UNC-4 represses CEH-12/HB9 to specify synaptic inputs to VA motor neurons in C. elegans (original) (raw)
Abstract
In Caenorhabditis elegans, VA and VB motor neurons arise as lineal sisters but synapse with different interneurons to regulate locomotion. VA-specific inputs are defined by the UNC-4 homeoprotein and its transcriptional corepressor, UNC-37/Groucho, which function in the VAs to block the creation of chemical synapses and gap junctions with interneurons normally reserved for VBs. To reveal downstream genes that control this choice, we have employed a cell-specific microarray strategy that has now identified _unc-4_-regulated transcripts. One of these genes, ceh-12, a member of the HB9 family of homeoproteins, is normally restricted to VBs. We show that expression of CEH-12/HB9 in VA motor neurons in unc-4 mutants imposes VB-type inputs. Thus, this work reveals a developmental switch in which motor neuron input is defined by differential expression of transcription factors that select alternative presynaptic partners. The conservation of UNC-4, HB9, and Groucho expression in the vertebrate motor circuit argues that similar mechanisms may regulate synaptic specificity in the spinal cord.
Keywords: C. elegans, synaptic specificity, unc-4, gap junctions, transcriptional repression, motor neuron
Animal movement is controlled by axial nerve cords in which interneurons synapse with motor neurons to regulate muscle activity. These connections are highly specific, as motor neurons innervate particular muscles as well as receive inputs from selected interneurons. Available evidence indicates that the architecture of these networks is largely imposed by genetic pathways in which neuronal fates are transcriptionally controlled (Arber et al. 2000; Glover 2000; Shirasaki and Pfaff 2002; Chen et al. 2003). This idea is especially well documented for motor circuit neurons that arise in the developing spinal cord (Shirasaki and Pfaff 2002). Here, distinct progenitor domains are delineated by reciprocal repression of unique combinations of homeodomain transcription factors that utilize the conserved corepressor protein, Groucho. In this “derepression” model, neuron identity is achieved by actively suppressing other fates (Muhr et al. 2001). For example, Pax6 and Olig2-mediated repression of the interneuron specifiers Nkx2.2 and Irx3 permits activation of the HB9 homeodomain transcription factor in the ventral motor neuron progenitor domain (Lee et al. 2004). HB9, in turn, specifies motor neuron fate by inhibiting expression of post-mitotic interneuron determinants (i.e., Chx10) (Thaler et al. 1999). The muscle-targeting specificity of motor neuron classes arising from this domain is defined by unique combinations of transcription factors (Sharma et al. 2000; Shirasaki and Pfaff 2002). Similarly, sensory neurons in local reflex circuits recognize post-synaptic motor neurons in mechanisms that depend on expression of specific transcriptional regulators in partner neurons (Arber et al. 2000; Chen et al. 2003, 2006). Inputs to vertebrate motor neurons from brain-derived interneurons are presumptively specific and are likely to depend on genetic pathways that also define motor neuron identity (Glover 2000). Although these observations provide strong support for the model that the specificity of motor circuit synapses is transcriptionally controlled, the precise pathways that define synaptic choice are poorly understood.
To simplify this problem, we adopted the nematode Caenorhabditis elegans because of its small, well-defined nervous system and facile genetics. In this animal, interneurons enter the ventral nerve cord from distal ganglia in head and tail regions to synapse with specific classes of motor neurons (Fig. 1). These connections include defined combinations of chemical synapses and gap junctions (White et al. 1986; Von Stetina et al. 2005). Mutants with “uncoordinated” or “Unc” movement phenotypes have been correlated with structural defects in this circuit. Of particular interest to the question of how transcription factors define the architecture of motor circuits is the finding that mutations in the unc-4 homeodomain gene alter inputs to VA class motor neurons (Miller et al. 1992; White et al. 1992). In wild-type animals, most VA and VB motor neurons arise as sister cells that adopt distinctive morphologies and synapse with separate sets of interneurons (Fig. 1; Sulston and Horvitz 1977; White et al. 1986). In unc-4 mutants, these cell lineages and morphological differences are preserved; VAs, however, are miswired with inputs from interneurons normally restricted to their VB sister cells. Thus, unc-4 explicitly controls synaptic choice but does not regulate other features of this motor circuit such as axonal outgrowth or process placement that could indirectly alter wiring specificity (White et al. 1992).
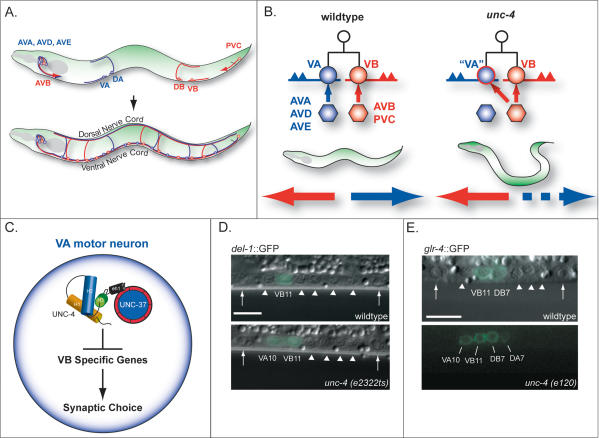
Figure 1.
VA synaptic inputs to VA motor neurons depend on UNC-4 repression of VB-specific genes. (A) Interneurons in head and tail ganglia extend axons into the ventral nerve cord to synapse with specific motor neurons. DA and VA class motor neurons receive inputs from interneurons AVA (gap junctions and chemical synapses), AVD, and AVE (chemical synapses) to comprise the “blue circuit,” whereas DB and VB motor neurons are connected to AVB (gap junctions) and PVC (chemical synapses) in the “red circuit.” VA and VB motor neurons innervate ventral muscles; DA and DB motor neurons extend commissural processes from the ventral side to synapse with dorsal muscles. (B) Most VA and VB motor neurons arise from a common progenitor but extend axons in opposite directions and are connected to separate sets of command interneurons; backward locomotion depends on the blue circuit and forward movement is driven by the red circuit. In unc-4 mutants, which cannot back, VA motor neurons are miswired with inputs (ectopic gap junction with AVB, chemical synapse from PVC) normally reserved for VB sister cells. (C) Model of UNC-4 action. The UNC-4 homeodomain protein and its corepressor, UNC-37/Groucho, specify VA inputs by blocking expression of VB genes. The UNC-4 eh1 domain interacts with UNC-37/Groucho. (D,E) VB genes are derepressed in VA motor neurons in unc-4 mutants. Lateral views of posterior ventral nerve cord of L2 larvae. (D) A GFP reporter for the DEG/ENaC subunit, del-1, is normally restricted to VB motor neurons in the L2 but is also expressed in VA motor neurons in unc-4 mutants. (E) _glr-4_∷GFP is expressed in DB and VB motor neurons in the wild-type and ectopically expressed in DA and VA motor neurons in unc-4. Arrowheads point to DIC images of motor neuron nuclei that do not express GFP. Arrows denote ventral nerve cord landmarks, P9.p and P10.p ectodermal blast cells. Anterior is to the left, ventral is down. Bars, 5 μm.
The UNC-4 homeodomain protein is expressed in VA motor neurons to prevent the adoption of VB-type inputs (Miller and Niemeyer 1995). This activity depends on physical interactions with UNC-37, the nematode homolog of Groucho. We have thus proposed that UNC-4 sustains VA inputs by turning off VB-specific genes (Pflugrad et al. 1997; Winnier et al. 1999). Here we describe the results of a neuron-specific microarray-based strategy to identify these transcripts. This approach has revealed VB-genes that are negatively regulated by unc-4 and unc-37 in VA motor neurons. We have confirmed that at least one of these genes, ceh-12/HB9, functions downstream from unc-4 to regulate synaptic choice.
CEH-12 is closely related to Mnx/HB9 homeodomain transcription factors, key determinants of motor neuron fate in flies, birds, and mammals (Arber et al. 1999; Thaler et al. 1999; Broihier and Skeath 2002). In C. elegans, ceh-12 is selectively detected in VB motor neurons in the ventral cord of wild-type animals but is also ectopically expressed in VAs in unc-4 and unc-37 mutants; we show that this misexpression of ceh-12 in the VAs is both necessary and sufficient to impose VB-type inputs. Thus, these results substantiate the model that normal interneuron synapses with VA motor neurons depend on UNC-4 repression of VB-specific genes (Winnier et al. 1999). Moreover, this work underscores the key role of transcriptional control in the establishment of functional motor neuron circuits and argues for the ancient evolutionary origin of this mechanism and the specific transcription factors that mediate it.
Results
The C. elegans motor circuit
UNC-4 is expressed in DA motor neurons in the embryo and in larval VA motor neurons that arise in the L1 stage (Miller and Niemeyer 1995). These “A-class” motor neurons share common morphological features, such as anteriorly directed axons and input from specific interneurons (AVA, AVD, AVE). In contrast, DB and VB or “B-class” motor neurons extend posterior axons and are connected to a different set of presynaptic command interneurons (AVB, PVC) (Fig. 1; White et al. 1986; Von Stetina et al. 2005). Most (nine out of 12) VA and VB motor neurons arise from a common progenitor cell; VA2–VA10 are sisters to VB3–VB11. VA motor neurons in this group are miswired with VB-type inputs in the mutant unc-4(e120) (Fig. 1B). In contrast, EM reconstruction of unc-4(e120) did not detect miswiring of VA motor neurons that do not have VB sisters (VA1, VA11, VA12) or of embryonic DA motor neurons. These results suggested that unc-4 is exclusively required to specify synaptic inputs to VA motor neurons that arise from a progenitor cell that also produces a VB motor neuron (White et al. 1992).
