Complete Nucleotide Sequence and Genetic Organization of the 210-Kilobase Linear Plasmid of Rhodococcus erythropolis BD2 (original) (raw)
Abstract
The complete nucleotide sequence of the linear plasmid pBD2 from Rhodococcus erythropolis BD2 comprises 210,205 bp. Sequence analyses of pBD2 revealed 212 putative open reading frames (ORFs), 97 of which had an annotatable function. These ORFs could be assigned to six functional groups: plasmid replication and maintenance, transport and metalloresistance, catabolism, transposition, regulation, and protein modification. Many of the transposon-related sequences were found to flank the isopropylbenzene pathway genes. This finding together with the significant sequence similarities of the ipb genes to genes of the linear plasmid-encoded biphenyl pathway in other rhodococci suggests that the ipb genes were acquired via transposition events and subsequently distributed among the rhodococci via horizontal transfer.
Linear DNA replicons occur in various organisms and viruses (3, 13). Their structural analyses revealed two distinct classes. The first one is characterized by covalently closed hairpin loops at each end and is found in the genus Borrelia and in the prophage N15. The linear elements of the second class (invertrons) carry characteristic terminal inverted repeats (TIRs) and proteins covalently bound to each 5′ end. Invertrons have been detected in several bacteriophages and viruses as well as in eukaryotic cells. Among bacteria, they occur in species of the genera Streptomyces (2), Rhodococcus (4, 6, 16), Mycobacterium (10), and Planobispora (14). So far, only two small linear plasmids, the 12-kb plasmid pSCL1 from Streptomyces clavuligerus and the 23-kb mycobacterial plasmid pCLP, and one megaplasmid of 350 kb (SCP1 from Streptomyces coelicolor A3) have been completely sequenced (10, 15, 18).
Rhodococcus erythropolis strain BD2 is a gram-positive isopropylbenzene (IPB) degrader and was found to cometabolize trichloroethene with dependence on IPB as an inducing substrate. Analysis of the IPB degradation pathway led to the finding that strain BD2 harbors a linear transmissible plasmid, pBD2, of approximately 210 kb carrying the ipb genes for IPB and trichloroethene oxidation and mediating arsenite and mercury resistance (4, 5). Linear plasmid pBD2 was sequenced to further characterize its genetic organization.
General features of pBD2.
To perform a complete sequence analysis, pBD2 linear DNA was isolated from R. erythropolis BD2 as described recently (4) and a cosmid library comprising 350 clones was generated by use of pWE15 (17). To map the cosmids on pBD2, hybridization studies with digoxigenin-labeled probes were performed under high-stringency conditions as described by Anderson and Young (1). The first cosmid was mapped with a probe of pMK34 carrying the ipb genes (8). Further rounds of hybridization with labeled end fragments of mapped cosmids led to the identification of seven cosmid clones covering 96% of pBD2. The cosmids were analyzed with respect to the presence of chromosomal contaminations and chimeric DNA, respectively. After assembly of the sequence generated from a small insert library of the selected cosmid DNAs in pTZr19 (12) and from pBD2 end fragments (cloning is described below), two gaps remained. One gap of 931 bp was closed by sequencing clones from a pDA71 small insert plasmid library (8). The other gap of 212 bp was closed by PCR using primers deduced from the sequence flanking the gap (P1, 5′-GAGCCACACAAACACCAG-3′, and P2, 5′-GCACGAAGTATGGCGAAC-3′).
Sequence data were analyzed with the software package (version 10.0) of the Genetics Computer Group (University of Wisconsin Biotechnology Center), and similarity searches were done by BLAST and FASTA.
The average GC content of the 210,205-bp plasmid pBD2 is 62.2%, which is lower than the average GC content of rhodococcal genomes (64 to 72%). Interestingly, the GC content was found to increase from the left to the right end. The region from the left end to 91 kb comprises 60.7% GC on average (region A), and the region from 91 kb to the right end contains 63.4% GC (region B).
