Toxin–antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci (original) (raw)
Abstract
Vancomycin-resistant enterococci (VRE) are common hospital pathogens that are resistant to most major classes of antibiotics. The incidence of VRE is increasing rapidly, to the point where over one-quarter of enterococcal infections in intensive care units are now resistant to vancomycin. The exact mechanism by which VRE maintains its plasmid-encoded resistance genes is ill-defined, and novel targets for the treatment of VRE are lacking. In an effort to identify novel protein targets for the treatment of VRE infections, we probed the plasmids obtained from 75 VRE isolates for the presence of toxin–antitoxin (TA) gene systems. Remarkably, genes for one particular TA pair, the mazEF system (originally identified on the Escherichia coli chromosome), were present on plasmids from 75/75 (100%) of the isolates. Furthermore, mazEF was on the same plasmid as vanA in the vast majority of cases (>90%). Plasmid stability tests and RT-PCR raise the possibility that this plasmid-encoded mazEF is indeed functional in enterococci. Given this ubiquity of mazEF in VRE and the deleterious activity of the MazF toxin, disruption of mazEF with pharmacological agents is an attractive strategy for tailored antimicrobial therapy.
Keywords: antibiotics, mazEF, axe-txe, relbE, VRE
Enterococci are the leading cause of surgical-site infections and the third leading cause of urinary tract and bloodstream infections (1, 2) and are implicated in bacterial endocarditis, intraabdominal infections, bacteremia, and meningitis (3). The intrinsic resistance of enterococci to cephalosporin and aminoglycoside antibiotics complicates treatment strategies. Historically, vancomycin has been effective in the management of enterococcal infections; however, after nearly 30 years of use, vancomycin-resistant enterococci (VRE) emerged in the mid-1980s. In a short time span, intensive care units in the U.S. have seen a dramatic increase in the percentage of enterococcal infections that are resistant to vancomycin, from 0.4% in 1989 to 25% in 1999 (3). In fact, in a recent study, 60% of Enterococcus faecium clinical isolates were resistant to vancomycin (4).
The genes encoding the elaborate protein systems that mediate vancomycin resistance generally reside on mobile genetic elements within the enterococci. In some cases, plasmids with the various vancomycin resistance gene clusters have been recovered from enterococci (5–8), although the prevalence of plasmid-encoded resistance in VRE has not been defined. Large plasmids often have intricate mechanisms by which they maintain themselves in the bacterial population. Some plasmids contain genes encoding postsegregational killing “plasmid addiction” systems (9, 10), in which the plasmid encodes a potent toxin and a labile antitoxin. Provided the plasmid is present, the antitoxin binds to and sequesters the toxin. However, if a plasmid-free daughter cell arises, the unstable antitoxin is degraded, and the toxic protein kills the bacterial cell from within.
If toxin–antitoxin (TA) systems are prevalent and functional on plasmids residing in pathogenic bacteria, this would present a novel opportunity for antibacterial therapy. For example, it has been suggested that small molecules that disrupt the TA interaction would free the toxin to kill the host (11–13). However, both the identity and prevalence of TA systems in pathogenic bacteria are unknown. In an effort to definitively link TA systems to VRE and to define new macromolecular targets for the treatment of these infections, we have searched for the presence of TA systems on plasmids isolated from VRE obtained from 75 different patients. Described herein is the discovery that TA genes are widespread in plasmids isolated from VRE, and surprisingly, the genes for one particular TA system that had previously been observed on the Escherichia coli chromosome (the mazEF system) appear on plasmids in 100% of the VRE strains. In addition, we show that in the vast majority of cases (>90%) the genes for vancomycin resistance are on the same plasmid as (and hence physically linked to) the genes for the mazEF TA system. Thus, TA systems appear to play a major role in enterococcal plasmid stability and are a critical component of the drug resistance of VRE. The identification herein of the commonality of a specific TA system presents an attractive target for tailored antimicrobial therapy.
Results
Plasmid-Encoded Vancomycin Resistance.
VRE were obtained from five different medical centers, 75 strains in total. The medical centers were: Carle Foundation Hospital (Urbana, IL), Memorial Medical Center (Springfield, IL), St. Mary's Hospital (Decatur, IL), Mount Sinai Hospital (Chicago, IL), and Barnes-Jewish Hospital (St. Louis, MO). Plasmid preparations from each strain were performed by an alkaline lysis method modified from standard protocols (14). PCR with primers complementary to vanA and vanB revealed that 75 of 75 of these strains had plasmids with the vanA gene cluster, and 7 of 75 also possessed plasmids harboring the vanB genes. Through strain typing using a PCR-based species identification method (15–19), it was determined that eight of the strains were Enterococcus faecalis, and 61 were E. faecium; six strains were not able to be typed by this method and are denoted as Enterococcus spp. A complete list of all strains and the features of their plasmids is in supporting information (SI) Tables 1–3.
Presence of TA Genes on Plasmids Isolated from VRE.
In an effort to define the prevalence of TA systems on plasmids isolated from VRE, PCR was used to probe the plasmid preparations from the 75 clinical isolates. Nondegenerate primers were designed such that each primer set would specifically amplify the genes of one TA system (9). These include three TA systems that have been previously observed on plasmids from Gram-positive bacteria [ω-ε-ζ (20), axe-txe (21), and par (22)], and seven that are representative of the major TA systems from Gram-negative bacteria [_parDE_ (23), ccdAB (24, 25), mazEF (26), relBE (27), higBA, (28), vagCD (29), and phd-doc (30)]. The exact primers used and their location on the TA genes are shown in SI Figs. 5–9 and Table 4. The par TA system utilizes a countertranscript RNA as the antitoxin, making it a “Type I” TA system (22). The other nine pairs are all “Type II” TA systems, meaning that both the toxin and antitoxin are proteins. The toxin components of these systems exert their antibacterial activity through a variety of mechanisms, including inhibition of DNA replication, inhibition of translation, and direct mRNA degradation (9).
