrast to P. pastoris oleate-induced cells, we observed a significant decrease in the number of large mature peroxisomes as early as 1 h after the shift of amine-induced cells to glucose/(−N) (compare Figure 5A with B). It was accompanied by the transient accumulation of GFP-SKL inside the vacuoles. Large mature peroxisomes disappeared almost completely after 2 h of glucose adaptation, and GFP-SKL fluorescence inside the vacuoles decreased significantly 3 h after the shift (Figure 5B). The kinetics of delivery of P. pastoris amine-induced peroxisomes to the vacuole was consistent with the high rates of AOX degradation in our biochemical studies under the same experimental setup (Figure 4C). However, these dramatic changes in peroxisome numbers occurred with no detectable changes in vacuolar morphology. The vacuoles remained perfectly round and peroxisomes could often be seen adjacent to the vacuolar membrane. Our observations are more consistent with the model that amine-induced peroxisomes enter the vacuole by macropexophagy. Interestingly, at each time point of pexophagy induction, a lot of vacuoles contained lumenal dot-like structures labeled with FM 4-64. However, these structures never colocalized with GFP-SKL dots. Therefore, we suggest that intravacuolar FM 4-64 dots constitute the microautophagic but not micropexophagic bodies. Together, these results indicate that amine-induced peroxisomes in P. pastoris are degraded by macropexophagy, which resembles the mechanism observed with methanol-induced cells shifted to ethanol medium, but it occurs with much higher rates of peroxisome delivery to the vacuole and vacuolar degradation.
We compared the pexophagy rates in P. pastoris WT, Δ_atg26_, and atg1-1 strains, 0 and 3 h after the shift from (−C)/methylamine to glucose/(−N) (Figure 6B). Peroxisomes disappeared from the P. pastoris WT strain, but some vacuolar GFP–SKL fluorescence was still present after 3 h of glucose adaptation. In contrast, the majority of amine-induced peroxisomes remained intact in the cytosol of the atg1-1 mutant. Furthermore, we never observed GFP-SKL in the vacuoles of the atg1-1 strain. Not surprisingly, the population of P. pastoris Δ_atg26_ cells had large mature peroxisomes, as in the atg1-1 mutant, as well as cells with vacuolar GFP-SKL (Figure 6B). These results suggest that the P. pastoris ATG26 gene is not essential for macropexophagy of peroxisomes induced by primary amines. The combined P. pastoris pexophagy data presented above demonstrate that the function of Atg26 protein and SG in pexophagy also depends on the peroxisome inducer, being essential for degradation of methanol- but not of oleate- and amine-induced peroxisomes in P. pastoris.
The P. pastoris ATG26, but Not the VAC8, Gene Is Essential for Macropexophagy of Methanol-induced Peroxisomes during Ethanol Adaptation
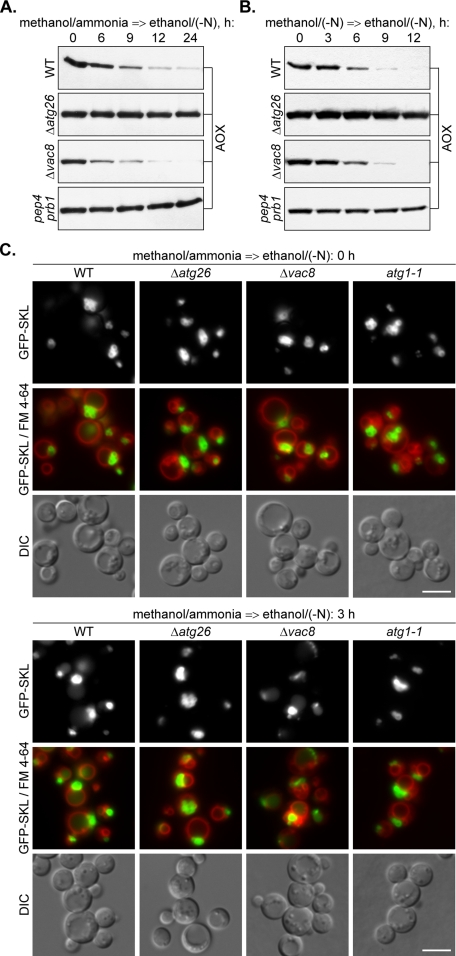
To determine whether the PpATG26 gene is essential for ethanol-induced macropexophagy of the P. pastoris methanol-induced peroxisomes under nitrogen starvation conditions, we induced the cells of the P. pastoris WT, Δ_atg26_, Δ_vac8_, and pep4 prb1 strains in methanol/ammonia or methanol/(−N) media and transferred them to ethanol/(−N) medium to induce pexophagy (Figure 7, A and B). Independent of the presence of nitrogen in methanol medium, the P. pastoris WT strain exhibited lower rates of peroxisomal AOX degradation in ethanol/(−N) than during adaptation of cells to glucose/(−N) (compare Figure 4, B and D, and Figure 7, A and B). However, both of them were consistent with the rates of classical micro- and macropexophagy described previously (Stasyk et al., 2003). Importantly, the levels of peroxisomal AOX in ethanol/(−N) medium remained completely stable in the cells of both P. pastoris pep4 prb1 and Δ_atg26_ strains, independent of the presence of nitrogen in methanol medium (Figure 7, A and B). These results indicate that the ATG26 gene is essential for both glucose- and ethanol-induced degradation of methanol-induced peroxisomes in P. pastoris. Surprisingly, the rates of AOX degradation in the P. pastoris Δ_vac8_ strain were indistinguishable from those found in the WT strain.
