Trigger Factor can antagonize both SecB and DnaK/DnaJ chaperone functions in Escherichia coli (original) (raw)
Abstract
Polypeptides emerging from the ribosome are assisted by a pool of molecular chaperones and targeting factors, which enable them to efficiently partition as cytoplasmic, integral membrane, or exported proteins. In Escherichia coli, the chaperones SecB, Trigger Factor (TF), and DnaK are key players in this process. Here, we report that, as with dnaK or dnaJ mutants, a secB null strain exhibits a strong cold-sensitive (Cs) phenotype. Through suppressor analyses, we found that inactivating mutations in the tig gene encoding TF fully relieve both the Cs phenotype and protein aggregation observed in the absence of SecB. This antagonistic effect of TF depends on its ribosome-binding and chaperone activities but unrelated to its peptidyl-prolyl cis/trans isomerase (PPIase) activity. Furthermore, in contrast to the previously known synergistic action of TF and DnaK/DnaJ above 30°C, a tig null mutation partially suppresses the Cs phenotype exhibited by a compromised DnaK/DnaJ chaperone machine. The antagonistic role of TF is further exemplified by the fact that the secB dnaJ double mutant is viable only in the absence of TF. Finally, we show that, in the absence of TF, more SecA and ribosomes are associated with the inner membrane, suggesting that the presence of TF directly or indirectly interferes with the process of cotranslational protein targeting to the Sec translocon.
Keywords: cold sensitivity, genetic suppression, nascent polypeptides, protein aggregation, protein export
It is known that de novo protein folding in Escherichia coli is mainly orchestrated by the chaperones Trigger Factor (TF), DnaK (Hsp70), and GroEL (Hsp60) (1, 2). The ribosome-bound TF is the first chaperone to interact cotranslationally with nascent polypeptides (3), and it is thought that the majority of the newly synthesized polypeptides can reach their native state in the cytoplasm without further help. Yet a substantial portion of cytoplasmic proteins need further co- and/or posttranslational assistance by either the DnaK or the GroEL chaperone machines for their correct folding. In contrast, nascent proteins destined to be secreted or inserted into the IM are prevented from premature folding. Whereas transmembrane segments in IM proteins are recognized by the signal recognition particle and directed cotranslationally to the Sec translocon, presecretory proteins are generally exported posttranslationally via the dedicated SecA/SecB pathway (4, 5). In this case, the SecB chaperone binds either co- and/or posttranslationally to unfolded polypeptides without specificity for signal sequences, maintains them in a protected yet unfolded state, and transfers them to the membrane-associated SecA (6–9).
The functional complementarities observed among the major cytoplasmic chaperones TF, DnaK, GroEL, and SecB suggest that they compete for the same pool of newly synthesized polypeptides. For example, the severe temperature-sensitive (Ts) phenotype and protein folding defect observed in the absence of both the DnaK and TF chaperones can be partially rescued by increasing the cellular levels of either SecB or GroEL/GroES (10–13). Consistent with this result, in the absence of the ribosome-bound TF, both DnaK and SecB interact with considerably shorter nascent polypeptide chains, and DnaK binds significantly more newly synthesized polypeptides (11, 14, 15). This functional overlap is further illustrated by peptide-binding scan analyses showing that TF, SecB, and DnaK can share potential binding sites present in nascent polypeptide substrates (16–18). Furthermore, it has been shown that DnaK promotes the export of several SecB-dependent proteins, and both GroEL and DnaK assist protein export in some cases as well (19–21). In contrast, TF does not generally assist the export of secretory proteins (22). Finally, in the absence of SecB, both DnaK and GroEL are up-regulated and vice versa, further supporting the notion that under certain conditions, these molecular chaperones may functionally substitute for each other (23).
Here, we provide additional evidence for such competition and interplay among molecular chaperones. First, we show that a secB null mutant exhibits a strong cold-sensitive (Cs) phenotype, and that this phenotype is most likely related to the resulting deficiency in protein export. Through a suppressor analysis, we next demonstrate that the absence of the TF chaperone largely relieves the _secB_- and, to a lesser extent, the _dnaJ_-dependent Cs phenotypes. Finally, we show that in the absence of TF, significantly more SecA and ribosomes are associated with the IM, suggesting that TF somehow interferes with cotranslational protein targeting to the translocon.
Results and Discussion
SecB Is Required for E. coli Growth at Low Temperature.