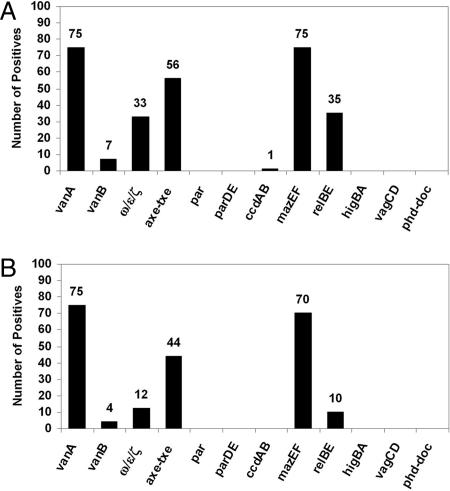
The results of this PCR analysis of the total DNA content isolated after plasmid preparation are displayed in Fig. 1A. Examination of these data reveals that TA systems are ubiquitous in the plasmids from these VRE isolates; in fact, there is at least one plasmid-encoded TA system in 75 of 75 (100%) of the samples. Most of the VRE isolates have multiple TA systems on their plasmids; the average number of plasmid-encoded TA systems for each VRE isolate was 2.7 (see SI Table 1).
Fig. 1.
PCR anaysis of VRE isolates. (A) The prevalence of genes encoding vancomycin resistance and plasmid stability systems as assessed by PCR analysis of the total plasmid preparation from 75 VRE isolates. (B) Analysis of the physical linkage between TA systems and vanA. Single plasmids containing the vanA gene cluster were isolated and probed by PCR with primers complementary to the genes encoding the various TA systems.
The genes for four TA systems are particularly prevalent and appear in multiple isolates: ω-ε-ζ (33/75, 44%), relBE (35/75, 47%), axe-txe (56/75, 75%), and mazEF (75/75, 100%). The prevalence of mazEF was particularly surprising, because this TA combination had been observed on the E. coli chromosome, with orthologs present on chromosomes of certain other bacteria (31). Also, the prevalence of axe-txe and relBE is of significance, because the axe-txe homologue, yefM-yoeB on the E. coli chromosome, is considered part of the relBE superfamily of TA systems (32, 33). Thus, the results of the PCR screen suggest that, in addition to mazEF, members of the relBE TA superfamily are quite abundant in VRE; in fact, only 7 of 75 clinical isolates in this study do not have plasmids with either axe-txe or relBE. See SI Table 1 for a complete list of the TA genes present in the total plasmid preparations from the individual VRE isolates. DNA sequencing was performed on ≈10% of all PCR products. In every case, these sequencing data confirmed the identity of the TA system, because the sequenced products had >95% sequence identity with their respective reference sequence (ω-ε-ζ, 96% identity; relBE, 99.7% identity; ccdAB, 99.1% identity; axe-txe, 99.9% identity; and mazEF, 98.8% identity; alignments are shown in SI Figs. 5–9).
Presence of TA Genes on Plasmids That Have Vancomycin-Resistance Genes.
Because many of the VRE strains contained multiple plasmids, the experiments described above gave no information about whether the TA systems resided on the same plasmids as the vancomycin resistance genes. Therefore, a determination was next made about the physical linkage of the vanA resistance cluster with the TA systems. Plasmids containing the vanA gene cluster were first isolated from each of the 75 VRE strains. This was performed one of two ways. First, all 75 of the clinical VRE isolates were subjected to conjugative mating (broth and filter matings; see Methods) with a plasmid-free recipient strain of E. faecium (BM4105-RF; RF, rifampin and fusidic acid) and subsequent selection on semisolid media containing vancomyin, rifampicin, and fusidic acid. Transconjugants were obtained for 33 of 75 of the isolates in this manner; plasmid preparation from these transconjugates followed by PCR with primers specific for the vanA gene was used to confirm the presence of the vanA gene cluster. For the 42 VRE strains that gave no transconjugates, the total plasmid content was subjected to agarose gel electrophoresis, and each band was extracted from the gel. PCR with primers for the vanA gene revealed the plasmids that harbored the vancomycin resistance genes.
With a single plasmid that harbors the vanA gene cluster now in hand for the 75 different VRE isolates, each of these plasmids was probed by PCR for the presence of the various TA systems, as above. A summary of these results is displayed in Fig. 1B, and the complete listing of individual strains is in SI Table 2. Although genes for the relBE (10/75; 13%) and ω-ε-ζ (12/75; 16%) TA systems were present on a relatively low percentage of the plasmids harboring the _vanA_-resistant gene cluster, the genes encoding the mazEF TA system were physically linked to the vancomycin resistance genes in a remarkable 93% (70/75) of the isolates, and axe-txe was present on 59% (44/75) of these _vanA_-containing plasmids. As before, DNA sequencing was used to confirm the identity of several of these PCR products.
Confirmation That TA Systems Reside on Plasmids in VRE.
To ensure that the observed results were not due to chromosomal contamination, cells from a plasmid-free strain of E. faecium (BM4105-RF) were taken through the alkaline lysis plasmid preparation protocol. Any DNA thus isolated was probed by PCR (as above) with primers specific for the various TA systems. In all cases, no amplification was observed. To further verify that a given TA locus resided on the plasmid (and not the chromosomal DNA) of VRE, two additional methods were used to eliminate contaminating chromosomal DNA in the plasmid preparations from the clinical VRE isolates before PCR analysis. First, after alkaline lysis preparation, the plasmid content from 15 of the VRE strains was treated with a “plasmid-safe” DNase, an enzyme that will digest linear DNA but not double-stranded supercoiled DNA. PCR amplification of the TA systems after this treatment closely matched the results of the initial PCR screen (see SI Table 5), suggesting that the TA loci reside on plasmid DNA. Second, after alkaline lysis preparation, the plasmid content from seven of the VRE strains was further purified by CsCl gradient ultracentrifugation. This highly purified plasmid DNA was then subjected to PCR analysis for the presence of the TA systems, and again the PCR results closely matched the results of the initial PCR screens (see SI Table 6). Finally, the flanking regions of the axe-txe and mazEF genes from three clinical isolates were sequenced. These DNA regions just outside the TA gene systems were found to be nearly identical to regions from two different enterococcal plasmids, pRUM and pPD1 (see SI Table 7 for DNA sequences obtained, results of BLASTN search with that sequence, and percent similarity to matched database DNA). Taken together, these data suggest the TA loci reside on plasmid DNA. For full experimental protocols for plasmid-safe DNase treatments, CsCI plasmid preparations, and sequencing, please see SI Text.