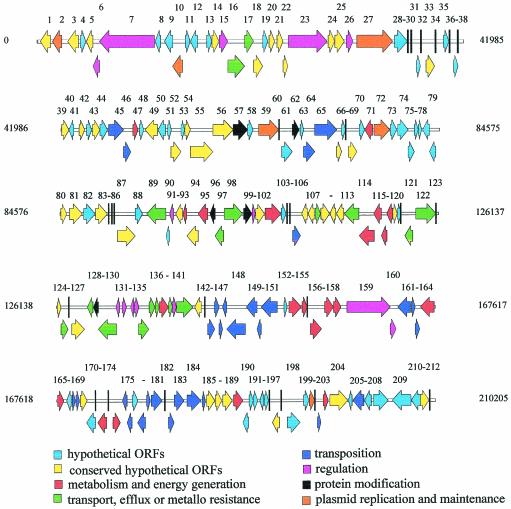
The coding region comprises 87.6% of the plasmid, and a total of 212 open reading frames (ORFs) were identified (Table 1), and 97 of them could be assigned to six functional groups: 7 code for plasmid maintenance and replication, 16 code for transport and metalloresistance, 23 code for catabolism, 32 code for transposition, 14 code for regulation, and 5 code for protein modification. Forty-seven ORFs code for conserved proteins, and 68 ORFs are hypothetical. Their locations on the plasmid are depicted in Fig. 1.
TABLE 1.
Predicted ORFs of pBD2
| ORF(s) | Nucleotide position ofa: | Function predicted by sequence similarityb | |
|---|---|---|---|
| Start codon | Stop codon | ||
| PBD2.001 | 1214 | 234 | Conserved hypothetical protein |
| PBD2.002 | 2352 | 1429 | Putative DNA uptake protein |
| PBD2.003 | 4008 | 2935 | Conserved hypothetical protein |
| PBD2.004 | 4200 | 4733 | Hypothetical protein |
| PBD2.005 | 5438 | 4806 | Conserved hypothetical protein |
| PBD2.006 | 6049 | 5435 | Putative regulatory protein |
| PBD2.007 | 11511 | 6046 | Putative regulatory protein |
| PBD2.008 | 12124 | 11564 | Hypothetical protein |
| PBD2.009 | 13298 | 12495 | Hypothetical protein |
| PBD2.010 | 14257 | 13295 | Putative ParA-family ATPase |
| PBD2.011-PBD2.013 | Hypothetical proteins | ||
| PBD2.014 | 17173 | 17880 | Conserved hypothetical protein |
| PBD2.015 | 17885 | 18721 | Putative regulatory protein |
| PBD2.016 | 18718 | 20409 | Putative type II/IV-secretion NTPase |
| PBD2.017 | 20409 | 21296 | TadB-like protein |
| PBD2.018 | 21293 | 22195 | Conserved hypothetical protein |
| PBD2.019 | 22198 | 22695 | Hypothetical protein |
| PBD2.020-PBD2.022 | Conserved hypothetical proteins | ||
| PBD2.023 | 24760 | 28539 | Putative regulator |
| PBD2.024 | 28662 | 29291 | Conserved hypothetical protein |
| PBD2.025 | 29321 | 30253 | Conserved hypothetical protein |
| PBD2.026 | 30459 | 31121 | A-factor receptor-like protein |
| PBD2.027 | 31489 | 35031 | Putative helicase |
| PBD2.028-PBD2.032 | Hypothetical proteins | ||
| PBD2.033 | 38316 | 39149 | Conserved hypothetical protein |
| PBD2.034-PBD2.038 | Hypothetical proteins | ||
| PBD2.039 | 41986 | 42816 | Conserved hypothetical |
| PBD2.040 | 43351 | 42821 | Hypothetical protein |
| PBD2.041 | 43947 | 44567 | Conserved hypothetical protein |
| PBD2.042 | 44796 | 45362 | Hypothetical protein |
| PBD2.043 | 45468 | 46268 | Conserved hypothetical protein |
| PBD2.044 | 46296 | 47138 | Hypothetical protein |
| PBD2.045 | 47277 | 48956 | Putative transposase |
| PBD2.046 | 48953 | 49759 | IS sequence |
| PBD2.047 | 49999 | 50580 | Putative hydrolase CbbY/CbbZ/GpH/YieH family |