Both A-class and B-class motor neurons are cholinergic and excitatory but exhibit separate locomotory functions. A-class motor neurons mediate backward movement, whereas B-class motor neurons are required for forward locomotion (Fig. 5, below; Supplementary Movie 1; Chalfie et al. 1985). The loss of normal inputs to VA motor neurons in unc-4(e120) blocks backward movement. Other unc-4 alleles display a similar phenotype (Fig. 5, below; Supplementary Movie 2; White et al. 1992). Movement may also be impaired by a reduction in synaptic vesicles at neuromuscular synapses of A-class motor neurons in unc-4 and unc-37 mutants (Lickteig et al. 2001).
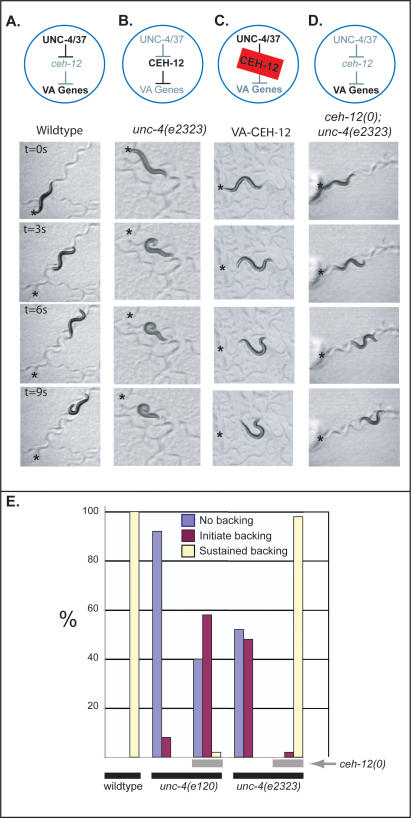
Figure 5.
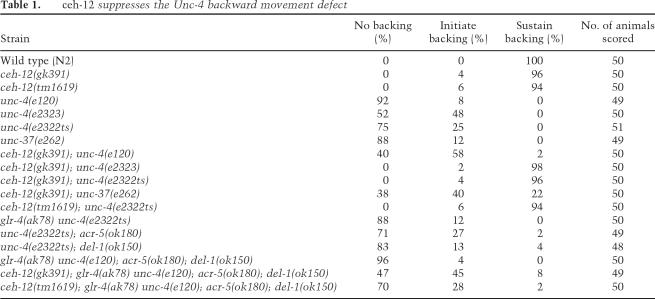
ceh-12 genetically interacts with unc-4 to control backward locomotion. Animals were touched on the head (asterisk) to stimulate backward locomotion. Models above movie panels indicate gene expression in VA motor neurons. (A) Wild-type animals back away for 9 sec following touch (Supplementary Movie 1). (B) Mutations in unc-4 or unc-37 derepress ceh-12 and block backward locomotion. Note that the unc-4(e2323) animal in the bottom panels coils in response to touch but cannot back (Supplementary Movie 2). (C) Transgenic expression of CEH-12 in VA motor neurons (VA-CEH-12) is sufficient to induce an Unc-4-like backing defect (Supplementary Movie 4). (D) ceh-12(0) restores backward movement to the hypomorphic allele, unc-4(e2323) (Supplementary Movie 5). (E) Quantification of Unc-4 backward movement defects reveals suppression by _ceh-12_-null alleles. Individual animals of each genotype were tapped on the head to evoke backward movement and scored according to three phenotypes: no backing (coil immediately upon tapping) (blue), initiate backing (but then stop) (purple), sustained backward movement (less than two body bends) (yellow). Note that the null allele unc-4(e120) is weakly suppressed by ceh-12(0), whereas the hypomorphic mutation, unc-4(e2323), is strongly suppressed by ceh-12(0) (i.e., shows sustained backward movement). All results are for ceh-12(gk391). Similar results were obtained for ceh-12(tm1619) (data not shown).
VA motor neuron-specific microarray profiles reveal candidate unc-4 target genes
We used the mRNA-tagging method to identify candidate _unc-4_-regulated genes in VA motor neurons (Fig. 2A; Roy et al. 2002). The _unc-4_p∷3XFlag-PAB-1 transgene (3FPAB) was crossed into an unc-37 mutant background and a mRNA-tagging profile generated for comparison to a wild-type VA motor neuron data set (Fig. 2B; S.E. Von Stetina, J.D. Watson, R.M. Fox, W.C. Spencer, K.L. Olszewski, P.J. Roy, S.K. Kim, and D.M. Miller III, in prep.). (For reasons that are unclear, the combination of 3FPAB with unc-4 alleles resulted in subviable animals that could not be profiled; see Materials and Methods.) Because UNC-4 functions with UNC-37/Groucho as a transcriptional repressor, target genes should be elevated in the unc-37 mutant data set (Fig. 1C). This prediction was confirmed by the enrichment of known _unc-4_-regulated genes, del-1 and glr-4 (Figs. 1D,E, 2C; Winnier et al. 1999). A total of 255 transcripts are significantly elevated in the unc-37 profile. Here we describe experiments confirming that one of these genes, ceh-12, is also negatively regulated by unc-4 and that it functions downstream from unc-4 to specify synaptic inputs to A-class motor neurons. A complete description of UNC-37/Groucho-regulated A-class motor neuron genes will be presented elsewhere.
Figure 2.
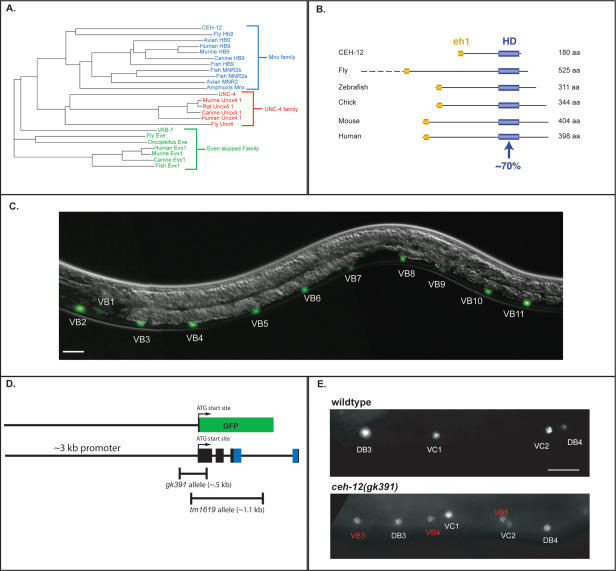
mRNA tagging identifies candidate unc-4 target genes. (A) An epitope-tagged mRNA-binding protein (3XFlag-PAB-1) is expressed in A-class motor neurons (red) using the unc-4 promoter. A-class motor neuron-specific transcripts are isolated by coimmunoprecipitation with anti-Flag. L2 larvae were utilized for this experiment because unc-4 function is required during this developmental stage (Miller et al. 1992). Artwork courtesy of Erik Jorgensen. (B, top) The _unc-4_∷3XFlag∷PAB-1 transgene is expressed in VA motor neurons in unc-37(e262). (Bottom) Closeup view of posterior ventral cord (boxed area in top) of mid-L2 larva shows anti-Flag staining (red) surrounding DAPI-stained (blue) nuclei of DA and VA motor neurons. Anterior is to the left, ventral is down. Bars: top, 10 μm; bottom, 2 μm. (C) Scatter plot of representative unc-37 (e262) microarray data set (red) versus the average of reference (wild-type) intensity values (green) for A-class motor neurons. Up-regulated genes (red) include del-l, glr-4, and ceh-12 (HB9 homeodomain protein).
ceh-12 encodes the nematode homolog of the HB9 homeodomain protein and regulates VB motor neuron- specific gene expression
Phylogenetic analysis indicates that ceh-12 is the closest C. elegans relative of the Mnx/HB9 family of homeodomain proteins (Fig. 3A). CEH-12 includes an N-terminal eh1 domain (Jimenez et al. 1997) that is also conserved in this location in other HB9 proteins (Fig. 3B). The presence of this potential Groucho interaction domain is consistent with the proposal that CEH-12 and other HB9 proteins are transcriptional repressors (William et al. 2003). The well-established and highly conserved role of HB9 proteins in motor neuron differentiation in other species (Arber et al. 1999; Thaler et al. 1999; Broihier and Skeath 2002) suggested to us that ceh-12 was also likely to regulate motor neuron fate in C. elegans.
Figure 3.
