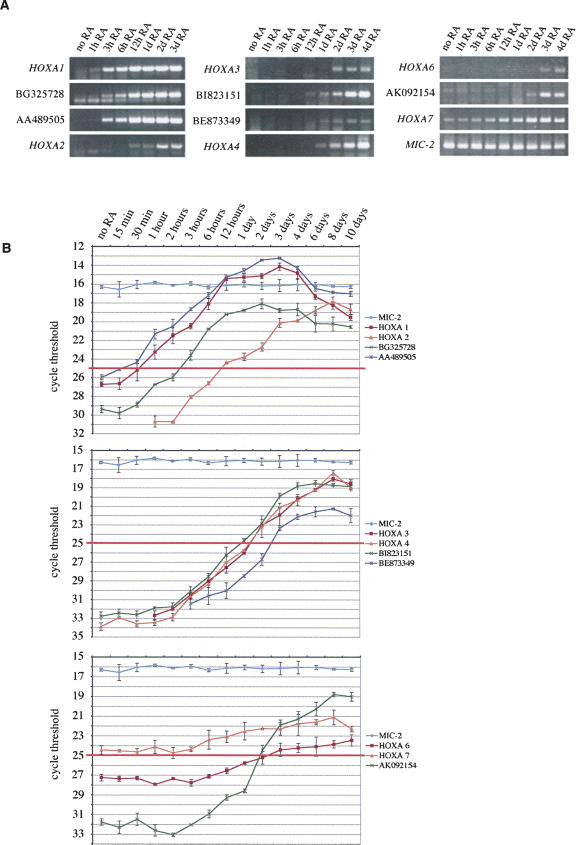
Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster (original) (raw)
Abstract
The ratio of noncoding to protein coding DNA rises with the complexity of the organism, culminating in nearly 99% of nonprotein coding DNA in humans. Nevertheless, a large portion of these regions is transcribed, creating the alleged paradox that noncoding RNA (ncRNA) represents the largest output of the human genome. Such a complex scenario may include epigenetic mechanisms where ncRNAs would be involved in chromatin regulation. We have investigated the intergenic, noncoding transcriptomes of mammalian HOX clusters. We show that “opposite strand transcription” from the intergenic spacer regions in the human HOXA cluster correlates with the activity state of adjacent HOXA genes. This noncoding transcription is regulated by the retinoic acid morphogen and follows the colinear activation pattern of the cluster. Opening of the cluster at sites of activation of intergenic transcripts is accompanied by changes in histone modifications and a loss of interaction with Polycomb group (PcG) repressive complexes. We propose that noncoding transcription is of fundamental importance for the opening and maintenance of the active state of HOX clusters.
Keywords: Noncoding transcription, epigenetics, HOX cluster, polycomb, chromatin
INTRODUCTION
The transcriptome of the mammalian genome is much larger than originally thought (Mattick 2001; Carninci et al. 2005). Detailed maps (5-nucleotide resolution) of transcriptional activity along human chromosomes using both cytoplasmic and nuclear poly(A)+ and poly(A)-RNA probes revealed that almost 90% of the nonrepetitive chromosome sequences are transcribed (Cheng et al. 2005). According to recent reports of the FANTOM-3 and RIKEN genome research groups, a large proportion of these would be classified as noncoding RNA (ncRNA) (Carninci et al. 2005). A major question that arises is, what are the functional role(s) of ncRNAs. In addition to a few ncRNA species that have been known for a long time (tRNA, rRNA, and spliceosomal RNAs), several new classes of these molecules have been described in recent years that seem to have predominantly, but not exclusively, an important impact on gene regulation either in cis or in trans (Bernstein and Allis 2005; Hüttendorfer et al. 2005). The factors that control ncRNA production are not known. Nevertheless, there is growing evidence that ncRNAs play a crucial role in epigenetic transmission of early determined transcription states, in particular in counteracting Polycomb group (PcG)-mediated repression.
The Polycomb group proteins, first described in Drosophila melanogaster, are part of a widely conserved transcriptional memory system that contributes to the epigenetic inheritance of cell identity (Orlando 2003; Ringrose and Paro 2004; Bantignies and Cavalli 2006). PcG proteins prevent changes in gene silencing programs and work as large complexes that act on nucleosome structure and the RNA–Polymerase II complex, leading to the inhibition of transcriptional activation (Levine et al. 2004). Recent genome-wide screens have identified PcG targets in human embryonic fibroblasts and in murine and human embryonic stem cells (Boyer et al. 2006; Bracken et al. 2006; Lee et al. 2006). These analyses indicate that the PcG controls most developmental regulators, including all four homeotic gene clusters (HOX).
PcG repression in Drosophila seems to be counteracted by intergenic transcription events. The expression of ncRNAs through PcG-specific epigenetic DNA elements (PcG response elements—PREs) correlates with the maintenance of the active state of homeotic PcG targets in the homeotic Bithorax complex (BX-C) (Rank et al. 2002; Schmitt et al. 2005; Sanchez-Elsner et al. 2006). Thus, anti-silencing by transcription could be a widespread and fundamental process used by cell memory systems to maintain the active state of developmentally regulated genes. Interestingly, the active portion of the human and mouse HOX clusters in primary fibroblasts is, in a very broad pattern, enriched for histone methylation marks usually associated with active regions, encompassing both transcription units and intergenic regions (Bernstein et al. 2005). The authors suggest that in the intergenic portions, representing methylated and transcriptional active chromatin domains, high levels of transcription occur that are maybe needed for the maintenance of HOX gene expression (Bernstein et al. 2005).
Homeotic gene clusters show a characteristic genomic organization in which the arrangement of transcription units on the chromosome mirrors their expression domains along the anterior–posterior axis of the developing embryo and often their timely regulated expression pattern during development (Duboule and Morata 1994; Krumlauf 1994; Zakany et al. 2001). Thus, HOX clusters show a particular spatially and timely coordinated colinearity. Activation of the cluster during development has been interpreted as a multistep process, involving the progressive (colinear) release of the coding units from heterochromatic silencing (Kondo et al. 1998). Retinoic acid (RA) is a well-characterized morphogen involved in this opening process. The addition of RA to tissue culture cells leads to the timely and spatially regulated activation of HOX genes (Boncinelli et al. 1991), but also generally triggers the onset of transcription of noncoding, intergenic RNAs (Cawley et al. 2004). Interestingly, RA induction seems to be counteracted by PcG proteins, antagonizing, e.g., Sonic hedgehog signaling (Bel-Vialar et al. 2000; Barna et al. 2002; Epping et al. 2005; Bracken et al. 2006).
We were interested in the identification of ncRNAs inside HOX clusters that might be used for the maintenance of the active state. Using a previously developed bioinformatic program (AntiHunter; http://bioinfo.crs4.it/AH2.0) capable of identifying opposite strand EST (OS-ESTs) clones directly from the output of dbEST BLAST searches (Lavorgna et al. 2004, 2005), we identified a strikingly large number of OS transcripts in the intergenic regions of human and mouse HOX clusters. Intergenic HOXA OS transcripts were analyzed by RT-PCR in human fetal and adult tissues, revealing characteristic expression profiles and tissue-specific isoforms of these ncRNAs. We show that in many cases OS-intergenic transcripts are coexpressed with adjacent coding HOX genes. Further, by RA induction of embryonic NT2D1 cells we show that expression of OS-RNAs is induced upon morphogen treatment, following the time-regulated colinearity of the _HOX_-sense transcripts. In some cases, OS-RNAs are activated even before neighboring HOX genes. Using immunoprecipitation of in vivo formaldehyde fixed chromatin (X-ChIP), we observed that onset of ncRNA activation in these cells is accompanied by distinct changes in the histone modification patterns and a loss of interaction with the EED/EZH2–PcG repressive complex (PRC2) at the noncoding transcription units. We hypothesize that OS transcription in noncoding, regulative regions is involved in the RA-induced activation process of mammalian HOX clusters, and prevents, as an anti-silencing mechanism, i.e., the re-repression of the cluster.
RESULTS
Opposite strand transcription units in mammalian HOX clusters
To identify intergenic transcripts we searched for noncoding mRNAs originating from the four mammalian HOX loci using the AntiHunter software tool (http://bioinfo.crs4.it/AH2.0) (Lavorgna et al. 2004, 2005). The HOX genes of each cluster are all transcribed in the same orientation. In order not to confuse intergenic transcripts with primary _HOX_-sense transcripts, e.g., for the human HOXC cluster, a long transcript containing _HOXC_5, _HOXC_6, and _HOXC_7 mRNAs has been described (Simeone et al. 1988), we concentrated our attention on noncoding intergenic RNAs transcribed from the “opposite strand” (OS).
