Selection and cloning of poly(rC)-binding protein 2 and Raf kinase inhibitor protein RNA activators of 2′,5′-oligoadenylate synthetase from prostate cancer cells (original) (raw)
Abstract
The antiviral and antitumor functions of RNase L are enabled by binding to the allosteric effectors 5′-phosphorylated, 2′,5′-linked oligoadenylates (2-5A). 2-5A is produced by interferon-inducible 2′,5′-oligoadenylate synthetases (OAS) upon activation by viral double-stranded RNA (dsRNA). Because mutations in RNase L have been implicated as risk factors for prostate cancer, we sought to determine if OAS activators are present in prostate cancer cells. We show that prostate cancer cell lines (PC3, LNCaP and DU145), but not normal prostate epithelial cells (PrEC), contain RNA fractions capable of binding to and activating OAS. To identify the RNA activators, we developed a cDNA cloning strategy based on stringent affinity of RNAs for OAS. We thus identified mRNAs for Raf kinase inhibitor protein (RKIP) and poly(rC)-binding protein 2 (PCBP2) that bind and potently activate OAS. In addition, human endogenous retrovirus (hERV) envelope RNAs were present in PC3 cells that bind and activate OAS. Analysis of several gene expression profiling studies indicated that PCBP2 RNA was consistently elevated in metastatic prostate cancer. Results suggest that OAS activation may occur in prostate cancer cells in vivo stimulated by cellular mRNAs for RKIP and PCBP2.
INTRODUCTION
Interferons (IFNs) are a group of cytokines that confer antiviral and antiproliferative effects by arming the cell with numerous defense mechanisms, including the 2′,5′-oligoadenylate synthetase (OAS)/RNase L system (1). Unusual 5′-phosphorylated, 2′,5′-oligoadenylates (2-5A) are produced from ATP by IFN-inducible OAS proteins in response to stimulation by dsRNA activators (2,3). To date, all naturally occurring RNA activators of RNase L that have been identified are dsRNA structures of various viral origins (4–8). RNA activation of OAS precedes and is required for the activation of RNase L. Latent RNase L, present in most mammalian cells, is a monomer lacking ribonuclease activity (9,10). However, upon binding 2-5A, RNase L dimerizes into a potent endoribonuclease that degrades single-stranded regions in viral and cellular RNAs after UpNp dinucleotide sequences (primarily UU and UA), and sequences within the L1 bulge of intact cellular 28S rRNA (10–13). The OAS/RNase L pathway is required for a complete innate antiviral response. Accordingly, mice lacking RNase L have a reduced ability to survive infections by any one of several different types of viruses, including Coxsackievirus B4, encephalomyocarditis virus and West Nile virus (14–16). Although transient activation of RNase L is compatible with cell survival, sustained activation is not (17–20). RNase L activity can result in apoptosis, an outcome that requires the MAP kinase, JNK, and is characterized by cytochrome c release from mitochondria and caspase activation (17,19). Prostate cancer cells deficient in RNase L are highly resistant to apoptosis by the RNase L activator, 2-5A, alone or in combination treatments with a topoisomerase I inhibitor and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) (18). While apoptosis eliminates virus-infected cells, a result consistent with the antiviral activity of RNase L, it can also function as a tumor suppressor mechanism. Interestingly, in several, but not all studies, mutations and variants of RNASEL were correlated with enhanced risk of prostate cancer (21–27). For instance, the common variant of RNase L, R462Q, was implicated in up to 13% of unselected prostate cancer cases (22). Recently, we identified a novel gammaretrovirus, XMRV, almost exclusively in prostate tumors homozygous for a reduced activity variant (R462Q) of RNase L (28). Those findings suggested that XMRV is susceptible to the OAS/RNase L pathway, thus providing evidence for an antiviral role of RNase L in humans.
IFN treatment of human cells results in transcription of a cluster of four OAS genes (OAS1, OAS2, OAS3 and OASL) located on chromosome 12q24.2, which encodes 8–10 latent isoforms of OAS, as a result of alternative splicing (29). Once activated, OAS proteins (excluding OASL), catalyze 2′-specific adenosine nucleotidyl transfer reactions with ATP as a substrate. However, the reason for multiple isoforms of OAS with essentially similar function remains unknown. OAS1 and OAS2 encoded proteins produce predominantly 2-5A trimer and longer oligomers, all of which activate RNase L (30). In contrast, OAS3 encoded proteins produce mostly 2-5A dimer, ppp5′A2′p5′A, which does not activate RNase L (31). Although dsRNAs, such as poly(I):poly(C), are potent activators of OAS, both dsRNA and ssRNA can bind to, and in some instances activate, OAS (32). Activation of OAS was proposed as a two-step process based upon the observation that the binding affinities of different in vitro selected ssRNA aptamers do not correlate with OAS activation potential and the elucidation of the crystal structure of porcine OAS1 (32,33).
Although viral dsRNAs potently activate OAS, IFN treatment by itself inhibits the growth of certain cancer cells, some of which may not be virus infected or may not be dependent on viral infections for cell growth (34). Unidentified nuclear RNA activators of OAS were detected in IFN-sensitive cancer cells but not normal cells, suggesting that these RNAs may play a role in cellular growth arrest (35). The recent identification of RNASEL as a prostate cancer susceptibility gene led us to examine the possible presence of RNA activators of OAS in prostate cancer cells (21). To identify such RNAs, we developed a cDNA cloning strategy based on high affinity of RNA for OAS. These efforts identified two RNA activators of OAS in prostate cancer cells encoding Raf kinase inhibitor protein (RKIP) (36) and poly(rC)-binding protein 2 (PCBP2) (37).
MATERIALS AND METHODS
Cell culture
Cell lines: LNCaP-FGC (ATCC Catalog no. CRL-1740), DU145 (ATCC Catalog no. HTB-81) and PC3 (ATCC Catalog no. CRL-1435) were grown in RPMI 1640 medium with 2 mM l-glutamine, fetal bovine serum 10%; 100 U/ml penicillin G and 100 μg/ml streptomycin. Normal prostate epithelial cells (PrEC) were obtained from Clonetics Corporation (San Diego, CA) and were maintained in prostate epithelial cell medium (PrEGM) supplemented with a mixture of various growth factors (SingleQuots) (Clonetics); and 10% fetal bovine serum.
Human recombinant OAS1 and RNase L: Escherichia coli (BL21-DE3 Novagen, EMD Biosciences, Inc., USA) was transformed with recombinant human His6-tagged OAS1 p42 cDNA in a modified pET9d plasmid (a gift from Rune Hartmann, University of Aarhus, Denmark) and grown overnight in Luria–Burtani (LB) broth containing 30 μg/ml kanamycin. An aliquot of 5 ml of the overnight culture was inoculated in 1 liter LB containing antibiotic and allowed to grow at 37°C until the OD600 reached 0.6. The culture was cooled on ice and human OAS was induced with 0.5 mM IPTG at 25°C for 12 h with constant shaking at 220 r.p.m.. The cells were harvested by centrifugation at 10 000 g for 10 min and washed with chilled phosphate-buffered saline (PBS). The cells were re-suspended in buffer A: [50 mM sodium phosphate buffer (pH 8.0), 300 mM sodium chloride, 5 mM magnesium chloride, 20 mM imidazole, 10% glycerol and 5 mM 2-mercaptoethanol] plus 0.1% NP-40 and EDTA-free complete protease inhibitor cocktail (Roche Diagnostics Corporation, Indianapolis, IN, USA) and lysed using a French press. Cell debris was removed by centrifugation at 20 000 g for 20 min and the lysate was loaded on Nickel(II) nitrilotriacetic acid (Ni2+-NTA) agarose HiTrap™ Chelating HP column (Amersham Biosciences) washed with buffer A and eluted with the same buffer containing 250 mM imidazole (buffer B). The eluate was dialyzed at least four times against buffer C: [10 mM HEPES (pH 7.4), 150 mM sodium chloride, 5 mM magnesium chloride, 5 mM 2-mercaptoethanol and 0.005% surfactant P20] and further purified with size exclusion chromatography with a Superdex HR16/60 column in buffer C. Human recombinant RNase L was expressed in Sf9 insect cells (Invitrogen) from a baculovirus vector and purified to homogeneity as described previously (38).
RNA isolation
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and DNA was digested with RNase-free DNase I (Ambion, Austin, TX, USA). Poly(A)+ RNA was isolated with the Oligotex mRNA Midi Kit (Qiagen). RNA concentrations were measured with the RIBOgreen quantification kit (Molecular Probes, Invitrogen).