The SecB chaperone is known to be critical for the processing of a subset of secretory proteins (24). One of the interesting phenotypes of the first secB mutants discovered was their inability to grow on LB agar plates (25). However, subsequent studies suggested that this phenotype may be due to a polar effect of the secB mutation on the downstream gpsA gene, whose product is involved in the biosynthesis of glycerol 3-phosphate, a precursor for phospholipid synthesis (26). To minimize such potential polar effects on gpsA gene expression, we generated an in-frame deletion/replacement of the secB gene by the cat gene. In agreement with Shimizu and colleagues (26), we found that the secB null mutant grew as well as the wild-type strain on LB at 37°C, independent of the absence or presence of plasmid-encoded GpsA (data not shown). This observation was confirmed by cotransduction experiments using a kanamycin-resistance cassette linked ≈40% to the secB gene (data not shown).
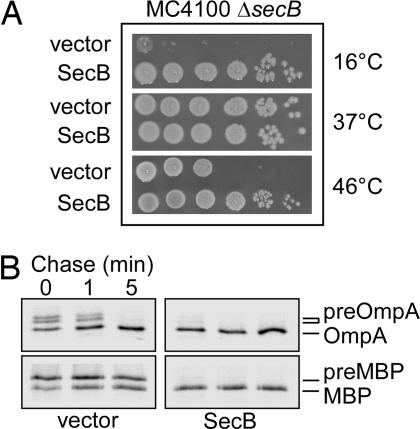
However, upon subsequent testing on LB agar plates, we found that the secB null mutant exhibited a strong bactericidal Cs phenotype below 23°C and a relatively weak Ts phenotype at temperatures above 45°C. Such phenotypes were not observed on M9 glucose minimal medium (data not shown). Subsequent complementation experiments established that a low-copy plasmid expressing SecB could fully rescue all of the observed growth phenotypes, whereas GpsA could not, even under high expression levels (Fig. 1A; data not shown). These results demonstrate that the strong Cs and weak Ts phenotypes are due to the absence of SecB. Consistent with the role of SecB in protein export, the secB null mutant exerted an export defect (Fig. 1B). The Cs phenotype of the secB null mutant most likely reflects the involvement of SecB in protein export. Indeed, a large number of mutations in components of the Sec translocon lead to a Cs phenotype (27), and it has been reported that at least one step in the export process is Cs (27). Thus, following a temperature downshift, export may be critically dependent on upstream chaperone “holdases,” such as SecB or DnaK, to maintain precursor proteins in a translocation-competent conformation and protect them from proteolysis.
Fig. 1.
SecB-dependent phenotypes. (A) The MC4100 Δ_secB_ strain was transformed with either the p29SEN vector or p29SEN-SecB. Fresh transformants were serially diluted 10-fold and spotted on LB-ampicillin agar plates in the presence of 10 μM IPTG inducer. Plates were incubated for either 18 h at 37 or 46°C or for 6 days at 16°C. (B) Pulse–chase experiments showing the processing of OmpA and maltose-binding protein (MBP) precursors in the MC4100 Δ_secB_ mutant in the presence of either p29SEN vector or p29SEN-SecB.
Mutations in various genes that code for molecular chaperones with a broad spectrum of substrate specificity generally result in increased steady-state levels of aggregated proteins (28). We found that the secB mutant strain indeed accumulated a significant level of aggregated proteins when compared with its isogenic parent, even when grown at the permissive temperature of 37°C (Fig. 2C). These results agree with the recent proteomic study of the aggregates accumulated in the absence of SecB (29).
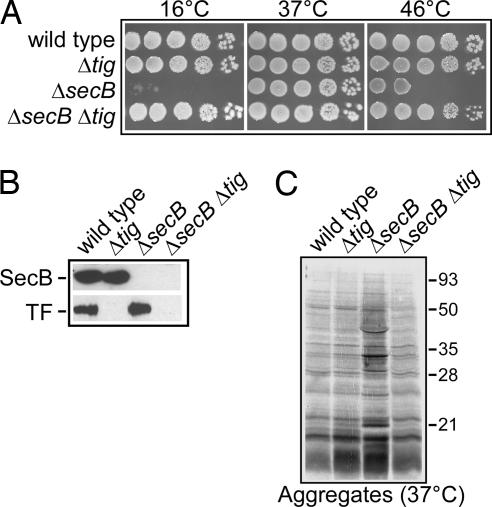
Fig. 2.
TF antagonizes SecB. (A) The deletion of the tig gene suppresses the Cs and Ts phenotypes of the Δ_secB_ mutant. MC4100 and its isogenic mutant derivatives were grown to mid-log phase, serially diluted 10-fold, spotted on LB agar plates, and incubated at the indicated temperatures. (B) Western blot analysis confirming the absence of SecB and/or TF in the mutant strains. (C) Protein aggregation in MC4100 and its isogenic mutant derivatives at the permissive temperature of 37°C. The aggregated proteins were separated by SDS/PAGE and visualized by silver staining.
The tig Null Mutation Fully Suppresses the Various Phenotypes of a secB Null Mutant.