Antibiotic Susceptibility of VRE Isolates.
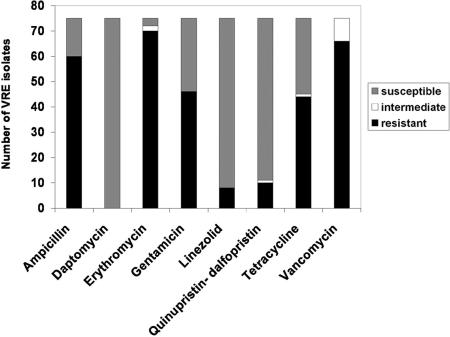
All 75 VRE strains were assessed for their susceptibility to common antibacterial agents. As expected, all strains were resistant to vancomycin, at the level of either full or intermediate resistance (according to National Committee for Clinical Laboratory Standards/Clinical and Laboratory Standards Institute standards; Fig. 2). The vast majority of the strains were also resistant to erythromycin and ampicillin, and a significant number were resistant to gentamicin and tetracycline. Only a handful of strains showed resistance to linezolid and quinupristin-dalfopristin, and all 75 isolates were susceptible to daptomycin. The results from this evaluation are displayed in Fig. 2, and the antibiotic resistance profile for each individual strain is in SI Table 3.
Fig. 2.
Antibiotic-resistance profiles of the 75 VRE isolates.
The mazEF System Confers Plasmid Stability in Enterococci.
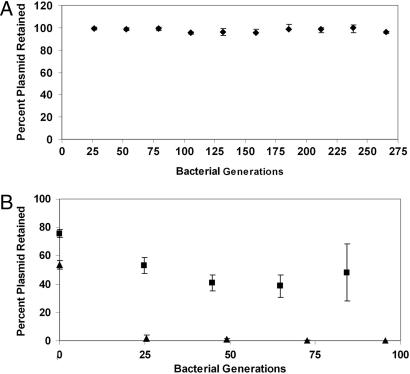
As discussed below, mazEF and homologous systems (parD and chpB) have been most extensively studied on their plasmid and chromosomal origins in enterobacteria (34–36). Because 100% of the VRE strains obtained for this study harbored genes for a plasmid-encoded mazEF TA system (Fig. 1A), we sought to determine whether this plasmid addiction system was indeed functional in enterococci. As a first step, plasmid stability was assessed with a transconjugate that harbors a single plasmid encoding both vanA and mazEF but not any of the other TA systems that were examined. This plasmid is designated pS345RF. This plasmid was found to be stable in the absence of vancomycin for 265 bacterial generations (Fig. 3A). However, this experiment does not conclusively show that mazEF is functional in VRE, because other undetected or unknown plasmid stability/maintenance systems may be present on this plasmid. To directly determine whether the mazEF genes are functional in enterococci, the mazEF loci identified in VRE was cloned into an unstable enterococcal vector (pAM401; ref. 37), and plasmid stability was measured in E. faecalis strain OG1X in the absence of antibiotic selection for the plasmid. This new plasmid, designated pAM401EF, was found to be considerably more stable than the parent vector (Fig. 3B).
Fig. 3.
Plasmid stability assays. (A) Plasmid pS345RF, which has mazEF as the only detectable TA gene system, is stable for 265 generations in enterococci in the absence of any antibiotic selection. Plasmid stability was determined by replica plating onto selective media. (B) Plasmid pAM401 is highly unstable in the enterococcal strain OG1X (▴) in the absence of antibiotic selection. When the mazEF loci is cloned into pAM401, the stability of the plasmid is increased (■). Error bars represent standard deviation from the mean.
The Transcripts for MazEF Are Synthesized in VRE.
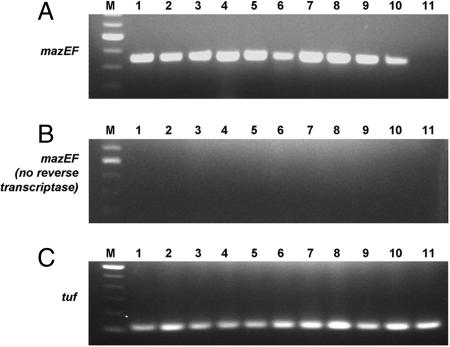
To confirm that the mRNA coding for the MazEF proteins is indeed being produced in VRE, we performed RT-PCR from total mRNA isolated from 10 of the VRE clinical isolates. As displayed in Fig. 4, all 10 of the isolates that had been shown to harbor mazEF did indeed produce the mazEF transcript, as assessed by RT-PCR (Fig. 4A, lanes 1–10). This same RT-PCR analysis was also performed on the total RNA isolated from the plasmid-free E. faecium strain BM4105-RF, and no mazEF transcript was detected (Fig. 4A, lane 11). To ensure the observed RT-PCR products were not due to DNA contamination, controls were conducted in which the reverse transcriptase enzyme was not added, but all other components (including thermostable DNA polymerase and primers for mazEF) were added. Analysis shows that no amplification is observed under these conditions (Fig. 4B). Finally, RT-PCR with primers for the enterococcal elongation factor (tuf, encoding EF-tu of enterococci) gave the expected result (Fig. 4C).
Fig. 4.
