Whole-genome ChIP–chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo (original) (raw)
Abstract
Genetic studies have identified numerous sequence-specific transcription factors that control development, yet little is known about their in vivo distribution across animal genomes. We determined the genome-wide occupancy of the dorsoventral (DV) determinants Dorsal, Twist, and Snail in the Drosophila embryo using chromatin immunoprecipitation coupled with microarray analysis (ChIP–chip). The in vivo binding of these proteins correlate tightly with the limits of known enhancers. Our analysis predicts substantially more target genes than previous estimates, and includes Dpp signaling components and anteroposterior (AP) segmentation determinants. Thus, the ChIP–chip data uncover a much larger than expected regulatory network, which integrates diverse patterning processes during development.
Keywords: Dorsoventral patterning, segmentation, gene regulatory networks, ChIP–chip
Systematic genetic studies of embryonic development have revealed a key role for sequence-specific transcription factors in axis formation and cell differentiation (St Johnston and Nusslein-Volhard 1992). However, it has been difficult to unravel the detailed regulatory interactions through which these transcription factors regulate development. One of the best-studied gene regulatory networks controls the dorsoventral (DV) patterning of the Drosophila embryo (see Stathopoulos et al. 2002). This network is initiated by a nuclear concentration gradient of the NF-κB transcription factor Dorsal, which is established by differential activation of the Toll receptor in ventral and ventrolateral regions of precellular embryos (for review, see Moussian and Roth 2005). The ensuing Dorsal gradient leads to the differential regulation of at least 50 target genes across the DV axis. Approximately 40 of these genes encode either sequence-specific transcription factors or components of the Notch, EGF, FGF, and Dpp signaling pathways, which further specify diverse cell fates across the DV axis (e.g., see Stathopoulos et al. 2002). More than 100 functional interconnections among these regulatory factors drive the initial phases of DV patterning (Levine and Davidson 2005).
At the core of the DV patterning network are Dorsal and the products encoded by two of its earliest target genes, Twist and Snail. Twist functions as a basic helix–loop–helix (bHLH) activator, while Snail is a zinc-finger repressor. At least half of the tissue-specific enhancers that are regulated by different levels of Dorsal also contain binding sites for Twist and/or Snail (Stathopoulos et al. 2002; Markstein et al. 2004). Our current understanding of the DV patterning network is based on decades of genetic screens (e.g., see Nusslein-Volhard et al. 1980), investigations of genetic interactions (e.g., see Simpson 1983; Kosman et al. 1991; Leptin 1991), lacZ reporter assays (e.g., see Jiang et al. 1991), in vitro DNA-binding assays (e.g., see Huang et al. 1993), and more recently, bioinformatics methods (Markstein et al. 2002, 2004; Papatsenko and Levine 2005). This network has served as a model for understanding axis formation, tissue specification, and cell shape changes during gastrulation (Ip and Gridley 2002). Nonetheless, the genome-wide physical contacts through which the relevant transcription factors interact with the genome and regulate gene expression in vivo are essentially unknown.
Here we determine the genome-wide occupancy of Dorsal, Twist, and Snail in the Drosophila embryo using chromatin immunoprecipitation coupled with microarray analysis (ChIP–chip). These assays accurately identify known DV enhancers and further identify several hundred additional potential target enhancers, a dramatic increase over previous estimates. We present evidence that a significant fraction of these enhancers is functional, and suggest that extensive integration occurs between diverse patterning networks, including those controlling DV patterning and segmentation.
Results and Discussion
Known enhancers are bound by Dorsal, Twist, and Snail
ChIP–chip assays were performed with antibodies directed against Dorsal, Twist, or Snail on Toll10b mutant embryos, aged 2–4 h. These embryos contain a constitutively activated form of the Toll receptor, which results in high levels of nuclear Dorsal protein and uniform expression of Twist and Snail throughout the embryo (e.g., see Stathopoulos et al. 2002). The high levels of Dorsal, Twist, and Snail cause all cells to form derivatives of the mesoderm at the expense of neurogenic and dorsal ectoderm. Thus, these embryos represent a uniform cell type with respect to DV fate.
The whole-genome ChIP–chip experiments reveal several hundred strong binding clusters of Dorsal, Twist, and Snail with up to 40-fold ChIP enrichment, most of which span regions of ∼1 kb in length. To identify the binding patterns of bona fide target enhancers of the Dorsal regulatory network, we first analyzed known enhancers. The 22 known enhancers fall into three classes—type 1, type 2, and type 3—based on which levels of nuclear Dorsal regulate their expression (Supplementary Table S1; see Biemar et al. 2006).