First, to filter out repetitive regions, AntiHunter runs RepeatMasker (http://www.repeatmasker.org) on the target sequence. Then, performing a BLAST search on the resulting sequence versus the EST database, the software identifies OS-ESTs with respect to annotated genes. The program gains independent information on the EST strand source by analyzing the splice junctions of the genomic regions matching a spliced EST, and the presence of a poly-A tail in the “3” of the annotated ESTs. Notably, as AntiHunter can tolerate a variable number of bases between an annotated gene and an anti-sense transcript, this can be useful in detecting anti-sense or opposite strand transcripts with respect to genes with only partially characterized 5′ and/or 3′ ends. It can also facilitate the detection of transcribed regulatory regions that originate from intergenic regions.
We identified several transcripts inside all four human and mouse HOX clusters, in many different tissues (both normal and malignant), from adult and fetal samples and from different cell lines (the complete output of the AntiHunter search is available upon request; e-mail: orlando@igb.cnr.it). Several other unspliced intergenic transcripts were found by the same method (data not shown). This in silico mapping revealed a potential overlap between some of the spliced OS-ESTs with _HOX_-sense transcripts. However, many spliced OS-RNAs do not show obvious overlap with HOX genes.
In vivo identification of intergenic OS-RNAs in the HOXA cluster
For in vivo studies on the OS-RNAs identified, we concentrated on the human HOXA cluster. In order to exclude database artifacts, we selected only correctly spliced OS-ESTs. As the sequences corresponding to the acceptor and donor splice sites are highly conserved (99.24% of introns have GT-AG at their 5′-3′ ends) (Lehner et al. 2002), they can be used to verify the DNA strand that is transcribed. Spliced OS-EST clones that do not follow the GT-AG rule probably result from misleading annotations of the EST sequences and were not further investigated.
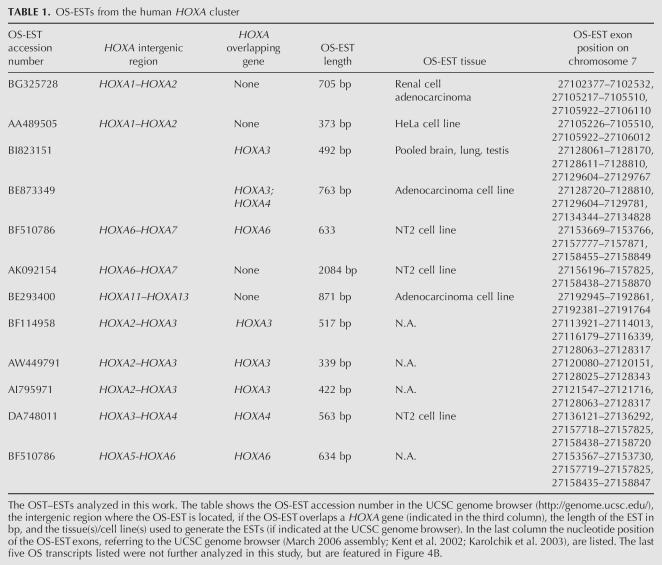
For reverse transcription PCR analysis we selected 6 OS-ESTs in the human HOXA cluster (see Table 1). These are located between HOXA1_–_A2 (OS-ESTs: BG325728, AA489505; see Fig. 1A), between HOXA3_–_A4 (OS-ESTs: BI823151, BE873349; see Fig. 1B), between HOXA6_–_A7 (OS-EST: AK092154; see Fig. 1C), and between HOXA11_–_A13 (OS-EST: BE293400; see Fig. 1D). RT-PCR on total RNA from seven adult tissues and embryonic placenta (from the MVP human normal adult tissue total RNA bank, Stratagene) showed that the OS transcripts are widely expressed and have a characteristic expression pattern (e.g., the transcripts between HOXA1 and HOXA2, between HOXA6 and HOXA7, and near HOXA11) that is often quite similar to that of nearby _HOX_-sense transcripts (Fig. 1). Thus, in many cases expression of HOXA OS-RNAs correlates with the active state of nearby HOXA genes. Some of these OS-ESTs are alternatively spliced, and part of much longer transcripts, e.g., combining exons of BG325728 with exons of BI823151, suggesting that anti-sense transcription starting from the 3′ end can expand far into the cluster (data not shown).
TABLE 1.
OS-ESTs from the human HOXA cluster
FIGURE 1.
Expression patterns of different HOXA genes and OS transcripts in adult human tissues and placenta. (A–D) The relative positions of HOXA (including alternative splice forms, marked by “#”) and OS transcripts are schematically drawn above and below the genomic line, respectively. Exons are indicated by black numbered rectangles. (E) Constitutive MIC-2 expression is shown as a control for the PCR, showing that similar amounts of cDNA were present in the RT reactions for each tissue. MIC-2 (CD99) encodes a T-cell surface anti-gene expressed in most human tissues except spermatozoa.
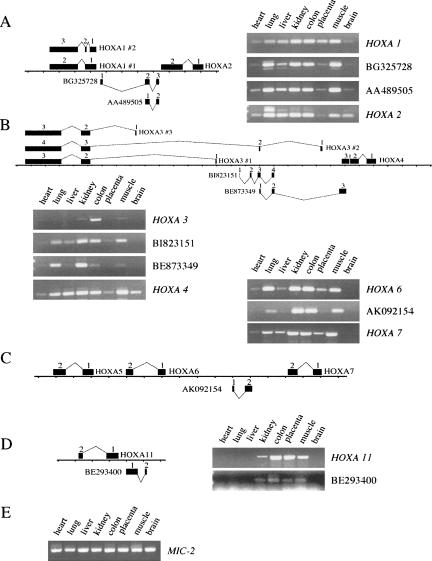
RT-PCR-analysis was repeated with total RNAs from six fetal tissues and analyzed by real-time PCR (Fig. 2). Again, we found tissue-specific expression patterns that in some cases resemble the expression profile of sense transcripts. Strikingly similar are, e.g., the patterns for HOXA6, AK092154, and HOXA7, which are mainly expressed in fetal ovary, kidney, and lung. The expression levels of the intergenic transcripts are comparable to those of _HOX_-sense transcripts. Expression levels of both types of transcripts vary between 50% (HOXA6 in fetal kidney) and below 1% of the level of the MIC-2 standard.
FIGURE 2.
Real-time RT-PCR analysis of different HOXA genes and OS transcripts in fetal human tissues. (A) Expression levels of the HOXA genes 1, 2, 3, 4, 6, and 7 and the ncRNAs BG325728, AA489505, BI823151, BE8733499, and AK092154 in human fetal tissues as percentages of MIC-2 expression. Note change of scale above the red line. (B) Expression levels of the HOXA genes 1, 2, 3, 4, and 7 and the ncRNAs BG325728, BI823151, BE8733499, and AK092154 as in A but with adjusted scale to visualize small peaks. Note change of scale above the red line. Primer pairs used have comparable amplification efficiencies. The mean values from at least two independent experiments are shown. Error bars indicate the standard deviation.
Induction of intergenic OS-RNAs by RA follows HOX colinearity
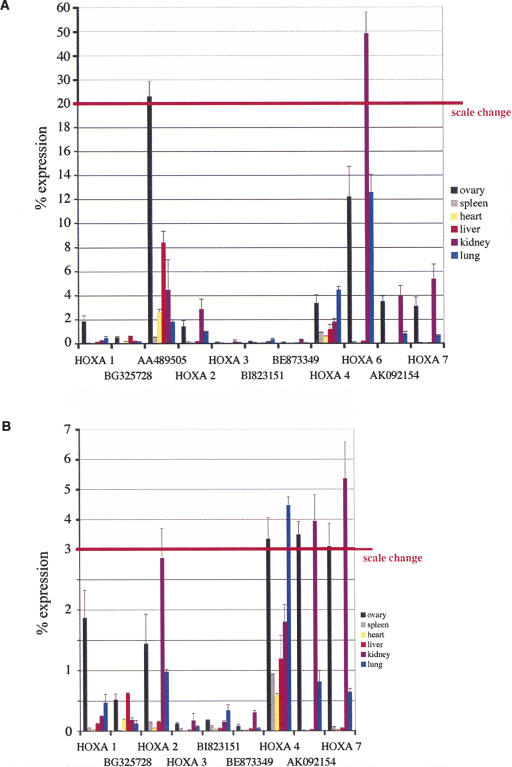
Progressive de-silencing of HOX clusters can be induced by RA. To investigate the expression timing of HOXA OS transcripts, we used the teratocarcinoma cell line NT2D1. These cells have been extensively used to study the expression of HOX genes after treatment with RA (Boncinelli et al. 1991; Simeone et al. 1991). Upon RA treatment the genes of the anterior half of the HOXA cluster start to be expressed in a sequential order that is colinear with their position in the cluster. HOXA1 is expressed first, followed by the progressive activation of the next 5′ genes until HOXA6, which is not found expressed before 48 h of morphogen induction. HOXA7 is active at low levels without RA treatment, but shows also increased expression levels after induction (Simeone et al. 1991).