Figure 7.
The P. pastoris ATG26 but not the VAC8 gene is essential for macropexophagy of methanol-induced peroxisomes during ethanol adaptation. The P. pastoris WT (GS200), Δ_atg26_ (Pdg3D), Δ_vac8_ (WDY53), and pep4 prb1 (SMD1163) cells were induced in methanol/ammonia (A) or methanol/(−N) (B) medium and transferred to ethanol/(−N) medium to induce pexophagy. At the indicated time points, the culture samples were collected and processed for immunoblotting with antibodies against peroxisomal AOX. (C) The P. pastoris WT (STN026), Δ_atg26_ (STN028), Δ_vac8_ (STN032), and atg1-1 (STN030) cells were induced in methanol/ammonia medium with FM 4-64 and transferred to ethanol/(−N) medium for 3 h. Pexophagy was monitored by fluorescence microscopy. Peroxisomes were labeled with GFP-SKL and vacuolar membranes with FM 4-64. Bar, 5 μm.
To confirm that methanol-induced peroxisomes are delivered to the vacuoles during ethanol adaptation of the P. pastoris Δ_vac8_ but not Δ_atg26_ strain, we labeled peroxisomes with the GFP–SKL fusion protein expressed under control of promoter of the P. pastoris AOX1 gene and the vacuolar membrane with FM 4-64 (Sakai et al., 1998). Cells of the P. pastoris WT, Δ_atg26_, Δ_vac8_, and atg1-1 strains were grown in methanol/ammonia medium and transferred to ethanol/(−N) for 3 h (Figure 7C). All four strains developed clusters of large peroxisomes in methanol medium, but only the cells of the WT and Δ_vac8_ strains showed peroxisome degradation. The diffuse vacuolar GFP–SKL fluorescence, typical for the WT and Δ_vac8_ cells after 3 h of ethanol adaptation, was absent in the atg1-1 and Δ_atg26_ mutants. Because vacuolar morphology in the WT strain did not undergo any changes in ethanol/(−N) medium (Figure 7C), we conclude that P. pastoris methanol-induced peroxisomes are delivered to the vacuole by macropexophagy. The P. pastoris ATG26 gene seemed to be essential for macropexophagy of methanol-induced peroxisomes independent of the presence of nitrogen in methanol and/or ethanol media. These data suggest an essential and specific role of sterol glucoside in the degradation of methanol-induced peroxisomes irrespective of the mode of pexophagy used by these peroxisomes. In contrast to PpAtg26, PpVac8 protein was nonessential for macropexophagy of methanol-induced peroxisomes during ethanol adaptation. Vac8 is the armadillo repeat protein essential for homotypic vacuole fusion, Cvt pathway, and piecemeal microautophagy of the nucleus but not macroautophagy in S. cerevisiae (Fleckenstein et al., 1998; Pan and Goldfarb, 1998; Wang et al., 1998; Scott et al., 2000; Roberts et al., 2003). It is required for micropexophagy but not for starvation-induced autophagy in P. pastoris (Chang et al., 2005). Our data indicate that the function of PpVac8 in degradation of methanol-induced peroxisomes might be micropexophagy specific.