RT-PCR analysis of 10 clinical VRE isolates (lanes 1–10) and a plasmid-free enterococcal strain (BM4105-RF) (lane 11). The isolates used were U563, C27282, C31582, C531926, S177, S234, S345, D1, SL278, and SL518 (lanes 1–10, respectively). (A) RT-PCR with primers complementary to mazEF indicates that this transcript is present in the clinical isolates but not in plasmid-free E. faecium. (B) Controls for DNA contamination, in which the reverse transcriptase is left out of the reaction mix, give no amplification of mazEF. (C) RT-PCR with primers complementary to the enterococci tuf gene (encoding EF-tu) gives the expected products. Lane M contains the 100-bp DNA ladder.
Discussion
Although the resistance of enterococci to vancomycin in hospital settings is a fairly recent phenomenon, VRE are now responsible for a large subset of nosocomial infections. This upward trend in resistance is alarming: VRE itself is now a major and largely untreatable infection, and VRE can pass the vancomycin resistance genes to the highly virulent methecillin-resistant Staphylococcus aureus(MRSA) (38, 39). Despite the high medical relevance of VRE, little is known about the nature of plasmid-encoded vancomycin resistance. Further, the mechanisms that allow these mobile genetic elements to be retained in enterococci even in the absence of antibiotic selection are not well understood.
TA systems have been identified on the chromosomes of many different bacteria. Although the exact function of these chromosomal TA systems is still unclear, in some cases, they are believed to be linked to a bacterial stress response (12, 40, 41). In addition to the chromosomal location of the TA genes, they have also been found on plasmids within certain (mostly Gram-negative) bacteria (9). On plasmids, the function of TA systems seems straightforward: to ensure faithful propagation of bacteria harboring the plasmid through the postsegregational killing of plasmid-free daughter cells. Thus, it has been speculated that disruption of the TA interaction with pharmacological agents would unveil the active toxin and kill the cell from within (11–13). However, many different TA systems have been described, and no systematic analyses have been performed to determine their prevalence in clinically important bacterial infections; it has thus been impossible to know which TA systems would be present and operational in the most untreatable infections. This lack of knowledge about the prevalence and identities of TA systems in clinically unmanageable bacteria has hampered compound development; indeed, to date, no small-molecule disruptors of any TA proteins have been reported.
The major findings of our survey of the plasmid content from 75 different VRE isolates are as follows: (i) Vancomycin resistance is plasmid encoded in our isolates; (ii) genes encoding TA systems are ubiquitous in plasmids isolated from VRE; (iii) mazEF, and to a lesser extent axe-txe, relBE, and ω-ε-ζ, are common in plasmids from VRE isolates. Whereas ω-ε-ζ had been previously observed on plasmids from Gram-positive Bacillus (42), Streptococcus (43), and Enterococcus (44), axe-txe and par are the only other TA systems that had been found on a plasmid from a Gram-positive bacterium, and axe-txe was from a single E. faecium isolate (as discussed below; ref. 21). The mazEF and relBE systems were both originally identified on the E. coli chromosome; relBE has also been found on a plasmid from E. coli (45), and the mazEF homologues, parD and pem, were identified on plasmids R1 and R100, respectively (34, 46). Thus, the finding that mazEF and relBE are common on plasmids from VRE is especially striking.
The mazEF TA system was originally discovered on the E. coli chromosome and has been well characterized genetically and biochemically (47–50). It consists of a 9-kDa antitoxin (MazE) that binds to and renders inactive the 12-kDa toxin (MazF). MazE is a labile protein that is degraded by the ClpA protease; thus, in the absence of the genes encoding mazEF, the cellular MazE concentration drops rapidly, leaving MazF to kill or inhibit the growth of the cell (51). The MazF toxin is an endoribonuclease that is specific for ACA sequences (52–54), and its degradation of mRNA in this manner is believed to lead to the observed bacterial cell death (55). Although mazEF orthologs with varying degrees of homology are present on other bacterial chromosomes (31), including three mazEF homologs on the E. faecalis V583 genome and one on the V583 plasmid pTEF1 (33), the PCR products investigated in this study show surprisingly strong sequence similarity to the mazEF genes from the E. coli K12 genome. Although its role on the E. coli chromosome is not entirely clear, mazEF appears to be a “stress-induced suicide module” (55), because it triggers cell death under conditions of amino acid starvation signals (41), antibiotic treatment (56), phage infection (57), thymine starvation (58), oxidative stress, and high temperatures (55). The role of the chromosomally encoded mazEF TA system in E. coli stress response has been reviewed extensively (11, 40, 55).
Because relBE is also believed to have originated from the E. coli chromosome and has been only rarely been found on plasmids (27, 59), it was also surprising to find that almost half of the VRE isolates were observed to have relBE on their plasmids. In E. coli, relBE appears to be part of a complex bacterial response to nutrient starvation (60). Importantly, a crystal structure of the RelBE protein–protein complex was recently solved (61).
The axe-txe TA system was initially identified from a plasmid isolated from a single VRE clinical isolate, and in this strain, axe-txe was not physically linked with the vanA resistance element (21). The prevalence of axe-txe within clinical VRE isolates had not previously been determined, but here we show that axe-txe is quite common on plasmids isolated from VRE (appearing in 75% of the isolates), and in most cases, these genes reside on the same plasmid as the vancomycin resistance. Similarly, ω-ε-ζ appeared quite frequently on the plasmids from VRE (44% of the isolates). Interestingly, the plasmid on which axe-txe was discovered, pRUM (21) was found to make up the sequences of the flanking regions of several axe-txe and mazEF genes from the VRE isolates (SI Table 7).