2-5A synthesis assays
2-5A synthesis reaction mixtures, prepared in DEPC-treated H2O, contained OAS buffer [20 mM HEPES (pH 7.6), 20 mM magnesium acetate, 20 mM potassium chloride and 1 mM EDTA], 10 mM ATP (Sigma-Aldrich Inc., USA) (pH 7.4), 2 μg/ml RNA and 20 μg/ml OAS. Samples were prepared on ice, mixed, centrifuged briefly and incubated at 37°C for the indicated time. Control reactions were prepared exactly as described above, except either without ATP, or without both ATP and OAS. 2-5A synthesis reactions were passed through a Centricon-10 filter (Millipore Corp.). 2-5A in each sample was measured with an RNase L-dependent FRET assay as described previously (38). To generate a standard curve, authentic trimeric 2-5A, diluted to final concentrations of 0.1, 0.3, 1, 3, 10, 30, 100 and 300 nM in DEPC-treated water was used. Fluorescence was measured at 5, 30, 60, 90 and 120 min with a Wallac 1420 fluorimeter (Perkin-Elmer LAS Inc., USA) (absorption 485 nm/emission 535 nm).
In vitro selection of the OAS-binding RNAs
All solutions were made with DEPC-treated H2O. The buffer in the OAS preparations was exchanged with OAS buffer without EDTA with a Centricon-10 filter. Ni2+-NTA agarose (Novagen, EMD Bioscience), equilibrated in OAS buffer without EDTA, was incubated with OAS for 30 min. The OAS–resin complex was washed with 500 μl of OAS buffer minus EDTA. OAS was repeatedly (5–7 times) added to the complex until the Ni2+-NTA agarose was saturated with OAS. PC3 Poly(A)+ RNA, at a molar ratio of 25:1 to bound OAS, was used in the in vitro selection assays. RNA was added to the OAS–resin complex and incubated for 15 min and mixed manually every 2 min. The sample was washed twice with 500 μl of OAS buffer minus EDTA. The RNA–OAS–resin complex was washed with 100 μl of the same buffer supplemented with 250 mM potassium chloride followed by two washes with 500 μl of OAS buffer minus EDTA. The RNA–OAS complex was removed from the resin by adding 250 μl strip buffer [20 mM Tris–HCl (pH 8.0), 100 mM sodium chloride and 100 mM EDTA] in DEPC-treated H2O. Imidazole was not used due to possible site-specific cleavage of RNAs (39). The supernatant was added to 250 μl of protein digestion buffer [50 mM Tris–HCl (pH 7.5), 5 mM calcium chloride and 0.5% SDS] and treated with 100 μg/ml Proteinase K (Promega Corp., Madison, WI, USA) at 37°C for 90 min. The samples were extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1), pH 6.6 (Ambion) and the selected RNAs were precipitated with 300 mM sodium acetate, pH 5.2, and 2.5 volumes of 100% ethanol with 10 μg glycogen as a carrier. The RNA was washed with 75% ethanol and air-dried. The pellet was dissolved in DEPC-treated H2O, purified and concentrated with the RNeasy® MinElute Cleanup Kit (Qiagen). OAS was monitored by loading aliquots of washes, beads or supernatant onto a 10% SDS–PAGE, and subsequent silver-staining (Bio-Rad Inc., Hercules, CA, USA). Control mock binding experiments were run in parallel without the addition of OAS to determine if RNA species capable of activating OAS were non-specifically binding to the Ni2+-NTA agarose. RNA isolated from the in vitro selection experiments with and without OAS were quantitated with the RIBOgreen quantification kit and analyzed for their ability to activate OAS with the 2-5A synthesis reactions. 2-5A levels in samples were quantified in FRET assays.
cDNAs preparation and cloning
Selected RNAs were annealed with either random hexamer or oligo(dT) primers, and cDNAs were obtained by reverse transcription. First-strand synthesis was performed for 1 h at 55°C with Superscript™ Choice System for cDNA Synthesis (Invitrogen). Second-strand synthesis was performed according to the manufacturer's instructions except without the addition of linkers. Three control reactions were carried out in parallel with each RT–PCR. The first control was without the addition of selected RNA; the second control was without reverse transcriptase; and the third control was without the addition of primers. The PCR products were cloned into pBluescript KS+. The recombinant clones were sequenced and identified using the BLAST site at www.ncbi.nih.gov. Primers specific for hERV env RNAs, as described previously (40), were used in PCR (Promega PCR Master Mix) with 2 μl of the cDNA produced from PC3 total or selected RNA (described above). Control reactions included PCR with cDNAs generated without RNA, PCR on cDNA with RNA recovered from mock in vitro selection assay, and PCR for GAPDH. Primers for PCR were as follows. GAPDH exon 8; forward d(CCAAGGCTGTGGGCAAGG), reverse d(TCCGACGCCTGCTTCACC), β-actin; forward d(GCCAGCTCACCATGGATGAT), reverse d(TTTAAGGTGTGCACTTTTATTC), PCBP2; forward d(CACCGGTGTGATTGAAGG), reverse d(TGCCAACTTCCTTTCCATG), RKIP; forward d(CACAATGTGATTTTATGGT), reverse d(TCTTCATTCAGGTTTCTAT). PCRs were as follows: 95°C, 4 min, [95°C, 30 s (annealing temperature 45 s), 72°C, 1 min] for 30 cycles, 72°C, 10 min. Annealing temperature for GAPDH was at 53°C; ERV3 env was 63°C; hERV 4-1 SU env was 55°C; hERV 4-1 TM env was 58°C; β-actin was 50°C; PCBP2 was 52°C and RKIP was 46°C. The selected cDNAs were cloned into pGEM®-T Easy (Promega Corp.).
Northern blots
Twenty micrograms of PC3, LNCaP, DU145 and PrEC RNA were separated on 1.2% agarose formaldehyde gels and transferred to Hybond-N+ (Amersham Pharmacia Biotech) membranes. The transferred membranes were hybridized with [α-32P]dCTP-labeled probes. cDNA probes specific for PCBP2, RKIP and GAPDH were generated using Prime-It® RmT Random Primer Labeling Kit (Stratagene). Hybridized membranes were exposed to film (Kodak BioMax XAR Film) for 12–48 h at −80°C. Blots were stripped in four washes of boiling 0.1× SSC [0.3 M sodium chloride and 0.03 M sodium citrate (pH 7.0)], 0.1% SDS for 5 min.
Preparation of RNAs
RNA was prepared from linearized plasmids by runoff T7 RNA polymerase transcription reactions as described previously (41) then resolved on a formaldehyde–agarose gel and eluted using a crush-and-soak method at 4°C. The RNA was extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1), pH 6.6 (Ambion), precipitated with 300 mM sodium acetate (pH 5.2) and 2.5 volume of 100% ethanol and dissolved in DEPC-treated H2O. Aliquots of the purified RNAs were run on formaldehyde gels to confirm RNA integrity. The RNAs used in subsequent assays were first denatured at 70°C and cooled at room temperature for 15 min to allow renaturation. The purified selected RNAs were used in 2-5A synthesis assays and surface plasmon resonance (SPR) assays. Poly(I):poly(C) was obtained from Amersham Bioscience.
Surface plasmon resonance
Kinetic characterization of OAS:RNA binding was performed with SPR on a Biocore 3000 (Biacore Inc., USA). The response units, RU, indicating the extent of binding, were monitored with time. For this purpose, NTA chips (Biacore Inc.) were saturated with Ni2+ solution to achieve a baseline shift of ≥40 response units (RU). The purified His6-tagged OAS1 p42 (200 ng/ml) in 10 mM HEPES (pH 7.4) containing 5 mM magnesium chloride, 150 mM sodium chloride, 10 μM EDTA and 0.005% surfactant P20 (SPR buffer; Biacore Inc.) was immobilized on Ni2+-NTA Biacore sensorchip at a flow rate of 10 μl/min for 1 min at 25°C to achieve a resonance response of 250–300 RU. An additional wash for 5 min at a flow rate of 20 μl/min was performed with buffer alone. The in vitro transcribed, gel-purified RNAs were dissolved in DEPC-treated water and freeze-dried. The freeze-dried RNAs were dissolved in SPR buffer to make a stock of 10 μM RNA. Different concentrations of RNAs (0.1, 0.5, 1, 5, 10, 25, 50 and 100 nM) were prepared and passed over the OAS immobilized sensorchip at a flow rate of 10 μl/min for 1 min while association was monitored. The dissociation was monitored with buffer alone for additional 5 min. Data normalization against buffer alone, analysis and fitting was performed with BIAevaluation™ software, version 3.2 (Biacore Inc.) with the option for simultaneous _k_a/_k_d calculation. Fitting of sensorgram data was carried out according to global fitting, and the _k_a and _k_d values were calculated with a 1:1 Langmuir model. In addition, these data were also used for independent _K_D calculations by Scatchard plot analysis.