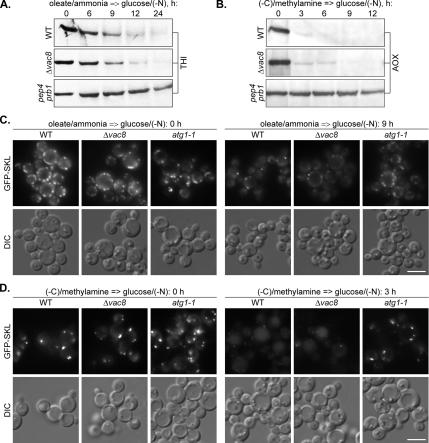
The P. pastoris VAC8 Gene Is Also Not Essential for Degradation of Oleate- and Amine-induced Peroxisomes
If the PpVac8 is really required only for the homotypic fusion of VSM during micropexophagy but not for the heterotypic fusion of pexophagosomes with the vacuole during macropexophagy (Dunn et al., 2005; this study), the P. pastoris Δ_vac8_ mutant could be a valuable tool to discriminate between the two pexophagy modes. To test this possibility and support our morphological observations on the mode of degradation of P. pastoris oleate- and amine-induced peroxisomes, we compared the degradation of peroxisomal THI and AOX in the P. pastoris WT, Δ_vac8_, and pep4 prb1 cells after the shift from oleate/ammonia (Figure 8A) and (−C)/methylamine (Figure 8B) to glucose/(−N). In contrast to the pep4 prb1 mutant with completely blocked pexophagy, the P. pastoris Δ_vac8_ strain exhibited essentially the same rates of degradation of oleate- and amine-induced peroxisomes as the WT strain. The biochemical data were confirmed by fluorescence microscopy of the P. pastoris WT, Δ_vac8_, and atg1-1 cells under the same experimental conditions (Figure 8, C and D). The majority of oleate- and amine-induced peroxisomes disappeared in the P. pastoris Δ_vac8_ mutant after 9 and 3 h of glucose adaptation, respectively, essentially as in the WT strain. At the same times, most of the oleate- and amine-induced peroxisomes remained intact in the cytosol of the atg1-1 mutant. These data clearly demonstrate that the Vac8 protein is not essential for degradation of the P. pastoris oleate- and amine-induced peroxisomes. Therefore, the phenotype of the P. pastoris Δ_vac8_ mutant is consistent with our conclusions (see above) that in this organism oleate- and amine-induced peroxisomes are delivered to the vacuole by macropexophagy.
Figure 8.
The P. pastoris VAC8 gene is not essential for degradation of oleate- and amine-induced peroxisomes. P. pastoris WT (GS200), Δ_vac8_ (WDY53), and pep4 prb1 (SMD1163) cells were induced in oleate/ammonia (A) or (−C)/methylamine (B) medium and transferred to glucose/(−N) medium to induce pexophagy. At the indicated time points, the culture samples were collected and processed for immunoblotting with antibodies against peroxisomal THI or AOX. (C) P. pastoris WT (STN018), Δ_vac8_ (STN024), and atg1-1 (STN022) cells were induced in oleate/ammonia medium and transferred to glucose/(−N) medium for 9 h. (D) P. pastoris WT (STN026), Δ_vac8_ (STN031), and atg1-1 (STN029) cells were induced in (−C)/methylamine medium and transferred to glucose/(−N) medium for 3 h. The progress of pexophagy was determined by fluorescence microscopy. Peroxisomes were labeled with GFP-SKL. Bar, 5 μm.
DISCUSSION
Oleate- and Amine-induced Peroxisomes Are Degraded by Macropexophagy in Yeast
Although peroxisome turnover has been investigated in several yeast species, it is unclear how selective it is in different model systems. Furthermore, very little is known regarding the mechanisms of pexophagy (Kiel et al., 2003; Farré and Subramani, 2004; Dunn et al., 2005). Knowledge of the selectivity and mechanisms of pexophagy in different model systems is necessary for a complete understanding of this process. Studies in P. pastoris have revealed that both micropexophagy and macropexophagy can selectively degrade peroxisomes (Farré and Subramani, 2004). In this study, we have addressed several important questions. First, we used different conditions for peroxisome induction in several model systems to address the selectivity of peroxisome turnover. Second, we have used biochemical and microscopy methods to study the rates and mode of pexophagy in each case. Third, we have defined both species-specific and peroxisome inducer-specific requirements for one pexophagy-specific protein, Atg26. Finally, we report that Vac8 is required for the micropexophagy, but not for the macropexophagy, machinery. These studies demonstrate the importance of several model systems in the elucidation of the functional role of genes involved in pexophagy.
Because degradation of oleate-induced peroxisomes is usually a slow process that needs to be monitored over prolonged periods of time (Hutchins et al., 1999; Nazarko et al., 2005b), we developed reliable assays to follow the degradation of oleate- and amine-induced peroxisomes of Y. lipolytica and P. pastoris in glucose medium without nitrogen, glucose/(−N). The use of medium lacking nitrogen avoids the problem of peroxisome dilution due to cell growth and division. Similar to degradation of oleate-induced peroxisomes in S. cerevisiae (Hutchins et al., 1999), degradation of Y. lipolytica and P. pastoris oleate- and amine-induced peroxisomes was highly selective despite the use of nitrogen starvation conditions. We provide two lines of evidence for the selective degradation of peroxisomes in glucose/(−N): 1) the Y. lipolytica WT cells maintained stable levels of mitochondria but not of oleate- or amine-induced peroxisomes (Figure 1); and 2) the P. pastoris Δ_atg26_ cells affected in pexophagy but not in autophagy maintained the stable levels of methanol-induced peroxisomes, in contrast to the WT cells (Figure 4, B and D). Therefore, the combined S. cerevisiae, Y. lipolytica, and P. pastoris data strongly suggest that the turnover of surplus peroxisomes in the vacuole in yeast occurs by a selective mechanism under both growth and starvation conditions. Similar results were observed previously for the transport of aminopeptidase I to vacuole in S. cerevisiae (Scott et al., 1996; Baba et al., 1997), which is mediated by the receptor protein Atg19 in both growing and starving cells (Leber et al., 2001; Scott et al., 2001). Thus, our results predict a receptor-mediated mechanism for pexophagy. Importantly, clofibrate-induced peroxisomes in rat hepatocytes were selectively degraded after administration of either normal or fasting diet to rats (Luiken et al., 1992). Therefore, not only the molecular machinery of pexophagy but also its selectivity toward superfluous peroxisomes under both normal and starvation conditions is conserved in lower and higher eukaryotes.