Thus, TA systems appear to be widespread on plasmids isolated from VRE. Further, all of those plasmids that contain the vanA gene cluster encode at least one TA system. In addition, the mazEF TA system is ubiquitous on plasmids residing in the Gram-positive pathogen VRE (appearing in 100% of the isolates). This high prevalence of mazEF suggests the encoded protein products as a logical target for tailored antibacterial therapy. To determine whether mazEF is functional in these VRE isolates, two sets of experiments were performed. First, the stability of a plasmid that contains mazEF as the only detectable TA system (plasmid pS345RF) was determined; this plasmid also harbors the vanA gene cluster. As shown in Fig. 3A, even in the absence of vancomycin, there is no detectable plasmid loss from enterococci over 265 generations of bacterial growth. To directly demonstrate mazEF functionality in enterococci, the mazEF loci were cloned into an unstable enterococcal vector and shown to exert a stabilizing effect on the plasmid in enterococci (Fig. 3B). Second, to determine whether the mazEF transcript is being synthesized by VRE, RT-PCR was performed on the total mRNA preparation from multiple VRE isolates. As shown in Fig. 4, the mazEF transcript is indeed being made whenever the mazEF genes are present. Taken together, the plasmid stability and RT-PCR results are consistent with a functional mazEF in the VRE clinical isolates.
Although our current study was limited to enterococci, given its prevalence in VRE it would not be surprising to find mazEF on plasmids within closely related bacteria, such as Streptococcus and Staphylococcus. In addition, because the mazEF family of TA systems was among the first plasmid addiction systems to be discovered and studied (the first TA system to be identified was ccdAB; refs. 25 and 34), much is known about the biochemistry of these proteins. Most important to drug design and discovery, there are crystal structures available for both MazE itself (62) and the MazE–MazF protein–protein complex (63). Given the available evidence suggesting that MazF lethality can initially be reversed by MazE, but that a “point of no return” exists from which bacterial cells cannot recover (51, 64, 65), the disruption of the MazE–MazF interaction with a small molecule should, over time, free the MazF toxin and lead to cell death (66). With the current discovery that this E. coli TA system has made the leap to plasmids and is ubiquitous in VRE, efforts to develop disruptors of the TA interaction can proceed with full confidence that mazEF is a relevant and important target for antibacterial therapy.
Methods
Plasmid DNA Isolation.
Plasmid DNA from all enterococcal strains and transconjugants was isolated by a modified alkaline lysis method (14). Cell pellets were initially incubated with lysozyme (20 mg/ml) in 25 mM Tris/10 mM EDTA (pH 8.0), containing 50 mM glucose at 37°C for 1 h. After phenol-chloroform extraction, plasmid DNA was precipitated by addition of isopropanol and pelleted by centrifugation. The isopropanol was gently removed, and 70% ethanol was added to the DNA pellet. The plasmid DNA was again pelleted by centrifugation, the supernatant was removed, and the pellet was resuspended in 100 μl of 10 mM Tris·HCl buffer, pH 8.5.
Total DNA Isolation.
Total DNA from enterococci was isolated and purified by an alkaline-lysis method modified from standard protocols (67). After phenol-chloroform extraction, total DNA was precipitated by adding 100 μl of a 1:1 mixture of 7.5 M NH4OAc and 100% ethanol on ice for 30 min. Total DNA was recovered by removing the supernatant after centrifugation and resuspending the pellet in 10 mM Tris·HCl buffer (pH 8.5) to a final volume of 100 μl.
Isolation of Plasmids Encoding Vancomycin Resistance.
Filter and broth matings were attempted with all clinical strains of VRE and recipient strains E. faecium BM4105-RF and BM4105-SS (SS, streptomycin and spectinomycin), as described with minor modifications (68, 69). E. faecium BM4105-RF and BM4105-SS are resistant to RF and SS, respectively. To 3 ml of fresh brain–heart infusion (BHI) broth (BBL, Sparks, MD), equal (1-ml) volumes of donor strain and recipient strain cultures (grown overnight in BHI broth at 37°C) were added and further incubated for 4 h. The mating mixture was plated on BHI agar plates containing rifampin (50 μg/ml), fusidic acid (20 μg/ml), and vancomycin (10 μg/ml) or streptomycin (200 μg/ml), spectinomycin (100 μg/ml), and vancomycin (10 μg/ml). After incubation at 37°C for up to 48 h, plates were checked for the absence (donor and recipient cultures only) or presence (mating mixtures) of colonies on triple selective media.
For filter matings, mating mixtures were filtered through a sterile membrane filter [pore size, 0.45 μm; MF-Millipore (Billerica, MA) membrane filter HAWP 2500]. The filters were incubated on BHI agar plates for 24 h at 37°C. After mating, the cells were resuspended in 1 ml of BHI broth and spread on BHI agar containing rifampin (50 μg/ml), fusidic acid (20 μg/ml), and vancomycin (10 μg/ml) or streptomycin (200 μg/ml), spectinomycin (100 μg/ml), and vancomycin (10 μg/ml). The plates were incubated at 37°C for 24–48 h and checked for the absence (donor and recipient cultures only) or presence (mating mixtures) of colonies on triple selective media.
Total plasmid DNA was isolated, as described above, from clinical VRE strains that repeatedly failed to mate with E. faecium BM4105-RF or -SS. This plasmid DNA was analyzed by agarose gel electrophoresis in 1% agarose and stained with ethidium bromide. Individual bands were extracted by using the QIAquick gel extraction kit (Qiagen, Valencia, CA). The vancomycin-resistance plasmid DNA was identified by PCR for the vanA gene.
PCR Analysis of Plasmids.
For all clinical VRE isolates, PCR amplification was performed from purified total DNA or from purified plasmid DNA. Oligodeoxynucleotide primer pairs were synthesized (Integrated DNA Technologies, Coralville, IA) that amplify the ddlE. faecalis and ddlE. faecium genes, the tuf gene, the groES gene, and a strongly conserved sequence in E. faecium, to identify clinical enterococci isolates at the genus and species level, as described (15–19). A primer pair derived from the vanA gene sequence was used to amplify specifically the vancomycin-resistance determinant in Enterococcus, as well as a pair of PCR primers specific for the vanB gene sequence (70). Based on the sequence alignment of the axe-txe, par, parDE, ccd, mazEF, relBE, higBA, vagCD, and phd-doc TA genes in the National Center for Biotechnology Information database, pairs of fixed primers specific to each TA system were designed to amplify internal fragments with sizes ranging from 456 bp to 1.04 kb. The sequence of primers to amplify the ω-ε-ζ gene system was identical to those reported (44). The sequences of all primers are listed in SI Table 4.