High-performance liquid chromatography (HPLC)
2-5A synthesis reaction mixtures were passed through a Centricon-10 filter (Millipore Corp.) to remove the protein and RNA before HPLC. Fifty microliter of a 1:10 dilution of clarified 2-5A synthesis reaction mixtures in 20 mM Tris–HCl (pH 7.5) were loaded onto a Dionex P100 analytical HPLC column, interfaced with Beckman System Gold HPLC system connected to a 32 Karat workstation, at a flow rate of 1 ml/min and eluted with a linear gradient of 0–750 mM NaCl over a period of 1 h. The chromatograms were analyzed with 32 Karat software (Beckman-Coulter).
In silico validation of RKIP and PCBP2 expression
We interrogated RKIP and PCBP2 expression in the ONCOMINE public microarray database (42), which contains transcription profiling of 35 different tumors and 14 177 microarrays profiling for these different tumors. We selected prostate cancer profiling studies from this database for expression analysis of RKIP and PCBP2.
RESULTS
RNA activators of OAS in prostate cancer cells
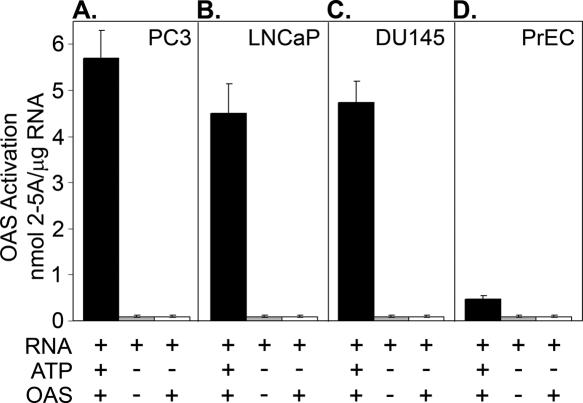
To monitor and compare RNA activators of OAS in prostate cancer and normal cells, we isolated RNA from three established human prostate cancer cell lines (PC3, LNCaP and DU145) and normal prostatic epithelial cells (PrEC). RNA was incubated with purified histidine-tagged human OAS1 (subsequently ‘OAS’) while conversion of ATP to 2-5A was determined by measuring activation of RNase L in FRET assays (Materials and Methods). 2-5A levels were calculated from relative fluorescence units, corresponding to RNA cleavage, when compared to a standard curve generated with known amounts of authentic 2-5A trimer. RNA quality and integrity was established by separation on RNA chips with an Agilent bioanalyzer (data not shown). Remarkably, amounts of 2-5A were 9- to 12-fold higher in OAS reactions containing RNA isolated from the three prostate cancer cell lines (PC3, LNCaP, DU145) in comparison to reactions with RNA isolated from normal prostate epithelial RNA (PrEC) (Figure 1). 2-5A synthesis was dependent on the presence of OAS, RNA and ATP. The highest level of OAS activation (2 h incubation at 37°C) was obtained with PC3 RNA (5.8 nmol 2-5A/μg total RNA), whereas the RNA isolated from LNCaP and DU145 cells produced 4.4 and 4.7 nmol 2-5A/μg total RNA, respectively. In contrast, RNA from the PrEC cells produced only ∼0.5 nmol 2-5A/μg RNA. Because PC3 RNA reproducibly showed the highest OAS activating potential, we used RNA from these cells to clone RNA activators of OAS.
Figure 1.
Activation of OAS by total RNA isolated from prostate cancer cell lines. Total RNA preparations from prostate cancer cell lines (A, B and C: PC3, LNCaP, DU145, respectively) or normal prostate epithelial cells (D, PrEC) were incubated with OAS and ATP at 37°C for 2 h and 2-5A was measured by FRET assay as described (Materials and Methods). Control reactions were without ATP and/or OAS. Linear regression analysis was used to fit the standard curve data points within 95% confidence limits.
RNA activators of OAS are present in the poly(A)+ fraction of PC3 cell RNA
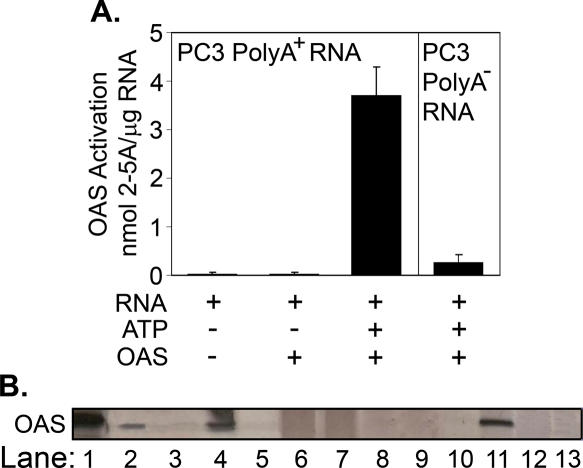
To determine whether the PC3 RNA responsible for OAS activation was mRNA, poly(A)+ and poly(A)− RNA fractions were compared (Figure 2). The poly(A)+ RNA fraction clearly contained the OAS activator(s) since it had a 15-fold higher specific activity in the OAS assay than the poly(A)− RNA (Figure 2A). Curiously, however, the specific activity of the poly(A)+ RNA was slightly lower than that of the total RNA, despite being enriched for the OAS-activating species. These results suggest that there may be RNA species inhibitory to OAS in the poly(A)+ RNA fraction.
Figure 2.
In vitro selection of poly(A)+ RNA species that bind OAS. (A) Activation of OAS with poly(A)+ and poly(A)− fractions of PC3 RNA. OAS reactions were at 37°C for 2 h. 2-5A levels were determined by FRET assay. (B) Binding and elution of OAS from Ni2+-NTA agarose as determined by SDS–PAGE and silver-staining of gel. Lane 1, purified OAS; lane 2, input OAS; lane 3, wash of OAS:Ni2+-NTA beads; lane 4, aliquot of the OAS:Ni2+-NTA beads; lane 5, effluent after loading PC3 poly(A)+ RNA; lanes 6 and 7, consecutive washes of RNA:OAS:Ni2+-NTA beads with buffer; lane 8, KCl wash of RNA:OAS:Ni2+-NTA beads; lanes 9 and 10, additional washes of RNA:OAS:Ni2+-NTA beads; lane 11, elution of RNA:OAS with EDTA; lane 12, post-elution wash of beads; and lane 13, proteinase digestion of eluted OAS to prepare bound RNA.
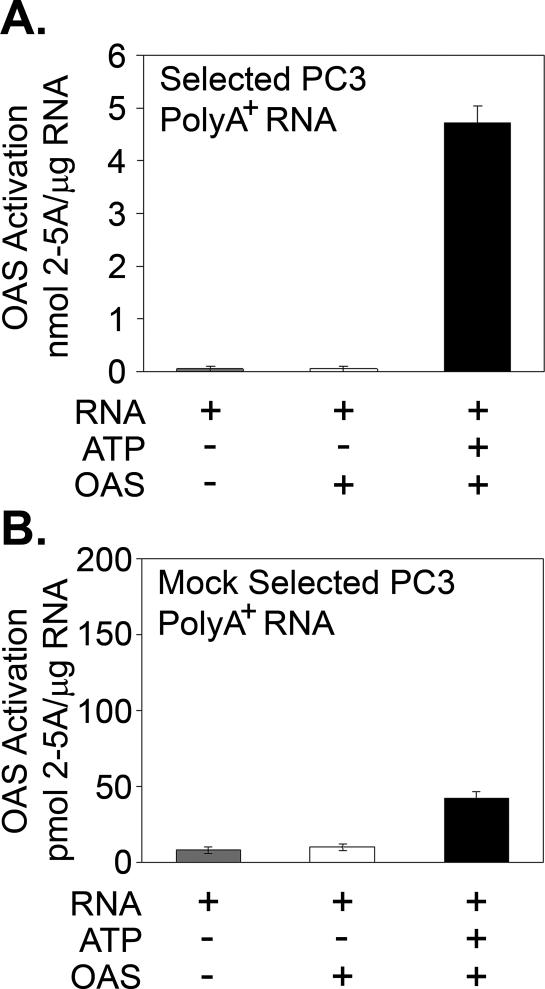
To isolate RNA species in the PC3 poly(A)+ RNA fraction that have high affinity for OAS, we developed an in vitro selection method (Materials and Methods). Binding and elution of OAS to and from the Ni2+-NTA beads during the RNA binding and washing procedures was monitored by gel electrophoresis and silver staining of the OAS (Figure 2B). These methods included a KCl wash to remove any RNA species that bind OAS with low affinities. The bound RNA, recovered after proteinase K digestion of the OAS, produced 4.8 nmol 2-5A/μg RNA when incubated with freshly added OAS, while <50 pmol 2-5A/μg RNA was produced by OAS from RNA recovered with Ni2+-NTA agarose alone (Figure 3).
Figure 3.