CEH-12 is the nematode HB9 homolog, is expressed in VB motor neurons, and specifies VB fate. (A) Phylogenetic tree comparing members of the Mnx/HB9, UNC-4, and Even-skipped homeodomain transcription factor families. CEH-12 is more closely related to members of the Mnx family than to other C. elegans homeodomain proteins. (B) Genomic structures of Mnx family homeodomain genes. CEH-12 contains a homeodomain (∼70% identical to other species) and conserved N-terminal eh-1 Groucho interaction domain. (C) A promoter-GFP reporter, _ceh-12_∷GFP, is expressed in all 11 VB motor neurons and in a single head neuron, RID; occasional weak GFP expression was observed in the pharyngeal/intestinal valve cell and in the excretory gland cell (data not shown). (D) Structure of _ceh-12_∷GFP and ceh-12 deletion alleles. _ceh-12_∷GFP was constructed using 2.8 kb of promoter sequence upstream of the predicted start site. The gk391 allele deletes 0.5 kb of ceh-12, including the predicted start site. The tm1619 allele is a 1.1-kb deletion that removes the start site and most of the homeodomain. Both alleles are predicted null mutations. (E, top) Immunostaining reveals VAB-7 expression in DB and VC motor neurons in wild-type adult animals. (Bottom) VAB-7 is detected ectopically in VB motor neurons in ceh-12(gk391) mutant adult. Similar results are seen with ceh-12(tm1619) (data not shown). Bars: C,E, 10 μm.
We confirmed that ceh-12 is expressed in C. elegans motor neurons by constructing a _ceh-12_∷GFP reporter gene (Fig. 3D). Three independent transgenic lines display GFP fluorescence exclusively in VB motor neurons in the ventral nerve cord (Figs. 3C, 4A). To determine if ceh-12 controls gene expression in these cells, we examined the ceh-12 deletion alleles tm1619 and gk391 (Fig. 3D) for regulation of known motor neuron markers. The VB reporters _del-1_∷GFP and _acr-5_∷GFP, the A-class marker _unc-4_∷GFP, and the A- and B-class label _acr-2_∷GFP were not affected by these ceh-12 alleles (Supplemental Material). Immunostaining, however, revealed that VAB-7/Even-skipped is derepressed in VB motor neurons in ceh-12 mutants (Fig. 3E). VAB-7/Eve is normally restricted to DB motor neurons in the ventral nerve cord during the L2 larval stage in which _ceh-12_∷GFP is expressed in VB motor neurons (VAB-7 is also expressed in VC motor neurons in the adult) (Fig. 3E; Esmaeili et al. 2002). Our results mirror similar transcriptional regulation in Drosophila, in which dHB9 represses Eve in selected motor neuron classes to prevent the adoption of the axonal trajectory of Eve+ motor neurons (Broihier and Skeath 2002). In C. elegans, however, ectopic VAB-7 in the VBs is not sufficient to impose the dorsally directed axonal outgrowth of DB motor neurons, as VB motor axons remain in the ventral nerve cord in ceh-12 mutants (Supplemental Material). The limited effect of ceh-12 mutants on VB motor neuron fate is consistent with the observation that these animals do not display obvious movement defects (Table 1; Supplemental Material; Supplementary Movie 3), and suggests that ceh-12 may regulate a specific subset of VB traits.
Figure 4.
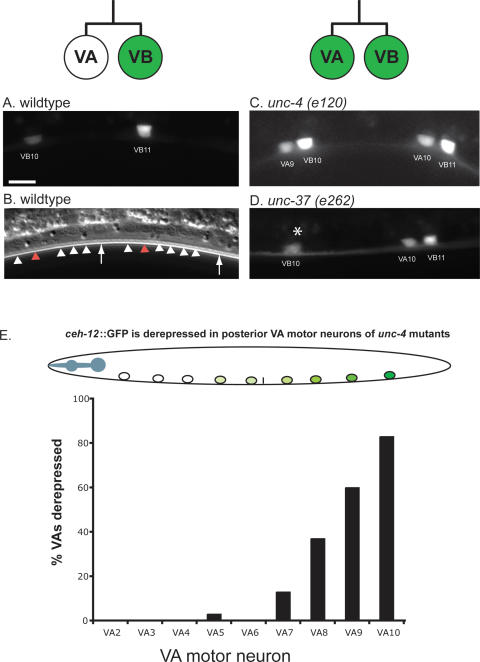
_ceh-12_∷GFP is negatively regulated by unc-4 and unc-37 in posterior VA class motor neurons. (A) Closeup view of wild-type posterior ventral cord showing expression of _ceh-12_∷GFP in VB motor neurons. Bar, 5 μm. (B) White arrowheads in wild-type DIC image point to motor neuron nuclei that do not express _ceh-12_∷GFP; red arrowheads denote VB10 and VB11 that express GFP in the top panel. Arrows identify landmark P9.p and P10.p ectoblasts. (C,D) Ectopic _ceh-12_∷GFP expression in VA motor neurons in unc-4 and unc-37 mutants. All are mid-L2 stage larvae. Anterior is to the left, ventral is down. Asterisk denotes gut autofluorescence. (E) _ceh-12_∷GFP is preferentially derepressed in posterior VA motor neurons in unc-4(e120). n = 30.
Table 1.
ceh-12 suppresses the Unc-4 backward movement defect
_ceh-12_∷GFP is negatively regulated by unc-4 and unc-37 in posterior A-class neurons
Our model predicts that UNC-4 target genes are normally expressed in VB motor neurons but turned off in the VAs by the combined activity of UNC-4 and UNC-37/Groucho (Fig. 1; Winnier et al. 1999). We have confirmed this prediction by showing that _ceh-12_∷GFP is normally expressed in VB motor neurons in the wild-type but is also detected in VA motor neurons in unc-4 and unc-37 mutants (Fig. 4C,D). This finding agrees with microarray results that detected significant elevation of the endogenous ceh-12 transcript in unc-37 mutant VA motor neurons (Fig. 2C). Surprisingly, ectopic _ceh-12_∷GFP expression is strongest for VA motor neurons in the posterior ventral nerve cord. For example, in unc-4(e120), 80% of VA10 neurons show _ceh-12_∷GFP, whereas only ∼15% of VA7 motor neurons are affected; ectopic _ceh-12_∷GFP was never detected (n = 30) in VA2 at the anterior end of the ventral nerve cord (Fig. 4E). A similar bias in _ceh-12_∷GFP expression was also observed for posteriorly located DA motor neurons in unc-4 and unc-37 mutants (data not shown). This finding is consistent with the elevation of ceh-12 transcript levels in microarray data generated from unc-4 mutant DA motor neurons using the MAPCeL approach (Fox et al. 2005; R.M. Fox and D.M. Miller, unpubl.). On the basis of these results, we conclude that ceh-12 is a strong candidate for a VB gene that is negatively regulated by the unc-4 pathway in posterior A-class motor neurons.
CEH-12 expression in VA motor neurons induces an Unc-4-like movement defect
If ceh-12 is sufficient to miswire VA motor neurons, then ectopic expression of CEH-12 in the VAs should result in a backward Unc defect. To test this idea, we used the unc-4 promoter to drive CEH-12 protein expression in A-class motor neurons. These “VA-CEH-12” animals are unable to execute backward movement when stimulated by head touch and instead coil dorsally, a phenotype that resembles that of unc-4 mutants (Fig. 5B,C; Supplemental Material; Supplementary Movie 4). Because CEH-12 is predicted to act as a transcriptional repressor, this behavior could have resulted from CEH-12-dependent repression of unc-4. However, _unc-4_∷GFP is expressed normally in A-class motor neurons in VA-CEH-12 animals (Supplemental Material). Thus, we propose that CEH-12 must be acting on other VA genes to induce the backward Unc phenotype (Fig. 5).
CEH-12 functions downstream from unc-4 and unc-37 to induce the Unc-4 movement defect
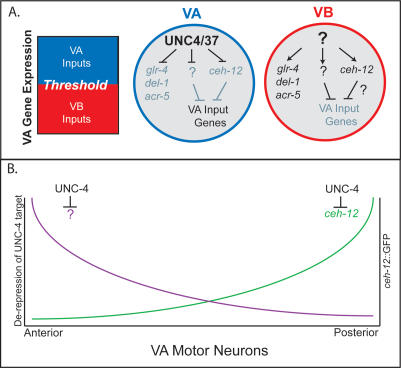
Above, we showed that ectopic expression of CEH-12 in VA motor neurons is sufficient to produce an Unc-4-like backward movement defect in VA-CEH-12 worms. If derepression of ceh-12 in unc-4 mutants is also responsible for this phenotype, then the loss of ceh-12 activity should result in improved backward locomotion for these animals. To test this possibility, we used the ceh-12 deletion alleles (Fig. 3D) in genetic experiments with unc-4 and unc-37 mutants to detect “suppression” of the Unc-4 phenotype. As shown in Figure 5E, ceh-12(0) affords weak but quantitatively measurable improvement of backward locomotion in the null allele, unc-4(e120). A similar effect was observed in double-mutant combinations of ceh-12(0) with unc-37(e262) (Table 1). Incomplete suppression of the Unc-4 phenotype by ceh-12(0) could be indicative of a second downstream pathway that is also derepressed in unc-4 mutants. This model is consistent with the observation that _ceh-12_∷GFP is preferentially expressed in posterior VA motor neurons in unc-4 mutants (Fig. 4E), although anterior VAs are also miswired. This result suggests that normal VA inputs are selectively restored to posterior VA motor neurons in ceh-12(0); unc-4(0) double mutants, whereas anterior VAs remain miswired due to the ectopic activity of a presumptive parallel pathway that functions in these cells (Fig. 7, below).