The 10 type 1 enhancers (associated with twi, sna, miR-1, htl, hbr, mes3, CG12177, ady43A, tin, and Phm) are activated by peak levels of Dorsal in the presumptive mesoderm, and are all constitutively activated in Toll10B mutant embryos (e.g., see Stathopoulos et al. 2002; Biemar et al. 2006). The ChIP–chip experiments identify strong binding peaks (greater than fivefold enrichment) of Dorsal, Twist, and Snail (DTS) within five of the 10 enhancers (twi, sna, miR-1, CG12177 and Phm) (Fig. 1A,B). Another three enhancers, those associated with htl, tin, and ady43A, show significant but lower (less than fivefold) binding peaks restricted to Twist and Snail (TS) binding. This observation is consistent with earlier studies indicating that these enhancers might be primarily activated by Twist (Papatsenko and Levine 2005). Hence, eight of the 10 known type 1 enhancers exhibit significant in vivo occupancy by Twist and Snail.
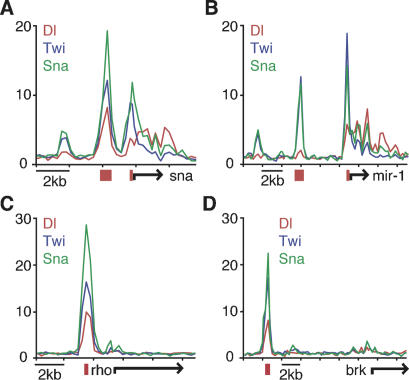
Figure 1.
Drosophila ChIP–chip identifies known enhancers of Dorsal, Twist, and Snail. Drosophila embryos, aged 2–4 h, from Toll10b mothers were used to perform ChIP using antibodies against Dorsal, Twist, and Snail. Most enhancers known to be regulated by these factors were successfully identified, including type 1 target genes sna (A), mir-1 (B), and type 2 target genes rho (C) and brk (D). The graphs show unprocessed ChIP enrichment ratios (_Y_-axis) for Dorsal (red), Twist (blue), and Snail (green), across chromosomal regions (_X_-axis). Gene model (black arrow) and the known enhancer (red) are shown below.
An even greater correspondence between known enhancers and in vivo occupancy is seen for the type 2 [sim, E(spl), vn, rho, vnd and _brk_] and type 3 enhancers (ths, sog, ind, dpp, zen and tld), which are regulated by intermediate and low levels of the Dorsal gradient, respectively (Fig. 1C,D). All 12 enhancers are silenced in Toll10B mutant embryos due to constitutive expression of the Snail repressor (e.g., see Biemar et al. 2006). Remarkably, every enhancer exhibits strong DTS or TS peaks with greater than fivefold enrichment in the ChIP–chip assays. Thus, ChIP–chip assays correctly identified 20 of the 22 known Dorsal target enhancers.
Genome-wide identification of putative target enhancers
Most known DV enhancers are associated with overlapping binding clusters of Dorsal, Twist, and Snail regardless of whether they mediate activation or repression. Moreover, 17 of the 20 binding clusters at known enhancers display greater than fivefold enrichment of Twist and/or Snail. Using these binding criteria, we identified 428 high-confidence DTS regions and 433 high-confidence TS regions across the genome (see also Supplemental Material).
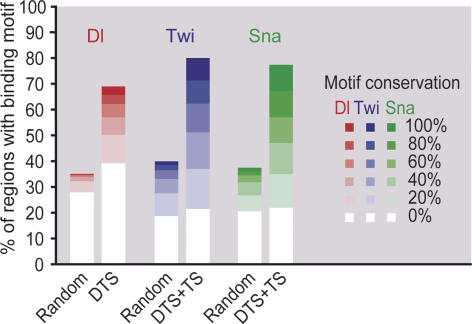
To confirm these regions through independent evidence, we performed sequence analysis on these regions using the known consensus binding motifs of Dorsal, Twist, and Snail. As expected, the identified regions are highly enriched in all three binding motifs (Fig. 2). Moreover, a large fraction of the motifs is conserved across the 12 sequenced Drosophila species providing evidence that the discovered regions are functionally important (Fig. 2). Finally, when motifs that are enriched in these regions were identified de novo, the known binding motifs can be rediscovered (Supplemental Material). Hence, the regions we identified represent putative target gene enhancers of the DV network.
Figure 2.