PCR amplification was performed in a final volume of 100 μl containing 1–3 μl of purified total DNA or plasmid DNA, 1× PCR buffer (20 mM Tris·HCl/50 mM KCl, pH 8.4), 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 0.5 μM each primer, and 2.5 units of Taq polymerase (Invitrogen, Carlsbad, CA). PCR was carried out in a DNA thermal cycler (PTC-200; MJ Research, Cambridge, MA), with an initial denaturation step (94°C, 3 min), 30 cycles of denaturation (94°C, 1 min), annealing (54°C, 1 min), and extension (72°C, 1 min 30 s), followed by a final extension step (72°C, 10 min). PCR amplification products were analyzed by agarose gel electrophoresis in 1% agarose and stained with ethidium bromide.
Sequencing Analysis.
Approximately 10% of the PCR products generated from custom primers for the TA systems, vancomycin-resistant determinants, and species-specific genes were submitted for DNA sequencing by the University of Illinois W. M. Keck Center for Comparative and Functional Genomics. Sequence data were analyzed by using BioEdit, version 7.0.4.1, computer software and the BLAST database to verify the identity of the PCR products and sequence identity to known genes (71).
Antibiotic Susceptibility Tests.
Susceptibility testing was performed in duplicate by using the microdilution broth method as outlined by the Clinical and Laboratory Standards Institute CLSI, formerly National Committee for Clinical Laboratory Standards (NCCLS) (72). Briefly, organisms were inoculated into broth and incubated at 37°C until the culture reached the turbidity of the 0.5 McFarland standard. The test medium used was cation-adjusted Mueller Hinton II Broth (BBL) for all organisms tested. For testing daptomycin susceptibility, the broth was supplemented with 50 mg/liter Ca2+. Antibiotic concentrations were prepared manually in the range of the minimum inhibitory concentration (MIC) interpretive standards for Enterococcus spp. and dispensed with a 125-μl Matrix Impact pipettor. The MIC was defined as the lowest concentration of each antimicrobial agent that resulted in ≥80% reduction in growth.
Plasmid Stability Tests.
The stability of the vancomycin-resistant plasmid in the transconjugant, S345RF, was studied in triplicate, as described, with some modifications (73). One hundred microliters of an overnight culture of the strain grown in BHI broth containing 10 μg/ml vancomycin was used to inoculate 10 ml of fresh BHI broth containing 10 μg/ml vancomycin and grown for 4 h. Dilutions of the culture were prepared in sterile H2O and plated onto nonselective BHI agar plates for viable counts. The 10−4 dilution of the culture was used to inoculate 10 ml of BHI broth without selection and grown overnight at 37°C. Dilutions and viable counts were made with the culture as before, and a 10−4 dilution of the culture was subcultured into 10 ml of fresh BHI broth. This process was repeated until ≈250–300 generations of growth were achieved. Viable-count plates containing 75–150 colonies were replica-plated onto BHI agar plates with and without 10 μg/ml vancomycin. The proportion of the population retaining the vancomycin-resistant plasmid compared with the total number of viable cells was calculated in this way. Plasmid DNA was isolated throughout the stability assay from selected colonies to confirm the maintenance or loss of the vancomycin-resistant plasmid.
The stability of the enterococcal vector pAM401 (with and without the mazEF loci cloned in at the XbaI restriction site) was assessed in triplicate. These experiments were performed in an analogous fashion to the experiments described with pS345RF (above), except chloramphenicol was used to select for cells that contained the plasmid, and the assay was continued for ≈75–95 generations of bacterial growth. The data points at bacterial generation zero in Fig. 3B represent the time at which the bacteria were introduced into media without antibiotic selection. Plasmid retention was determined by replica plating; generations from overnight growth on the antibiotic-free plates were not included in the bacterial generation count (x axis) and could account for the appearance of a mixed (plasmid and plasmid-free) population at generation zero.
RNA Extraction and RT-PCR Analysis.
To 10 ml of fresh BHI broth, 10 μl of a culture grown overnight in BHI broth was added and further incubated until ≈1 × 109 cells were present. RNA was extracted by using the RNeasy Mini Kit for Isolation of Total RNA from Bacteria and treated with the RNase-Free DNase set (Qiagen). RT-PCR was performed by using the SuperScript One-Step RT-PCR with Platinum Taq kit (Invitrogen). Primers used for RT-PCR are as listed in SI Table 1. Intragenic primers used for mRNA mazEF expression were designed from the E. coli K12 mazEF sequence, and the control primers were based on the tuf gene specific to enterococcal species (17). Processed RNA (100 ng) was used in RT-PCR, as well as PCRs with Platinum TaqDNA polymerase (Invitrogen) to detect DNA contamination. RT-PCR was performed in a total reaction volume of 50 μl in a DNA thermal cycler (PTC-200; MJ Research), with an initial cycle at 50°C for 30 min for cDNA synthesis and then one cycle at 94°C for 2 min for predenaturation. PCR amplification began immediately after cDNA synthesis and predenaturation with 35 cycles of denaturation (94°C, 30 s), annealing (54°C, 45 s), and extension (72°C, 1 min), followed by a final extension step (72°C, 10 min). RT-PCR amplification products were analyzed by agarose gel electrophoresis in 1% agarose and stained with ethidium bromide.
Supplementary Material
Supporting Information
Acknowledgments
This work was supported by National Institutes of Health Grant NIHGMS R01-GM68385 and Office of Naval Research Grant N00014-02-1-0390. E.M.M. was partially supported by a National Institutes of Health Cell and Molecular Biology Training grant. We thank Cubist Pharmaceuticals (Lexington, MA) for the gift of daptomycin, Pfizer (New York, NY) for the gift of linezolid, and King Pharmaceuticals (Bristol, TN) for the gift of quinupristin-dalfopristin. We also thank Prof. Y. Ike (Gunma University Graduate School of Medicine, Gunma, Japan) for the BM4105-RF and -SS strains and Prof. K. Weaver (University of South Dakota, Vermillion, SD) for the OG1X strain. Finally, we thank the bacterial laboratories at Carle Foundation Hospital (Urbana, IL), Memorial Medical Center (Springfield, IL), St. Mary's Hospital (Decatur, IL), Mount Sinai Hospital (Chicago, IL), and Barnes-Jewish Hospital (St. Louis, MO) for the VRE isolates.