Figure 7.
Model of UNC-4 function to regulate input to VA class motor neurons. (A) A-type synaptic inputs are imposed when VA gene expression exceeds a specific threshold. In VA motor neurons, the UNC-4/UNC-37(Groucho) complex blocks expression of VB genes (glr-4, del-1, acr-5) that do not affect synaptic choice and at least two other VB genes, ceh-12 and an unknown target (?), that repress VA input genes. VB motor neurons do not express UNC-4, and therefore the level of VA gene expression is not sufficient to specify A-type synapses to these cells. (B) UNC-4 target gene expression is biased along the A/P axis. ceh-12 is preferentially repressed by unc-4 in posterior VA motor neurons, whereas an as-yet-unidentified unc-4 target gene (?) regulates synaptic choice anteriorly.
The proposal that ceh-12 functions downstream from unc-4 in parallel to at least one additional partially redundant pathway is also consistent with our finding that the weak or hypomorphic unc-4 alleles, e2323 and e2322ts, are strongly suppressed by ceh-12(0). For example, unc-4(e2323) animals are unable to sustain backward movement, whereas almost all (98%) of ceh-12(0); unc-4(e2323) animals readily execute reverse locomotion when touched on the head (Fig. 5D,E; Table 1; Supplementary Movie 5). In this case, we propose that the residual function of these hypomorphic unc-4 alleles prevents full derepression of target genes, and therefore limits their consequent negative effects on putative downstream VA input genes. In this situation, subsequent elimination of ceh-12 function in posterior VA motor neurons allows net VA motor neuron activity throughout the cord to exceed a threshold required for the restoration of backward locomotion (Fig. 7, below).
Candidate genes that could function in parallel to ceh-12 include del-1 (DEG/ENaC channel subunit), acr-5 (nicotinic ACh receptor subunit) and glr-4 (ionotrophic glutamate receptor), VB genes that we have previously shown are negatively regulated in VA motor neurons by unc-4 and unc-37 (Fig. 1; Winnier et al. 1999). As cell surface proteins and ion channel components, DEL-1, ACR-5, and GLR-4 are plausible synaptic determinants. However, null alleles of these loci have no effect on the Unc-4 phenotype either alone or in combination with ceh-12 (Table 1). Furthermore, ceh-12 does not regulate the expression of these ion channel components in VB motor neurons (Supplemental Material). These genetic results rule out a role for del-1, acr-5, and glr-4 as unc-4 target genes that regulate synaptic choice. unc-4 may repress these downstream genes to prevent expression of other VB traits in VA motor neurons.
UNC-4 and CEH-12 regulate the specificity of gap junctions between command interneurons and motor neurons
The wiring diagram of the C. elegans nervous system was originally deduced from reconstruction of serial sections photographed in the electron microscope (White et al. 1986). A partial EM reconstruction of unc-4(e120) revealed the miswiring defect of selected VA motor neurons (VA2, VA3, VA10). Aberrant gap junctions with AVB are particularly prominent, and are placed directly on the VA motor neuron soma, a location adjacent to the cell nucleus that is also characteristic of the usual AVB gap junctions with DB and VB motor neurons (Fig. 6A; White et al. 1992). We have now confirmed these results with a GFP-tagged marker that allows visualization in the light microscope of gap junctions between AVB command interneurons and specific ventral nerve cord motor neurons.
Figure 6.
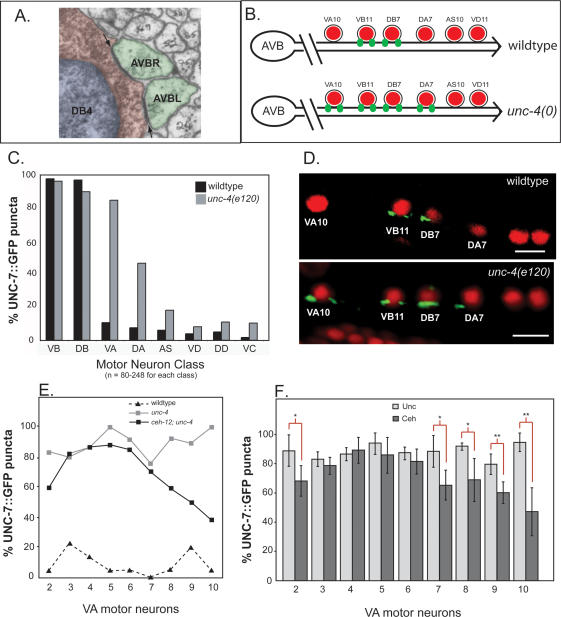
Gap junctions between command interneuron AVB and specific motor neurons are visualized with UNC-7S∷GFP. (A) Electron micrograph (courtesy of John White and Michael Nonet) depicting electron-dense gap junction synapses (arrows) between AVBL and AVBR interneuron processes (green) and the cell soma (red) of DB4 in the ventral nerve cord. The nucleus of DB4 is pseudocolored blue. (B) Posterior ventral nerve cord showing gap junctions (green puncta) between AVB and soma of specific motor neurons (red) in wild-type (top) and unc-4 (bottom) mutants. (C) Quantification of UNC-7S∷GFP puncta on specific motor neuron classes in wild-type and unc-4(e120). UNC-7S∷GFP puncta are consistently observed adjacent to DB and VB motor neurons. Most (∼80%) VA motor neurons (VA2–VA10) are miswired with UNC-7S∷GFP puncta in unc-4(e120); DA motor neurons may also be affected (45%) (see DA7 in B). UNC-7S∷GFP puncta are rarely associated with other motor neuron classes (AS, VD, DD, VC). (D) Confocal images of UNC-7S∷GFP puncta (green) adjacent to motor neuron nuclei stained with DAPI (red). Lateral view of adult; anterior is to the left, ventral is down. Bars, 5 μm. (E,F) Rescue of the AVB wiring defect in ceh-12; unc-4 is biased to posterior VA motor neurons. UNC-7S∷GFP puncta were scored for VA2–VA10. (E) Fraction of VAs with AVB gap junctions in wild-type (black triangles), unc-4(e120) (gray boxes), and ceh-12(0); unc-4(e120) (black boxes). (n = 18–45 for each VA in each genotype.) (F) Fraction of VAs with AVB gap junctions were averaged for unc-4(e120, e2322ts, e2323) and unc-37(e262) mutants (Unc light-gray bars) and compared with the average of ceh-12; unc-4 and ceh-12; unc-37 double mutants (Ceh, dark-gray bars). A statistically significant decrease in UNC-7S∷GFP puncta is noted for Ceh animals in VA2 (P = 0.032), VA7 (P = 0.021), VA8 (P = 0.021), VA9 (P = 0.008), and VA10 (P = 0.002). (*) P ≤ 0.05; (**) P ≤ 0.01.
Invertebrate gap junctions are assembled from innexins, modular subunit proteins that function similarly to the vertebrate gap junction connexin proteins despite the absence of obvious sequence homologies (Phelan and Starich 2001). The recently discovered pannexins, the apparent evolutionary descendents of the innexins, constitute a second group of vertebrate gap junction proteins (Bruzzone et al. 2003; Panchin 2005). The C. elegans genome includes 25 innexin genes with distinct spatial and temporal patterns of expression (Starich et al. 2001). The unc-7 gene encodes at least two innexin isoforms, UNC-7L and UNC-7S (T.A. Starich, J. Xi, I.M. Skerrett, B.J. Nicholson, and J.E. Shaw, in prep.). AVB command interneurons express UNC-7S, and a GFP-tagged UNC-7S construct is detected in distinct puncta along the ventral nerve cord. In wild-type animals, most DB and VB motor neurons are marked with UNC-7S∷GFP puncta adjacent to the cell soma, whereas other motor neurons are rarely stained (Fig. 6C,D). This result is consistent with the motor circuit wiring diagram derived from EM reconstruction in which gap junctions with AVB interneurons are prominently located adjacent to cell soma of B-class motor neurons (Fig. 6A; White et al. 1986). Moreover, UNC-7S∷GFP rescues the forward movement defect of unc-7 mutants, thereby suggesting that UNC-7S∷GFP puncta correspond to functional gap junctions in the nervous system (T.A. Starich, J. Xi, I.M. Skerrett, B.J. Nicholson, and J.E. Shaw, in prep.). On the basis of these results, we conclude that UNC-7S∷GFP is a reliable marker of AVB gap junctions with specific ventral cord motor neurons.
In unc-4(e120) mutants, ectopic UNC-7S∷GFP puncta are consistently observed adjacent to the cell soma of nine VA motor neurons (VA2–VA10) (Fig. 6C,D). This result both confirms and extends the results of the original EM reconstruction of unc-4(e120). This study was limited to portions of the anterior and posterior ventral cord where AVB gap junctions were detected for VA2, VA3, and VA10; miswiring of VA4–VA9 was inferred from the Unc-4 backward movement defect that appears to result from VA dysfunction throughout the intervening body region (White et al. 1992).