We observed that suppressors of the secB Cs phenotype appear spontaneously at an approximate frequency of 10−4 at 14°C. To identify recessive mutations able to suppress the Cs phenotype, we performed a random transposon mutagenesis of the secB mutant, simultaneously selecting for both drug resistance and colony formation at 14°C. Interestingly, we found that a large proportion of the insertional mutations that suppressed the secB Cs phenotype were either in the tig structural gene or affected its expression (34 of 123).
To ensure that the suppressive effect was indeed due to the resulting loss of TF function, we engineered a complete deletion of the tig gene and found that it was also capable of completely suppressing the secB Cs phenotype (Fig. 2A). The absence of TF and SecB was confirmed by Western blot analysis (Fig. 2B). As expected, the secB Cs phenotype was restored following expression of plasmid-encoded TF (Fig. 3C). The efficient suppression of the secB Cs phenotype by the tig null mutation was also confirmed in the MG1655 strain background (data not shown). Subsequent testing showed that deletion of tig additionally rescued the weak Ts phenotype of the secB mutant at 46°C (Fig. 2A).
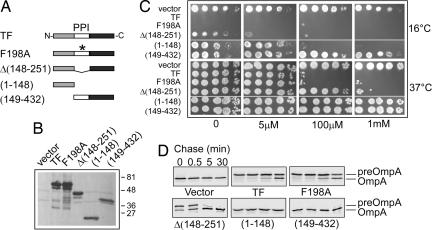
Fig. 3.
TF domain requirement. (A) Schematic representation of the various p29SEN-based TF constructs used in this work. Light gray, N-terminal domain; white, PPIase domain; black, C domain. (B) Steady-state levels of the TF constructs as judged by Western blot analysis using anti-TF antibodies. (C) Growth of the MC4100 Δ_secB_ Δ_tig_ double mutant transformed with the various p29SEN-based TF constructs. Fresh transformants were serially diluted and spotted on LB-ampicillin agar plates with or without IPTG. (D) Pulse–chase experiments showing the processing of OmpA precursor in the MC4100 Δ_secB_ Δ_tig_ double mutant transformed with the various p29SEN-based TF constructs in the presence of 100 μM IPTG.
It has been previously shown that TF can retard protein export, because a tig mutation significantly accelerated the export of several Sec-dependent proteins (22, 30). In agreement with these studies, we observed that deletion of tig suppressed the need for SecB in assisting preOmpA export (Fig. 3D). In addition, the tig null allele also exerted a positive effect on bacterial growth in the presence of compromised sec alleles in either the secA or secY genes. Nevertheless, the suppression was much less efficient than the one seen with the secB null mutant [supporting information (SI) Fig. 6].
As shown above and also reported by Baars et al. (29), a secB mutation led to an increased accumulation of aggregated proteins in the cytoplasm (Fig. 2C). Interestingly, we found that the tig deletion efficiently suppressed protein aggregation in the absence of SecB (Fig. 2C). In contrast, increasing TF levels at an otherwise permissive temperature of growth completely blocked the growth of the secB null mutant and intensified protein aggregation (Fig. 3C; data not shown). These results are in agreement with the observations that TF markedly delays protein folding relative to translation (31) and with the detrimental effect on cell growth resulting from TF overexpression in the absence of DnaK or DnaJ (13). Taken together, the results suggest that TF likely exerts its negative effect on bacterial growth by interacting for inappropriately long periods of time with newly synthesized polypeptides when other chaperones or targeting factors are limiting.
TF Antagonistic Effect Relies on Its Ribosome-Binding but Not on Its PPIase Activity.
TF is principally composed of three domains. The N-terminal domain mediates ribosome-binding and participates, together with the C-terminal domain, in TF chaperone function (13, 32, 33). The central domain possesses a PPIase activity, which is not required for TF chaperone function in vivo (13, 34, 35). We asked which of the TF structural domains are necessary for its antagonistic effect in the absence of SecB (Fig. 3). As expected, plasmid-encoded wild-type TF fully restored the Cs phenotype seen in the absence of SecB. The TF-F198A mutant with inactivated PPIase activity (34) behaved like wild-type TF (Fig. 3 C and D). Although less severe, a similar effect was observed when the complete PPIase domain was deleted (Fig. 3 C and D). In contrast, we found that the TF N-terminal domain was absolutely essential for its antagonistic effect at 16°C (Fig. 3 C and D). However, by itself, the N-terminal domain did not affect bacterial growth at permissive temperatures, protein aggregation, or preOmpA processing, even under maximal inducer concentrations (Fig. 3 C and D; data not shown).
TF also Antagonizes the DnaK/DnaJ Chaperone Machine at Low Temperatures.