Abbreviations
VRE
vancomycin-resistant enterococci
TA
toxin–antitoxin
RF
rifampin and fusidic acid
SS
streptomycin and spectinomycin
BHI
brain–heart infusion.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Richards MJ, Edwards JR, Culver DH, Gaynes RP. Infect Control Hosp Epidemiol. 2000;21:510–515. doi: 10.1086/501795. [DOI] [PubMed] [Google Scholar]
- 2.Shepard BD, Gilmore MS. Microbes Infect. 2002;4:215–224. doi: 10.1016/s1286-4579(01)01530-1. [DOI] [PubMed] [Google Scholar]
- 3.Koch S, Hufnagel M, Huebner J. Exp Opin Biol Ther. 2004;4:1519–1531. doi: 10.1517/14712598.4.9.1519. [DOI] [PubMed] [Google Scholar]
- 4.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 5.Tomita H, Pierson C, Lim SK, Clewell DB, Ike Y. J Clin Microbiol. 2002;40:3326–3333. doi: 10.1128/JCM.40.9.3326-3333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomita H, Tanimoto K, Hayakawa S, Morinaga K, Ezaki K, Oshima H, Ike Y. J Bacteriol. 2003;185:7024–7028. doi: 10.1128/JB.185.23.7024-7028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasman H, Villadsen AG, Aarestrup FM. Microb Drug Resist. 2005;11:178–184. doi: 10.1089/mdr.2005.11.178. [DOI] [PubMed] [Google Scholar]
- 8.Johnsen PJ, Simonsen GS, Olsvik O, Midtvedt T, Sundsfjord A. Microb Drug Resist. 2002;8:161–170. doi: 10.1089/107662902760326869. [DOI] [PubMed] [Google Scholar]
- 9.Hayes F. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes K, Rasmussen PB, Molin S. Proc Natl Acad Sci USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelberg-Kulka H, Sat B, Reches M, Amitai S, Hazan R. Trends Microbiol. 2004;12:66–71. doi: 10.1016/j.tim.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Gerdes K, Christensen SK, Lobner-Olesen A. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 13.DeNap JBC, Hergenrother PJ. Org Biomol Chem. 2005;3:959–966. doi: 10.1039/b500182j. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 15.Cheng S, McCleskey FK, Gress MJ, Petroziello JM, Liu R, Namdari H, Beninga K, Salmen A, DelVecchio VG. J Clin Microbiol. 1997;35:1248–1250. doi: 10.1128/jcm.35.5.1248-1250.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutka-Malen S, Evers S, Courvalin P. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ke D, Picard FJ, Martineau F, Menard C, Roy PH, Ouellette M, Bergeron MG. J Clin Microbiol. 1999;37:3497–3503. doi: 10.1128/jcm.37.11.3497-3503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng LJ, Hsueh PR, Wang YH, Lin HM, Luh KT, Ho SW. J Clin Microbiol. 2001;39:3326–3331. doi: 10.1128/JCM.39.9.3326-3331.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velasco D, Perez S, Pena F, Dominguez MA, Cartelle M, Molina F, Moure R, Villanueva R, Bou G. Diagn Microbiol Infect Dis. 2004;49:151–156. doi: 10.1016/j.diagmicrobio.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Camacho AG, Misselwitz R, Behlke J, Ayora S, Welfle K, Meinhart A, Lara B, Saenger W, Welfle H, Alonso JC. Biol Chem. 2002;383:1701–1713. doi: 10.1515/BC.2002.191. [DOI] [PubMed] [Google Scholar]
- 21.Grady R, Hayes F. Mol Microbiol. 2003;47:1419–1432. doi: 10.1046/j.1365-2958.2003.03387.x. [DOI] [PubMed] [Google Scholar]
- 22.Weaver KE, Weaver DM, Wells CL, Waters CM, Gardner ME, Ehli EA. J Bacteriol. 2003;185:2169–2177. doi: 10.1128/JB.185.7.2169-2177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Pogliano J, Helinski DR, Konieczny I. Mol Microbiol. 2002;44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- 24.Van Melderen L. Int J Med Microbiol. 2002;291:537–544. doi: 10.1078/1438-4221-00164. [DOI] [PubMed] [Google Scholar]
- 25.Ogura T, Hiraga S. Proc Natl Acad Sci USA. 1983;80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelberg-Kulka H, Glaser G. Annu Rev Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- 27.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. Proc Natl Acad Sci USA. 2001;98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian QB, Ohnishi M, Tabuchi A, Terawaki Y. Biochem Biophys Res Commun. 1996;220:280–284. doi: 10.1006/bbrc.1996.0396. [DOI] [PubMed] [Google Scholar]
- 29.Pullinger GD, Lax AJ. Mol Microbiol. 1992;6:1631–1643. doi: 10.1111/j.1365-2958.1992.tb00888.x. [DOI] [PubMed] [Google Scholar]
- 30.Jensen RB, Gerdes K. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 31.Mittenhuber G. J Mol Microbiol Biotechnol. 1999;1:295–302. [PubMed] [Google Scholar]
- 32.Anantharaman V, Aravind L. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey DP, Gerdes K. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bravo A, de Torrontegui G, Diaz R. Mol Gen Genet. 1987;210:101–110. doi: 10.1007/BF00337764. [DOI] [PubMed] [Google Scholar]
- 35.Santos Sierra S, Giraldo R, Diaz Orejas R. FEMS Microbiol Lett. 1998;168:51–58. doi: 10.1111/j.1574-6968.1998.tb13254.x. [DOI] [PubMed] [Google Scholar]
- 36.Kamphuis MB, Bonvin AM, Monti MC, Lemonnier M, Munoz-Gomez A, van den Heuvel RH, Diaz-Orejas R, Boelens R. J Mol Biol. 2006;357:115–126. doi: 10.1016/j.jmb.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 37.Wirth R, An FY, Clewell DB. J Bacteriol. 1986;165:831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferber D. Science. 2003;302:1488. doi: 10.1126/science.302.5650.1488. [DOI] [PubMed] [Google Scholar]