UNC-7S∷GFP puncta are similarly misplaced on VA class motor neurons in other unc-4 alleles and in the Groucho mutant, unc-37(e262) (Fig. 6F; Supplementary Table 1). These results establish that the AVB-to-VA gap junction defect is a component of the Unc-4-null phenotype and confirm the essential role of UNC-37/Groucho in UNC-4 function. Surprisingly, we also observed UNC-7S∷GFP puncta on DA class motor neurons (Fig. 6B–D) and on VA11 (Supplementary Table 2) in unc-4 and unc-37 mutants (VA1 and VA12 were not scored) (see Materials and Methods). The incomplete penetrance of this effect (<50%) and the limited number of DA motor neurons examined in the original EM reconstruction of unc-4(e120) may explain why these gap junctions with AVB were not previously observed (White et al. 1992).
ceh-12 function is required for miswiring of posterior VA motor neurons with AVB gap junctions
To determine if derepression of ceh-12 is required for the creation of ectopic AVB gap junctions with VA motor neurons, we examined the distribution of UNC-7S∷GFP puncta in ceh-12(0); unc-4(e120) double mutants. Whereas the frequency at which UNC-7S∷GFP puncta are placed next to VA motor neurons in the anterior ventral cord is comparable to that of unc-4(e120) (i.e., 90%–95%), a significant reduction is noted for posterior VA motor neurons in ceh-12(0); unc-4(e120) animals (e.g., 30% for VA10) (Fig. 6E). This effect is confirmed by similar results showing a posterior bias for ceh-12(0) rescue of the AVB gap junction miswiring defect in two additional unc-4 alleles and in unc-37(e262) (Fig. 6F). The finding here that ceh-12 is selectively required for the Unc-4 gap junction defect in the posterior ventral cord is congruent with the observation that _ceh-12_∷GFP is preferentially derepressed in posterior VA motor neurons in unc-4 mutants (Fig. 4E). Although not directly scored by the UNC-7S∷GFP assay, restoration of normal inputs (e.g., gap junctions and chemical synapses with AVA) to posterior VAs is also likely given the partial suppression of backward movement in ceh-12(0); unc-4(e120) animals (Fig. 5E). We also note a statistically significant (p = 0.032) reduction of UNC-7S∷GFP puncta on the anteriorly located VA2 motor neuron in ceh-12(0) doubles with unc-4 and unc-37 alleles. Because we do not observe ectopic _ceh-12_∷GFP expression in VA2 in unc-4 or unc-37 mutants (Fig. 4) this result could be indicative of an additional non-cell-autonomous role for CEH-12, as also reported for HB9 in Drosophila motor neuron specification (Broihier and Skeath 2002).
Discussion
Transcription factor cascades define the structure of the vertebrate motor circuit by regulating the differentiation of specific neurons that contribute to this network (Shirasaki and Pfaff 2002). A striking feature of these pathways is the frequent use of negative gene regulation to produce distinct fates between neurons generated from adjacent progenitor domains (Lee and Pfaff 2001; Muhr et al. 2001). Here we show that a similar mechanism of repression involving conserved transcriptional components distinguishes the fates of C. elegans motor neurons born as sisters from a common mother cell (Fig. 1). Our results also offer a strikingly new finding, an explicit link between this biological strategy and the choice of presynaptic partners, a developmental decision of critical importance to motor neuron function. Below we present a model of transcriptional regulation of synaptic specificity in C. elegans and discuss the possibility that related schemes may also define wiring in the vertebrate spinal cord.
A transcriptional switch regulates synaptic choice in the C. elegans motor circuit
C. elegans mutants in the unc-4 homeodomain gene display a strong backward movement defect that results from the miswiring of VA class motor neurons with inputs normally reserved for VB motor neurons (Fig. 1; Miller et al. 1992; White et al. 1992). Intriguingly, other aspects of VA cell fate (i.e., axon trajectory and process placement) are unchanged, suggesting that UNC-4 functions to control only the synaptic fate of this cell type. Here we show that this change in synaptic specificity depends in part on misexpression of the VB-specific transcription factor, CEH-12/HB9, in VA motor neurons (Fig. 7). Normally, UNC-4 functions with UNC-37/Groucho to block ceh-12/HB9 expression in the VAs. Because HB9 is also believed to function as a transcriptional repressor in other organisms (Broihier and Skeath 2002; William et al. 2003), we propose that ectopic CEH-12/HB9 in unc-4 and unc-37 mutants triggers miswiring by turning off genes that specify VA inputs. It is possible that ectopic CEH-12/HB9 also activates VB genes that drive the creation of VB-type inputs. Our results provide strong genetic evidence for at least one additional pathway downstream from UNC-4 that functions in parallel to CEH-12/HB9. The relative contributions of these pathways to VA input specificity are biased along the anterior–posterior (A/P) axis with ectopic CEH-12 selectively driving the creation of VB inputs to posterior VA motor neurons in unc-4 mutants and the presumptive parallel pathway imposing VB inputs to anterior VAs. Finally, a third set of VB genes, glr-4, del-1, and acr-5 are negatively regulated by unc-4 but have no detectable role in the VA miswiring defect. These cell surface proteins and ion channel components could be indicative of physiologically important differences in the excitability or signaling capacity of VA versus VB motor neurons. In the future, it will be interesting to determine if ectopic ceh-12 expression contributes to the observed depletion of synaptic vesicles in unc-4 mutant neurons (Lickteig et al. 2001).
Although ceh-12 is required for the imposition of VB-type inputs to posterior VA motor neurons in unc-4 mutants, inputs to most VB motor neurons apparently do not depend on ceh-12 activity. Two lines of evidence support this conclusion. First, ceh-12 knock-out mutants do not show an obvious forward movement defect as would be expected if VB motor neurons were miswired (Table 1; Supplemental Material; Supplementary Movie 3). Second, the elimination of ceh-12 activity in these mutants does not perturb the creation of gap junctions between most VBs and AVB command interneurons (Supplementary Table 3). These data are consistent with the proposal suggested above that ceh-12 functions in parallel to a redundant pathway in VB motor neurons that is sufficient to retain VB-type inputs (Fig. 7).
Transcriptional regulation by UNC-4 versus CEH-12/HB9 in VA motor neurons results in gap junctions with different sets of command interneurons
We describe in this work the use of a GFP-tagged UNC-7S marker protein for visualizing gap junctions between specific neuron pairs in the C. elegans motor circuit (T.A. Starich, J. Xi, I.M. Skerrett, B.J. Nicholson, and J.E. Shaw, in prep.). This assay has provided an unprecedented opportunity to score gap junction specificity in the light microscope in multiple animals and in a variety of different mutant backgrounds.
These experiments indicate that the innexin, UNC-7S, is expressed in AVB command interneurons for assembly into gap junctions with B-class motor neurons. Genetic and physiological data suggest that these gap junctions are likely to be heterotypic, and also include the innexin UNC-9 (T.A. Starich, J. Xi, I.M. Skerrett, B.J. Nicholson, and J.E. Shaw, in prep.). The ectopic gap junctions between AVB and A-class motor neurons that appear in unc-4 mutants may have a similar subunit composition, as unc-9 is the most abundant innexin transcript expressed in A-class motor neurons (Fox et al. 2005; S.E. Von Stetina, J.D. Watson, R.M. Fox, W.C. Spencer, K.L. Olszewski, P.J. Roy, S.K. Kim, and D.M. Miller III, in prep.). It follows that UNC-9 is also a likely candidate for assembly into gap junctions between VA and AVA command interneurons in wild-type animals (Fig. 1; White et al. 1986). Gap junctions with AVB tend to be located on the motor neuron soma (Fig. 6), whereas gap junctions with AVA are more often distributed along the length of the motor neuron partner (White et al. 1986). Thus, unc-4 may orchestrate the assembly of UNC-9 into gap junctions at particular locations within A-class motor neurons and with selected presynaptic partners. Although gap junctions have been previously thought to provide a largely developmental role in the generation of neural networks in higher vertebrates, recent evidence suggests that these “electrical” synapses are also important for neural function in adult nervous systems (Bennett and Zukin 2004). This view is consistent with ultrastructural and immunochemical data showing that gap junctions are widely distributed in the mature mammalian brain and spinal cord (Rash et al. 2000). As the mechanisms that control the specificity of gap junction assembly in the vertebrate CNS are unknown (Hestrin and Galarreta 2005), the discovery of downstream genes that regulate gap junction placement in C. elegans could provide targets for molecular studies in more complex nervous systems. Moreover, the joint regulation by unc-4 (or ceh-12) of the specificity of chemical and electrical synapse formation (White et al. 1992) is indicative of a common nexus for pathways controlling the assembly of both types of synapses.