To study degradation of Y. lipolytica and P. pastoris amine-induced peroxisomes, two primary amines, methylamine and ethylamine, were used for peroxisome induction as the sole sources of carbon and nitrogen. These carbon-limited conditions allowed us to follow pexophagy with the same peroxisomal markers, as after oleate or methanol induction, and did not influence the selectivity of pexophagy. Selective degradation of Y. lipolytica and P. pastoris peroxisomes after the transfer of cells from (−C)/methylamine, (−C)/ethylamine, or methanol/(−N) medium to glucose/(−N) is also consistent with selective degradation of P. pastoris and H. polymorpha peroxisomes after addition of solid glucose to stationary phase cultures in methanol/ammonia medium (Tuttle et al., 1993). However, to our knowledge, this is the first study on degradation of peroxisomes induced solely by primary amines in yeast.
Degradation of both oleate- and amine-induced peroxisomes was dependent on the components of general autophagic machinery, such as PpAtg1 (Stromhaug et al., 2001; Mukaiyama et al., 2002) and YlTrs85 (Nazarko et al., 2005a) as well as on the vacuolar proteases A and B (Tuttle and Dunn, 1995). Although it was not possible to distinguish between macro- and micropexophagy of oleate-induced peroxisomes in glucose/(−N) with S. cerevisiae (Hutchins et al., 1999) and both macropexophagy and microautophagy of methanol-induced peroxisomes were observed in methanol/glucose/(−N) with H. polymorpha (Monastryska et al., 2004), we could easily exclude micropexophagy as the mechanism of turnover for oleate- and amine-induced peroxisomes in Y. lipolytica and P. pastoris. A characteristic feature of micropexophagy in P. pastoris is the development of VSMs that engulf the whole cluster of methanol-induced peroxisomes (Dunn et al., 2005). However, we never observed 1) clustering of oleate- and amine-induced peroxisomes and 2) invagination or septation of vacuolar membranes around oleate- and amine-induced peroxisomes during glucose adaptation in either Y. lipolytica (Figure 2) or P. pastoris (Figure 5). Oleate- and amine-induced peroxisomes were degraded individually in glucose/(−N) with no visible changes in vacuolar membrane curvature at the sites of peroxisome uptake. Therefore, we conclude that peroxisomes induced by oleate or primary amines are degraded by macropexophagy in yeast. It is worthwhile to point out that clofibrate-induced peroxisomes in rat hepatocytes were degraded by macropexophagy after incubation of cells in the medium without amino acids in the presence of cycloheximide (Luiken et al., 1992). Furthermore, mouse liver peroxisomes induced by phthalate esters were also degraded by macropexophagy after discontinuation of treatment with phthalate esters (Iwata et al., 2006). These results demonstrate that macropexophagy occurs in mammals. This increases the importance of yeast macropexophagy models. Here, we present two new macropexophagy conditions in two model yeast strains. The P. pastoris pexophagy model is particularly attractive because it allows comparative studies on macropexophagy of methanol-, oleate-, and amine-induced peroxisomes.