We analyzed the expression of HOXA genes and intergenic OS-ESTs in both RA-treated and control cells by RT-PCR. The same OS-EST transcripts were chosen, localized between HOXA1_–_A2, HOXA3_–_A4, and HOXA6_–_A7, as in the coexpression study shown in Figure 1. Total RNA was extracted from NT2D1 cells after 0, 1, 3, and 12 h and 1, 2, 3, and 4 d of RA treatment. Results are shown in Figure 3A. We found HOXA genes activated sequentially by RA treatment as previously described (Simeone et al. 1991). Strikingly, also the OS-EST expression follows this temporal colinearity. OS-ESTs start to be transcribed at the same time or even shortly before the neighboring HOX genes, i.e., BG325728 and AA489505 together with HOXA1 and before HOXA2, BI82315 before HOXA3 and HOXA4, and AK092154 before HOXA6. HOXA7 is indeed active in these cells without RA treatment, although the level of its transcription clearly increases upon morphogen treatment. Thus, RA-induced intergenic transcription in the HOXA6–A7 region appears to be related to HOXA6 activation. To get a more refined picture, we repeated this analysis using real-time PCR, also including shorter (15 and 30 min) and longer periods (6, 8, and 10 d) of RA treatment. Figure 3B shows the results that closely resemble the conventional RT-PCR (Fig. 3A). The graphs show the cycle threshold value (CT) from two independent RT-PCR reactions plotted against the length of RA treatment. The threshold cycle value is directly correlated to the amount of template present before PCR amplification. The more template that is present, the lower the number of cycles (the threshold cycle) needed to measure a significant amplification. In most cases, with the exception of HOXA2, HOXA3, and BE873349, very low expression levels of _HOX_-sense transcripts and OS-RNAs also could be detected without RA treatment. We believe that this represents a basal transcription level that might be characteristic for genes that are developmentally regulated, and thus prone to respond fast to activating signals. A low degree of “leakiness,” detectable only with such sensitive methods as real-time PCR, might be the consequence. Only HOXA7 levels are high enough to result in a visible band on a gel stained with ethidium bromide. The threshold cycle number that corresponds to a visible band seems to be around 25 (Fig. 3B, red line). The response to RA induction can be observed when the curves in Figure 3B start to rise. Clearly, both OS-EST transcripts in the HOXA1–2 region are induced before HOXA2. In addition, AA489505 seems to get activated slightly before HOXA1 (after 15 min). HOXA1, AA489505, and BG325728 expression shows a peak around day 2 and goes down later, whereas HOXA2 seems not to reach the peak until day 8. Thus, HOXA2 clearly responds much slower to RA treatment than HOXA1 and the intergenic transcripts >3′. In the region containing HOXA3 and HOXA4 the situation is much more difficult to interpret as all four transcripts analyzed show very similar behavior. Nevertheless, both OS-RNAs are activated together with HOXA3 and HOXA4, BI823151 slightly earlier than both sense transcripts. AK092154 is induced after 3 h of RA treatment, whereas HOXA6 responds after 6–12 h (showing, though, a much higher starting expression level). AK092154 passes a CT of 25 after 2 d, whereas HOXA6 does not reach this level before day 3, as can be seen also in the gel (Fig. 3A). Again, the intergenic transcript responds much faster to RA treatment than the _HOX_-sense transcript. HOXA7 is expressed at “visible” levels from the beginning, as expected, but also shows a response to RA treatment after 6 h, leading to a significant increase in expression.
FIGURE 3.
RT-PCR analysis of HOXA genes and OS transcripts in NT2D1 cells. (A) NT2D1 cells were treated with retinoic acid (RA) for 1, 3, 6, and 12 h and 1, 2, 3, and 4 d. Total RNA was extracted from treated and untreated (no RA) cells and analyzed by RT-PCR. Note that HOXA7 is constitutively expressed in NT2 cells. Constitutive MIC-2 expression is shown as the loading control. (B) Analysis of HOXA and ncRNA transcription by real-time RT-PCR. NT2D1 cells were treated with retinoic acid (RA) for 15 and 30 min; 1, 3, 6, and 12 h; and 1, 2, 3, 4, 6, 8, and 10 d. Total RNA was extracted from treated and untreated (no RA) cells and analyzed by RT and PCR amplification on a LightCycler. The cycle threshold numbers of at least two independent experiments are plotted against RA-incubation time. The cycle threshold numbers are reciprocally correlated to the amount of template at the start of linear amplification. MIC-2 expression does not change upon RA treatment (light blue line at threshold cycle 16). The red line indicates threshold cycle 25, which corresponds approximately to a visible amplification product in an ethidium bromide-stained gel. Primer pairs used have comparable amplification efficiencies. The mean values from at least two independent experiments are shown. Error bars indicate the standard deviation.
RA response elements, CpG elements, and OS-transcripts—A bioinformatic analysis
RA is thought to activate transcription of HOX genes by interacting with members of the nuclear RA receptor family (RAR α, β, γ), and the Retinoid X receptor family (RXR α, β, γ). Heterodimers of both receptors bind to recognition sites near the genes regulated by RA, known as retinoid response elements (RAREs). In the absence of ligand, receptors are potent repressors, recruiting histone deacetylases and co-repressor complexes. Binding of RA leads to the displacement of repressive complexes, the targeting of histone acetyl transferases and coactivators, and finally, the activation of the HOX targets (Xu et al. 1999; Gronemeyer and Miturski 2001; McKenna and O'Malley 2002). As mentioned before, HOX genes are repressed by PcG complexes, most likely by counteracting RA activation (Bel-Vialar et al. 2000; Barna et al. 2002; Epping et al. 2005; Bracken et al. 2006). In Drosophila, reactivation of PcG-repressed genes is accompanied by intergenic transcription events in intergenic, regulative DNA elements encompassing large regions of the homeotic loci (Bae et al. 2002; Drewell et al. 2002; Rank et al. 2002; Schmitt et al. 2005). In mammalian HOX clusters, such regions, important for the activation of homeotic genes, most likely would contain RAREs. Thus, in order to see if the sites of OS transcription would run through RAREs, we have screened the part of the HOXA cluster activated by RA in NT2 cells (HOXA1–HOXA7) for potential RAREs using the Matinspector tool (Werner 2000) by Genomatix (http://www.genomatix.de).
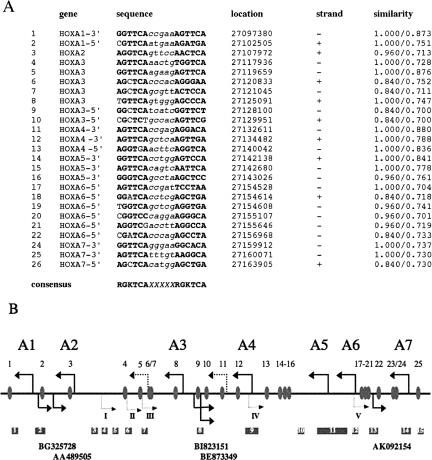
We identified 25 potential sequences of the DR5 class (see Fig. 4A), some of which have been described before (Mainguy et al. 2003, and references therein). Only two of these RAREs have been functionally tested in vivo (RAREs 1 and 13) (Mainguy et al. 2003); the others we consider thus as “potential” RARES, and their position has been analyzed with respect to the HOX coding units and the OS transcripts identified (Fig. 4B). Interestingly, the intergenic regions containing OS-transcripts, HOXA1–2, HOXA3–4, and HOXA6–7, also show potential RAREs. Upon activation of OS transcription, these presumptive retinoid-sensitive regions would be transcribed. Other predicted RAREs reside in the HOXA3 gene body (RAREs 4–8), in the intergenic region between HOXA4 and HOXA5 (RAREs 13–16), 3′ of HOXA1 and 5′ of HOXA7. Two clusters of potential RAREs reside in 3′ of HOXA3 and 5′ of HOXA6. This clustering of potential RAREs is significantly above the average number of sites to be expected in a random sequence with the same base composition (see Materials and Methods). Both regions are also characterized by several OS transcripts. The regions between HOXA2 and HOXA4 and between HOXA6 and HOXA7 contain additional OS transcripts that we have identified by AntiHunter, but did not choose to further analyze due to the partial overlap with protein coding HOXA transcripts (see Table 1). Thus, in these regions many predicted RAREs would also be transcribed by activated OS transcripts.