Enrichment of Dorsal-, Twist-, and Snail-binding motifs in bound regions. Sequences from DTS and TS regions with greater than fivefold ChIP enrichment were searched for the presence of the known Dorsal (red), Twist (blue), or Snail (red) motifs, and the evolutionary conservation of these motifs across the 12 sequenced Drosophila genomes was determined (as percent of branch length within the phylogenetic tree). The graph shows the fraction of regions at each conservation cutoff (0%, 20%, 40%, 60%, 80%, and 100%). As a control, the same analysis was performed with regions of identical length that were randomly distributed among intronic and intergenic regions. Using a χ2 test, the motif enrichment and evolutionary conservation was highly significant for Dorsal (p < 10−148), Twist (p < 10−228), and Snail (p < 10−306).
Identification of new DV enhancers
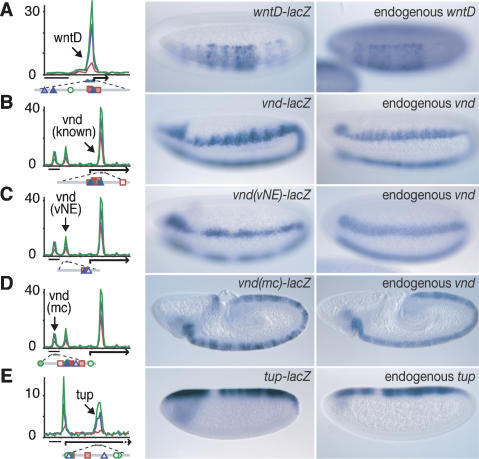
To show that newly identified regions indeed function as enhancers in vivo, we selected putative enhancers of primary DV genes; i.e., those genes that are expressed as localized stripes across the DV axis (Stathopoulos et al. 2002; Biemar et al. 2006). In addition to the 22 known DV enhancers, we identified 47 new putative enhancers, some of which appear to regulate the same gene (Supplementary Table S1). By attaching the genomic sequence to a lacZ reporter and expressing the construct in transgenic embryos, we show that seven of these enhancers are bona fide DV enhancers and that regulation by multiple enhancers occurs (Fig. 3; Supplemental Material).
Figure 3.
Identification of novel DV enhancers. ChIP–chip data identified enhancers for the DV genes wntD (A), vnd (B–D), and tup (E). The left column shows the ChIP enrichment ratios of Dorsal (red), Twist (blue), and Snail (green). Bar, 2 kb. The tested enhancer (arrow) is shown with transcription factor motifs below (Dorsal site, red square; Twist site, blue triangle; Snail site, green circle; shading corresponds to their conservation across the 12 sequenced Drosophila genomes). The pattern of enhancer_-_driven lacZ expression in transgenic embryos (middle column) resembles that of the respective endogenous genes (right column). Unexpectedly, in addition to the previously known vnd enhancer (B), two regulatory regions (C,D) were identified that drive lacZ expression in subsets of the endogenous vnd expression pattern: The region in C drives early expression in the vNE, while the region in D drives later expression in the medial column (mc). Embryos are oriented anterior to the left, dorsal is up.
The wntD gene is expressed in portions of the presumptive mesoderm where it mediates feedback inhibition of Toll signaling (Ganguly et al. 2005; Gordon et al. 2005). A cluster of DTS-binding peaks was identified in the 5′-flanking region, and the corresponding genomic DNA fragment mediates lacZ expression in the same region of the mesoderm as the endogenous gene (Fig. 3A). Similar results were obtained with the DTS-binding cluster located in the 5′-flanking region of mes5/mdr49 (Supplemental Material).
The vnd locus contains a well-documented intronic enhancer that mediates expression in the neurogenic ectoderm and recapitulates the spatial and temporal expression pattern of the endogenous gene (Stathopoulos et al. 2002; Markstein et al. 2004). The ChIP–chip analysis detected this enhancer but also revealed two novel clusters further upstream (Fig. 3B). When tested for lacZ reporter activity, these novel genomic sequences directed lacZ expression in a pattern resembling that of the endogenous gene over different time periods: One directs early vnd expression in the presumptive ventral neurogenic ectoderm (vNE) (Fig. 3C) while the other directs later expression in the medial column (mc) (Fig. 3D) of the developing nervous system. All three enhancers contain evolutionarily conserved binding sites for Dorsal, Twist, and Snail, suggesting that the enhancers are not redundant but may function to fine-tune the vnd expression pattern. Overlapping enhancer activity was also observed for multiple miR-1 enhancers (Supplemental Material). Overall, as many as a third of all DV genes have multiple binding clusters, and thus might be subject to similar regulatory control.
Several of the occupied regions are associated with Dpp target genes expressed in the dorsal ectoderm. When the tup and pnr intronic sequences are tested in transgenic embryos, both fragments function as authentic enhancers and direct localized expression in the dorsal ectoderm, comparable to the endogenous tup and pnr expression patterns (Fig. 3E; Supplemental Material). These results suggest that the Dorsal patterning network directly regulates the expression of Dpp target genes (see below).