OAS activation by poly(A)+ RNA fraction of PC3 cells. (A) Activation of OAS with the OAS-selected PC3 poly(A)+ RNA. (B) Lack of OAS activation with non-specifically selected RNA isolated from PC3 cells on Ni2+-NTA beads without the addition of His6-tagged OAS. 2-5A levels were determined by FRET assays. The same concentration of RNA (2 μg/ml) was used in (A) and (B). OAS reactions were at 37°C for 2 h.
Cloning and identification of PC3 mRNAs that activate OAS
To clone and identify the RNA species that bind OAS, we performed reverse transcription using the PC3 poly(A)+ RNA fraction isolated by in vitro selection. Typically, RNA(s) that bind to and activate OAS are double-stranded or have a defined secondary structure. Therefore, reverse transcription at lower temperatures may not allow efficient cDNA synthesis. Accordingly, the temperature of the first-strand synthesis reaction was performed at 55°C. To maximize the amount of clones produced, separate reverse transcription reactions were performed with either random hexamer or oligo(dT) primers. Using this strategy, only two clones were obtained from ∼16 rounds of selection and cloning. The two clones were sequenced and identified as partial cDNAs for RKIP and PCBP2. The RKIP cDNA, 368 nt in length, was from the 3′-UTR [nt 1138–1506; GenBank accession no. NM_002567] whereas the PCBP2 cDNA, 72 nt in length, was from the 5′ region of the ORF (356–428 nt, translation start site at 351 nt; GenBank accession no. AB188306). The RKIP clone was produced using oligo(dT) primed reverse transcription, while the PCBP2 encoding clone was produced using random hexamer primers.
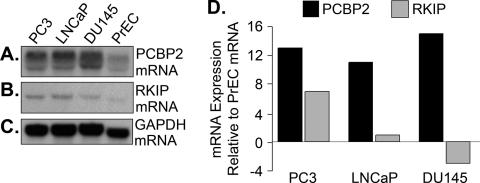
The level of expression of the selected RNAs in the prostate cancer cell lines and PrEC was determined in northern blots. Levels of PCBP2 mRNA were highly elevated (>10-fold) in the prostate cancer cells (PC3, LNCaP and DU145) compared with the normal (PrEC) cells after normalizing for levels of GAPDH mRNA (Figure 4). The PCBP2 cDNA probe identified a doublet produced from alternative splicing. However, while normalized levels of RKIP mRNA were elevated in the PC3 cells (6-fold), they were not elevated in the LNCaP and DU145 cells.
Figure 4.
Levels of RKIP and PCBP2 mRNAs in prostate cancer cell lines and normal prostate epithelial cells. (A), (B) and (C): Northern blot of PCBP2, RKIP and GAPDH mRNAs, respectively. (D) Relative levels of PCBP2 and RKIP mRNAs (each normalized to GAPDH mRNA levels) in PC3, LNCaP and DU145 cells expressed as a ratio of the same RNAs in PrEC cells (also normalized for GAPDH mRNA). Relative amounts of RNA were estimated using NIH image software (version 1.61).
hERV env RNAs bind OAS
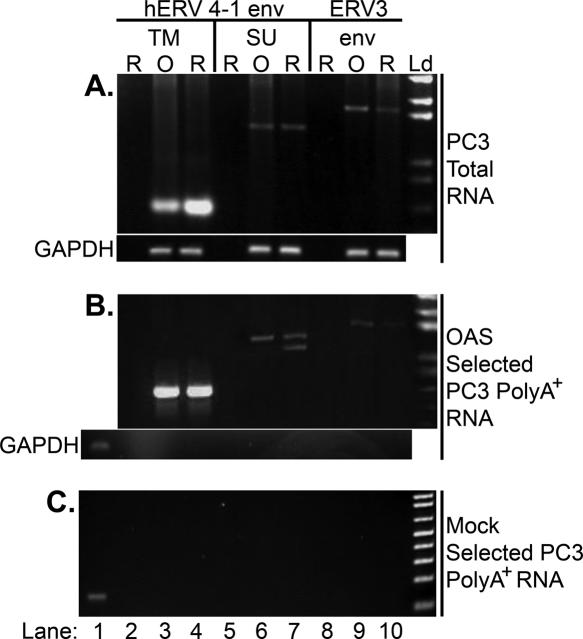
A report of the presence of hERV RNAs in prostate cancer cells, and the recognition that viral RNAs often activate OAS, led us to examine if such RNAs were capable of binding and activating OAS (40). Therefore, RT–PCR was performed for hERV env RNAs with PC3 poly(A)+ RNA fractions isolated by OAS-selection. PCR primers were specific for the env regions of hERV 4-1 transmembrane ™ (599 bp) (nt 7501–8099; GenBank accession no. M10976), hERV 4-1 surface subunit (SU) (1349 bp) (nt 6211–7559; GenBank accession no. M10976) and ERV3 (1745 bp) (nt 786–2530; GenBank accession no. M12140), previously identified in prostate cancer (40). RNA initiated PCR performed with both total RNA and OAS-selected RNA from PC3 cells showed the presence of hERV 4-1 TM and SU and ERV3 env RNAs (Figure 5A and B). No PCR products were obtained when GAPDH primers were used on cDNA prepared from the OAS-selected RNA indicating that GAPDH mRNA is a suitable non-specific RNA control (Figure 5B, lower panel). In a mock selection, performed in the absence of OAS, these hERV cDNAs were not amplified (Figure 5C). Primers specific for GAPDH mRNA was performed on PC3 cDNA as a positive control (Figure 5C, lane 1). These findings show that the different hERV env RNAs are present in PC3 cells and that these RNA bind OAS.
Figure 5.
HERV env RNAs bind to OAS. (A) hERV 4-1 TM, hERV 4-1 env SU and ERV3 env RNAs in total RNA from PC3 cells as determined by RT–PCR. Reverse transcription was with random hexamer primer (lanes labeled ‘R’) or oligo(dT) primer (lanes labeled ‘O’) and PCR was with primers specific for hERV 4-1 env TM (lanes 2–4), hERV 4-1 env SU (lanes 5–7) or ERV3 env (lanes 8–10). Lanes 2, 5 and 8 were negative controls without RNA; lower gel, RT–PCR with GAPDH-specific primers as a loading control. (B) RT–PCR was performed as described for (A) except with OAS-selected RNA from PC3 cells. Bottom gel, lane 1, PCR with GAPDH-specific primers on cDNA prepared from PC3 cells as a control; lanes 2–10, PCR with GAPDH-specific primers did not amplify cDNA prepared from the OAS-selected RNA. (C) Lack of hERV 4-1 env TM (lanes 3 and 4), hERV 4-1 env SU (lanes 6 and 7) and ERV3 env (lanes 9 and 10) in cDNA prepared from the mock selected PC3 poly(A)+ RNA with Ni2+-NTA beads lacking OAS. The amount of RNA for (A), 1 μg; for (B), ∼350 ng; and for (C), ∼100 ng as determined by the ribogreen RNA quantification kit. Equal amounts (2 μl) of cDNA was used in each RT reaction. Lane 1, PCR with GAPDH-specific primers on cDNA prepared from PC3 cells as a control. Ld, DNA ladder (1 kb Plus DNA Ladder; Invitrogen).
OAS-binding affinities of the selected RNAs
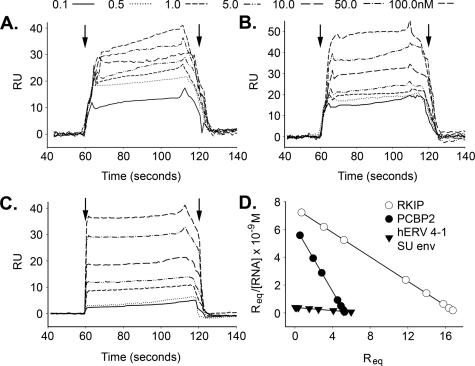
To determine affinities of the selected RNAs for OAS, SPR was performed on a Biacore 3000 instrument (Materials and Methods). Representative sensograms for RKIP, PCBP2 and hERV 4-1 SU env RNAs show concentration-dependent steady-state binding to OAS with rapid association/dissociation rates (Figure 6A–C). Global data analyses with BIAevaluation™ software (version 3.2; Biacore Inc.) were used to determine the dissociation constants (_K_D) and the association and dissociation rates (_k_a and _k_d, respectively) (Table 1). Scatchard analyses with SigmaPlot™ (version 2000) produced similar _K_D values (Figure 6D and data not shown). The quality of the fit was determined by χ2-values, as well as from the magnitude and distribution of the residuals (Table 1) (43). The _K_D values are in the subnanomolar to nanomolar range. The highest and lowest affinities (0.9 and 18 nM) were obtained for PCBP2 and hERV 4-1 SU RNAs, respectively (Table 1). Although there are considerable differences in the _k_a and _K_D for different RNAs, the off rates, _k_d, are very similar. There was no significant binding of the RNAs to the Ni2+-NTA chip alone (data not shown).