FIGURE 4.
In silico analysis of retinoid response elements and CpG islands (RAREs) in the human HOXA cluster. Potential consensus sites for RAR–RXR heterodimers of the DR5 class were identified using the Matinspector software. The 25 RAREs found are shown in A. In bold, conserved nucleotides are indicated (see the consensus at the bottom of the figure); in italics, the 5mer spacer region. In addition, the chromosomal location (according to the UCSC genome browser, assembly March 2006), the strand and the values for core and matrix similarities (from the Matinspector analysis) (Werner 2000) are shown. (B) The location of the potential RAREs in the HOXA cluster (gray ovals) is shown. The first exon of each HOX gene is indicated by a thick black arrow. The two black dotted arrows refer to alternative exons of HOXA3. Arrows below indicate the first exons of the OS transcripts particularly featured in this report. Dotted arrows refer to mapped OS transcripts, not further analyzed in this study (I=BF114958, II=AW449791, III=AI795971, IV=DA748011, V=BF510786; see Table 1). Below CpG islands are shown (dark gray squares).
In addition, the CpG islands in this region of the HOXA cluster, as mapped by the UCSC Genome Browser (Kent et al. 2002; Karolchik et al. 2003), are shown in Figure 4B. In most (but not all) cases the presumptive transcription start sites of HOX genes and OS transcripts reside in or near CpG islands. Very striking is the CpG island between HOXA1 and BG325728 (No. 2 in Fig. 4B), which is centered in the region where the promoters of both transcription units most likely reside, suggesting a bidirectional promoter region. Bidirectional promoters are usually associated with CpG islands, whereas only 60% of unidirectional promoters show this association in mammals (Engström et al. 2006).
Taken together, these data suggest that many regions of the HOXA cluster with in silico-predicted RAREs are transcribed by OS transcripts during the activation of the cluster, which might have an important role in the RA-induced activation process and the maintenance of the activated state. Transcription through such RARE-containing regions further downstream of the respective OS transcription start might help to displace repressive complexes, to change the epigenetic state of the chromatin environment, and to recruit coactivators and chromatin remodeling factors.
Changes in histone methylation patterns and PcG presence upon RA treatment of NT2D1 cells
Using immunoprecipitation of formaldehyde fixed chromatin (X-ChIP) with antibodies specific for modified histone variants and three PcG proteins, Bracken and coworkers recently showed that repressive methylation marks and PcG proteins are present at inactive HOXA genes in NT2 cells and that they disappear upon activation by RA (Bracken et al. 2006).
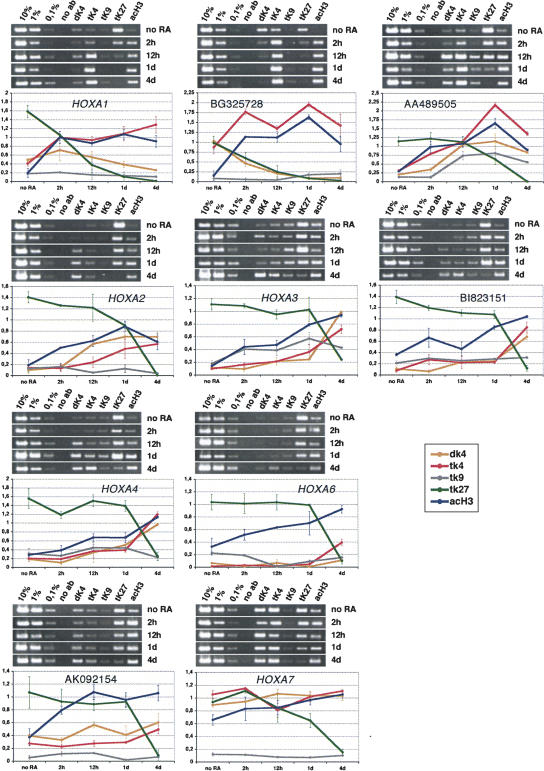
In order to see if intergenic regions containing OS transcripts show similar behavior to HOX coding genes, we examined histone H3 methylation patterns and PcG protein interactions in NT2D1 cells treated with RA for 2 h, 12 h, 1 d, and 4 d. The cells were cross-linked, chromatin was prepared and used first for immunoprecipitation with sera against histone H3 dimethylated at lysine 4 (H3K4me2), histone H3 trimethylated at lysine 4 (H3K4me3), histone H3 trimethylated at lysine 9 (H3K9me3), histone H3 trimethylated at lysine 27 (H3K27me3), and acetylated histone H3 (H3K9/18ac; this sera recognizes histone H3 acetylated at lysine 9 and/or 18) (see also Breiling et al. 2001).
Di- and trimethylation of lysine 4 of histone H3 is considered a mark for transcriptional activity, as well as acetylation of lysines 9 and 18 of this histone. Trimethylation of K4 has been found to be specific for transcriptional active genes of all four HOX clusters in promonocytic U937 cells (Guenther et al. 2005). In contrast, trimethylation of lysines 9 and 27 of histone H3 are mostly repressive marks that are used for interactions with silencing complexes like those of the PcG of proteins (for a summarizing review on histone modifications, see Peterson and Laniel 2004). Immunopurified DNA was analyzed by PCR, using primer pairs specific for the coding portion of HOXA genes and OS transcripts under investigation. Primers of ∼400 base pairs were designed that contained the start of the first exon of the respective transcription units (see Fig. 1 for a schematic representation of the regions). In some cases, when no primer pair that amplifies well under the established conditions (see Materials and Methods) was found, primers around the second exon were chosen (HOXA3, HOXA4, HOXA6). For BI823151 and BE873349, a primer pair containing the third exon of BI823151 and the first exon of BE873349 was chosen. Similar results as presented below were obtained with a primer pair containing the first exon of BI823151 (data not shown).
As shown in Figure 5, we observe clear changes in histone modifications that correlate with the timely and colinear activation of the respective genes. In general, levels of di- and trimethylation of K4 as well as H3-acetylation increase, whereas trimethylation of lysine 27 of H3 is reduced in a reciprocal manner. Trimethylation of lysine 9 of histone H3 is usually a characteristic of silent heterochromatic regions, but it also has been found to be enriched in active genes (Vakoc et al. 2005). As we did not observe significant changes in H3K9me3 upon activation, we conclude that this methylation mark might play a dual role in _HOXA_-transcriptional regulation. This is in accordance with previous findings that showed that lysine 9 dimethylation of histone H3 is found at very low levels in promoter regions of HOXB genes, and no increase of these is observed upon gene activation by RA (Chambeyron and Bickmore 2004). All the OS transcripts under investigation show a similar profile. BG325728 shows a very striking behavior, which already shows a high level of lysine 4 methylation without RA treatment, and which might indicate that this region is already prone to transcription before morphogen treatment.
FIGURE 5.
Histone modifications in the HOXA cluster. ChIP analysis of histone modifications of untreated NT2D1 cells (no RA) and of cells treated for 2 h, 12 h, 1 d, and 4 d with retinoic acid (RA). Chromatin was precipitated with the antisera indicated above the gel photographs in each panel (dK4=histone H3 dimethylated at lysine 4, tK4=histone H3 trimethylated at lysine 4, tK9=histone H3 trimethylated at lysine 9, tK27=histone H3 trimethylated at lysine 27, acH3=acetylated histone H3). Immunopurified DNA was analyzed by PCR using primer pairs near or in the first exon of the indicated transcription units. PCR reactions with 10%, 1%, and 0.1% of the input DNA were loaded to determine the linear range of amplification and to use for normalization during quantification (see Materials and Methods). Below the gel photographs a quantification of two independent ChIP experiments is shown. The values on the y-axis represent the intensities of immunoprecipitated DNA bands as a percentage of the intensities of the input DNA bands. The graphs show the changes in histone modifications after the different periods (untreated, 2 h, 12 h, 1 d, and 4 d) of retinoic acid treatment (dK4=orange 4, tK4=red, tK9=gray, tK27=green, acH3=dark blue). The mean values from at least two independent experiments are shown. Error bars indicate the standard deviation.