Connections between the DV and the anterioposterior (AP) patterning networks
We noticed that many of the new DTS/TS clusters are associated with AP genes involved in segmentation (Supplementary Table S2). Although classical genetic studies argue that AP and DV patterning of the early embryo are controlled by separate maternal genetic programs (for review, see St Johnston and Nusslein-Volhard 1992), it is conceivable that the expression of AP target genes is modulated by the DV network. Indeed, DV modulation of segmentation gene expression has been observed previously (e.g., see Carroll et al. 1987; Liaw and Lengyel 1993; Rothe et al. 1994; Gao and Finkelstein 1998).
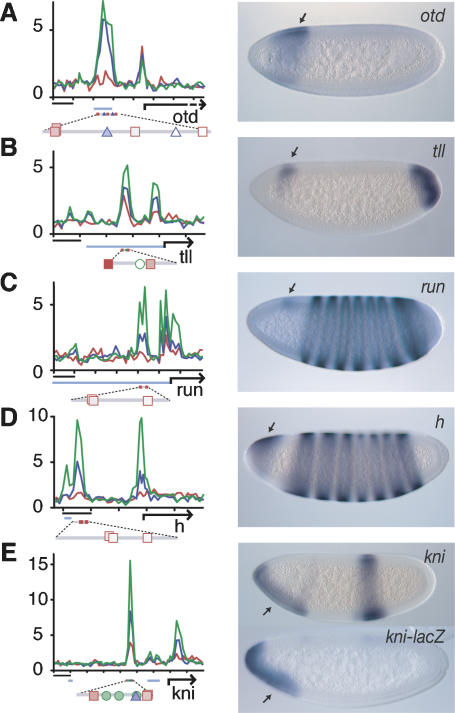
The gap gene orthodenticle (otd) is expressed in two stripes across the AP axis in the early embryo. The anterior stripe shows diminished expression on the ventral side (Fig. 4A). Previous studies identified a 5′ enhancer that recapitulates the normal expression pattern, including Dorsal-dependent suppression in ventral regions (Gao and Finkelstein 1998). ChIP–chip identified a strong DTS cluster within the limits of this enhancer (Fig. 4A). A similar DV bias in the expression pattern was found for the gap gene tailless (tll) (Fig. 4B) and the pair-rule genes runt and hairy (Fig. 4C,D). In each case, the regions identified by ChIP–chip overlap or map close to known regulatory regions (Pankratz et al. 1990; Butler et al. 1992; Liaw and Lengyel 1993) and contain several Dorsal-binding motifs (Fig. 4B–D).
Figure 4.
Regulation of AP genes by Dorsal, Twist, and Snail. ChIP–chip binding data of Dorsal (red), Twist (blue), and Snail (green) at the loci of AP-regulated genes are shown in the left column (fold enrichment is indicated on the _Y_-axis): otd (A), tll (B), run (C), h (D), and kni (E). Bar, 2 kb. The presence of binding motifs and their conservation across the 12 sequenced Drosophila genomes at the occupied regions are indicated below the graphs (Dorsal site, red square; Twist site, blue triangle; Snail site, green circle). Many identified regions overlap with previously identified regulatory regions (light blue). Association with, and hence regulation by, Dorsal, Twist, and Snail may be responsible for the DV modulation exhibited by these AP genes, as shown in the right column (arrows). Embryos are oriented anterior to the left, dorsal is up.
At the gap gene knirps, a DTS-binding cluster was found in a region distinct from the known Bicoid-dependent enhancer (Fig. 4E; Pelegri and Lehmann 1994). This newly identified genomic region functions as a bona fide enhancer directing expression in the anteroventral domain like endogenous knirps (Fig. 4E). Thus, the ChIP–chip analysis identified novel AP regulatory regions modulated by DV activity.
In summary, many segmentation genes contain DTS/TS-binding clusters, and at least some of these regions modulate gene expression across the DV axis, particularly in anterior regions of the embryo. We conclude that the Dorsal gradient does not only regulate primary DV target genes, but rather appears to fine-tune a large number of genes that do not contribute to DV axis formation themselves, at least based on their known genetic function.
Broad integration of patterning networks
Many DTS/TS-binding clusters are also found at genes encoding signal transduction components (Supplementary Table S3). Analysis of the network formed by these pathways suggests that the Dorsal gradient controls the expression of many target genes by multiple regulatory pathways (Fig. 5).
Figure 5.