| PBD2.048 | 50731 | 51189 | Hypothetical protein |
| PBD2.049 | 52759 | 51329 | Conserved hypothetical protein |
| PBD2.050 | 53650 | 52799 | Hypothetical protein |
| PBD2.051 | 53950 | 54312 | Hypothetical protein |
| PBD2.052 | 54309 | 55418 | Conserved hypothetical protein |
| PBD2.053 | 55439 | 55801 | Hypothetical protein |
| PBD2.054-PBD2.056 | Conserved hypothetical proteins | ||
| PBD2.057 | 61274 | 62917 | Putative peptidase |
| PBD2.058 | 62936 | 63544 | Hypothetical protein |
| PBD2.059 | 64185 | 66446 | Putative DNA translocase |
| PBD2.060 | 66479 | 66808 | Hypothetical protein |
| PBD2.061 | 66805 | 67956 | Hypothetical protein |
| PBD2.062 | 67946 | 68740 | Putative type 4 peptidase |
| PBD2.063 | 68872 | 69237 | Hypothetical protein |
| PBD2.064 | 69234 | 70493 | Putative DNA integrase/recombinase |
| PBD2.065 | 70490 | 72976 | Putative DNA integrase/recombinase |
| PBD2.066 | 72973 | 73524 | Conserved hypothetical protein |
| PBD2.067 | 73528 | 73947 | Hypothetical protein |
| PBD2.068 | 73984 | 74307 | Hypothetical protein |
| PBD2.069 | 74336 | 75229 | Conserved hypothetical protein |
| PBD2.070 | 75493 | 76104 | Hypothetical protein |
| PBD2.071 | 76977 | 76126 | Putative PAPS reductase |
| PBD2.072 | 77154 | 78938 | Putative septum site-determining protein (MinD) |
| PBD2.073-PBD2.079 | Hypothetical proteins | ||
| PBD2.080 | 84576 | 85223 | Conserved hypothetical protein |
| PBD2.081 | 85513 | 86955 | Conserved hypothetical protein |
| PBD2.082 | 87086 | 88336 | Hypothetical protein |
| PBD2.083 | 88407 | 89702 | Conserved hypothetical protein |
| PBD2.084 | 89832 | 90089 | Hypothetical protein |
| PBD2.085 | 90184 | 90444 | Hypothetical protein |
| PBD2.086 | 90445 | 90747 | PemK-like growth inhibitor/PICK> |
| PBD2.087 | 90776 | 92746 | Conserved hypothetical protein |
| PBD2.088 | 92746 | 93543 | Hypothetical protein |
| PBD2.089 | 96124 | 93977 | Putative cadmium resistance protein (CadA) |
| PBD2.090 | 96509 | 96117 | Hypothetical protein |
| PBD2.091 | 96965 | 96537 | Putative ArsR family regulator |
| PBD2.092 | 97942 | 97178 | Conserved hypothetical protein |
| PBD2.093 | 98406 | 97987 | Putative Rieske protein |
| PBD2.094 | 99668 | 98403 | Conserved hypothetical protein |
| PBD2.095 | 100726 | 99665 | Putative metallo-oxido-reductase |
| PBD2.096 | 101526 | 100972 | Putative lipoprotein signal peptidase |
| PBD2.097 | 102455 | 101505 | Putative efflux protein |
| PBD2.098 | 102524 | 104554 | Putative copper export protein |
| PBD2.099 | 104657 | 105568 | Putative peptidase of M23/37 family |
| PBD2.100 | 105601 | 105960 | Putative regulator |
| PBD2.101 | 105961 | 106911 | Conserved hypothetical protein |