Interplay and functional complementarities among cytoplasmic chaperones in both protein folding and export pathways are particularly well illustrated by the SecB and the DnaK/DnaJ chaperone machines (19–21, 36). Although TF functions synergistically with the DnaK/DnaJ chaperone machine in the folding of newly synthesized proteins (13–15), as observed for the secB mutant, the dnaKdnaJ mutants are more sensitive to high levels of TF compared with the wild-type isogenic strain. This finding suggested to us that physiological levels of TF may also antagonize the DnaK/DnaJ chaperone machine at low temperatures. Because the DnaK/DnaJ chaperone machine is essential at temperatures below 18°C (37), we chose to explore this hypothesis. Because the deletion of dnaK leads to an increased expression of all heat shock proteins and to the rapid accumulation of extragenic suppressors that enable bacterial growth (37), we decided to concentrate on the role of TF in the absence of DnaJ, the major DnaK cochaperone.
In contrast to the dnaK deletion, the dnaJ null mutation still allows basal DnaK activity through two other DnaK cochaperones, namely CbpA and DjlA, resulting in a DnaK-dependent Cs phenotype at 14°C (38). We introduced the dnaJ null mutation into the wild-type parent and its isogenic tig mutant derivative and tested its effect on bacterial growth at various temperatures. In sharp contrast to the previously seen synergistic effect of the tig and dnaJ mutations at high temperature (ref. 13; Fig. 4A), deletion of tig was beneficial at 14°C when the DnaK/DnaJ machine was partially compromised (Fig. 4A). Similar results were observed in both the MG1655 and Mph42 strain backgrounds (data not shown). However, in contrast to the very efficient suppression of the secB Cs phenotype, suppression of the dnaJ Cs phenotype by the tig mutation occurred in a much narrower temperature range, as we did not observe efficient suppression at 12°C. This difference likely reflects the plethora of other cellular functions in which the DnaK/DnaJ chaperone machine participates (39).
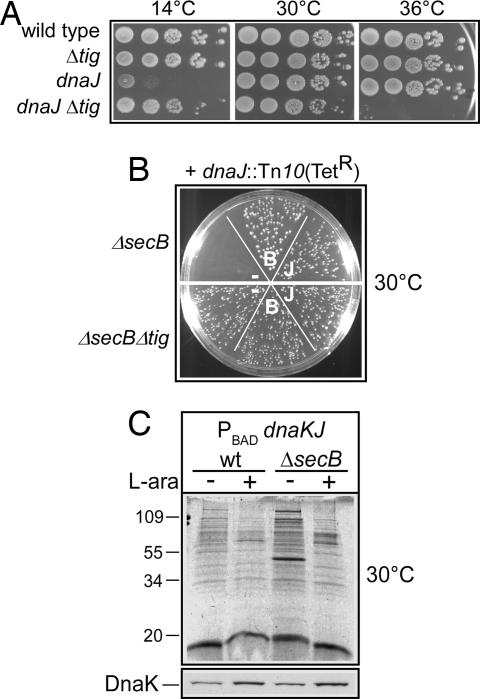
Fig. 4.
TF antagonizes the DnaK/DnaJ chaperone machine at low temperatures. (A) Growth of MC4100 and its isogenic Δ_tig_, dnaJ::Tn_10-42_, and Δ_tig dnaJ_::Tn_10-42_ mutant derivatives following incubation on LB agar plates at the indicated temperatures. (B) The dnaJ::Tn_10-42_ mutant allele was introduced by phage P1 transduction into either the Δ_secB_ or Δ_secB_ Δ_tig_ mutant derivatives in the presence of the various p29SEN-based constructs. −, vector; B, SecB; J, DnaJ. The results of a representative P1 transduction experiment performed at 30°C on LB-tetracycline-ampicillin agar plates supplemented with 10 μM IPTG are shown. (C) Intracellular protein aggregation at 30°C after a 3-h depletion of DnaK/DnaJ in the presence (wt) or in the absence (Δ_secB_) of chromosomally encoded SecB. + indicates the presence of 0.5% l-arabinose, and − indicates the absence of l-arabinose and the presence of 0.4% glucose. Western blot analysis of the resulting steady-state levels of DnaK after the 3-h depletion is shown. The nature of the aggregates is shown in SI Table 1 and Fig. 7.
The secB dnaJ Double Mutant Is Synthetically Lethal only in the Presence of TF.
Next we asked whether TF plays a beneficial or detrimental role for bacterial growth in the absence of both the SecB and DnaK/DnaJ chaperones. To do this, we performed phage P1-mediated transduction experiments as follows: A P1 lysate was grown on a dnaJ::Tn_10_(TetR) mutant strain and used to infect either the single secB or the double secB tig mutant. We found that although we could easily construct the secB tig dnaJ triple mutant at 30 or 33°C, we were unable to construct the secB dnaJ double mutant in the tig+ background (Fig. 4B; data not shown). As a control, we showed that the presence of either plasmid-encoded SecB or DnaJ permitted the introduction of the dnaJ mutant allele in the tig+ background. However, upon prolonged incubation (2–3 days) of the transductants, we noticed the occasional appearance of slowly growing secB dnaJ::TetR transductants, very likely due to the accumulation of unknown suppressor mutations (data not shown). The synthetic lethality between secB and dnaJ was also confirmed in the absence of DnaK by cotransduction experiments using a Δ_dnaKdnaJ_::KanR allele (data not shown).