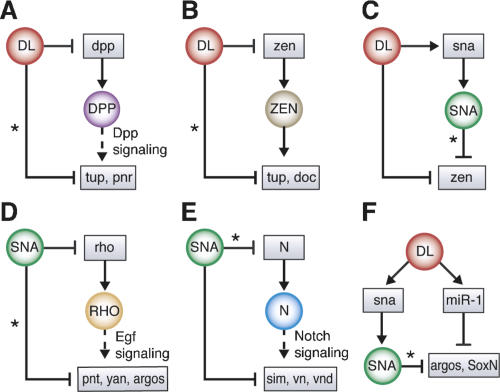
Integration of new putative target genes into the Dorsal network. Many newly identified putative target genes of Dorsal, Twist, and Snail are genes that were previously thought to be induced further downstream in the DV patterning network. Thus, Dorsal appears to regulate many target genes both directly and indirectly (a configuration also known as a feed-forward motif) in the suppression of dorsal ectodermal fate (A–C) and neurectodermal fate (D–F). Newly identified connections are marked by a star. Examples of such configurations are shown for target genes of Dpp signaling (A), which are also directly regulated by Dorsal; target genes of the transcription factor Zen (B), which are also directly regulated by Dorsal; target gene suppression by Dorsal (C), which can also be mediated by Snail; targets of EGF signaling (D), which are regulated by Snail at multiple levels; targets of Notch signaling (E), which are also regulated by Snail at multiple levels; and suppression of target genes by Snail or through induction of microRNAs (F). The regulatory relationship (activation, arrow; repression, orthogonal bar) of new connections was inferred based on the tissue-specific expression patterns of the target genes (Stathopoulos et al. 2002; Biemar et al. 2006).
Dorsal directly represses Dpp expression in the mesoderm and neuroectoderm, leading to localized Dpp signaling in the dorsal ectoderm (Huang et al. 1993). Dpp activates a variety of genes, including tup and pnr. Our accurate identification of intronic tup and pnr enhancers suggests that these genes are directly regulated by the Snail repressor (Fig. 3E), in addition to indirect regulation by the Dorsal gradient via Dpp signaling (Fig. 5A). zen is another well-known target gene of Dorsal in the dorsal ectoderm, and its product, a homeodomain transcription factor, functions synergistically with Dpp signaling (e.g., see Xu et al. 2005). Target genes of Zen also appear to be subject to additional regulation by the Dorsal gradient (Fig. 5B). In the dorsal ectoderm, Dorsal may regulate gene expression by two mechanisms: direct repression (Kirov et al. 1993), and indirect repression via Snail (Fig. 5C).
Similar network configurations regulate gene expression in the neuroectoderm. High levels of Dorsal repress the expression of rho via Snail in the mesoderm (e.g., see Zinzen et al. 2006), thereby blocking EGF signaling in Toll10b mutant embryos. Our ChIP–chip data suggest that the Dorsal network regulates additional genes encoding EGF signaling components as well as EGF target genes such as pnt, aop/yan, and argos (Fig. 5D). In the case of Notch signaling, it is known that the Dorsal network represses Notch target genes such as sim in Toll10B mutant embryos through Snail (e.g., see Markstein et al. 2004). The Dorsal network may also regulate Notch signaling more directly, by suppressing genes encoding components of the signaling pathway including Notch itself (Fig. 5E).
Although repression of neuroectodermal target genes is likely to occur predominantly through Snail, Dorsal also induces the expression of a number of microRNAs in Toll10b mutant embryos, including miR-1 (Biemar et al. 2005; Sokol and Ambros 2005). We found that some of the neuroectodermal genes repressed by Snail are also predicted targets of these microRNAs (Fig. 5F). Hence, there may be multiple tiers of repression in the DV system, similar to the activities of the gap repressors in the AP system (e.g., see Clyde et al. 2003).
In summary, the present ChIP–chip study revealed an unexpectedly broad distribution of binding peaks for Dorsal, Twist, and Snail in the genome, and suggests extensive integration of the Dorsal regulatory network with additional patterning processes, such as Dpp signaling in the dorsal ectoderm and segmentation across the AP axis. In addition to the observed tight correlation between binding peaks and known enhancers, two lines of evidence suggest that a significant fraction of the newly identified regions is functional: First, the bound regions are highly enriched in evolutionarily conserved Dorsal, Twist, and Snail sequence motifs; and, second, several of the identified enhancers were experimentally confirmed by lacZ reporter gene expression in transgenic embryos. Thus, while genetic studies identified core sets of regulators for each developmental process in Drosophila, we found that gene regulation integrates information more widely from several different systems. It is likely that integration of diverse patterning processes will also apply to mammalian development, including stem cell differentiation.