| PBD2.102 | 107029 | 108786 | Putative cytochrome _aa_3 oxidase UE I |
| PBD2.103 | 108816 | 109355 | Hypothetical protein |
| PBD2.104 | 109769 | 109488 | Conserved hypothetical protein |
| PBD2.105 | 109768 | 110046 | Putative transposase |
| PBD2.106 | 110070 | 110912 | Putative transposase |
| PBD2.107 | 111671 | 111081 | Conserved hypothetical protein |
| PBD2.108 | 112450 | 111740 | Conserved hypothetical protein |
| PBD2.109 | 112568 | 113122 | Putative copper resistance protein (CopC) |
| PBD2.110-PBD2.112 | Conserved hypothetical proteins | ||
| PBD2.113 | 117355 | 115742 | Putative apolipoprotein-_N_-acyltransferase (CutE) |
| PBD2.114 | 118983 | 117352 | Putative cytochrome c biogenesis protein (ResB) |
| PBD2.115 | 119786 | 118980 | Putative _c_-type cytochrome biogenesis protein (CcdA) |
| PBD2.116 | 120427 | 119783 | Putative thioredoxin |
| PBD2.117 | 121062 | 120424 | Putative thioredoxin |
| PBD2.118 | 121113 | 121556 | Conserved hypothetical protein |
| PBD2.119 | 121575 | 121991 | Hypothetical protein |
| PBD2.120 | 122259 | 121921 | Conserved hypothetical protein |
| PBD2.121 | 123261 | 122383 | Putative heavy metal transporter |
| PBD2.122 | 123626 | 125815 | Putative membrane transport protein |
| PBD2.123 | 125842 | 126093 | Hypothetical protein |
| PBD2.124 | 126138 | 126554 | Conserved hypothetical protein |
| PBD2.125 | 126551 | 127327 | Putative permease |
| PBD2.126 | 127406 | 127675 | Conserved hypothetical protein |
| PBD2.127 | 127729 | 129096 | Conserved hypothetical protein |
| PBD2.128 | 130061 | 129459 | Putative cadmium resistance protein (CadD) |
| PBD2.129 | 130635 | 130111 | Putative lipoprotein signal peptidase |
| PBD2.130 | 132647 | 130632 | Putative cadmium resistance protein (CadA) |
| PBD2.131 | 132957 | 132577 | Putative ArsR-family regulator |
| PBD2.132 | 133263 | 133721 | Putative MerR-family regulator |
| PBD2.133 | 134563 | 134159 | Putative MerR-family regulator |
| PBD2.134 | 134634 | 135059 | Putative ArsR-family regulator |
| PBD2.135 | 135056 | 136153 | Putative oxyanion translocation protein (ArsB) |
| PBD2.136 | 136181 | 136834 | Putative arsenate reductase (ArsC) |
| PBD2.137 | 136876 | 137292 | Putative arsenate reductase (ArsC) |
| PBD2.138 | 137334 | 138320 | Putative thioredoxin reductase (TrxB) |
| PBD2.139 | 138348 | 138758 | Putative arsenate reductase (ArsC) |
| PBD2.140 | 138819 | 139226 | Putative trans-acting repressor (ArsD) |
| PBD2.141 | 139243 | 141000 | Putative arsenite ATPase catalytic subunit (ArsA) |
| PBD2.142 | 141927 | 141190 | Conserved hypothetical protein |
| PBD2.143 | 142325 | 142588 | Hypothetical protein |
| PBD2.144-PBD2.146 | IS sequences | ||
| PBD2.147-PBD2.149 | Putative transposases | ||