Because of the TF-dependent synthetic lethality of secB and dnaJ (or dnaKdnaJ) mutant alleles, we placed the dnaKdnaJ operon under the control of an l-arabinose inducible promoter. This chromosomally encoded construct enabled us to observe some of the potential phenotypes of the otherwise synthetic lethal combination of the secB and dnaKdnaJ mutant alleles. Under these conditions, the resulting depletion of both DnaK and DnaJ during bacterial growth at permissive temperature resulted in a severe accumulation of aggregated proteins, uniquely in the absence of SecB (Fig. 4C). Mass spectrometry analysis revealed that such aggregates mainly comprise known DnaK substrates, as well as previously unrelated secretory proteins (SI Table 1 and Fig. 7). Thus, these results are in full agreement with the observed secB dnaK synthetic lethality and further illustrate the critical role played by the DnaK/DnaJ chaperone machine (most likely during protein export) as a back-up system in the absence of SecB and in the impeding presence of TF (19–21).
TF Modulates the Association of both SecA and Ribosomes with the IM.
In the absence of SecB, and perhaps also DnaK/DnaJ, nascent polypeptides of some secretory proteins form partially folded intermediates in the cytoplasmic space, which cannot be efficiently translocated through the IM via the general Sec machinery (5). Upon a temperature downshift, this phenotype may be exacerbated, owing to the compromised Sec translocon function (27). The absence of ribosome-bound TF in the tig mutant may relieve this phenotype by allowing premature cotranslational interaction of SecA with nascent polypeptides of secretory proteins, as well as the subsequent SecA-dependent targeting of the ribosome nascent chain complex to the Sec translocon (40), thus bypassing the need for cytoplasmic chaperones.
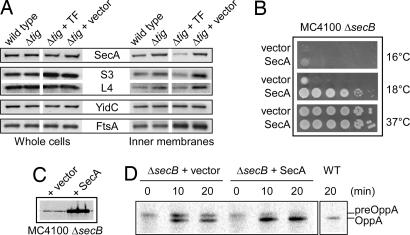
To address this possibility, we isolated IM vesicles from both the wild-type strain and its isogenic tig mutant grown at 37°C (Fig. 5A) and 16°C (SI Fig. 8) and compared the levels of the peripheral membrane protein SecA to those of the integral membrane proteins YidC and/or SecE. We found that significantly more SecA (≥30% after quantification with the ChemiDoc Bio-Rad chemiluminescent detection system) was associated with IMs in the absence of TF. As a control, the peripheral membrane protein FtsA (41) was not affected by the absence of TF (Fig. 5A). In contrast, the overexpression of TF significantly decreased (by >2-fold) the levels of SecA at the IM (Fig. 5A).
Fig. 5.
TF modulates both SecA and ribosome association with IMs. (A) Western blot analysis of either whole-cell or IM preparations showing SecA, L4, S3, YidC, and FtsA levels from cells grown at 37°C. The MC4100 wild type and its Δ_tig_ mutant derivative, as well as the Δ_tig_ mutant transformed with either p29SEN vector or p29SEN-TF are shown. Expression of TF from p29SEN-TF was induced with 100 μM IPTG for 1 h. Comparison of cytoplasmic and whole membrane fractions observed under such conditions is shown in SI Fig. 9. (B) Complementation of the Cs phenotype of MC4100 Δ_secB_ by p29SEN vector or p29SEN-SecA. Fresh transformants were serially diluted 10-fold and spotted on LB-ampicillin agar plates in the presence of 100 μM IPTG and incubated at the indicated temperatures. (C) SecA expression levels in these transformants at 37°C after 2-h induction with 100 μM IPTG. (D) Suppression of the OppA export defect of MC4100 Δ_secB_ by p29SEN vector or p29SEN-SecA at 23°C in the presence of 100 μM IPTG.
Next, we compared the composition of the whole membrane to the cytoplasmic fractions and found that without TF ≈20% of total SecA was found in the membrane fraction. This percentage also dropped by a factor of 2 when TF was overexpressed (see SI Fig. 9). The relevance for such an increase in membrane-bound SecA in the absence of TF was further emphasized by the fact that overexpression of SecA [≈18-fold with 100 μM isopropyl β-d-thiogalactoside (IPTG)] appreciably rescued the Cs phenotype of the secB mutant down to 18°C (Fig. 5 B and C), as well as the export of OppA, a SecB-dependent secretory protein (Fig. 5D). However, in contrast to the full suppression of the secB Cs phenotype at 14°C exerted by the tig null mutation, the overexpression of SecA only partially improved bacterial growth (Fig. 5B). These results suggest that SecA is not the sole limiting factor and that potentially other factors participate in protein export, especially at very low temperatures, in the absence of SecB and/or DnaK/DnaJ.