Materials and methods
ChIP–chip experiments
The ChIP–chip protocol was developed based on protocols from the Young and Maschat laboratories (Chanas et al. 2004; Lee et al. 2006; see Supplemental Material). Cross-linking of the embryos was performed for 5 min in a 5% formaldehyde/hexane solution (Toth and Biggin 2000). The Cy5-labeled amplified immunoprecipated DNA and the Cy3-labeled amplified input DNA were hybridized to whole-genome Drosophila arrays using the CGH protocol and CGH reagents provided by Agilent Technologies.
Drosophila whole-genome array design
A set of 11 arrays was designed to cover the entire non-repeat-masked euchromatic regions of Drosophila melanogaster. This was achieved by 446,931 features (60mers) that were spaced on average ∼280 base pairs (bp) from each other. Eight-hundred-sixty-six control spots (oligos from desert regions, the middle of long exons, Arabidopsis thaliana, and repetitive sequences) were added to each array. The array was printed by Agilent Technologies.
Drosophila stocks
ChIP assays were performed on 2–4-h Toll10b embryos (Stathopoulos et al. 2002). P-element transformations and in situ hybridizations were performed with the yw67 wild-type strain as described (e.g., see Stathopoulos et al. 2002).
Antibodies and beads
Polyclonal antibodies were raised against the HIS-purified (1) N-terminal 50 amino acids of Dorsal in rabbits, (2) full-length Twist in rats, and (3) full-length Snail in guinea pigs (Zinzen et al. 2006). Antibodies were preabsorbed overnight against ∼12–24-h embryos (Dorsal), or against overnight collections of dorsalized embryos derived from the gd7/FM3 strain (Stathopoulos et al. 2002). The antibodies were then incubated with magnetic Dynabeads coated with protein A (Dorsal, Snail) or protein G (Twist) overnight.
Data processing
Processing of the raw data through an error model and identification of bound regions were performed as previously described for yeast (Pokholok et al. 2005). To identify DTS and TS regions, the union of Twist and Snail regions was determined and regions with an at least fivefold enrichment of either Twist or Snail were selected. Regions with at least twofold enrichment of Dorsal qualified as DTS regions and the remaining regions were called TS regions.
Gene assignment
First, the bound regions located within 2 kb from known gene models were assigned to that gene. The remaining binding regions were then assigned to the gene with the closest transcription start site.
Motif analysis
The binding motifs for Dorsal, Twist, and Snail used for the analysis were obtained from Markstein et al. (2002, 2004), Papatsenko and Levine (2005), Zinzen et al. (2006), Jaspar (MA0022, MA0023, MA0086), and Transfac (M00043, M00120, M00044, M00060). The motifs were matched to the genome of D. melanogaster and the orthologous regions of 11 additional sequenced drosophilid genomes. Conservation was assessed by the relative total branch length (substitutions per neutrally evolving site) of the phylogenetic tree connecting D. melanogaster with the other species.
Cloning, injection, and visualization of enhancers
DNA fragments were PCR-amplified from genomic DNA (for primers, see Supplemental Material), T/A-cloned into the Promega pGEM T-Easy vector, and subcloned via NotI into the gypsy-insulated pCaSpeR vector E2G (Markstein et al. 2004). Each construct was introduced into the germline by microinjection and three to eight independent transgenic lines were tested. Embryos were collected and fixed according to standard methods, hybridized with digoxygenin-labeled antisense RNA probes against lacZ or the endogenous gene, and visualized colorimetrically (Jiang et al. 1991).
Acknowledgments
We thank J. Love for microarray experimental support, F. Lewitter and B. Yuan for help in the Drosophila whole-genome microarray design, E. Herbolsheimer for bioinformatics support, X. Li for technical advice, and M. Markstein, F. Biemar, and Young laboratory members for discussions. The work was supported by NIH grants HG002668 and GM069676 (to R.A.Y.), GM46638 (to M.L.), and a grant by the Moore Foundation and a postdoctoral fellowship by Schering (to A.S.). R.A.Y. consults for Agilent Technologies.