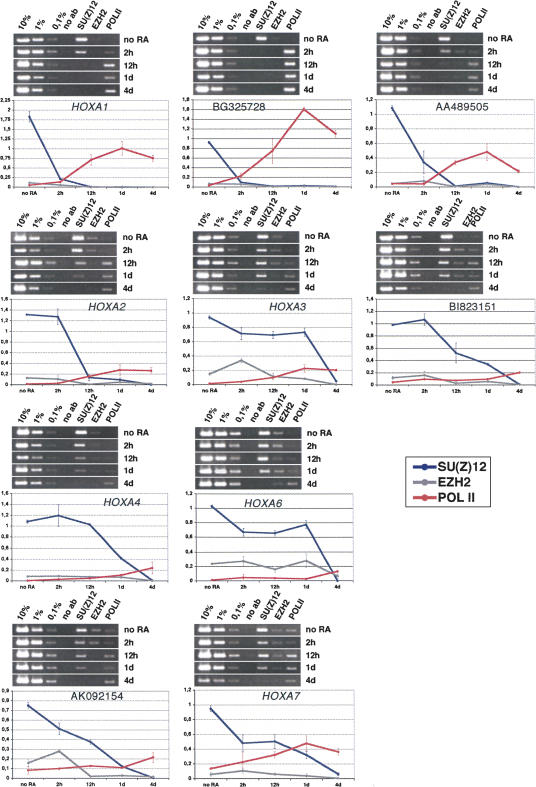
To monitor the presence of PRC2 components, we repeated the ChIP analysis using antibodies specific for SU(Z)12, EZH2, and as a mark for activated transcription, RNA-polymerase II (POL II). SU(Z)12 is part of the EED–EZH2 complex (the Polycomb repressive complex 2/PRC2), which also contains the histone methyltransferase Enhancer of Zeste 2 (EZH2), responsible for the PcG-specific trimethylation of lysine 27 of histone H3 (Cao et al. 2001; Cao and Zhang 2004). We found a clear correlation of the H3K27me3 modification and SU(Z)12 and EZH2 presence, indicating, that all the genes under investigation, in particular the OS transcripts, are repressed by the EED–EZH2 complex (Fig. 6). The levels observed for EZH2 are generally very low. The antibody against EZH2, although successfully used for X-ChIP (Caretti et al. 2004), seems to perform rather poorly under the conditions of our protocol. Nevertheless, the results for this protein clearly show the same tendency as those observed for SU(Z)12, i.e., a distinct reduction after RA treatment. Thus, upon activation of HOX genes by RA, the interaction with the SU(Z)12–EZH2 complex is lost, paralleled by methylation of lysine 4 and acetylation of histone H3. In addition, we observe increasing levels of RNA-polymerase II upon RA treatment.
FIGURE 6.
PRC2 components in the HOXA cluster. PCR analysis with cross-linked and immunoprecipitated DNA from untreated NT2D1 cells and cells treated for 2 h, 12 h, 1 d, and 4 d with retinoic acid (RA). Chromatin was precipitated with the antisera against SUZ(Z)12, EZH2, and POLII (as indicated above the gel photographs in each panel). PCR reactions with 10%, 1%, and 0.1% of the input DNA are loaded to determine the linear range of amplification and to use for normalization during quantification. Below the gel photographs quantitative analyses of the ethidium bromide-stained bands from two independent experiments are shown. The values on the y-axis represent the intensities of immunoprecipitated DNA bands as a percentage of the intensities of the input DNA bands (SU(Z)12=dark blue, EZH2=gray, POLII=red). The mean values from at least two independent experiments are shown. Error bars indicate the standard deviation.
In a study of the mouse HOXD4 locus, Rastegar and coworkers have shown that activation of HOXD4 by RA is preceded by chromatin opening (H3 lysine 4 methylation and H3 and H4 acetylation) at a 3′ enhancer region containing a RARE. Upon RA induction POL II is recruited first to the enhancer element and only later to the HOXD4 P1 promoter, suggesting that in this system a transcription event through an intergenic regulative element, slightly preceding the activation of the major transcription unit, also takes place during the activation process (Rastegar et al. 2004). In some cases we find the polymerase present before we actually observe a transcript (e.g., HOXA2, HOXA3, BI823151, AK092154, or HOXA6), although a clear increase in protein levels was observed when transcription was activated. Nevertheless, low levels of POL II have also been reported for repressed promoters from the Drosophila bithorax complex (Breiling et al. 2001, 2004; Dellino et al. 2004), indicating that PcG repression creates a chromatin environment that inhibits the correct engagement and progressive movement of the polymerase and associated proteins but not their binding.
Cis or trans effects of intergenic transcripts?
An important question is whether the transcripts, themselves, that we have identified have a role in HOX gene activation. In a first attempt, we analyzed whether or not the OS transcripts we have identified are evolutionarily conserved. In collaboration with the FANTOM-3 and RIKEN genome research groups we have performed sequence comparisons between human and mouse genomes, which showed that the HOX clusters show sequence identity of >75%, and that both human and mouse HOX clusters code for a large number of OS transcripts (Carninci et al. 2005; Engström et al. 2006). Nevertheless, although some of these ESTs occupy corresponding locations, none of them, including the OS-RNAs examined in this study, show conservation across species. The same holds true for OS-RNAs we identified near the promoter regions of dispersed HOX loci (Engström et al. 2006).
Nevertheless, we have tested the hypothesis that intergenic HOXA OS-RNAs may act in trans. To this aim, we chose a region between HOXA3 and HOXA4 in which two OS-RNAs were identified (BI823151 and BE873349). We transfected nontreated NT2 cells with in vitro-transcribed sense and the respective anti-sense RNA of BE873349. No HOX gene expression could be detected in these cells within 2 d after transfection (data not shown). The same negative result was obtained when the BE873349 transcript, under control of the CMV promoter, was constitutively expressed in undifferentiated NT2D1 cells. In both cases we could not observe any induction effect (data not shown). We thus conclude that the intergenic transcripts we have identified have no direct role in HOX gene activation, which is sustained by the finding that these transcripts are not conserved. We cannot exclude the possibility that additional factors are needed for successful activation, and that some targeting by OS transcripts (maybe others than the ones analyzed by us) is needed for this. So far, though, we favor the idea that it is the transcription event per se that is of importance and not the transcript itself.
DISCUSSION
Intergenic transcription and epigenetic regulation at homeotic clusters
From our analysis we conclude that the HOXA cluster shows a highly complex profile of intergenic, noncoding transcripts. Computational analysis of all other human and murine HOX clusters shows an as intriguing picture. RT-PCR analysis performed in fetal and adult tissues revealed a correlation between the expression of HOXA OS-RNAs and transcription of HOX genes that follows the same colinear pattern. In addition we show that the expression of HOXA OS-RNAs is regulated by the RA morphogen and that it correlates with the loss of by PcG proteins, suggesting that it might have a role in gene regulation.
This is not restricted to the HOX clusters. We have also recently mapped noncoding transcripts near many dispersed human and mouse homeotic loci (Engström et al. 2006). In general, a large portion of the mammalian genome can give rise to transcripts from both strands, often forming coregulated sense–anti-sense pairs (Katayama et al. 2005). The in vivo binding sites for transcription factors like SP1, cMYC, and p53, as identified by ChIP on CHIP experiments, are localized in a large number of genomic regions that do not always fit with the positions of protein coding genes (Cawley et al. 2004). In order to correlate the binding profile of these factors with regulated transcription, RNA profiles in differentiating pluripotent human germ cells were investigated. This led to the identification of a variety of genomic regions that produce ncRNAs, most of them surrounding the transcription units of inducible, protein-coding mRNAs (Cawley et al. 2004; Kampa et al. 2004). Intergenic, noncoding transcription is also characteristic of loci harboring multigene families like the globin and immunoglobulin genes (Ashe et al. 1997; Gribnau et al. 2000; Bolland et al. 2004). In particular, it has been proposed that intergenic transcription in the globin gene cluster may contribute to the opening of large chromatin domains necessary for the proper developmental regulation of the globin genes (Gribnau et al. 2000). Similar conclusions were drawn for anti-sense transcription events observed throughout the IghV region in early B cells and VDJ recombination (Bolland et al. 2004). More recent data suggests, though, that the situation is more complicated. Intergenic transcription in the human β-globin locus is increased in cells deficient in the RNAseIII-family protein Dicer, a component of the RNA interference machinery (Hausecker and Proudfoot 2005). Thus, intergenic transcription events also could be indicative of RNAi-related silencing processes, where they are the source of Dicer-dependent small RNA molecules that might be used for the targeting of silencing complexes (Hausecker and Proudfoot 2005). Taken together, transcription of intergenic (and most likely noncoding) regions containing regulatory DNA elements seems to be a developmentally controlled event necessary for the reorganization and maintenance of active or repressive chromatin structures. We thus propose that these regions play a role in the epigenetic regulation of the cluster.
In fact, we show that the transcription units of HOXA coding genes and OS-RNAs before RA induction are targets of the PcG complex PRC2 and are characterized by PcG-specific histone modification patterns, suggesting that the repressed state of these ncRNAs is maintained—as one of the HOX coding genes—by PcG complexes. Interestingly, we found several potential RAREs predicted in the intergenic regions coding for OS transcripts. This suggests that these regions have some features of regulative, epigenetic DNA elements and that transcription through them might have an important role in RA-induced activation and maintenance, counteracting PcG repression.