It is known that ribosomes specifically interact with the Sec translocon and that one of the main contacts between the ribosome and the translocon occurs in the neighborhood of ribosomal protein L23 (42–44). The L23 protein is located near the polypeptide tunnel exit and also serves as a TF docking site (32, 45). Thus, the presence of L23-bound TF may somehow sterically interfere with the association of the ribosome with the translocon. By comparing the levels of ribosomal proteins to those of YidC and/or SecE in IM vesicles from both the wild-type strain and its isogenic tig mutant grown at 37°C (Fig. 5A) or 16°C (SI Fig. 8), we found that, comparable to SecA, significantly more ribosomal proteins associated with IMs in the absence of TF, whereas TF overexpression reduced the amount of ribosomal proteins at the IM (Fig. 5A). Furthermore, we also compared whole membrane to cytoplasmic fractions and found that without TF ≈4% of total S3 was found in the membrane fraction, and that this percentage dropped, similarly to SecA, when TF was overexpressed (SI Fig. 9).
Taken together, all of the above results suggest that in the absence of TF, SecB (and perhaps DnaK)-dependent protein export is facilitated by increased concentrations of active SecA and ribosomes at the Sec translocon (see model in SI Fig. 10). Whether the observed increased ribosome levels at the IM are due directly to the absence of TF steric hindrance or indirectly due to a more efficient SecA-dependent targeting of the ribosome to the IM remains to be determined.
Materials and Methods
Bacterial Strains.
Genetic analyses were carried out in the E. coli MG1655 (46), Mph42 (47), and MC4100 (48, 49) genetic backgrounds. GP108 is MC4100 dnaJ::Tn_10-42_(TetR) (49). The MC4100 tig-13::Tn_10_(KanR) mutant has a miniTn_10_(KanR) cassette inserted 86 nucleotides upstream of the tig initiation codon (this work).
The deletion of the tig gene was carried out as described by Yu and colleagues (50), except that the CmR cassette was amplified from plasmid pKD3 (51). The Δ_tig_::CmR allele was then moved by bacteriophage P1 transduction into either the MC4100 or MG1655 strain backgrounds, and the CmR cassette was subsequently removed by using the FLP recombinase (51). The in-frame deletion/replacement of the 468-nt secB gene by the 639-nt cat gene from plasmid pKD3 was similarly carried out. After recombination into the bacterial chromosome, we noted that the resulting cat gene had undergone a fortuitous deletion of two guanines 25-nt upstream of the initiation codon and 48-nt downstream of the transcription start of the secB P2 promoter. The strain GP495 is an Mph42 derivative in which the endogenous dnaKdnaJ promoters are replaced by the PBAD promoter, together with the upstream araC and kanamycin resistance genes. In this case, the level of expression of DnaK in the presence of 0.5% l-arabinose inducer is comparable to that of DnaK from its native promoter at 30°C. Higher expression levels could not be achieved by using this conditional strain (data not shown). Strain GP502 is GP495 Δ_secB_::cat.
Genetic Selections.
To select for loss-of-function mutants suppressing the Δ_secB_ Cs phenotype, the MC4100 Δ_secB_ Cs mutant was grown at 37°C in LB broth and subjected to transposon mutagenesis by using the miniTn_10_(KanR) from bacteriophage λNK1316 as described by Kleckner and colleagues (52). Approximately 30,000 independent KanR clones thusly obtained at 37°C were pooled and selected as colony formers at 14°C for 8 days. To ensure that the suppressive effect was due to a single transposition event, the KanR alleles of individual colonies were back-crossed into MC4100 Δ_secB_ by P1 transduction and rechecked for the suppression of the Cs phenotype at 14°C. The exact localization of the miniTn_10_(KanR) insertions in the E. coli chromosome was determined by inverse-PCR as described (53).
Isolation of Aggregated Proteins.
Aggregated proteins were isolated essentially as described by Tomoyasu and colleagues (54), except that all volumes of solutions were increased 10-fold. To monitor protein aggregation upon DnaK and DnaJ depletion, overnight cultures of strains GP495 and GP502 grown at 30°C in LB supplemented with 0.5% l-arabinose were diluted 1:100 in the same medium and grown at 30°C to an OD600 of 0.4. Cells were pelleted, resuspended in prewarmed LB supplemented either with 0.4% glucose or 0.5% l-arabinose, and grown for 3 h at 30°C. Identical densities of cells were then pelleted, and aggregated proteins were isolated.