Footnotes
References
- Biemar F., Zinzen R., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Zinzen R., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Sementchenko V., Manak J.R., Levine M.S., Manak J.R., Levine M.S., Levine M.S. Spatial regulation of microRNA gene expression in the Drosophila embryo. Proc. Natl. Acad. Sci. 2005;102:15907–15911. doi: 10.1073/pnas.0507817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemar F., Nix D.A., Piel J., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Nix D.A., Piel J., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Piel J., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Sementchenko V., Bell I., Manak J.R., Levine M.S., Bell I., Manak J.R., Levine M.S., Manak J.R., Levine M.S., Levine M.S. Comprehensive identification of Drosophila dorsal–ventral patterning genes using a whole-genome tiling array. Proc. Natl. Acad. Sci. 2006;103:12763–12768. doi: 10.1073/pnas.0604484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler B.A., Soong J., Gergen J.P., Soong J., Gergen J.P., Gergen J.P. The Drosophila segmentation gene runt has an extended cis-regulatory region that is required for vital expression at other stages of development. Mech. Dev. 1992;39:17–28. doi: 10.1016/0925-4773(92)90022-c. [DOI] [PubMed] [Google Scholar]
- Carroll S.B., Winslow G.M., Twombly V.J., Scott M.P., Winslow G.M., Twombly V.J., Scott M.P., Twombly V.J., Scott M.P., Scott M.P. Genes that control dorsoventral polarity affect gene expression along the anteroposterior axis of the Drosophilaembryo. Development. 1987;99:327–332. doi: 10.1242/dev.99.3.327. [DOI] [PubMed] [Google Scholar]
- Chanas G., Lavrov S., Iral F., Cavalli G., Maschat F., Lavrov S., Iral F., Cavalli G., Maschat F., Iral F., Cavalli G., Maschat F., Cavalli G., Maschat F., Maschat F. Engrailed and polyhomeotic maintain posterior cell identity through cubitus-interruptus regulation. Dev. Biol. 2004;272:522–535. doi: 10.1016/j.ydbio.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Clyde D.E., Corado M.S., Wu X., Pare A., Papatsenko D., Small S., Corado M.S., Wu X., Pare A., Papatsenko D., Small S., Wu X., Pare A., Papatsenko D., Small S., Pare A., Papatsenko D., Small S., Papatsenko D., Small S., Small S. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. 2003;426:849–853. doi: 10.1038/nature02189. [DOI] [PubMed] [Google Scholar]
- Ganguly A., Jiang J., Ip Y.T., Jiang J., Ip Y.T., Ip Y.T. Drosophila WntD is a target and an inhibitor of the Dorsal/Twist/Snail network in the gastrulating embryo. Development. 2005;132:3419–3429. doi: 10.1242/dev.01903. [DOI] [PubMed] [Google Scholar]
- Gao Q., Finkelstein R., Finkelstein R. Targeting gene expression to the head: The Drosophilaorthodenticle gene is a direct target of the Bicoid morphogen. Development. 1998;125:4185–4193. doi: 10.1242/dev.125.21.4185. [DOI] [PubMed] [Google Scholar]
- Gordon M.D., Dionne M.S., Schneider D.S., Nusse R., Dionne M.S., Schneider D.S., Nusse R., Schneider D.S., Nusse R., Nusse R. WntD is a feedback inhibitor of Dorsal/NF-κB in Drosophila development and immunity. Nature. 2005;437:746–749. doi: 10.1038/nature04073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.D., Schwyter D.H., Shirokawa J.M., Courey A.J., Schwyter D.H., Shirokawa J.M., Courey A.J., Shirokawa J.M., Courey A.J., Courey A.J. The interplay between multiple enhancer and silencer elements defines the pattern of decapentaplegic expression. Genes & Dev. 1993;7:694–704. doi: 10.1101/gad.7.4.694. [DOI] [PubMed] [Google Scholar]
- Ip Y.T., Gridley T., Gridley T. Cell movements during gastrulation: Snail dependent and independent pathways. Curr. Opin. Genet. Dev. 2002;12:423–429. doi: 10.1016/s0959-437x(02)00320-9. [DOI] [PubMed] [Google Scholar]
- Jiang J., Kosman D., Ip Y.T., Levine M., Kosman D., Ip Y.T., Levine M., Ip Y.T., Levine M., Levine M. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes & Dev. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- Kirov N., Zhelnin L., Shah J., Rushlow C., Zhelnin L., Shah J., Rushlow C., Shah J., Rushlow C., Rushlow C. Conversion of a silencer into an enhancer: Evidence for a co-repressor in dorsal-mediated repression in Drosophila. EMBO J. 1993;12:3193–3199. doi: 10.1002/j.1460-2075.1993.tb05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D., Ip Y.T., Levine M., Arora K., Ip Y.T., Levine M., Arora K., Levine M., Arora K., Arora K. Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science. 1991;254:118–122. doi: 10.1126/science.1925551. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Johnstone S.E., Young R.A., Johnstone S.E., Young R.A., Young R.A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptin M. twist and snail as positive and negative regulators during Drosophilamesoderm development. Genes & Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- Levine M., Davidson E.H., Davidson E.H. Gene regulatory networks for development. Proc. Natl. Acad. Sci. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw G.J., Lengyel J.A., Lengyel J.A. Control of tailless expression by bicoid, dorsal and synergistically interacting terminal system regulatory elements. Mech. Dev. 1993;40:47–61. doi: 10.1016/0925-4773(93)90087-e. [DOI] [PubMed] [Google Scholar]