Figure 6.
Sensograms for kinetic analysis of OAS binding with RNAs. Net SPR signals as a function of time with different levels (A) RKIP, (B) PCBP2 and (C) hERV 4-1 SU env RNAs injected to Ni2+-NTA sensor chips (Biacore Inc.) containing immobilized His6-tagged OAS. Arrows indicate time of injection of RNA and subsequent wash with buffer alone to monitor dissociation of the RNA. (D) Scatchard plot for binding of RKIP, PCBP2 and hERV 4-1 SU env RNAs with OAS using SPR data. RU, response units; _R_eq, response unit at equilibrium.
Table 1.
Kinetic parameters of RNA interactions with OAS determined by BIAcore analysis in comparison to OAS activation determined by FRET assays
| RNA | _K_D (M) | _k_a (M−1 s−1) | _k_d (s−1) | χ2 | OAS activation (nmol 2-5A/μg RNA) |
|---|---|---|---|---|---|
| PCBP2 | 8.58E−10 | 1.23E+4 | 1.06E−5 | 0.064 | 20.6 ± 4.2 |
| hERV 4-1 TM env | 8.55E−10 | 1.83E+4 | 1.56E−5 | 4.89 | 6.2 ± 0.6 |
| ERV3 env | 8.03E−9 | 3.63E+3 | 2.92E−5 | 2.77 | 22.5 ± 3.1 |
| Poly(I):poly(C) | 7.76E−9 | 1.25E+3 | 9.68E−6 | 0.252 | 37.4 ± 5.3 |
| RKIP | 2.29E−9 | 4.94E+3 | 1.13E−5 | 0.67 | 15.4 ± 2.8 |
| β-Actin | 1.23E−9 | 9.09E+3 | 1.12E−5 | 0.519 | 3.3 ± 1.3 |
| hERV 4-1 SU env | 1.81E−8 | 5.52E+2 | 1E−5 | 3.7 | 16.6 ± 2.5 |
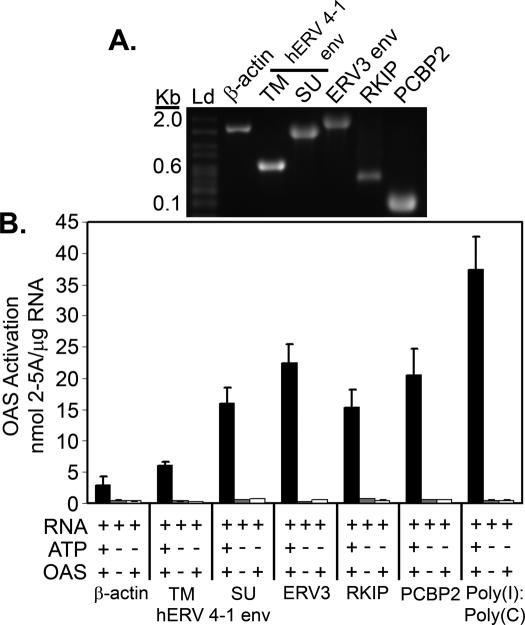
OAS-selected RNAs potently activate the catalytic function of OAS
To measure the abilities of the RNAs to activate OAS, they were produced by in vitro transcription and gel-purified to remove contaminants (Materials and Methods). To confirm RNA size and integrity, RNA species were analyzed by formaldehyde gel electrophoresis (Figure 7A). 2-5A produced in OAS reactions was measured by the ability to activate RNase L using FRET. The OAS-selected RNA species with the highest activating abilities were ERV3 env and PCBP2 RNAs, producing 22.5 and 21 nmol 2-5A/μg RNA, respectively. HERV 4-1 SU env, RKIP RNA and hERV 4-1 TM env produced ∼16, 15.5 and 6 nmol 2-5A/μg RNA, respectively. In comparison, poly(I):poly(C) and β-actin RNA produced 37.5 and 3 nmol 2-5A/μg RNA, respectively (Figure 7B and Table 1). As expected, no 2-5A was produced in control mixtures lacking OAS and/or ATP. Therefore, all of the OAS-selected RNAs, with the exception of the hERV 4-1 TM RNA, approached or exceeded half the activity of poly(I):poly(C) and were several-fold more potent than β-actin RNA.
Figure 7.
Activation of OAS by selected RNA species. (A) In vitro transcribed and gel- purified RNAs (as indicated) analyzed by formaldehyde gel electrophoresis. (B) Activation of OAS was determined with the purified in vitro transcribed RNAs (as indicated). 2-5A levels were measured by FRET. Assays were in the presence or absence of OAS and ATP (as indicated). Reactions were at 37°C for 2 h.
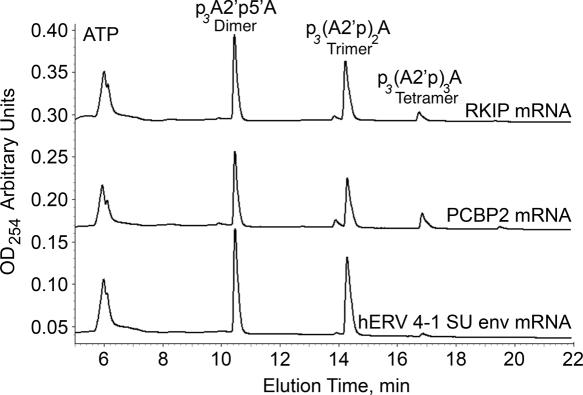
2-5A oligomer size distribution
To determine amounts of different 2-5A oligomers generated in the OAS reactions, HPLC was performed (Figure 8 and Table 2). Representative HPLC profiles of 2-5A produced with RKIP, PCBP2 and hERV 4-1 SU env RNAs as OAS activators are shown (Figure 8). Poly(I):poly(C) and β-actin RNA produced the largest and least amounts of functional 2-5A (trimer and longer species) (ppp(A2′p)_n_A, n = 2 to ≥ 4) (Table 2). The relative abilities of the different RNAs to cause trimeric or longer oligomers of 2-5A to be synthesized were in agreement with RNase L activation data (Figure 7 and Table 2).
Figure 8.
2-5A oligomer distribution as determined by HPLC of OAS reaction products using as activator, RKIP, PCBP2 and hERV 4-1 SU env RNAs as indicated. Reactions were at 37°C for 2 h. The _y_-axis, optical density at 254 nm, is translated upward for the clarity of data. Positions of 2-5A dimer p3A2′p5′A, trimer p3(A2′p)2A and tetramer p3(A2′p)3A are indicated.
Table 2.
Percentage of 2-5A oligomers produced with different RNAs
| RNA | ATP (%) | p3(A2′p)A (% dimer) | p3(A2′p)A2 (% trimer) | p3(A2′p)A ≥3 (% tetramer and longer) |
|---|---|---|---|---|
| Poly(I):poly(C) | 13.5 | 30.5 | 36.3 | 19.6 |
| PCBP2 | 36.6 | 28.2 | 27.2 | 7.7 |
| RKIP | 44.5 | 26.5 | 25.5 | 3.5 |
| hERV 4-1 SU env | 38.1 | 30.6 | 30.2 | 1 |
| hERV 4-1 TM env | 44.8 | 31.4 | 22.6 | 1.2 |
| ERV3 env | 30 | 34 | 35 | 6 |
| β-Actin | 64.8 | 32.7 | 2.4 | 0 |
Levels of RKIP and PCBP2 RNAs in human prostate and prostate tumors
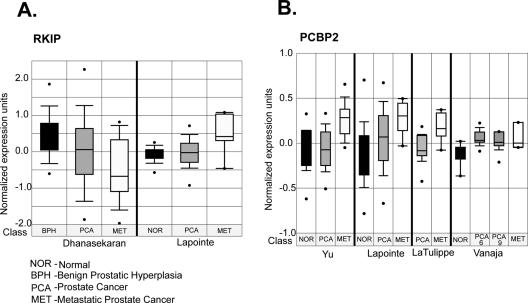
To compare levels of RKIP and PCBP2 RNA in normal prostate and prostate cancer, we interrogated the expression of these genes in prostate tumor expression profiling studies from several different research groups using the ONCOMINE expression profiling database (42). ONCOMINE is an oncogenomic database of 14 177 microarrays that profiled 35 different types of tumors from which gene expression results can be mined by a web-based platform. RKIP showed variable patterns of RNA expression in two independent studies. Although Dhanasekaran et al. (44) showed downregulation RKIP in localized and metastatic prostate cancer, Lapointe et al. (45) showed upregulation of RKIP RNA in lymph node metastasis, but not in primary tumors (Figure 9A). In contrast, data from four independent prostate cancer profiling studies indicated upregulation of PCBP2 expression in prostate cancer, particularly in metastatic tumors (Figure 9B). These data show overexpression of PCBP2 in aggressive prostate tumors in agreement with our findings from the prostate cancer cell lines (Figure 4).