As already mentioned, noncoding transcription has been linked to derepression of PcG targets in Drosophila. Several intergenic transcripts have been identified in the homeotic Bithorax complex (BX-C), which corresponds to the central part of the mammalian HOX clusters (Bae et al. 2002; Drewell et al. 2002; Rank et al. 2002; Schmitt et al. 2005). As in mammals, spatial expression of BX-C genes along the anterior–posterior axis of the embryo reflects their 3′-5′ order in the chromosome. Systematic RNA in situ hybridization experiments highlighted the presence of several putatively independent noncoding transcripts spanning most intergenic regulatory regions of the BX-C. Interestingly, the expression domains of these ncRNAs are restricted to the same tissues in which adjacent BX-C genes are expressed. The expression of some of these ncRNAs through epigenetic DNA elements, PREs, correlates with the maintenance of long-term memory of BX-C gene expression (Rank et al. 2002; Schmitt et al. 2005). Some of these transcripts apparently have a direct role in gene activation (Sanchez-Elsner et al. 2006). They are bound by the HMT ASH1 and serve to recruit this protein to the PRE. Transgenic expression of these transcripts is able to activate the target gene, regulated by the PRE from which they stem (Sanchez-Elsner et al. 2006). In our case, we have no indication that the OS transcripts we have identified in the human HOXA cluster have a similar function. Neither the transfection of NT2 cells with OS-RNA, nor the expression of noncoding transcripts in trans had an effect on HOX gene activation. Nevertheless, there are many more noncoding transcripts in the HOX clusters, and some of these—not among those we have analyzed in more detail—might show trans effects. Importantly though, the OS transcripts in the HOX clusters show no conservation across species (Engström et al. 2006), which argues against a direct role of these transcripts despite the fact that HOX clusters are among the most conserved loci in mammals. Thus, we suggest that the transcription event in cis may have a mechanistic function in the activation and maintenance of the cluster. In this context the identification of ncRNA and PcG protein binding sites may be diagnostic of the presence of PRE-like DNA elements in the mammalian HOX clusters.
The role of ncRNAs in the regulation of HOX clusters
What is then the mechanistic role of intergenic transcription? A naturally occurring case of loss of function of such a kind of transcription has been reported again from the globin gene cluster, resulting in the failure of proper activation of the adult _δ_- and γ-globin genes (Calzolari et al. 1999). Gribnau and colleagues showed that the production of intergenic transcripts is necessary for chromatin remodeling and maintenance of the active state of adult globin genes (Gribnau et al. 2000). Our results show that in fetal and adult tissues intergenic OS transcripts and nearby HOXA genes are co-expressed. Moreover, upon RA-induced activation of the HOXA cluster in NT2D1 cells, we observed the persistence of the production of OS-RNAs together with HOX gene transcription. This also suggests that in the case of HOX clusters, noncoding transcription correlates with progressive and long-term expression of coding mRNAs, most likely preventing re-repression of these genes (anti-silencing).
Elegant work from Wendy Bickmore's laboratory showed that RA-induced activation of the mouse HOXB cluster (in embryonic stem cells and embryos) appears to involve a primary step in which the cluster is decondensed. Gene activation occurs only subsequently (Chambeyron and Bickmore 2004; Chambeyron et al. 2005). Decondensation is accompanied by changes in the higher-order chromatin structure and in histone modifications. Nevertheless, changes in histone modifications alone, e.g., induced by treatment with the histone deacetylase inhibitor trichostatin A, which leads to hyperacetylation of histone tails (an active mark), are not sufficient to induce gene activation and structural changes (Chambeyron and Bickmore 2004). Thus, the activation of HOX clusters is a multilayered process, involving activator binding (RAR/RXR), chromatin remodeling and decondensation, changes in histone marks, and higher-order structural changes. We propose that intergenic transcription would be part of the decondensation process during HOX gene activation.
Using ChIP analysis we show that upon RA induction specific changes in histone modifications occur in the “coding” regions of the intergenic OS transcripts that we have identified. These changes are accompanied by a loss of interaction with the EED/EZH2–PcG complex, which otherwise appears to control these regions. Thus, our data indicate that an activating rearrangement of chromatin in parallel with a reduced contact with the PcG complexes takes place near the promoter regions of HOX genes and in nearby intergenic, transcribed regions containing retinoid response elements. An attractive hypothesis would be that RA-triggered intergenic transcription may be fundamental for the decondensation of the cluster, setting the stage for the activation of HOX genes and, most importantly, preventing re-repression of the region.
Further studies are needed to show that by abolishing transcription through regulative regions activation or maintenance of nearby protein coding genes is disturbed. This is, though, complicated by the fact that the actual promoter structure of these transcription units is very complex (Carninci et al. 2005). It was shown in Drosophila that artificially induced transcription through an intergenic Polycomb response element prevents the establishment of PcG repression (Schmitt et al. 2005). Thus, gene activation and anti-silencing by nonprotein coding transcription units seem to be fundamental aspects of cellular memory systems in many species. Our results suggest a dynamic process conserved in Drosophila and mammals occurring at homeotic gene clusters, which involves the production of ncRNA and the displacement of PcG proteins, resulting in an epigenetic switch of gene activity, and has a potential impact on the higher-order structure of the region.
MATERIALS AND METHODS
Treatment of NT2D1 cells with RA
The human embryonic carcinoma cell line NT2D1 was maintained in Dulbecco's modified minimal essential medium supplemented with 10% fetal bovine serum, 20 mM HEPES Buffer, 2 mM Glutamine, 200 U/mL Penicillin, and 200 μg/mL Streptomycin (all from Invitrogen) in a humidified atmosphere of 5% CO2 in air. NT2/D1 cells were induced to differentiate with 10 μM all-trans retinoic acid (RA, Sigma). Cells were seeded at a density of 106 cells/75 cm2 tissue culture flask and re-fed every 24 h with fresh medium containing RA.
RNA samples
Adult total RNA samples (heart, lung, liver, kidney, colon, placenta, muscle, and brain) were gifts from Massimo Cocchia (TIGEM), and derive from the MVP human normal adult tissue total RNA bank from Stratagene. Fetal total RNA samples (ovary, heart, and liver from a week 15 Meckel Syndrome abortion; spleen, kidney, and lung from a week 20 Down Syndrome abortion) were obtained from Lucio Nitsch (bank of fetal biological samples, Telethon). Total RNA from NT2 tissue culture cells and mouse embryos was prepared using the Trizol reagent (Invitrogen).
RT-PCRs
Reverse transcription was performed with M-MLV Reverse Transcriptase (Ambion) using DNAse treated total RNA as template (1–2 μg per reaction) and a 16mer polydT primer in the supplied buffer or using the oligo-(dT)18 RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas). The cDNA (200 ng) was PCR-amplified using EuroTaq DNA polymerase (Euroclone) in the supplied buffer. PCR conditions: 1 cycle: 94°C×1 min; 35 cycles: 94°C×1 min/60°C×1 min/72°C×1 min; 1 cycle: 72°C×5 min.
Real-time RT-PCR was performed using the QuantiTect Reverse Transcription Kit by Quiagen. One microgram of total RNA was processed in 20 μL reactions according to the manufacturer's specifications, using the primer mix provided, or using 20 pmol of strand-specific primers. One microliter of the cDNA was used for a 20μL PCR reaction using the QuantiTect SYBR Green PCR Kit by Quiagen and a DNA engine Opticon 2 lightcycler by MJ Research. PCR conditions: 1 cycle: 95°C×15 min; 45 cycles: 95°C×15 sec/60°C×30 sec/72°C×1 min, read; melting curve 65°C to 95°C, read every 1°C. Cycle threshold numbers for each amplification were measured with the OPTICON monitor software provided by MJ Research. For this assay, primer pairs of equal efficiency were used (the graphs obtained by plotting CT values against template concentration result in parallel lines; thus, the slopes of the mean regression lines drawn through the data points are similar; data not shown).