PpVac8 Is Required for the Micropexophagy, but Not for the Macropexophagy, Machinery
Recently, it was reported that PpVac8 is required for glucose-induced micropexophagy of methanol-induced peroxisomes but not for starvation-induced autophagy (Chang et al., 2005). Here, we show for the first time that PpVac8 is not essential for ethanol-induced macropexophagy of methanol-induced peroxisomes (Figure 7). Furthermore, PpVac8 is also not essential for macropexophagy of oleate- and amine-induced peroxisomes under glucose adaptation (Figure 8). Therefore, our results place PpVac8 in a new category of proteins specifically required for one of the two pexophagy pathways, i.e., for micropexophagy. Recently, it was shown that the H. polymorpha Atg25 is a macropexophagy-specific protein required for glucose-induced macropexophagy but not for starvation-induced microautophagy of methanol-induced peroxisomes (Monastyrska et al., 2005). Currently, the requirement in Vac8 is the only known difference between the P. pastoris macro- and micropexophagy machineries. It was previously shown that PpVac8 localizes to perivacuolar spots and also to vacuolar membranes or VSMs during macro- or micropexophagy of methanol-induced peroxisomes, respectively (Ano et al., 2005). PpVac8 is not required for the formation of either the VSM (Chang et al., 2005) or the MIPA (Dunn et al., 2005). Instead, PpVac8 might be required for the last stage of micropexophagy, the homotypic fusion of the VSM, completing sequestration of the cluster of methanol-induced peroxisomes from the cytosol (Dunn et al., 2005). Indeed, S. cerevisiae Vac8 is essential for homotypic vacuole fusion (Veit et al., 2001; Wang et al., 2001), but it is dispensable for macroautophagy of bulk cytosol (Scott et al., 2000). Therefore, the persistence of macropexophagy of methanol-, oleate-, and amine-induced peroxisomes in the P. pastoris Δ_vac8_ strain (this study) is consistent with the continuation of macroautophagy of bulk cytosol in P. pastoris and S. cerevisiae Δ_vac8_ mutants (Scott et al., 2000; Chang et al., 2005). These results strongly suggest that heterotypic fusion of autophagosome/pexophagosome with the vacuole does not require the assistance of the Vac8 protein in yeast. The latter function might be mediated by an orthologue of the H. polymorpha Atg25 coiled-coil protein that has been shown to be required for the fusion of pexophagosomes with the vacuole but not for the homotypic vacuole fusion (Monastyrska et al., 2005).
Methanol-induced Peroxisomes Have a Specific Requirement of Sterol Glucoside for Pexophagy in P. pastoris
Ugt51/Atg26 has recently been suggested to be a unique Atg protein, because its catalytic activity is essential for macro- and micropexophagy of methanol-induced peroxisomes but not for starvation-induced autophagy in P. pastoris, or the Cvt pathway in S. cerevisiae (Oku et al., 2003; Stasyk et al., 2003). Our data (Figures 1 and 3) show that macropexophagy of both oleate- and amine-induced peroxisomes is not affected in the Y. lipolytica mutant lacking SG. Furthermore, we did not observe any difference in the degradation of peroxisomes induced by methylamine (C1 compound with a similar metabolism to methanol) or ethylamine (C2 compound) in the Y. lipolytica atg26 mutant (Figure 1B and Supplemental Figure 1B). This suggests that the difference in the role of sterol glucosyltransferase observed in Y. lipolytica and P. pastoris (Stasyk et al., 2003) is not directly related to metabolic differences between Y. lipolytica oleate- and P. pastoris methanol-induced peroxisomes. Therefore, we conclude that SG has a species-specific function in pexophagy in that it is required for pexophagy in P. pastoris but not in Y. lipolytica. Supporting this idea, recent work, submitted while this manuscript was being reviewed, shows that in S. cerevisiae Atg26 is also not involved in the degradation of peroxisomes (Cao and Klionsky, 2007).
However, P. pastoris Atg26 is not essential for macropexophagy of both oleate- and amine-induced peroxisomes in glucose/(−N), but increases its efficiency (Figures 4, A and C, and 6). At the same time, the PpATG26 gene is essential for macropexophagy of methanol-induced peroxisomes in ethanol/(−N) (Figure 7). Therefore, SG also has a peroxisome inducer-specific function in P. pastoris pexophagy, being specifically essential for degradation of methanol- but not of oleate- and amine-induced peroxisomes. Recently, it was shown that synthesis of SG by PpAtg26 at the PAS is a prerequisite of MIPA formation. Furthermore, it was suggested that the conversion of sterol into SG is required for maturation and elongation of the PAS into the MIPA (Yamashita et al., 2006). SG formation might be also required for maturation and elongation of the PAS into a pexophagosome. However, it is unclear what the difference is between formation of pexophagosomes around methanol-, oleate-, and amine-induced peroxisomes and why ergosterol conversion is specifically essential for sequestration of methanol-induced peroxisomes. The simplest explanation would be that the larger size of methanol-induced peroxisomes relative to oleate- and amine-induced peroxisomes might result in the enhanced SG requirement for macropexophagy of methanol-induced peroxisomes (Supplemental Figure 2). However, the size of the MIPA that is needed for micropexophagic sequestration of a cluster of methanol-induced peroxisomes is much smaller than the size of a single pexophagosome around methanol-induced peroxisome, but synthesis of SG is still essential for MIPA formation as well. The other possibility is that the differences in protein composition of peroxisomal membrane in methanol-, oleate- and amine-induced cells may account for the diverse requirement of SG for subsequent pexophagy. Further studies are required to clarify the role of SG in macropexophagy of peroxisomes induced by different substrates in P. pastoris.
ACKNOWLEDGMENTS
We thank Dr. Marilyn Farquhar (University of California, San Diego, San Diego, CA) for the use of the electron microscope facility. This work was supported by National Institutes of Health Grants DK-41737 and GM-069373 (to S.S.).