Figure 9.
(A) RKIP and (B) PCBP2 expression in prostate tumors from publicly available cancer microarray studies. Box plots of expression of these genes in published prostate cancer profiling studies from independent groups (44,45,55–57). Expression array analyses of multiple cancer microarray datasets were collected and analyzed, and statistical significance was calculated. The data were collected from gene expression array database ONCOMINE (www.oncomine.org) (42). Dots represent the minimum and maximum values. Class represents the type of tissue in which the expression was measured. Numbers in the study of Vanaja et al. are the Gleason scores (57).
DISCUSSION
RNA activators of OAS in prostate cancer cells
Here we provide the first identification of cellular RNA species that activate OAS. Prostate cancer cells, especially from prostate cancer metastasis, are enriched in RNAs capable of activating OAS compared with normal prostate epithelial cells. OAS activation usually leads to stimulation of RNase L. Because sustained RNase L activity causes apoptosis, these findings are consistent with reports of mutations in RNase L in prostate cancer (21,22,25,26). Alternatively, defects in apoptosis pathways could render prostate cancer cells resistant to RNase L activity. Most of the RNAs studied here (except for hERV4-1 TM) potently activated OAS, with 40–60% of the activity obtained with poly(I):poly(C) (Figure 7). Our data show there is no clear correlation between RNA size, binding affinity and OAS activation (Table 1 and Figure 7). For instance, ERV3 RNA has a 10-fold lower _K_D than PCBP2 RNA, yet both activate OAS to the same extent. RKIP and β-actin RNAs have similar _K_D's, yet RKIP RNA is 5-fold more active. These findings support a prior report that suggested RNA binding per se is insufficient for OAS activation (32). Analysis of the crystal structure of porcine OAS1 led to a model suggesting that activation of OAS is a two-step process in which RNA binding induces conformation changes in the catalytic domain (33).
RKIP and PCBP2 mRNAs potently activate OAS
The two cellular mRNAs capable of binding and activating OAS encode RKIP and PCBP2. There were no obvious secondary structure or sequence motif similarities between these two RNAs (data not shown). RKIP, a member of the phosphatidylethanolamine binding protein family (PEBP-2), is an inhibitor of Raf kinase (46) [reviewed in (36)]. In addition to inhibiting the Raf-MEK-ERK cascade, RKIP also modulates G-protein and NF-κB signaling (47,48). RKIP suppresses prostate cancer metastasis by inhibiting angiogenesis and cell invasion and by promoting apoptosis (49,50). Enhancing RKIP levels in metastatic prostate cancer cells decreased invasion, whereas decreasing RKIP expression in non-metastatic prostate cancer cells increased their ability to invade (49). In contrast, RKIP does not affect growth of prostate cancer cells in vitro or the ability to form colonies in soft agar. RKIP expression is rapidly induced upon chemotherapeutic drug treatment in human prostate and breast cancer cells, sensitizing the cells to apoptosis while decreased the expression of RKIP confers resistance to chemotherapeutic agents (50). A recent study showed that RKIP expression was a prognostic marker for prostate cancer, with low expression of RKIP correlating with a high rate of recurrence based on prostate-specific antigen levels (51). These studies suggest that RKIP may sensitize prostate cancer cells to apoptosis by inhibiting the Raf/MEK/ERK and NF-κB survival signaling pathways. It is tempting to speculate an apoptotic function for RKIP mRNA, by way activating the OAS/RNase L pathway, that is completely independent of the encoded protein. However, there was no consistent difference in levels of RKIP mRNA in the three prostate cancer cell lines and normal prostate epithelial cells (Figure 4). In addition, there were variable levels of RKIP mRNA in prostate cancer in two microarray studies (44,45). Therefore, RKIP mRNA does not appear to be responsible for the higher OAS activation obtained with total RNA from prostate cancer cell lines compared to normal prostate epithelial cells (Figure 1).
PCBP2 (also known as hnRNP E2 and αCP-2) is a member of a family of host proteins with triplicate KH RNA-binding domains and poly(C)-binding activity. PCBP2 has a range of cellular functions including mRNA stabilization and control of both translational initiation and transcription (37). Interestingly, PCBP2 binds to a conserved UC-rich region containing a CCCUCCC motif in the 3′-UTR of the androgen receptor mRNA, suggesting a role for PCBP2 in modulating AR mRNA stability and/or translation (52,53). In addition, PCPB2 is an essential host factor in poliovirus replication playing roles in both translation initiation and viral RNA synthesis (54). Perhaps OAS and RNase L affect translation or stability of PCBP2 mRNA and thus indirectly regulate both virus replication and androgen receptor synthesis. The former activity is consistent with the antiviral activity of the OAS/RNase L pathway, while the latter is consistent with the proposed role of RNase L in prostate cancer pathogenesis. In this regard, PCBP2 expression was higher in all of the prostate cancer cell lines that we examined compared with normal prostate epithelial cells. Furthermore, PCBP2 RNA levels were elevated in localized and/or metastatic prostate cancer from four independent microarray studies (45,55–57) (Figure 9A). These investigations suggest that PCBP2 is a potential marker for aggressive forms of prostate cancer and that it could regulate androgen receptor synthesis.
hERV RNAs bind and activate OAS
We identified three hERV env RNAs that bind with high affinity and activate OAS. hERVs are descendants of exogenous retroviruses that integrated into genomic DNA of host germ line cells and account for ∼8–9% of the total human genome (58–60). Many functions for hERVs have been proposed, including roles in tumorigenesis, autoimmune diseases and neurological disorders (61–65). In addition, the fusogenic properties of a hERV-W envelope protein is utilized for the formation of the placental syncytiotrophoblast layer (66,67). hERV transcripts are found in various human tissues (68), including prostate cancer cells specifically, but lacking in normal prostatic tissues (40). However, the pathobiology of hERV RNAs is poorly understood.
It is likely that the OAS/RNase L pathway suppresses retroviral infections. For instance, overexpression of OAS (69) or RNase L (70) in cultured cells impairs HIV replication, and suppression of RNase L activity by expression of an RNase L inhibitor (RLI, ABCE1, HP68) enhances HIV growth in culture (71). OAS is activated by HIV-1 TAR RNA (8). In addition, we identified a novel gammaretrovirus, XMRV, almost exclusively in prostate cancer patients homozygous for a reduced activity variant of RNase L (28). The fact that some hERV RNAs are capable of activating OAS suggests that their role in prostate cancer may be more protective than destructive by initiating an antiproliferative mechanism in cancer cells. Nevertheless, further studies are clearly required to establish the roles of these RNA activators of OAS in the RNA biology of prostate cancer. Our findings show that OAS activators are enriched in prostate cancer. Furthermore, these results demonstrate that both viral and cellular RNA can possess the ability to bind and activate OAS.