- 39.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. Science. 2003;302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 40.Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. Trends Biochem Sci. 2005;30:672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 41.Aizenman E, Engelberg-Kulka H, Glaser G. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ceglowski P, Boitsov A, Chai S, Alonso JC. Gene. 1993;136:1–12. doi: 10.1016/0378-1119(93)90441-5. [DOI] [PubMed] [Google Scholar]
- 43.Zielenkiewicz U, Ceglowski P. J Bacteriol. 2005;187:6094–6105. doi: 10.1128/JB.187.17.6094-6105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnsen PJ, Osterhus JI, Sletvold H, Sorum M, Kruse H, Nielsen K, Simonsen GS, Sundsfjord A. Appl Environ Microbiol. 2005;71:159–168. doi: 10.1128/AEM.71.1.159-168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gronlund H, Gerdes K. J Mol Biol. 1999;285:1401–1415. doi: 10.1006/jmbi.1998.2416. [DOI] [PubMed] [Google Scholar]
- 46.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Zhang Y, Inouye M. J Biol Chem. 2003;278:32300–32306. doi: 10.1074/jbc.M304767200. [DOI] [PubMed] [Google Scholar]
- 48.Lah J, Marianovsky I, Glaser G, Engelberg-Kulka H, Kinne J, Wyns L, Loris R. J Biol Chem. 2003;278:14101–14111. doi: 10.1074/jbc.M209855200. [DOI] [PubMed] [Google Scholar]
- 49.Marianovsky I, Aizenman E, Engelberg-Kulka H, Glaser G. J Biol Chem. 2001;276:5975–5984. doi: 10.1074/jbc.M008832200. [DOI] [PubMed] [Google Scholar]
- 50.Lah J, Simic M, Vesnaver G, Marianovsky I, Glaser G, Engelberg-Kulka H, Loris R. J Biol Chem. 2005;280:17397–17407. doi: 10.1074/jbc.M501128200. [DOI] [PubMed] [Google Scholar]
- 51.Pedersen K, Christensen SK, Gerdes K. Mol Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. Mol Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 53.Munoz-Gomez AJ, Santos-Sierra S, Berzal-Herranz A, Lemonnier M, Diaz-Orejas R. FEBS Lett. 2004;567:316–320. doi: 10.1016/j.febslet.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. J Biol Chem. 2005;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 55.Engelberg-Kulka H, Hazan R, Amitai S. J Cell Sci. 2005;118:4327–4332. doi: 10.1242/jcs.02619. [DOI] [PubMed] [Google Scholar]
- 56.Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H. J Bacteriol. 2001;183:2041–2045. doi: 10.1128/JB.183.6.2041-2045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hazan R, Engelberg-Kulka H. Mol Genet Genom. 2004;272:227–234. doi: 10.1007/s00438-004-1048-y. [DOI] [PubMed] [Google Scholar]
- 58.Sat B, Reches M, Engelberg-Kulka H. J Bacteriol. 2003;185:1803–1807. doi: 10.1128/JB.185.6.1803-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gotfredsen M, Gerdes K. Mol Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 60.Wilson DN, Nierhaus KH. Nat Struct Mol Biol. 2005;12:282–284. doi: 10.1038/nsmb0405-282. [DOI] [PubMed] [Google Scholar]
- 61.Takagi H, Kakuta Y, Okada T, Yao M, Tanaka I, Kimura M. Nat Struct Mol Biol. 2005;12:327–331. doi: 10.1038/nsmb911. [DOI] [PubMed] [Google Scholar]
- 62.Loris R, Marianovsky I, Lah J, Laeremans T, Engelberg-Kulka H, Glaser G, Muyldermans S, Wyns L. J Biol Chem. 2003;278:28252–28257. doi: 10.1074/jbc.M302336200. [DOI] [PubMed] [Google Scholar]
- 63.Kamada K, Hanaoka F, Burley SK. Mol Cell. 2003;11:875–884. doi: 10.1016/s1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 64.Amitai S, Yassin Y, Engelberg-Kulka H. J Bacteriol. 2004;186:8295–8300. doi: 10.1128/JB.186.24.8295-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolodkin-Gal I, Engelberg-Kulka H. J Bacteriol. 2006;188:3420–3423. doi: 10.1128/JB.188.9.3420-3423.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Engelberg-Kulka H, Sat B, Reches M, Amitai S, Hazan R. Trends Microbiol. 2004;12:66–71. doi: 10.1016/j.tim.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 67.Dutka-Malen S, Leclercq R, Coutant V, Duval J, Courvalin P. Antimicrob Agents Chemother. 1990;34:1875–1879. doi: 10.1128/aac.34.10.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christie PJ, Korman RZ, Zahler SA, Adsit JC, Dunny GM. J Bacteriol. 1987;169:2529–2536. doi: 10.1128/jb.169.6.2529-2536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clewell DB, An FY, White BA, Gawron-Burke C. J Bacteriol. 1985;162:1212–1220. doi: 10.1128/jb.162.3.1212-1220.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Depardieu F, Perichon B, Courvalin P. J Clin Microbiol. 2004;42:5857–5860. doi: 10.1128/JCM.42.12.5857-5860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 72.National Committee for Clinical Laboratory Standards. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Wayne, PA: National Committee for Clinical Laboratory Standards; 2003. [Google Scholar]
- 73.Simpson AE, Skurray RA, Firth N. J Bacteriol. 2003;185:2143–2152. doi: 10.1128/JB.185.7.2143-2152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information