Abbreviations used:
AOX
alcohol oxidase
Atg
autophagy-related
Cvt
cytoplasm-to-vacuole targeting
EYFP
enhanced yellow fluorescent protein
MIPA
micropexophagic membrane apparatus
PAS
preautophagosomal structure
PiB
phosphate buffer
SG
sterol glucoside
THI
thiolase
VSM
vacuolar sequestering membrane.
Footnotes
REFERENCES
- Ano Y., Hattori T., Kato N., Sakai Y. Intracellular ATP correlates with mode of pexophagy in Pichia pastoris. Biosci. Biotechnol. Biochem. 2005;69:1527–1533. doi: 10.1271/bbb.69.1527. [DOI] [PubMed] [Google Scholar]
- Baba M., Osumi M., Scott S. V., Klionsky D. J., Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends R. J., Faber K. N., Kram A. M., Kiel J. A., van der Klei I. J., Veenhuis M. A stretch of positively charged amino acids at the N terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J. Biol. Chem. 2000;275:9986–9995. doi: 10.1074/jbc.275.14.9986. [DOI] [PubMed] [Google Scholar]
- Barth G, Gaillardin C. Yarrowia lipolytica. In: Wolf K, editor. Nonconventional Yeasts in Biotechnology. Berlin, Germany: Springer; 1996. pp. 313–388. [Google Scholar]
- Bellu A. R., Kiel J. A. Selective degradation of peroxisomes in yeasts. Microsc. Res. Tech. 2003;61:161–170. doi: 10.1002/jemt.10325. [DOI] [PubMed] [Google Scholar]
- Cao Y., Klionsky D. J. Atg26 is not involved in autophagy-related pathways in Saccharomyces cerevisiae. Autophagy. 2007;3:e1–e4. doi: 10.4161/auto.3371. [DOI] [PubMed] [Google Scholar]
- Chang T., Schroder L. A., Thomson J. M., Klocman A. S., Tomasini A. J., Stromhaug P. E., Dunn W. A., Jr PpATG9 encodes a novel membrane protein that traffics to vacuolar membranes, which sequester peroxisomes during pexophagy in Pichia pastoris. Mol. Biol. Cell. 2005;16:4941–4953. doi: 10.1091/mbc.E05-02-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J. M., Barringer K. J., Hessler A. Y., Madden K. R. Pichia pastoris as a host system for transformations. Mol. Cell. Biol. 1985;5:3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J. M., Russell K. A. Transformation. In: Higgins D. R., Cregg J., editors. Methods in Molecular Biology, Pichia Protocols. Totowa, NJ: Humana Press; 1998. pp. 27–40. [Google Scholar]
- Dunn W. A., Jr, Cregg J. M., Kiel J.A.K.W., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Farré J. C., Subramani S. Peroxisome turnover by micropexophagy: an autophagy-related process. Trends Cell Biol. 2004;14:515–523. doi: 10.1016/j.tcb.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Fleckenstein D., Rohde M., Klionsky D. J., Rudiger M. Yel013p (Vac8p), an armadillo repeat protein related to plakoglobin and importin alpha is associated with the yeast vacuole membrane. J. Cell Sci. 1998;111:3109–3118. doi: 10.1242/jcs.111.20.3109. [DOI] [PubMed] [Google Scholar]
- Gunkel K., van der Klei I. J., Barth G., Veenhuis M. Selective peroxisome degradation in Yarrowia lipolytica after a shift of cells from acetate/oleate/ethylamine into glucose/ammonium sulfate-containing media. FEBS Lett. 1999;451:1–4. doi: 10.1016/s0014-5793(99)00513-x. [DOI] [PubMed] [Google Scholar]
- Harding T. M., Hefner-Gravink A., Thumm M., Klionsky D. J. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J. Biol. Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Hutchins M. U., Veenhuis M., Klionsky D. J. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 1999;112:4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- Iwata J., Ezaki J., Komatsu M., Yokota S., Ueno T., Tanida I., Chiba T., Tanaka K., Kominami E. Excess peroxisomes are degraded by autophagic machinery in mammals. J. Biol. Chem. 2006;17:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- Kawamata T., Kamada Y., Suzuki K., Kuboshima N., Akimatsu H., Ota S., Ohsumi M., Ohsumi Y. Characterization of a novel autophagy-specific gene, ATG29. Biochem. Biophys. Res. Commun. 2005;30:1884–1889. doi: 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- Kiel J. A., Komduur J. A., van der Klei I. J., Veenhuis M. Macropexophagy in Hansenula polymorpha: facts and views. FEBS. Lett. 2003;14:1–6. doi: 10.1016/s0014-5793(03)00794-4. [DOI] [PubMed] [Google Scholar]
- Kim J., Kamada Y., Stromhaug P. E., Guan J., Hefner-Gravink A., Baba M., Scott S. V., Ohsumi Y., Dunn W. A., Jr, Klionsky D. J. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001;16:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Cregg J. M., Dunn W. A., Jr, Emr S. D., Sakai Y., Sandoval I. V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Komduur J. A. Haren, The Netherlands: University of Groningen; 2004. Molecular aspects of peroxisome degradation in Hansenula polymorpha. [Google Scholar]