Acknowledgments
These studies were supported by a grant from the National Institutes of Health, National Cancer Institute (CA103943 R.H.S.) and a Molecular Medicine Fellowship from Cleveland State University and the Cleveland Clinic Foundation (R.J.M.). We thank Rune Hartmann, University of Aarhus, Denmark, for the gift of human hexahistidine-OAS1 p42 cDNA. Funding to pay the Open Access publication charges for this article was provided by grant CA103943 from the National Institutes of Health, National Cancer Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stark G.R., Kerr I.M., Williams B.R., Silverman R.H., Schreiber R.D. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Hovanessian A.G., Brown R.E., Kerr I.M. Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature. 1977;268:537–540. doi: 10.1038/268537a0. [DOI] [PubMed] [Google Scholar]
- 3.Kerr I.M., Brown R.E. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc. Natl Acad. Sci. USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharp T.V., Raine D.A., Gewert D.R., Joshi B., Jagus R., Clemens M.J. Activation of the interferon-inducible (2′–5′) oligoadenylate synthetase by the Epstein–Barr virus RNA, EBER-1. Virology. 1999;257:303–313. doi: 10.1006/viro.1999.9689. [DOI] [PubMed] [Google Scholar]
- 5.Mordechai E., Kon N., Henderson E.E., Suhadolnik R.J. Activation of the interferon-inducible enzymes, 2′,5′-oligoadenylate synthetase and PKR by human T-cell leukemia virus type I Rex-response element. Virology. 1995;206:913–922. doi: 10.1006/viro.1995.1014. [DOI] [PubMed] [Google Scholar]
- 6.Gribaudo G., Lembo D., Cavallo G., Landolfo S., Lengyel P. Interferon action: binding of viral RNA to the 40-kilodalton 2′–5′-oligoadenylate synthetase in interferon-treated HeLa cells infected with encephalomyocarditis virus. J. Virol. 1991;65:1748–1757. doi: 10.1128/jvi.65.4.1748-1757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai S.Y., Patel R.C., Sen G.C., Malhotra P., Ghadge G.D., Thimmapaya B. Activation of interferon-inducible 2′–5′ oligoadenylate synthetase by adenoviral VAI RNA. J. Biol. Chem. 1995;270:3454–3461. doi: 10.1074/jbc.270.7.3454. [DOI] [PubMed] [Google Scholar]
- 8.Maitra R.K., McMillan N.A., Desai S., McSwiggen J., Hovanessian A.G., Sen G., Williams B.R., Silverman R.H. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology. 1994;204:823–827. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]
- 9.Zhou A., Molinaro R.J., Malathi K., Silverman R.H. Mapping of the human RNASEL promoter and expression in cancer and normal cells. J. Interferon Cytokine Res. 2005;25:595–603. doi: 10.1089/jir.2005.25.595. [DOI] [PubMed] [Google Scholar]
- 10.Dong B., Silverman R.H. 2-5A-dependent RNase molecules dimerize during activation by 2–5A. J. Biol. Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 11.Wreschner D.H., McCauley J.W., Skehel J.J., Kerr I.M. Interferon action—sequence specificity of the ppp(a2′p)na-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 12.Iordanov M.S., Paranjape J.M., Zhou A., Wong J., Williams B.R., Meurs E.F., Silverman R.H., Magun B.E. Activation of p38 mitogen-activated protein kinase and c-Jun NH2-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R, and alternative pathways. Mol. Cell. Biol. 2000;20:617–627. doi: 10.1128/mcb.20.2.617-627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Floyd-Smith G., Slattery E., Lengyel P. Interferon action: RNA cleavage pattern of a (2′–5′)oligoadenylate—dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 14.Zhou A., Paranjape J., Brown T.L., Nie H., Naik S., Dong B., Chang A., Trapp B., Fairchild R., Colmenares C., et al. Interferon action and apoptosis are defective in mice devoid of 2′–5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samuel M.A., Whitby K., Marri A., Williams B.R.G., Silverman R.H., Diamond M.S. PKR and RNASEL contribute to protection against lethal west nile virus infection by controlling early spread in the periphery and viral replication in neurons. J. Virol. 2006;80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flodstrom-Tullberg M., Hultcrantz M., Stotland A., Maday A., Tsai D., Fine C., Williams B., Silverman R., Sarvetnick N. RNase l and double-stranded RNA-dependent protein kinase exert complementary roles in islet cell defense during coxsackievirus infection. J. Immunol. 2005;174:1171–1177. doi: 10.4049/jimmunol.174.3.1171. [DOI] [PubMed] [Google Scholar]
- 17.Rusch L., Zhou A., Silverman R.H. Caspase-dependent apoptosis by 2′,5′-oligoadenylate activation of RNase l is enhanced by IFN-β. J. Interferon Cytokine Res. 2000;20:1091–1100. doi: 10.1089/107999000750053762. [DOI] [PubMed] [Google Scholar]
- 18.Malathi K., Paranjape J.M., Ganapathi R., Silverman R.H. HPC1/RNASEL mediates apoptosis of prostate cancer cells treated with 2′,5′-oligoadenylates, topoisomerase I inhibitors, and tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 2004;64:9144–9151. doi: 10.1158/0008-5472.CAN-04-2226. [DOI] [PubMed] [Google Scholar]
- 19.Li G., Xiang Y., Sabapathy K., Silverman R.H. An apoptotic signaling pathway in the interferon antiviral response mediated by RNase l and c-Jun NH2-terminal kinase. J. Biol. Chem. 2004;279:1123–1131. doi: 10.1074/jbc.M305893200. [DOI] [PubMed] [Google Scholar]
- 20.Castelli J.C., Hassel B.A., Maran A., Paranjape J., Hewitt J.A., Li X.L., Hsu Y.T., Silverman R.H., Youle R.J. The role of 2′–5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 1998;5:313–320. doi: 10.1038/sj.cdd.4400352. [DOI] [PubMed] [Google Scholar]
- 21.Carpten J., Nupponen N., Isaacs S., Sood R., Robbins C., Xu J., Faruque M., Moses T., Ewing C., Gillanders E., et al. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nature Genet. 2002;30:181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 22.Casey G., Neville P.J., Plummer S.J., Xiang Y., Krumroy L.M., Klein E.A., Catalona W.J., Nupponen N., Carpten J.D., Trent J.M., et al. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nature Genet. 2002;32:581–583. doi: 10.1038/ng1021. [DOI] [PubMed] [Google Scholar]
- 23.Downing S.R., Hennessy K.T., Abe M., Manola J., George D.J., Kantoff P.W. Mutations in ribonuclease L gene do not occur at a greater frequency in patients with familial prostate cancer compared with patients with sporadic prostate cancer. Clin. Prostate Cancer. 2003;2:177–180. doi: 10.3816/cgc.2003.n.027. [DOI] [PubMed] [Google Scholar]
- 24.Maier C., Haeusler J., Herkommer K., Vesovic Z., Hoegel J., Vogel W., Paiss T. Mutation screening and association study of RNASEL as a prostate cancer susceptibility gene. Br. J. Cancer. 2005;92:1159–1164. doi: 10.1038/sj.bjc.6602401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rennert H., Bercovich D., Hubert A., Abeliovich D., Rozovsky U., Bar-Shira A., Soloviov S., Schreiber L., Matzkin H., Rennert G., et al. A novel founder mutation in the rnasel gene, 471delaaag, is associated with prostate cancer in ashkenazi jews. Am. J. Hum. Genet. 2002;71:981–984. doi: 10.1086/342775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rennert H., Zeigler-Johnson C.M., Addya K., Finley M.J., Walker A.H., Spangler E., Leonard D.G., Wein A., Malkowicz S.B., Rebbeck T.R. Association of susceptibility alleles in ELAC2/HPC2, RNASEL/HPC1, and MSR1 with prostate cancer severity in European American and African American men. Cancer Epidemiol. Biomarkers Prev. 2005;14:949–957. doi: 10.1158/1055-9965.EPI-04-0637. [DOI] [PubMed] [Google Scholar]
- 27.Wiklund F., Jonsson B.A., Brookes A.J., Stromqvist L., Adolfsson J., Emanuelsson M., Adami H.O., Augustsson-Balter K., Gronberg H. Genetic analysis of the RNasel gene in hereditary, familial and sporadic prostate cancer. Clin. Cancer Res. 2004;10:7150–7156. doi: 10.1158/1078-0432.CCR-04-0982. [DOI] [PubMed] [Google Scholar]
- 28.Urisman A., Molinaro R.J., Fischer N., Plummer S.J., Casey G., Klein E.A., Malathi K., Magi-Galluzzi C., Tubbs R.R., Ganem D., et al. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Rebouillat D., Hovanessian A.G. The human 2′,5′-oligoadenylate synthetase family: interferon-induced proteins with unique enzymatic properties. J. Interferon Cytokine Res. 1999;19:295–308. doi: 10.1089/107999099313992. [DOI] [PubMed] [Google Scholar]
- 30.Marie I., Blanco J., Rebouillat D., Hovanessian A.G. 69-kDa and 100-kDa isoforms of interferon-induced (2′–5′)oligoadenylate synthetase exhibit differential catalytic parameters. Eur. J. Biochem. 1997;248:558–566. doi: 10.1111/j.1432-1033.1997.t01-1-00558.x. [DOI] [PubMed] [Google Scholar]
- 31.Rebouillat D., Hovnanian A., Marie I., Hovanessian A.G. The 100-kDa 2′,5′-oligoadenylate synthetase catalyzing preferentially the synthesis of dimeric pppA2′p5′A molecules is composed of three homologous domains. J. Biol. Chem. 1999;274:1557–1565. doi: 10.1074/jbc.274.3.1557. [DOI] [PubMed] [Google Scholar]