- Markstein M., Markstein P., Markstein V., Levine M.S., Markstein P., Markstein V., Levine M.S., Markstein V., Levine M.S., Levine M.S. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Zinzen R., Markstein P., Yee K.P., Erives A., Stathopoulos A., Levine M., Zinzen R., Markstein P., Yee K.P., Erives A., Stathopoulos A., Levine M., Markstein P., Yee K.P., Erives A., Stathopoulos A., Levine M., Yee K.P., Erives A., Stathopoulos A., Levine M., Erives A., Stathopoulos A., Levine M., Stathopoulos A., Levine M., Levine M. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–2394. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- Moussian B., Roth S., Roth S. Dorsoventral axis formation in the Drosophila embryo—Shaping and transducing a morphogen gradient. Curr. Biol. 2005;15:R887–R899. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Lohs-Schardin M., Sander K., Cremer C., Lohs-Schardin M., Sander K., Cremer C., Sander K., Cremer C., Cremer C. A dorso–ventral shift of embryonic primordia in a new maternal-effect mutant of Drosophila. Nature. 1980;283:474–476. doi: 10.1038/283474a0. [DOI] [PubMed] [Google Scholar]
- Pankratz M.J., Seifert E., Gerwin N., Billi B., Nauber U., Jackle H., Seifert E., Gerwin N., Billi B., Nauber U., Jackle H., Gerwin N., Billi B., Nauber U., Jackle H., Billi B., Nauber U., Jackle H., Nauber U., Jackle H., Jackle H. Gradients of Kruppel and knirps gene products direct pair-rule gene stripe patterning in the posterior region of the Drosophila embryo. Cell. 1990;61:309–317. doi: 10.1016/0092-8674(90)90811-r. [DOI] [PubMed] [Google Scholar]
- Papatsenko D., Levine M., Levine M. Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proc. Natl. Acad. Sci. 2005;102:4966–4971. doi: 10.1073/pnas.0409414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegri F., Lehmann R., Lehmann R. A role of polycomb group genes in the regulation of gap gene expression in Drosophila. Genetics. 1994;136:1341–1353. doi: 10.1093/genetics/136.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Walker K., Rolfe P.A., Herbolsheimer E., Rolfe P.A., Herbolsheimer E., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Rothe M., Wimmer E.A., Pankratz M.J., Gonzalez-Gaitan M., Jackle H., Wimmer E.A., Pankratz M.J., Gonzalez-Gaitan M., Jackle H., Pankratz M.J., Gonzalez-Gaitan M., Jackle H., Gonzalez-Gaitan M., Jackle H., Jackle H. Identical transacting factor requirement for knirps and knirps-related gene expression in the anterior but not in the posterior region of the Drosophila embryo. Mech. Dev. 1994;46:169–181. doi: 10.1016/0925-4773(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Simpson P. Maternal–zygotic gene interactions during formation of the dorsoventral pattern in Drosophila embryos. Genetics. 1983;105:615–632. doi: 10.1093/genetics/105.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol N.S., Ambros V., Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes & Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., Nusslein-Volhard C., Nusslein-Volhard C. The origin of pattern and polarity in the Drosophilaembryo. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Van Drenth M., Erives A., Markstein M., Levine M., Van Drenth M., Erives A., Markstein M., Levine M., Erives A., Markstein M., Levine M., Markstein M., Levine M., Levine M. Whole-genome analysis of dorsal–ventral patterning in the Drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- Toth J., Biggin M.D., Biggin M.D. The specificity of protein–DNA crosslinking by formaldehyde: In vitro and in drosophila embryos. Nucleic Acids Res. 2000;28:e4. doi: 10.1093/nar/28.2.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Kirov N., Rushlow C., Kirov N., Rushlow C., Rushlow C. Peak levels of BMP in the Drosophila embryo control target genes by a feed-forward mechanism. Development. 2005;132:1637–1647. doi: 10.1242/dev.01722. [DOI] [PubMed] [Google Scholar]
- Zinzen R.P., Senger K., Levine M., Papatsenko D., Senger K., Levine M., Papatsenko D., Levine M., Papatsenko D., Papatsenko D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr. Biol. 2006;16:1358–1365. doi: 10.1016/j.cub.2006.05.044. [DOI] [PubMed] [Google Scholar]