- Komduur J. A., Veenhuis M., Kiel J. A. The Hansenula polymorpha PDD7 gene is essential for macropexophagy and microautophagy. FEMS Yeast Res. 2003;3:27–34. doi: 10.1016/s1567-1356(02)00135-6. [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leber R., Silles E., Sandoval I. V., Mazon M. J. Yol082p, a novel CVT protein involved in the selective targeting of aminopeptidase I to the yeast vacuole. J. Biol. Chem. 2001;276:29210–29217. doi: 10.1074/jbc.M101438200. [DOI] [PubMed] [Google Scholar]
- Liu H., Tan X., Russell K. A., Veenhuis M., Cregg J. M. PER3, a gene required for peroxisome biogenesis in Pichia pastoris, encodes a peroxisomal membrane protein involved in protein import. J. Biol. Chem. 1995;270:10940–10951. doi: 10.1074/jbc.270.18.10940. [DOI] [PubMed] [Google Scholar]
- Liu H., Tan X., Veenhuis M., McCollum D., Cregg J. M. An efficient screen for peroxisome-deficient mutants of Pichia pastoris. J. Bacteriol. 1992;174:4943–4951. doi: 10.1128/jb.174.15.4943-4951.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luers G. H., Advani R., Wenzel T., Subramani S. The Pichia pastoris dihydroxyacetone kinase is a PTS1-containing, but cytosolic, protein that is essential for growth on methanol. Yeast. 1998;14:759–771. doi: 10.1002/(SICI)1097-0061(19980615)14:8<759::AID-YEA275>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Luers G. H., Thiele S., Schad A., Volkl A., Yokota S., Seitz J. Peroxisomes are present in murine spermatogonia and disappear during the course of spermatogenesis. Histochem. Cell Biol. 2006;125:693–703. doi: 10.1007/s00418-005-0114-9. [DOI] [PubMed] [Google Scholar]
- Luiken J. J., van den Berg M., Heikoop J. C., Meijer A. J. Autophagic degradation of peroxisomes in isolated rat hepatocytes. FEBS. Lett. 1992;304:93–97. doi: 10.1016/0014-5793(92)80596-9. [DOI] [PubMed] [Google Scholar]
- Matsuura A., Tsukada M., Wada Y., Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Meiling-Wesse K., Epple U. D., Krick R., Barth H., Appelles A., Voss C., Eskelinen E. L., Thumm M. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- Monastryska I., Sjollema K., van der Klei I. J., Kiel J. A., Veenhuis M. Microautophagy and macropexophagy may occur simultaneously in Hansenula polymorpha. FEBS Lett. 2004;568:135–138. doi: 10.1016/j.febslet.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Monastyrska I., Kiel J.A.K.W., Krikken A. M., Komduur J. A., Veenhuis M., van der Klei I. J. The Hansenula polymorpha ATG25 gene encodes a novel coiled-coil protein that is required for macropexophagy. Autophagy. 2005;1:92–100. doi: 10.4161/auto.1.2.1832. [DOI] [PubMed] [Google Scholar]
- Mukaiyama H., Baba M., Osumi M., Aoyagi S., Kato N., Ohsumi Y., Sakai Y. Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell. 2004;15:58–70. doi: 10.1091/mbc.E03-05-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaiyama H., Oku M., Baba M., Samizo T., Hammond A. T., Glick B. S., Kato N., Sakai Y. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells. 2002;7:75–90. doi: 10.1046/j.1356-9597.2001.00499.x. [DOI] [PubMed] [Google Scholar]
- Nazarko T. Y., Huang J., Nicaud J. M., Klionsky D. J., Sibirny A. A. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy. 2005a;1:37–45. doi: 10.4161/auto.1.1.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko T. Y., Nicaud J. M., Sibirny A. A. Observation of the Yarrowia lipolytica peroxisome-vacuole dynamics by fluorescence microscopy with a single filter set. Cell Biol. Int. 2005b;29:65–70. doi: 10.1016/j.cellbi.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Noda T., Suzuki K., Ohsumi Y. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–235. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- Oku M., Warnecke D., Noda T., Muller F., Heinz E., Mukaiyama H., Kato N., Sakai Y. Peroxisome degradation requires catalytically active sterol glucosyltransferase with a GRAM domain. EMBO J. 2003;22:3231–3241. doi: 10.1093/emboj/cdg331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Goldfarb D. S. YEB3/VAC8 encodes a myristylated armadillo protein of the Saccharomyces cerevisiae vacuolar membrane that functions in vacuole fusion and inheritance. J. Cell Sci. 1998;111:2137–2147. doi: 10.1242/jcs.111.15.2137. [DOI] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., Oapos;Toole E., Winey M., Goldfarb D. S. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y., Koller A., Rangell L. K., Keller G. A., Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J. Cell Biol. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki T., Zahringer U., Warnecke D. C., Fahl A., Knogge W., Heinz E. Sterol glycosides and cerebrosides accumulate in Pichia pastoris, Rhynchosporium secalis and other fungi under normal conditions or under heat shock and ethanol stress. Yeast. 2001;18:679–695. doi: 10.1002/yea.720. [DOI] [PubMed] [Google Scholar]