- 32.Hartmann R., Norby P.L., Martensen P.M., Jorgensen P., James M.C., Jacobsen C., Moestrup S.K., Clemens M.J., Justesen J. Activation of 2′–5′ oligoadenylate synthetase by single-stranded and double-stranded RNA aptamers. J. Biol. Chem. 1998;273:3236–3246. doi: 10.1074/jbc.273.6.3236. [DOI] [PubMed] [Google Scholar]
- 33.Hartmann R., Justesen J., Sarkar S.N., Sen G.C., Yee V.C. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′–5′-oligoadenylate synthetase. Mol. Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 34.Rosenblum M.G., Yung W.K., Kelleher P.J., Ruzicka F., Steck P.A., Borden E.C. Growth inhibitory effects of interferon-beta but not interferon-alpha on human glioma cells: correlation of receptor binding, 2′,5′-oligoadenylate synthetase and protein kinase activity. J. Interferon Res. 1990;10:141–151. doi: 10.1089/jir.1990.10.141. [DOI] [PubMed] [Google Scholar]
- 35.Hubbell H.R., Sheetz P.C., Iogal S.S., Brodsky I., Kariko K., Li S.W., Suhadolnik R.J., Sobol R.W. Heterogeneous nuclear RNA from hairy cell leukemia patients activates 2′,5′-oligoadenylate synthetase. Anticancer Res. 1991;11:1927–1932. [PubMed] [Google Scholar]
- 36.Keller E.T., Fu Z., Brennan M. The biology of a prostate cancer metastasis suppressor protein: Raf kinase inhibitor protein. J. Cell Biochem. 2005;94:273–278. doi: 10.1002/jcb.20169. [DOI] [PubMed] [Google Scholar]
- 37.Makeyev A.V., Liebhaber S.A. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thakur C.S., Xu Z., Wang Z., Novince Z., Silverman R.H. A convenient and sensitive fluorescence resonance energy transfer assay for RNase L and 2′,5′ oligoadenylates. Methods Mol. Med. 2005;116:103–113. doi: 10.1385/1-59259-939-7:103. [DOI] [PubMed] [Google Scholar]
- 39.Ushijima K., Gouzu H., Hosono K., Shirakawa M., Kagosima K., Takai K., Takaku H. Site-specific cleavage of tRNA by imidazole and/or primary amine groups bound at the 5′-end of oligodeoxyribonucleotides. Biochim. Biophys. Acta. 1998;1379:217–223. doi: 10.1016/s0304-4165(97)00101-3. [DOI] [PubMed] [Google Scholar]
- 40.Wang-Johanning F., Frost A.R., Jian B., Azerou R., Lu D.W., Chen D.T., Johanning G.L. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer. 2003;98:187–197. doi: 10.1002/cncr.11451. [DOI] [PubMed] [Google Scholar]
- 41.Kao C., Rudisser S., Zheng M. A simple and efficient method to transcribe rnas with reduced 3′ heterogeneity. Methods. 2001;23:201–205. doi: 10.1006/meth.2000.1131. [DOI] [PubMed] [Google Scholar]
- 42.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M. Oncomine: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myszka D.G. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr. Opin. Biotechnol. 1997;8:50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- 44.Dhanasekaran S.M., Barrette T.R., Ghosh D., Shah R., Varambally S., Kurachi K., Pienta K.J., Rubin M.A., Chinnaiyan A.M. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 45.Lapointe J., Li C., Higgins J.P., van de Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl Acad. Sci. USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeung K., Seitz T., Li S., Janosch P., McFerran B., Kaiser C., Fee F., Katsanakis K.D., Rose D.W., Mischak H., et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 47.Kroslak T., Koch T., Kahl E., Hollt V. Human phosphatidylethanolamine-binding protein facilitates heterotrimeric G protein-dependent signaling. J. Biol. Chem. 2001;276:39772–39778. doi: 10.1074/jbc.M106991200. [DOI] [PubMed] [Google Scholar]
- 48.Yeung K.C., Rose D.W., Dhillon A.S., Yaros D., Gustafsson M., Chatterjee D., McFerran B., Wyche J., Kolch W., Sedivy J.M. Raf kinase inhibitor protein interacts with NF-κB-inducing kinase and TAK1 and inhibits NF-κB activation. Mol. Cell. Biol. 2001;21:7207–7217. doi: 10.1128/MCB.21.21.7207-7217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu Z., Smith P.C., Zhang L., Rubin M.A., Dunn R.L., Yao Z., Keller E.T. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J. Natl Cancer Inst. 2003;95:878–889. doi: 10.1093/jnci/95.12.878. [DOI] [PubMed] [Google Scholar]
- 50.Chatterjee D., Bai Y., Wang Z., Beach S., Mott S., Roy R., Braastad C., Sun Y., Mukhopadhyay A., Aggarwal B.B., et al. RKIP sensitizes prostate and breast cancer cells to drug-induced apoptosis. J. Biol. Chem. 2004;279:17515–17523. doi: 10.1074/jbc.M313816200. [DOI] [PubMed] [Google Scholar]
- 51.Fu Z., Kitagawa Y., Shen R., Shah R., Mehra R., Rhodes D., Keller P.J., Mizokami A., Dunn R., Chinnaiyan A.M., et al. Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate. 2006;66:248–256. doi: 10.1002/pros.20319. [DOI] [PubMed] [Google Scholar]
- 52.Yeap B.B., Voon D.C., Vivian J.P., McCulloch R.K., Thomson A.M., Giles K.M., Czyzyk-Krzeska M.F., Furneaux H., Wilce M.C., Wilce J.A., et al. Novel binding of hur and poly(C)-binding protein to a conserved UC-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J. Biol. Chem. 2002;277:27183–27192. doi: 10.1074/jbc.M202883200. [DOI] [PubMed] [Google Scholar]
- 53.Yeap B.B., Wilce J.A., Leedman P.J. The androgen receptor mRNA. Bioessays. 2004;26:672–682. doi: 10.1002/bies.20051. [DOI] [PubMed] [Google Scholar]
- 54.Walter B.L., Parsley T.B., Ehrenfeld E., Semler B.L. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 2002;76:12008–12022. doi: 10.1128/JVI.76.23.12008-12022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu Y.P., Landsittel D., Jing L., Nelson J., Ren B., Liu L., McDonald C., Thomas R., Dhir R., Finkelstein S., et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 56.LaTulippe E., Satagopan J., Smith A., Scher H., Scardino P., Reuter V., Gerald W.L. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 57.Vanaja D.K., Ballman K.V., Morlan B.W., Cheville J.C., Neumann R.M., Lieber M.M., Tindall D.J., Young C.Y. PDLIM4 repression by hypermethylation as a potential biomarker for prostate cancer. Clin. Cancer Res. 2006;12:1128–1136. doi: 10.1158/1078-0432.CCR-05-2072. [DOI] [PubMed] [Google Scholar]
- 58.Li W.H., Gu Z., Wang H., Nekrutenko A. Evolutionary analyses of the human genome. Nature. 2001;409:847–849. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- 59.Leib-Mosch C., Brack-Werner R., Werner T., Bachmann M., Faff O., Erfle V., Hehlmann R. Endogenous retroviral elements in human DNA. Cancer Res. 1990;50:5636S–5642S. [PubMed] [Google Scholar]
- 60.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 61.Yolken R.H., Torrey E.F. Viruses, schizophrenia, and bipolar disorder. Clin. Microbiol. Rev. 1995;8:131–145. doi: 10.1128/cmr.8.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yi J.M., Lee W.H., Kim H.M., Kim H.S. Identification of new endogenous retroviral sequences belonging to the HERV-W family in human cancer cells. Intervirology. 2001;44:333–338. doi: 10.1159/000050067. [DOI] [PubMed] [Google Scholar]
- 63.Sutkowski N., Conrad B., Thorley-Lawson D.A., Huber B.T. Epstein–Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity. 2001;15:579–589. doi: 10.1016/s1074-7613(01)00210-2. [DOI] [PubMed] [Google Scholar]
- 64.Perl A. Role of endogenous retroviruses in autoimmune diseases. Rheum. Dis. Clin. North Am. 2003;29:123–143. doi: 10.1016/s0889-857x(02)00098-4. vii. [DOI] [PubMed] [Google Scholar]
- 65.Marguerat S., Wang W.Y., Todd J.A., Conrad B. Association of human endogenous retrovirus K-18 polymorphisms with type 1 diabetes. Diabetes. 2004;53:852–854. doi: 10.2337/diabetes.53.3.852. [DOI] [PubMed] [Google Scholar]
- 66.Mi S., Lee X., Li X., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.Y., Edouard P., Howes S., et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 67.Blond J.L., Lavillette D., Cheynet V., Bouton O., Oriol G., Chapel-Fernandes S., Mandrand B., Mallet F., Cosset F.L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seifarth W., Frank O., Zeilfelder U., Spiess B., Greenwood A.D., Hehlmann R., Leib-Mosch C. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 2005;79:341–352. doi: 10.1128/JVI.79.1.341-352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schroder H.C., Suhadolnik R.J., Pfleiderer W., Charubala R., Muller W.E. (2′–5′)Oligoadenylate and intracellular immunity against retrovirus infection. Int. J. Biochem. 1992;24:55–63. doi: 10.1016/0020-711x(92)90229-t. [DOI] [PubMed] [Google Scholar]
- 70.Maitra R.K., Silverman R.H. Regulation of human immunodeficiency virus replication by 2′,5′-oligoadenylate-dependent RNase L. J. Virol. 1998;72:1146–1152. doi: 10.1128/jvi.72.2.1146-1152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martinand C., Montavon C., Salehzada T., Silhol M., Lebleu B., Bisbal C. RNase l inhibitor is induced during human immunodeficiency virus type 1 infection and down regulates the 2-5A/RNase L pathway in human T cells. J. Virol. 1999;73:290–296. doi: 10.1128/jvi.73.1.290-296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]