UNC-4 regulates downstream pathways that function regionally along the A/P axis to control synaptic inputs to VA motor neurons
Our findings indicate that ceh-12 conspires with at least one additional pathway in VA motor neurons to control input specificity (Fig. 7). unc-4 regulation of ceh-12 is restricted to VA motor neurons in the posterior region of the ventral nerve cord (Fig. 4E). Because anterior VA motor neurons are also miswired in unc-4 mutants (Fig. 6), we have proposed that the presumptive downstream pathway functioning in parallel to ceh-12 may be selectively derepressed in anterior VAs (Fig. 7). Other _unc-4_-regulated genes should be represented in our microarray profile of unc-37 mutant VA motor neurons (Fig. 2). One plausible candidate in this data set that could function in parallel to ceh-12 is cog-1, the C. elegans homolog of the homeodomain transcription factor, Nkx6. In Drosophila, dHB9 and Nkx6 act together in ventrally projecting motor neurons to repress dorsal motor neuron traits (Broihier et al. 2004). COG-1 regulates a similar decision in the C. elegans nervous system by preventing ASER sensory neurons from adopting characteristics normally reserved for ASEL (Chang et al. 2003). Potential COG-1 interactions with CEH-12 are suggested by the observation that _cog-1_∷GFP is also expressed in VA and VB motor neurons (J. Schneider and D.M. Miller, unpubl.). cog-1 and other candidate unc-4 target genes in the microarray data set that function in parallel to ceh-12 may be revealed by RNA interference (RNAi) tests currently underway to detect genes that enhance _ceh-12_-dependent suppression of the Unc-4 phenotype (i.e., improved backward locomotion). Conversely, RNAi of transcripts that are depleted in the unc-37 microarray data set (Fig. 2C) and therefore potentially repressed by ectopic ceh-12 should result in an Unc-4 like movement defect if these genes are required for specifying VA-type inputs.
CEH-12 and HB9 specify motor neuron fate
Our results, showing that ceh-12 preserves VB motor neuron fate by repressing VAB-7/Eve, parallels earlier observations that HB9 regulates motor neuron differentiation in flies, birds, and mammals. In Drosophila, dHB9 is expressed in a subset of ventrally projecting motor neurons where it represses the dorsal motor neuron determinant, Eve, and blocks the adoption of a dorsal axon trajectory. Eve, in turn, opposes ventral fates in dorsal motor neurons by reciprocally repressing dHB9 in a Groucho-dependent mechanism (Broihier and Skeath 2002). Interestingly, HB9 is also restricted to ventrally projecting motor neurons in the vertebrate spinal cord where it acts to prevent expression of markers for interneurons arising from the adjacent V2 progenitor domain. In this case, ectopic expression of HB9 in V2 neuroblasts is sufficient to drive expression of motor neuron markers as well as impose motor neuron-like morphological characteristics (i.e., ventral axonal projections) (Arber et al. 1999; Thaler et al. 1999). This dual function of HB9 to block as well as activate expression of motor neuron-specific traits is similar to our finding that CEH-12 inhibits VA motor neuron differentiation while simultaneously promoting a specific VB trait. Together, these observations suggest that the key role of HB9 function in motor neuron differentiation is evolutionarily ancient. In this regard, we note that the UNC-4 homolog, UNCX4.1, is strongly expressed in the V3 neural progenitor domain immediately adjacent to the MN region in which HB9 resides (Mansouri et al. 1997). It will be interesting to determine if UNCX4.1 functions in the V3 domain to block HB9 expression.
Materials and methods
Strains and genetics
Nematode strains were maintained at 20°C–25°C using standard culture techniques (Brenner 1974). ceh-12 deletion alleles, gk391 and tm1619, were obtained from the C. elegans Knock-Out Consortium and the National Bioresource Project, respectively, and outcrossed six times. The wild-type strain was N2. All other strains used in this study are listed in the Supplemental Material.
Construction of unc-4∷3XFlag∷PAB-1 plasmid and transgenic line
PCR was used to amplify the 3XFlag fragment from p3XFlag-CMV7.1 (Sigma) flanked by KpnI and XhoI sites. The 1XFlag in pPRSK9 (_myo-3_∷Flag∷PAB-1) (Roy et al. 2002) was exchanged for the 3XFlag to generate pSV14. The 3.2-kb 3XFlag∷PAB-1 fragment was subcloned from pSV14 into the vector pSL1180 (Amersham) using KpnI and SacI to create pSV15. pSV9-TOPO contains the 3-kb PCR-amplified unc-4 promoter ligated into pCR2.1-TOPO (Invitrogen). The unc-4 promoter was subcloned into pSV15 to generate pSV16 at flanking NheI and KpnI sites. The unc-119 minigene from plasmid MM051 (Maduro and Pilgrim 1995) was ligated into pSV16 to make it suitable for microparticle bombardment (see below). The resulting plasmid (pSV17) contains the unc-119 minigene in the opposite orientation to the _unc-4_∷3XFlag∷PAB-1 fragment. The transgenic line, NC694, was generated by microparticle bombardment with pSV17 as described (Fox et al. 2005). Expression of the Flag epitope in A-class motor neurons was confirmed by immunostaining (Roy et al. 2002). pSV17 expression in unc-4 mutants resulted in slow-growing animals with a synthetic Unc phenotype. Therefore, unc-37(e262) animals (NC714), which were healthy and did not show additional movement defects resulting from the pSV17 plasmid, were used for microarray profiling experiments to identify candidate unc-4 target genes.
RNA isolation, amplification, and hybridization
The isolation of A-class-specific RNAs by mRNA tagging was performed as described (Roy et al. 2002). A detailed protocol is available on request. RNA was amplified using a modified Eberwine method, as described (Fox et al. 2005), with the exception that 25 ng of RNA was used as starting material.
Data analysis
The commercially available Affymetrix C. elegans GeneChip was used for all experiments. Profiles of VA motor neurons from unc-37(e262) mutants (Pflugrad et al. 1997) were compared with reference microarray data from wild-type VAs (S.E. Von Stetina, J.D. Watson, R.M. Fox, W.C. Spencer, K.L. Olszewski, P.J. Roy, S.K. Kim, and D.M. Miller III, in prep.). RMA normalization and SAM analysis were performed as described (Fox et al. 2005) with the following modification: A two-class unpaired analysis identified transcripts that differ by ≥1.7 times from the reference at a false discovery rate (FDR) of ≤5%. Because the penetrance of the transgene (_unc-4_∷3XFlag∷PAB-1) in DA motor neurons is higher in unc-37 mutants than in wild type, the initial list of significantly up-regulated transcripts (395) was filtered to remove genes normally enriched in wild-type DA motor neurons (Fox et al. 2005). The final unc-37 target list contains 255 significantly up-regulated genes and 331 transcripts with reduced intensity values relative to wild type.
Annotation of data sets
Data sets were annotated using a Perl script that searched WormBase release WS140 (http://ws140.wormbase.org; Fox et al. 2005).
BLAST and phylogenetic analysis
A BLAST search using the CEH-12 protein sequence reveals that the Drosophila Hb9 protein is the highest scoring nonnematode protein, suggesting that CEH-12 is a member of the Mnx/HB9 family. Phylogenetic analysis confirms this idea. The protein sequences for Mnx/HB9 family members, UNC-4 family members, and Even-skipped family members were entered into the EMBL-EBI ClustalW Web-based server (http://www.ebi.ac.uk/clustalw) and output as a Phlyogram Guidetree (Fig. 3).
Construction of ceh-12∷GFP reporter lines
A total of 2.8 kb of promoter sequence upstream of the predicted ceh-12 start was obtained by PCR amplification of genomic DNA and cloned into pCR2.1-TOPO (Stratagene). This fragment was subcloned into pPD95.75 (a gift from Andy Fire) to generate _ceh-12_∷GFP. The unc-119 minigene from MM051 (Maduro and Pilgrim 1995) was cloned into _ceh-12_∷GFP as a coselectable marker for biolistic transformation. Three independent transgenic lines (Supplemental Material) were created by microparticle bombardment, as described (Fox et al. 2005).
ceh-12 cDNA generation and transgenic expression of CEH-12
An RT–PCR reaction using 6 μg of mixed-stage N2 total RNA was used to generate ceh-12 cDNA. This 543-base-pair (bp) fragment was gel-purified (Qiagen) and cloned into pCR2.1-TOPO (Stratagene) to create C12ORF-TOPO and used to generate pSV47 (unc-4 promoter∷CEH-12∷unc-54 3′ untranslated region). pSV47 was injected with pJER1 (_myo-3_∷dsRed2) and pCG9 (_acr-5_∷YFP) into N2 animals; five “VA-CEH-12” lines were obtained (Supplemental Material) and analyzed for Unc-4 defects.
PCR detection of ceh-12 deletion alleles
Single-worm PCR was performed as described (Plasterk 1995). ceh-12 il and ceh-12 ir primers were used to detect the ceh-12 (gk391) deletion, and F33D11.4 IL and F33D11.4 IR primers were used to detect the ceh-12 (tm1619) allele. Information on deletion endpoints for can be found at http://www.wormbase.org. Primer sequences are listed in the Supplemental Material.
Movement assays
For each strain tested, 50 L4 hermaphrodites were grown overnight at 20°C [or 25°C for the temperature-sensitive allele, _unc-4(e2322ts)_]. Samples were coded with unrelated names by a nonscorer to ensure that the study was performed blind. The animal was tapped on the head with a platinum wire a maximum of three times. Backward movement was scored as: unable to back (does not move or coils immediately), initiates backing (begins sinusoidal motion backward before stopping or coiling), or sustains backward locomotion (performs backward movement for at least two sinusoidal waves).