- Scott S. V., Guan J., Hutchins M. U., Kim J., Klionsky D. J. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S. V., Hefner-Gravink A., Morano K. A., Noda T., Ohsumi Y., Klionsky D. J. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S. V., Nice D. C., 3rd, Nau J. J., Weisman L. S., Kamada Y., Keizer-Gunnink I., Funakoshi T., Veenhuis M., Ohsumi Y., Klionsky D. J. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- Stasyk O. V., Nazarko T. Y., Stasyk O. G., Krasovska O. S., Warnecke D., Nicaud J. M., Cregg J. M., Sibirny A. A. Sterol glucosyltransferases have different functional roles in Pichia pastoris and Yarrowia lipolytica. Cell Biol. Int. 2003;27:947–952. doi: 10.1016/j.cellbi.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Stasyk O. V., Stasyk O. G., Mathewson R. D., Farré J. C., Nazarko V. Y., Krasovska O. S., Subramani S., Cregg J. M., Sibirny A. A. Atg28, a novel coiled-coil protein involved in autophagic degradation of peroxisomes in the methylotrophic yeast Pichia pastoris. Autophagy. 2006;2:30–38. doi: 10.4161/auto.2226. [DOI] [PubMed] [Google Scholar]
- Straub M., Bredschneider M., Thumm M. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J. Bacteriol. 1997;179:3875–3883. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug P. E., Bevan A., Dunn W. A., Jr GSA11 encodes a unique 208-kDa protein required for pexophagy and autophagy in Pichia pastoris. J. Biol. Chem. 2001;276:42422–42435. doi: 10.1074/jbc.M104087200. [DOI] [PubMed] [Google Scholar]
- Tuttle D. L., Dunn W. A., Jr Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J. Cell Sci. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- Tuttle D. L., Lewin A. S., Dunn W. A., Jr Selective autophagy of peroxisomes in methylotrophic yeasts. Eur. J. Cell Biol. 1993;60:283–290. [PubMed] [Google Scholar]
- Veit M., Laage R., Dietrich L., Wang L., Ungermann C. Vac8p release from the SNARE complex and its palmitoylation are coupled and essential for vacuole fusion. EMBO J. 2001;20:3145–3155. doi: 10.1093/emboj/20.12.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. X., Catlett N. L., Weisman L. S. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J. Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. X., Kauffman E. J., Duex J. E., Weisman L. S. Fusion of docked membranes requires the armadillo repeat protein Vac8p. J. Biol. Chem. 2001;276:35133–35140. doi: 10.1074/jbc.M103937200. [DOI] [PubMed] [Google Scholar]
- Warnecke D., Erdmann R., Fahl A., Hube B., Muller F., Zank T., Zahringer U., Heinz E. Cloning and functional expression of UGT genes encoding sterol glucosyltransferases from Saccharomyces cerevisiae, Candida albicans, Pichia pastoris, and Dictyostelium discoideum. J. Biol. Chem. 1999;274:13048–13059. doi: 10.1074/jbc.274.19.13048. [DOI] [PubMed] [Google Scholar]
- Waterham H. R., de Vries Y., Russel K. A., Xie W., Veenhuis M., Cregg J. M. The Pichia pastoris PER6 gene product is a peroxisomal integral membrane protein essential for peroxisome biogenesis and has sequence similarity to the Zellweger syndrome protein PAF-1. Mol. Cell. Biol. 1996;16:2527–2536. doi: 10.1128/mcb.16.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer E. A., Luers G. H., Faber K. N., Wenzel T., Veenhuis M., Subramani S. Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J. Biol. Chem. 1996;271:18973–18980. doi: 10.1074/jbc.271.31.18973. [DOI] [PubMed] [Google Scholar]
- Yamashita S., Oku M., Wasada Y., Ano Y., Sakai Y. PI4P-signaling pathway for the synthesis of a nascent membrane structure in selective autophagy. J. Cell Biol. 2006;173:709–717. doi: 10.1083/jcb.200512142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Rayapuram N., Subramani S. The control of peroxisome number and size during division and proliferation. Curr. Opin. Cell Biol. 2005;17:376–383. doi: 10.1016/j.ceb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Zwart K., Veenhuis M., van Dijken J. P., Harder W. Development of amine oxidase-containing peroxisomes in yeasts during growth on glucose in the presence of methylamine as the sole source of nitrogen. Arch. Microbiol. 1980;126:117–126. doi: 10.1007/BF00511216. [DOI] [PubMed] [Google Scholar]