SecA and Ribosome Association with IMs.
Bacteria were grown in LB medium at 37 or 16°C to an OD600 of ≈1.0, and they were used to prepare whole-cell extracts and IM vesicles as described (55). Whole-cell extracts (≈14 μg) and IM fractions (≈4 μg) were separated on SDS/PAGE gels, transferred to nitrocellulose membranes, and probed with antibodies raised against SecA (D. Belin, Centre Médical Universitaire, Geneva, Switzerland), SecE, YidC, L4, L6 (J. Luirink, Vrije Universiteit, Amsterdam, The Netherlands), FtsA (C. Hale, Case Western Reserve University, Cleveland, OH), and S3 (I. Iost, Ecole Normale Supérieure, Paris, France).
Pulse–Chase Analysis, Steady-State Protein Expression Levels, and Plasmid Constructs.
Details are given in the SI Materials and Methods.
Supplementary Material
Supporting Information
Acknowledgments
We thank Drs. Joen Luirink, Isabelle Iost, Dominique Belin, Kazuei Igarashi, Cynthia Hale, and Jon Beckwith for the gift of strains and/or antibodies; Eirene Markenscoff Papadimitriou for help with some experiments; and Drs. Joen Luirink and Tassos Economou for their useful advice during the course of our work. This work was supported by the Swiss National Foundation (SNF) and the Canton of Geneva (SNF-3100-065403), a European Molecular Biology Organization long-term fellowship (to R.S.U.), and a Centre National de la Recherche Scientifique-ATIP grant (to P.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Bukau B, Deuerling E, Pfund C, Craig EA. Cell. 2000;101:119–122. doi: 10.1016/S0092-8674(00)80806-5. [DOI] [PubMed] [Google Scholar]
- 2.Young JC, Agashe VR, Siegers K, Hartl FU. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 3.Lill R, Crooke E, Guthrie B, Wickner W. Cell. 1988;54:1013–1018. doi: 10.1016/0092-8674(88)90116-x. [DOI] [PubMed] [Google Scholar]
- 4.Luirink J, Sinning I. Biochim Biophys Acta. 2004;1694:17–35. doi: 10.1016/j.bbamcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Veenendaal AK, van der Does C, Driessen AJ. Biochim Biophys Acta. 2004;1694:81–95. doi: 10.1016/j.bbamcr.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 7.Patel CN, Smith VF, Randall LL. Protein Sci. 2006;15:1379–1386. doi: 10.1110/ps.062141006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Randall LL, Hardy SJ. Cell Mol Life Sci. 2002;59:1617–1623. doi: 10.1007/PL00012488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vrontou E, Economou A. Biochim Biophys Acta. 2004;1694:67–80. doi: 10.1016/j.bbamcr.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Vorderwulbecke S, Kramer G, Merz F, Kurz TA, Rauch T, Zachmann-Brand B, Bukau B, Deuerling E. FEBS Lett. 2004;559:181–187. doi: 10.1016/S0014-5793(04)00052-3. [DOI] [PubMed] [Google Scholar]
- 11.Ullers RS, Luirink J, Harms N, Schwager F, Georgopoulos C, Genevaux P. Proc Natl Acad Sci USA. 2004;101:7583–7588. doi: 10.1073/pnas.0402398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, et al. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 13.Genevaux P, Keppel F, Schwager F, Langendijk-Genevaux PS, Hartl FU, Georgopoulos C. EMBO Rep. 2004;5:195–200. doi: 10.1038/sj.embor.7400067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 15.Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU. Cell. 1999;97:755–765. doi: 10.1016/s0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- 16.Knoblauch NT, Rudiger S, Schonfeld HJ, Driessen AJ, Schneider-Mergener J, Bukau B. J Biol Chem. 1999;274:34219–34225. doi: 10.1074/jbc.274.48.34219. [DOI] [PubMed] [Google Scholar]
- 17.Patzelt H, Rudiger S, Brehmer D, Kramer G, Vorderwulbecke S, Schaffitzel E, Waitz A, Hesterkamp T, Dong L, Schneider-Mergener J, et al. Proc Natl Acad Sci USA. 2001;98:14244–14249. doi: 10.1073/pnas.261432298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wild J, Altman E, Yura T, Gross CA. Genes Dev. 1992;6:1165–1172. doi: 10.1101/gad.6.7.1165. [DOI] [PubMed] [Google Scholar]
- 20.Wild J, Rossmeissl P, Walter WA, Gross CA. J Bacteriol. 1996;178:3608–3613. doi: 10.1128/jb.178.12.3608-3613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altman E, Kumamoto CA, Emr SD. EMBO J. 1991;10:239–245. doi: 10.1002/j.1460-2075.1991.tb07943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HC, Bernstein HD. J Biol Chem. 2002;277:43527–43535. doi: 10.1074/jbc.M205950200. [DOI] [PubMed] [Google Scholar]