RT-PCR primers (expected amplicon length cDNA versus genomic DNA is indicated):
- BG325728U 5′-CCTCCCCACCGTTCAATG-3′ 211 bp/2897 bp
- BG325728L: 5′-TGCTCCCTACCTTCCCTC-3′
- AA489505U 5′-AAACCAGCCATAGTCCCCACACTC-3′ 281 bp/691 bp
- AA489505L 5′-TTCAACCCCCTCCCCCATAAATCC-3′
- BI823151U 5′-GAAACGCCAAGACATAGAAAAC-3′ 236 bp/675 bp
- BI823151L 5′-GTTGGAGACAATTCCTGGTTC-3′
- BE873349U 5′-GGCGAAGAGTTTAGGAGCAG-3′ 240 bp/1030 bp
- BE873349L 5′-GCAGAACTTCTCCAAGCCAG-3′
- AK092154U 5′-CCACGCACCTATTCCCCC-3′ 222 bp/816 bp
- AK092154L 5′-GCTTTGCAACTCGCAACC-3′
- BE305073U 5′-TTCTTTCGCTTCCTCCAAAC-3′ 126 bp/544 bp
- BE305073L 5′-TCTTCTGGAGCTGAGGACTC-3′
- HOXA1U 5′-GCCGTACTCTCCAACTTTC-3′ 221 bp/673 bp
- HOXA1L 5′-CTCGCCTCAATACATTCACC-3′
- HOXA2U 5′-CACTTGTCTCTCAGTCAAATCC-3′ 252 bp/892 bp
- HOXA2L 5′-AGAAAACCGCACTTCTGCC-3′
- HOXA3U 5′-GCGATCTACGGTGGCTACC-3′ 770 bp/2100 bp
- HOXA3L 5′-GCTGCGACTTGGAGACTGG-3′
- HOXA4U 5′-CATTCTCCGGTTCTGAAACCAGATC-3′ 224 bp/774 bp
- HOXA4L 5′-TGTACCCCTGGATGAAGAAGATCC-3′
- HOXA6U 5′-TGGGCTGCGTGGAATTGATGAGC-3′ 250 bp/1380 bp
- HOXA6L 5′-GATGCAGCGCATGAACTCCTGCG-3′
- HOXA7U 5′-TCATTCCTCCTCGTCTTCC-3′ 351 bp/1296 bp
- HOXA7L 5′-GCCAATTTCCGCATCTACC-3′
- HOXA11U 5′-AACCAGATTTTGACTTGACGATCAGTG-3′ 444 bp/1900 bp
- HOXA11L 5′-ATTTCTATAGCACCGTGGGCAG-3′
- MIC2U 5′-TCTCCATGTCCACCTCCCCT-3′ 365 bp/n.d.
- MIC2L 5′-ACCCAGTGCTGGGGATGACT-3′
Statistical test for RARE enrichment in the HOXA cluster
In order to test if the enrichment of RAREs predicted by Matinspector (www.genomatrix.de) in several regions of the HOXA cluster is significant, we created random sequences with the same base content of the eight regions of the cluster we scanned for RAREs (3′-HOXA1, HOXA1–HOXA2, HOXA2–HOXA3, HOXA3–HOXA4, HOXA4–HOXA5, HOXA5–HOXA6, HOXA6–HOXA7, and 5′-HOXA7). These were created based on the wild-type sequences using the shuffleseq tool on the EMBOSS Web page (http://bioweb.pasteur.fr/docs/EMBOSS/shuffleseq.html). Twenty random sequences for each region were analyzed manually for RAREs of the DR5 type with Matinspector, and for the wild-type sequences. The average number of hits for each region was calculated and compared to the wild-type situation. In two cases (HOXA2–HOXA3 and HOXA6–HOXA7) the sequences showed an enrichment of RAREs that is 2 SD above the average distribution in random sequences, which we considered significant (the output of the statistical test is available upon request; e-mail: orlando@igb.cnr.it).
ChIP analysis
NT2D1 cells were seeded in large cell culture dishes and grown for 4–5 d to confluency. They were either not treated with RA or treated with RA 2 h, 12 h, 1 d, or 4 d before fixation. Cells were fixed in cell culture dishes by adding fixation solution directly to the medium. Cells were fixed at room temperature for 10 min. Fixed cells were washed once with PBS, scratched of the plates, and transferred into falcon tubes. Chromatin was then prepared as described previously (Breiling et al. 2004).
The antibody against POL II derives from Covance (8WG16), against SU(Z)12 from Abcam (ab 12,201), and the antibody against EZH2 was obtained from Zymed (36–6300). Antisera to H3 di- or trimethylated at K4 and trimethylated at K9 were produced as described (Breiling et al. 2004). Antisera against acetylated histone H3 were produced as described (Breiling et al. 2001). Antibodies against trimethylated K9 and K27 were obtained from T. Jenuwein.
PCR analysis was performed as described (Breiling et al. 2004). Primer pairs:
- HOXA1–I 5′-CGTCTGCTTCCTGATTTAACG-3′
- hOXA1–II 5′-CCATTGGCTGACACCGTCAC-3′
- BG325728–I 5′-CCGATTTGGAGTGCTGGAGC-3′
- BG325728–II 5′-CACAGCCCCACGGTCTCCC-3′
- AA489505–I 5′-CCTGAACTGTTCATGGCATCC-3′
- AA489505–II 5′-CTGCGGCGGCGGAGTGTG-3′
- HOXA2–I 5′-GAAAAGATGTCAGGCACTCAG-3′
- HOXA2–II 5′-CTCGCCCCCACGTACTCC-3′
- HOXA3–I 5′-CGGGCTGAGGCGGCTGTG-3′
- HOXA3–II 5′-TTGAGAGTAAGATGGATTTGCG-3′
- BI823151–I 5′-CGCTGGTTTTGGGGTAGG-3′
- BI823151–II 5′-AAAGCAAGCATTCACATTCTCC-3′
- HOXA4–I 5′-CAGAACCCCGAAATAGAAGGC-3′
- HOXA4–II 5′-GTACCCCTGGATGAAGAAGATC-3′
- HOXA6–I 5′-TCCCCACAAAAACACGACCTG-3′
- HOXA6–II 5′-TAGCGGTTGAAGTGGAACTCC-3′
- AK092154–I 5′-CAAGAATAGGCACCCCAGACC-3′
- AK092154–II 5′-AATCGGAGAGACTACCTGTAG-3′
- HOXA7–I 5′-GGGTTTGGTGTAAATCTGGGG-3′
- HOXA7–II 5′-CTCGTCCGTCTTGTCGCAGG-3′
PCR-Gels were quantified as described (Breiling et al. 2004) with the QuantityOne software (Biorad) and the results of two to three independent experiments were plotted in diagrams as percentage of the input (normalized against the input).
Transfection and expression of ncRNAs
The cDNA for BE873349 was cloned into the pCMV–Sport6 vector, which allows strand-specific expression from the T7 or SP6 promoter, respectively. Sense and anti-sense RNA were transcribed in vitro using the Megascript T7 and SP6 kit (Ambion). Four micrograms of each transcript were then used for transfecting NT2 cells with the TransMessenger system (Qiagen), according to the manufacturer's specifications. Cells were harvested after 0, 12, 24, 36, and 48 h, and total RNA was prepared and retrotranscribed as described above. In parallel, the coding sequence of BE873349 was cloned in both orientations into the plasmid pcDNA3.1, which allows constitutive expression under the control of the CMV promoter. NT2 cells were transiently transfected with the liposomal transfection reagent Metafectene (Biontex Laboratories) according to the manufacturer's specifications. After 0, 12, 24, 36, and 48 h total RNA was prepared and retrotranscribed as described.
ACKNOWLEDGMENTS
We thank Lucio Nitsch for providing RNA from human fetal tissues (bank of fetal biological samples, Telethon), Piero Carninci and the FANTOM-3 and RIKEN genome research groups, and Antonio Simeone and Vincenzo Zappavigna for stimulating discussions. This work was supported by grants from Fondazione Telethon (TCP00094), La Compagnia di San Paolo, Ministero dell'Istruzione, dell'Università e della Ricerca FIRB-MIUR, to V.O.