| PBD2.150 | 148521 | 148048 | IS sequence |
| PBD2.151 | 150275 | 148518 | Putative transposase |
| PBD2.152 | 151023 | 151370 | Hypothetical protein |
| PBD2.153 | 151549 | 152931 | IPB dioxygenase, ISP large subunit (IpbA1) |
| PBD2.154 | 153013 | 153576 | IPB dioxygenase, ISP small subunit (IpbA2) |
| PBD2.155 | 153585 | 153908 | IPB dioxygenase, ferredoxin (IpbA3) |
| PBD2.156 | 153905 | 155143 | IPB dioxygenase, ferredoxin reductase (IpbA4) |
| PBD2.157 | 155469 | 156422 | IPC dioxygenase (IpbC) |
| PBD2.158 | 156460 | 157272 | IPB dihydrodiol dehydrogenase (IpbB) |
| PBD2.159 | 157899 | 162674 | Putative sensor kinase (IpbS) |
| PBD2.160 | 162671 | 163300 | Putative response regulator (IpbT) |
| PBD2.161-PBD2.163 | Putative transposases | ||
| PBD2.164 | 167494 | 165956 | Putative medium-chain acyl-CoA ligase (AlkK) |
| PBD2.165 | 167618 | 168337 | Putative enoyl-CoA-hydratase |
| PBD2.166 | 169139 | 168636 | Hypothetical protein |
| PBD2.167 | 169256 | 169615 | Putative transposase |
| PBD2.168 | 169702 | 170064 | Putative transposase |
| PBD2.169 | 170140 | 170937 | Conserved hypothetical protein |
| PBD2.170 | 171854 | 170871 | Hypothetical protein |
| PBD2.171 | 172136 | 171861 | Hypothetical protein |
| PBD2.172 | 173109 | 172120 | Putative quinone oxidoreductase |
| PBD2.173 | 173590 | 173327 | Putative transposase |
| PBD2.174 | 173839 | 174663 | HOMODA hydrolase (IpbD) |
| PBD2.175 | 175431 | 174904 | IS sequence |
| PBD2.176 | 175976 | 175428 | IS sequence |
| PBD2.177 | 176027 | 176641 | Hypothetical protein |
| PBD2.178 | 177462 | 176599 | Putative transposase |
| PBD2.179 | 177629 | 178018 | IS sequence |
| PBD2.180-PBD2.185 | Putative transposases | ||
| PBD2.186-PBD2.188 | Conserved hypothetical proteins | ||
| PBD2.189 | 187441 | 188487 | Putative _N_-formylglutamate aminohydrolase |
| PBD2.190-PBD2.195 | Hypothetical proteins | ||
| PBD2.196 | 192561 | 191812 | Conserved hypothetical protein |
| PBD2.197-PBD2.199 | Hypothetical proteins | ||
| PBD2.200 | 195961 | 196566 | Putative exonuclease X |
| PBD2.201 | 196600 | 196884 | IS sequence |
| PBD2.202 | 196887 | 197252 | IS sequence |
| PBD2.203 | 197556 | 198101 | Putative acetyltransferase |
| PBD2.204 | 198263 | 200308 | Conserved hypothetical protein |
| PBD2.205 | 200766 | 200392 | Hypothetical protein |
| PBD2.206 | 202095 | 200878 | Putative transposase |
| PBD2.207-PBD2.210 | Hypothetical proteins | ||
| PBD2.211 | 208479 | 209462 | Conserved hypothetical protein |
| PBD2.212 | 209541 | 209783 | Hypothetical protein |
FIG. 1.
Physical map of the linear plasmid pBD2. Putative ORFs are grouped into six functional groups. Black bars represent ORFs smaller than 312 bp.