- 23.Muller JP. J Bacteriol. 1996;178:6097–6104. doi: 10.1128/jb.178.21.6097-6104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumamoto CA, Beckwith J. J Bacteriol. 1983;154:253–260. doi: 10.1128/jb.154.1.253-260.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumamoto CA, Beckwith J. J Bacteriol. 1985;163:267–274. doi: 10.1128/jb.163.1.267-274.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu H, Nishiyama K, Tokuda H. Mol Microbiol. 1997;26:1013–1021. doi: 10.1046/j.1365-2958.1997.6392003.x. [DOI] [PubMed] [Google Scholar]
- 27.Pogliano KJ, Beckwith J. Genetics. 1993;133:763–773. doi: 10.1093/genetics/133.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gragerov A, Nudler E, Komissarova N, Gaitanaris GA, Gottesman ME, Nikiforov V. Proc Natl Acad Sci USA. 1992;89:10341–10344. doi: 10.1073/pnas.89.21.10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baars L, Ytterberg AJ, Drew D, Wagner S, Thilo C, Van Wijk KJ, de Gier JW. J Biol Chem. 2006;281:10024–10034. doi: 10.1074/jbc.M509929200. [DOI] [PubMed] [Google Scholar]
- 30.Bowers CW, Lau F, Silhavy TJ. J Bacteriol. 2003;185:5697–5705. doi: 10.1128/JB.185.19.5697-5705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agashe VR, Guha S, Chang HC, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM. Cell. 2004;117:199–209. doi: 10.1016/s0092-8674(04)00299-5. [DOI] [PubMed] [Google Scholar]
- 32.Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. Nature. 2002;419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- 33.Merz F, Hoffmann A, Rutkowska A, Zachmann-Brand B, Bukau B, Deuerling E. J Biol Chem. 2006;81:31963–31971. doi: 10.1074/jbc.M605164200. [DOI] [PubMed] [Google Scholar]
- 34.Kramer G, Patzelt H, Rauch T, Kurz TA, Vorderwulbecke S, Bukau B, Deuerling E. J Biol Chem. 2004;279:14165–14170. doi: 10.1074/jbc.M313635200. [DOI] [PubMed] [Google Scholar]
- 35.Scholz C, Stoller G, Zarnt T, Fischer G, Schmid FX. EMBO J. 1997;16:54–58. doi: 10.1093/emboj/16.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullers RS, Luirink J, Harms N, Schwager F, Georgopoulos C, Genevaux P. Proc Natl Acad Sci USA. 2004;101:7583–7588. doi: 10.1073/pnas.0402398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bukau B, Walker GC. J Bacteriol. 1989;171:2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genevaux P, Schwager F, Georgopoulos C, Kelley WL. J Bacteriol. 2001;183:5747–5750. doi: 10.1128/JB.183.19.5747-5750.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bukau B, Horwich AL. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 40.Karamyshev AL, Johnson AE. J Biol Chem. 2005;280:37930–37940. doi: 10.1074/jbc.M509100200. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez M, Valencia A, Ferrandiz MJ, Sander C, Vicente M. EMBO J. 1994;13:4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitra K, Frank J. FEBS Lett. 2006;580:3353–3360. doi: 10.1016/j.febslet.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies KU. EMBO J. 2000;19:1900–1906. doi: 10.1093/emboj/19.8.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zito CR, Oliver D. J Biol Chem. 2003;278:40640–40646. doi: 10.1074/jbc.M308025200. [DOI] [PubMed] [Google Scholar]
- 45.Blaha G, Wilson DN, Stoller G, Fischer G, Willumeit R, Nierhaus KH. J Mol Biol. 2003;326:887–897. doi: 10.1016/s0022-2836(02)01436-5. [DOI] [PubMed] [Google Scholar]
- 46.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 47.Michaelis S, Hunt JF, Beckwith J. J Bacteriol. 1986;167:160–167. doi: 10.1128/jb.167.1.160-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casadaban MJ. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 49.Genevaux P, Schwager F, Georgopoulos C, Kelley WL. Genetics. 2002;162:1045–1053. doi: 10.1093/genetics/162.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Datsenko KA, Wanner BL. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kleckner N, Bender J, Gottesman S. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 53.Genevaux P, Bauda P, DuBow MS, Oudega B. Arch Microbiol. 1999;172:1–8. doi: 10.1007/s002030050732. [DOI] [PubMed] [Google Scholar]
- 54.Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Mol Microbiol. 2001;40:397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- 55.De Vrije T, Tommassen J, de Kruijff B. Biochim Biophys Acta. 1987;900:63–72. doi: 10.1016/0005-2736(87)90278-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information