Footnotes
REFERENCES
- Ashe, H.L., Monks, J., Wijgerde, M., Fraser, P., Proudfoot, N.J. Intergenic transcription and transinduction of the human β-globin locus. Genes & Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, E., Calhoun, V.C., Levine, M., Lewis, E.B., Drewell, R.A. Characterization of the intergenic RNA profile at abdominal-A and abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 2002;99:16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies, F., Cavalli, G. Cellular memory and dynamic regulation of Polycomb group proteins. Curr. Opin. Cell Biol. 2006;18:275–283. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Barna, M., Merghoub, T., Costoya, J.A., Ruggero, D., Branford, M., Bergia, A., Samori, B., Pandolfi, P.P. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev. Cell. 2002;3:499–510. doi: 10.1016/s1534-5807(02)00289-7. [DOI] [PubMed] [Google Scholar]
- Bel-Vialar, S., Core, N., Terranova, R., Goudot, V., Boned, A., Djabali, M. Altered retinoic acid sensitivity and temporal expression of Hox genes in polycomb-M33-deficient mice. Dev. Biol. 2000;224:238–249. doi: 10.1006/dbio.2000.9791. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Allis, C.D. RNA meets chromatin. Genes & Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bernstein, B.E., Kamal, M., Lindblad-Toh, K., Bekiranov, S., Bailey, D.K., Huebert, D.J., McMahon, S., Karlsson, E.K., Kulbokas, E.J., Gingeras, T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bolland, D.J., Wood, A.L., Johnston, C.M., Bunting, S.F., Morgan, G., Chakalova, L., Fraser, P.J., Corcoran, A.E. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 2004;5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- Boncinelli, E., Simeone, A., Acampora, D., Mavilio, F. Hox gene activation by retinoic acid. Trends Genet. 1991;7:329–334. doi: 10.1016/0168-9525(91)90423-n. [DOI] [PubMed] [Google Scholar]
- Boyer, L.A., Plath, K., Zeitlinger, J., Brambrink, T., Medeiros, L.A., Lee, T.I., Levine, S.S., Wernig, M., Tajonar, A., Ray, M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken, A.P., Dietrich, N., Pasini, D., Hansen, K.H., Helin, K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling, A., Turner, B.M., Bianchi, M.E., Orlando, V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- Breiling, A., O'Neill, L.P., D'Eliseo, D., Turner, B.M., Orlando, V. Epigenome changes in active and inactive polycomb-group-controlled regions. EMBO Rep. 2004;5:976–982. doi: 10.1038/sj.embor.7400260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolari, R., McMorrow, T., Yannoutsos, N., Langeveld, A., Grosveld, F. Deletion of a region that is a candidate for the difference between the deletion forms of hereditary persistence of fetal hemoglobin and δβ-thalassemia affects β- but not γ-globin gene expression. EMBO J. 1999;18:949–958. doi: 10.1093/emboj/18.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, R., Zhang, Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED–EZH2 complex. Mol. Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Cao, R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., Jones, R.S., Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2001;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Caretti, G., Di Padova, M., Micales, B., Lyons, G.E., Sartorelli, V. Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes & Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M.C., Maeda, N., Oyama, R., Ravasi, T., Lenhard, B., Wells, C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cawley, S., Bekiranov, S., Ng, H.H., Kapranov, P., Sekinger, E.A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Chambeyron, S., Bickmore, W.A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes & Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron, S., Da Silva, N.R., Lawson, K.A., Bickmore, W.A. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- Cheng, J., Kapranov, P., Drenkow, J., Dike, S., Brubaker, S., Patel, S., Long, J., Stern, D., Tammana, H., Helt, G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Dellino, G.I., Schwartz, Y.B., Farkas, G., McCabe, D., Elgin, S.C., Pirrotta, V. Polycomb silencing blocks transcription initiation. Mol. Cell. 2004;13:887–893. doi: 10.1016/s1097-2765(04)00128-5. [DOI] [PubMed] [Google Scholar]
- Drewell, R.A., Bae, E., Burr, J., Lewis, E.B. Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 2002;99:16853–16858. doi: 10.1073/pnas.222671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule, D., Morata, G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Engström, P.G., Suzuki, H., Ninomiya, N., Akalin, A., Sessa, L., Lavorgna, G., Brozzi, A., Luzi, L., Tan, S.L., Yang, L., et al. Complex loci in human and mouse genomes. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020047. e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epping, M.T., Wang, L., Edel, M.J., Carlee, L., Hernandez, M., Bernards, R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Gribnau, J., Diderich, K., Pruzina, S., Calzolari, R., Fraser, P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- Gronemeyer, H., Miturski, R. Molecular mechanisms of retinoid action. Cell. Mol. Biol. Lett. 2001;6:3–52. [PubMed] [Google Scholar]
- Guenther, M.G., Jenner, R.G., Chevalier, B., Nakamura, T., Croce, C.M., Canaani, E., Young, R.A. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausecker, D., Proudfoot, N.J. Dicer-dependent turnover of intergenic transcripts from the human β-globin gene cluster. Mol. Cell. Biol. 2005;25:9724–9733. doi: 10.1128/MCB.25.21.9724-9733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttendorfer, A., Schattner, P., Polacek, N. Noncoding RNAs: Hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Kampa, D., Cheng, J., Kapranov, P., Yamanaka, M., Brubaker, S., Cawley, S., Drenkow, J., Piccolboni, A., Bekiranov, S., Helt, G., et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31:5154. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama, S., Tomaru, Y., Kasukawa, T., Waki, K., Nakanishi, M., Nakamura, M., Nishida, H., Yap, C.C., Suzuki, M., Kawai, J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., Haussler, D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, T., Zakany, J., Duboule, D. Control of colinearity in AbdB genes of the mouse HoxD complex. Mol. Cell. 1998;1:289–300. doi: 10.1016/s1097-2765(00)80029-5. [DOI] [PubMed] [Google Scholar]
- Krumlauf, R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Lavorgna, G., Sessa, L., Guffanti, A., Lassandro, L., Casari, G. AntiHunter: Searching BLAST output for EST antisense transcripts. Bioinformatics. 2004;20:583–585. doi: 10.1093/bioinformatics/btg460. [DOI] [PubMed] [Google Scholar]
- Lavorgna, G., Triunfo, R., Santoni, F., Orfanelli, U., Noci, S., Bulfone, A., Zanetti, G., Casari, G. AntiHunter 2.0: Increased speed and sensitivity in searching BLAST output for EST antisense transcripts. Nucleic Acids Res. 2005;33:W665–W668. doi: 10.1093/nar/gki448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.I., Jenner, R.G., Boyer, L.A., Guenther, M.G., Levine, S.S., Kumar, R.M., Chevalier, B., Johnstone, S.E., Cole, M.F., Isono, K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner, B., Williams, G., Campbell, R.D., Sanderson, C.M. Antisense transcripts in the human genome. Trends Genet. 2002;18:63–65. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- Levine, S.S., King, I.F., Kingston, R.E. Division of labor in Polycomb group repression. Trends Biochem. Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- McKenna, N.J., O'Malley, B.W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Mainguy, G., In der Rieden, P.M., Berezikov, E., Woltering, J.M., Plasterk, R.H., Durston, A.J. A position-dependent organisation of retinoid response elements is conserved in the vertebrate Hox clusters. Trends Genet. 2003;19:476–479. doi: 10.1016/S0168-9525(03)00202-6. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. Noncoding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2001;211:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando, V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. doi: 10.1016/s0092-8674(03)00157-0. [DOI] [PubMed] [Google Scholar]
- Peterson, C.L., Laniel, M.A. Histones and histone modifications. Curr. Biol. 2004;14:546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Rank, G., Prestel, M., Paro, R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 2002;22:8026–8034. doi: 10.1128/MCB.22.22.8026-8034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar, M., Kobrossy, L., Kovacs, E.N., Rambaldi, I., Featherstone, M. Sequential histone modifications at Hoxd4 regulatory regions distinguish anterior from posterior embryonic compartments. Mol. Cell. Biol. 2004;24:8090–8103. doi: 10.1128/MCB.24.18.8090-8103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose, L., Paro, R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner, T., Gou, D., Kremmer, E., Sauer, F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Schmitt, S., Prestel, M., Paro, R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes & Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone, A., Pannese, M., Acampora, D., D'Esposito, M., Boncinelli, E. At least three human homeoboxes on chromosome 12 belong to the same transcription unit. Nucleic Acids Res. 1988;16:5379–5390. doi: 10.1093/nar/16.12.5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone, A., Acampora, D., Nigro, V., Faiella, A., D'Esposito, M., Stornaiuolo, A., Mavilio, F., Boncinelli, E. Differential regulation by retinoic acid of the homeobox genes of the four Hox loci in human embryonal carcinoma cells. Mech. Dev. 1991;33:215–227. doi: 10.1016/0925-4773(91)90029-6. [DOI] [PubMed] [Google Scholar]
- Vakoc, C.R., Mandat, S.A., Olenchock, B.A., Blobel, G.A. Histone H3 lysine 9 methylation and HP1 gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Werner, T. Computer-assisted analysis of transcription control regions. Matinspector and other programs. Methods Mol. Biol. 2000;132:337–349. doi: 10.1385/1-59259-192-2:337. [DOI] [PubMed] [Google Scholar]
- Xu, L., Glass, C.K., Rosenfeld, M.G. Coactivator and corepressor complexes in nuclear receptor function. Cur. Opin. Genet. Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Zakany, J., Kmita, M., Alarcon, P., de la Pompa, J.L., Duboule, D. Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell. 2001;106:207–217. doi: 10.1016/s0092-8674(01)00436-6. [DOI] [PubMed] [Google Scholar]