Construction of unc-7S∷GFP
The unc-7 locus encodes two isoforms: a long form (UNC-7L; accession no. Q03412), and a shorter form (UNC-7S; T.A. Starich and J.E. Shaw, unpubl.). UNC-7S and UNC-7L proteins differ only in the sequences of their predicted intracellular N termini. To generate a construct that expresses UNC-7S exclusively, a 8.6-kb region flanked by a SalI site located in Exon 2 of _unc-7_L (nucleotide 14,412 of cosmid R07D5) and a BamHI site downstream from the unc-7 locus (nucleotide 5788 of R07D5) was cloned into pBluescript (Stratagene). This genomic segment includes unc-7S promoter/regulatory elements. An in-frame GFP sequence with a translational stop was inserted at a SalI site in the last exon of unc-7, resulting in the predicted deletion of the C-terminal 17 amino acids of UNC-7S in the final expressed product. _unc-7S_∷GFP and _col-19_∷GFP were coinjected and transgenic animals were identified by _col-19_∷GFP expression in the adult hypodermis (Abrahante et al. 1998). UNC-7S∷GFP puncta were diffuse in the wild-type background but localized to tight ventral cord puncta in unc-7 mutants; these animals were also partially rescued for the Unc-7 forward movement defect (T.A. Starich, J. Xi, I.M. Skerrett, B.J. Nicholson, and J.E. Shaw, in prep.). We interpret these results to mean that UNC-7S∷GFP assembles into functional gap junctions that restore locomotory activity to unc-7 mutants.
UNC-7S∷GFP antibody staining and analysis
UNC-7S∷GFP transgenics were methanol-formaldehyde fixed as described (Finney and Ruvkun 1990) and incubated with monoclonal GFP antibodies (Quantum Biotechnologies) and goat anti-mouse IgG-Cy3. The ventral cord motor neurons between the retrovesicular ganglion (RVG) at the anterior end of the cord and the posterior preanal ganglion (PAG) of L4 and adult animals were scored for the association of anti-GFP puncta. (Motor neurons in the RVG and PAG were not scored [e.g., VA1 and VA12] because they are difficult to identify unambiguously in these ganglia.) Results were recorded for each motor neuron of each type in this interval (VA2–VA11, VB3–VB11, DA2–DA7, DB3–DB7, DD2–DD5, VD3–VD11, AS2–AS10, VC1–VC6). All scoring was performed “blind”; genotypes of each sample were unknown to the individuals performing the experiment until after scoring was completed.
The percentages of UNC-7S∷GFP puncta were averaged from three unc-4 alleles (e120, e2322ts, e2323) and unc-37(e262) (Unc) and compared with the averages from ceh-12; unc-4 and ceh-12; unc-37 animals (Ceh). A two-tailed _t_-test was performed to determine if the average UNC-7S∷GFP puncta of Unc was significantly different versus the average of Ceh.
Microscopy
VAB-7 immunostaining was performed as described (Esmaeili et al. 2002). Images were obtained in a Zeiss Axioplan compound microscope equipped with a Hammamatsu Orca camera. Confocal images in Figure 6 were obtained in an Olympus FV1000 equipped with a UV laser for DAPI excitation and pseudocolored for anti-GFP (green) and DAPI (red).
Acknowledgments
We thank John White, Oliver Hobert, Piali Sengupta, Bruce Appel, and members of the Miller laboratory for comments on the manuscript; Villu Maricq for generously providing the _glr-4_∷GFP strain and glr-4(ak78); Bob Barstead and Gary Moulder (C. elegans Knockout Consortium, OMRF, Oklahoma City, OK) for acr-5(ok180) and del-1(ok150); Mark Edgley (C. elegans Knock-Out Consortium, Vancouver, Canada) for ceh12(gk391); Shohei Mitani (National Bioresource Project, Tokyo, Japan) for ceh-12(tm1619); Julie Ahringer for VAB-7 antibody; Sean Schaeffer for help with confocal microscopy; and John Mote, Braden Boone, and Shawn Levy of the VMSR for help with microarray experiments. This work was supported by NIH grants R01 NS26115, R21 MH77302, and P01 DK58212 (to D.M.M.), F31 NS046293 (to R.M.F.), F31 NS043068 (to S.E.V.), T32 HD07502 (to R.M.F. and S.E.V.), GM56367 (to J.E.S.), and P30 CA68485, P60 DK20593, P30 DK58404, HD15052, P30 EY08126, and P01 HL6744. Additional support for the microarray experiments was provided by DK58749 (A.L. George). Funding was also provided by MMF 3183923802 (to J.E.S.).
Footnotes
References
- Abrahante J.E., Miller E.A., Rougvie A.E., Miller E.A., Rougvie A.E., Rougvie A.E. Identification of heterochronic mutants in Caenorhabditis elegans. Temporal misexpression of a collagen∷green fluorescent protein fusion gene. Genetics. 1998;149:1335–1351. doi: 10.1093/genetics/149.3.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S., Han B., Mendelsohn M., Smith M., Jessell T.M., Sockanathan S., Han B., Mendelsohn M., Smith M., Jessell T.M., Sockanathan S., Mendelsohn M., Smith M., Jessell T.M., Sockanathan S., Smith M., Jessell T.M., Sockanathan S., Jessell T.M., Sockanathan S., Sockanathan S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- Arber S., Ladle D.R., Lin J.H., Frank E., Jessell T.M., Ladle D.R., Lin J.H., Frank E., Jessell T.M., Lin J.H., Frank E., Jessell T.M., Frank E., Jessell T.M., Jessell T.M. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–498. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- Bennett M.V., Zukin R.S., Zukin R.S. Electrical coupling and neuronal synchronization in the mammalian brain. Neuron. 2004;41:495–511. doi: 10.1016/s0896-6273(04)00043-1. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broihier H.T., Skeath J.B., Skeath J.B. Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron. 2002;35:39–50. doi: 10.1016/s0896-6273(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Broihier H.T., Kuzin A., Zhu Y., Odenwald W., Skeath J.B., Kuzin A., Zhu Y., Odenwald W., Skeath J.B., Zhu Y., Odenwald W., Skeath J.B., Odenwald W., Skeath J.B., Skeath J.B. Drosophila homeodomain protein Nkx6 coordinates motoneuron subtype identity and axonogenesis. Development. 2004;131:5233–5242. doi: 10.1242/dev.01394. [DOI] [PubMed] [Google Scholar]
- Bruzzone R., Hormuzdi S.G., Barbe M.T., Herb A., Monyer H., Hormuzdi S.G., Barbe M.T., Herb A., Monyer H., Barbe M.T., Herb A., Monyer H., Herb A., Monyer H., Monyer H. Pannexins, a family of gap junction proteins expressed in brain. Proc. Natl. Acad. Sci. 2003;100:13644–13649. doi: 10.1073/pnas.2233464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Sulston J.E., White J.G., Southgate E., Thomson J.N., Brenner S., Sulston J.E., White J.G., Southgate E., Thomson J.N., Brenner S., White J.G., Southgate E., Thomson J.N., Brenner S., Southgate E., Thomson J.N., Brenner S., Thomson J.N., Brenner S., Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J. Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Johnston R.J., Jr., Hobert O., Johnston R.J., Jr., Hobert O., Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes & Dev. 2003;17:2123–2137. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.H., Hippenmeyer S., Arber S., Frank E., Hippenmeyer S., Arber S., Frank E., Arber S., Frank E., Frank E. Development of the monosynaptic stretch reflex circuit. Curr. Opin. Neurobiol. 2003;13:96–102. doi: 10.1016/s0959-4388(03)00006-0. [DOI] [PubMed] [Google Scholar]
- Chen A.I., de Nooij J.C., Jessell T.M., de Nooij J.C., Jessell T.M., Jessell T.M. Graded activity of transcription factor Runx3 specifies the laminar termination pattern of sensory axons in the developing spinal cord. Neuron. 2006;49:395–408. doi: 10.1016/j.neuron.2005.12.028. [DOI] [PubMed] [Google Scholar]
- Esmaeili B., Ross J.M., Neades C., Miller D.M., III, Ahringer J., Ross J.M., Neades C., Miller D.M., III, Ahringer J., Neades C., Miller D.M., III, Ahringer J., Miller D.M., III, Ahringer J., Ahringer J. The C. elegans even-skipped homologue, vab-7,specifies DB motoneurone identity and axon trajectory. Development. 2002;129:853–862. doi: 10.1242/dev.129.4.853. [DOI] [PubMed] [Google Scholar]
- Finney M., Ruvkun G., Ruvkun G. The unc-86gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Fox R.M., Von Stetina S.E., Barlow S.J., Shaffer C., Olszewski K.L., Moore J.H., Dupuy D., Vidal M., Miller D.M., III, Von Stetina S.E., Barlow S.J., Shaffer C., Olszewski K.L., Moore J.H., Dupuy D., Vidal M., Miller D.M., III, Barlow S.J., Shaffer C., Olszewski K.L., Moore J.H., Dupuy D., Vidal M., Miller D.M., III, Shaffer C., Olszewski K.L., Moore J.H., Dupuy D., Vidal M., Miller D.M., III, Olszewski K.L., Moore J.H., Dupuy D., Vidal M., Miller D.M., III, Moore J.H., Dupuy D., Vidal M., Miller D.M., III, Dupuy D., Vidal M., Miller D.M., III, Vidal M., Miller D.M., III, Miller D.M., III A gene expression fingerprint of C. elegans embryonic motor neurons. BMC Genomics. 2005;6:42. doi: 10.1186/1471-2164-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.C. Development of specific connectivity between premotor neurons and motoneurons in the brain stem and spinal cord. Physiol. Rev. 2000;80:615–647. doi: 10.1152/physrev.2000.80.2.615. [DOI] [PubMed] [Google Scholar]
- Hestrin S., Galarreta M., Galarreta M. Electrical synapses define networks of neocortical GABAergic neurons. Trends Neurosci. 2005;28:304–309. doi: 10.1016/j.tins.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Jimenez G., Paroush Z., Ish-Horowicz D., Paroush Z., Ish-Horowicz D., Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes & Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Pfaff S.L., Pfaff S.L. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat. Neurosci. 2001;4 (Suppl.):1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Jurata L.W., Funahashi J., Ruiz E.C., Pfaff S.L., Jurata L.W., Funahashi J., Ruiz E.C., Pfaff S.L., Funahashi J., Ruiz E.C., Pfaff S.L., Ruiz E.C., Pfaff S.L., Pfaff S.L. Analysis of embryonic motoneuron gene regulation: Derepression of general activators function in concert with enhancer factors. Development. 2004;131:3295–3306. doi: 10.1242/dev.01179. [DOI] [PubMed] [Google Scholar]
- Lickteig K.M., Duerr J.S., Frisby D.L., Hall D.H., Rand J.B., Miller D.M., III, Duerr J.S., Frisby D.L., Hall D.H., Rand J.B., Miller D.M., III, Frisby D.L., Hall D.H., Rand J.B., Miller D.M., III, Hall D.H., Rand J.B., Miller D.M., III, Rand J.B., Miller D.M., III, Miller D.M., III Regulation of neurotransmitter vesicles by the homeodomain protein UNC-4 and its transcriptional corepressor UNC-37/groucho in Caenorhabditis elegans cholinergic motor neurons. J. Neurosci. 2001;21:2001–2014. doi: 10.1523/JNEUROSCI.21-06-02001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M., Pilgrim D., Pilgrim D. Identification and cloning of unc-119,a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A., Yokota Y., Wehr R., Copeland N.G., Jenkins N.A., Gruss P., Yokota Y., Wehr R., Copeland N.G., Jenkins N.A., Gruss P., Wehr R., Copeland N.G., Jenkins N.A., Gruss P., Copeland N.G., Jenkins N.A., Gruss P., Jenkins N.A., Gruss P., Gruss P. Paired-related murine homeobox gene expressed in the developing sclerotome, kidney, and nervous system. Dev. Dyn. 1997;210:53–65. doi: 10.1002/(SICI)1097-0177(199709)210:1<53::AID-AJA6>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Miller D.M., III, Niemeyer C.J., Niemeyer C.J. Expression of the unc-4homeoprotein in Caenorhabditis elegans motor neurons specifies presynaptic input. Development. 1995;121:2877–2886. doi: 10.1242/dev.121.9.2877. [DOI] [PubMed] [Google Scholar]
- Miller D.M., Shen M.M., Shamu C.E., Burglin T.R., Ruvkun G., Dubois M.L., Ghee M., Wilson L., Shen M.M., Shamu C.E., Burglin T.R., Ruvkun G., Dubois M.L., Ghee M., Wilson L., Shamu C.E., Burglin T.R., Ruvkun G., Dubois M.L., Ghee M., Wilson L., Burglin T.R., Ruvkun G., Dubois M.L., Ghee M., Wilson L., Ruvkun G., Dubois M.L., Ghee M., Wilson L., Dubois M.L., Ghee M., Wilson L., Ghee M., Wilson L., Wilson L. C. elegans unc-4gene encodes a homeodomain protein that determines the pattern of synaptic input to specific motor neurons. Nature. 1992;355:841–845. doi: 10.1038/355841a0. [DOI] [PubMed] [Google Scholar]
- Muhr J., Andersson E., Persson M., Jessell T.M., Ericson J., Andersson E., Persson M., Jessell T.M., Ericson J., Persson M., Jessell T.M., Ericson J., Jessell T.M., Ericson J., Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Panchin Y.V. Evolution of gap junction proteins—The pannexin alternative. J. Exp. Biol. 2005;208:1415–1419. doi: 10.1242/jeb.01547. [DOI] [PubMed] [Google Scholar]
- Pflugrad A., Meir J.Y., Barnes T.M., Miller D.M., III, Meir J.Y., Barnes T.M., Miller D.M., III, Barnes T.M., Miller D.M., III, Miller D.M., III The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4to govern motor neuron identity in C. elegans. Development. 1997;124:1699–1709. doi: 10.1242/dev.124.9.1699. [DOI] [PubMed] [Google Scholar]
- Phelan P., Starich T.A., Starich T.A. Innexins get into the gap. Bioessays. 2001;23:388–396. doi: 10.1002/bies.1057. [DOI] [PubMed] [Google Scholar]
- Plasterk R.H. Reverse genetics: From gene sequence to mutant worm. Methods Cell Biol. 1995;48:59–80. doi: 10.1016/s0091-679x(08)61383-7. [DOI] [PubMed] [Google Scholar]
- Rash J.E., Staines W.A., Yasumura T., Patel D., Furman C.S., Stelmack G.L., Nagy J.I., Staines W.A., Yasumura T., Patel D., Furman C.S., Stelmack G.L., Nagy J.I., Yasumura T., Patel D., Furman C.S., Stelmack G.L., Nagy J.I., Patel D., Furman C.S., Stelmack G.L., Nagy J.I., Furman C.S., Stelmack G.L., Nagy J.I., Stelmack G.L., Nagy J.I., Nagy J.I. Immunogold evidence that neuronal gap junctions in adult rat brain and spinal cord contain connexin-36 but not connexin-32 or connexin-43. Proc. Natl. Acad. Sci. 2000;97:7573–7578. doi: 10.1073/pnas.97.13.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P.J., Stuart J.M., Lund J., Kim S.K., Stuart J.M., Lund J., Kim S.K., Lund J., Kim S.K., Kim S.K. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Sharma K., Leonard A.E., Lettieri K., Pfaff S.L., Leonard A.E., Lettieri K., Pfaff S.L., Lettieri K., Pfaff S.L., Pfaff S.L. Genetic and epigenetic mechanisms contribute to motor neuron pathfinding. Nature. 2000;406:515–519. doi: 10.1038/35020078. [DOI] [PubMed] [Google Scholar]
- Shirasaki R., Pfaff S.L., Pfaff S.L. Transcriptional codes and the control of neuronal identity. Annu. Rev. Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Starich T., Sheehan M., Jadrich J., Shaw J., Sheehan M., Jadrich J., Shaw J., Jadrich J., Shaw J., Shaw J. Innexins in C. elegans. Cell Commun. Adhes. 2001;8:311–314. doi: 10.3109/15419060109080744. [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Horvitz H.R., Horvitz H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Thaler J., Harrison K., Sharma K., Lettieri K., Kehrl J., Pfaff S.L., Harrison K., Sharma K., Lettieri K., Kehrl J., Pfaff S.L., Sharma K., Lettieri K., Kehrl J., Pfaff S.L., Lettieri K., Kehrl J., Pfaff S.L., Kehrl J., Pfaff S.L., Pfaff S.L. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23:675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- Von Stetina S.E., Treinin M., Miller D.M., III, Treinin M., Miller D.M., III, Miller D.M., III The motor circuit. Int. Rev. Neurobiol. 2005;69:125–167. doi: 10.1016/S0074-7742(05)69005-8. [DOI] [PubMed] [Google Scholar]
- White J.G., Southgate E., Thomson J.N., Brenner S., Southgate E., Thomson J.N., Brenner S., Thomson J.N., Brenner S., Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;B314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- White J.G., Southgate E., Thomson J.N., Southgate E., Thomson J.N., Thomson J.N. Mutations in the Caenorhabditis elegans unc-4gene alter the synaptic input to ventral cord motor neurons. Nature. 1992;355:838–841. doi: 10.1038/355838a0. [DOI] [PubMed] [Google Scholar]
- William C.M., Tanabe Y., Jessell T.M., Tanabe Y., Jessell T.M., Jessell T.M. Regulation of motor neuron subtype identity by repressor activity of Mnx class homeodomain proteins. Development. 2003;130:1523–1536. doi: 10.1242/dev.00358. [DOI] [PubMed] [Google Scholar]
- Winnier A.R., Meir J.Y., Ross J.M., Tavernarakis N., Driscoll M., Ishihara T., Katsura I., Miller D.M., III, Meir J.Y., Ross J.M., Tavernarakis N., Driscoll M., Ishihara T., Katsura I., Miller D.M., III, Ross J.M., Tavernarakis N., Driscoll M., Ishihara T., Katsura I., Miller D.M., III, Tavernarakis N., Driscoll M., Ishihara T., Katsura I., Miller D.M., III, Driscoll M., Ishihara T., Katsura I., Miller D.M., III, Ishihara T., Katsura I., Miller D.M., III, Katsura I., Miller D.M., III, Miller D.M., III UNC-4/UNC-37-dependent repression of motor neuron-specific genes controls synaptic choice in Caenorhabditis elegans. Genes & Dev. 1999;13:2774–2786. doi: 10.1101/gad.13.21.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]