The deduced proteins of 20 of the pBD2 ORFs, are similar to proteins encoded by other linear plasmids; for example, PBD2.001 shows significant similarities of 73 and 54%, respectively, to 201L1 of the linear plasmid pHG201 from Rhodococcus opacus MR11 (7) and to FIR1 of linear plasmid pFiD188 from Rhodococcus fascians D188 (11). PBD2.002 exhibits a similarity of 54% to FIR2 of pFiD188. Eighteen ORFs (PBD2.014 to PBD2.018, PBD2.020 to PBD2.025, PBD2.027, PBD2.052, PBD2.054 to PBD2.056, PBD2.064, and PBD2.065) show significant similarities to ORFs of the 350-kb linear plasmid SCP1 from S. coelicolor A3(2) (20).
The protein products of six ORFs within region A—PBD2.002, PBD2.010, PBD2.027, PBD2.059, PBD2.072, PBD2.086, and the ORF located close to the right terminus, PBD2.200—show similarities to proteins implicated in plasmid maintenance and DNA processing (Table 1). In addition, all of the ORFs conserved on linear plasmids and nearly none of the potential transposon functions were detected within these regions. It is tempting to speculate that several of these hypothetical proteins are implicated in plasmid maintenance, especially those which are conserved and widely distributed among linear plasmids. Taken together, these findings suggest that region A and the right pBD2 terminus are essential for plasmid maintenance and DNA processing.
Plasmid dynamics in the region encoding resistance and catabolic functions.
Within the high-GC region, region B, a cluster of conserved genes mediating pBD2-encoded arsenite resistance was detected. Ten kilobases downstream of the resistance functions, the ipb gene cluster, encoding the three subunits of the IPB dioxygenase, the 3-isopropylcatechol dioxygenase, and the IPB dihydrodiol dehydrogenase, was found (ORFs PBD2.153 to PBD2.158) (Table 1). The deduced proteins are 94 to 100% similar to the analogous proteins of a linear-plasmid-encoded biphenyl (BPH) degradation pathway in Rhodococcus sp. strain RHA1 and the BPH degradation pathway in Rhodococcus sp. strain I1 (16) (accession no. CAA06877). The deduced proteins of two ORFs downstream of the ipb genes, PBD2.159 and PBD2.160, show 62 to 80% similarities to two-component signal transduction systems of the BPH degradation pathways in Rhodococcus sp. strain M5 and R. erythropolis TA421 (9) (accession no. AB014348). The close association together with the very high similarities suggests that this potential two-component regulatory system represents the IPB pathway regulatory system. The product of ORF PBD2.174, located 10 kb downstream of the ipb cluster, is identical to the 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase (EtbD1) in Rhodococcus sp. strain RHA1 (19). Therefore, PBD2.174 is designated ipbD.
Taken together, the similarities of the key enzymes and the regulators of the IPB degradative pathway genes in R. erythropolis BD2 and the linear-plasmid-encoded functions of BPH degradation pathways indicate that the ipb and bph operons have been distributed among gram-positive soil bacteria via linear-plasmid-mediated horizontal gene transfer.
The ipb structural and regulatory genes are flanked by a total of 22 ORFs showing significant similarities to insertion sequences, integrases, and transposases (Fig. 1; Table 1). This high number of transposon-related ORFs in the close vicinity of the ipb genes indicates that the ipb genes could have been acquired via transposition events. Furthermore, this suggests that this part of the plasmid has undergone a high frequency of dynamic rearrangements.
Structural characteristics of the pBD2 termini.
The presence of terminal proteins bound to the 5′ ends of pBD2 was demonstrated by λ exonuclease, exonuclease III, and mung bean nuclease treatment and gel retardation analyses under proteolytic and nonproteolytic conditions according to the protocol of Kalkus et al. (6). The two terminal fragments, a 1.5-kb _Kpn_I and a 3.8-kb _Bam_HI fragment, were cloned from pBD2 DNA isolated under proteolytic conditions. Additionally, a 4.9-kb _Eco_RI/_Not_I fragment with a 1-kb overlap of the _Kpn_I fragment was cloned from pBD2.
It is apparent from sequence analyses of the pBD2 termini that pBD2 does not contain long TIRs and that the similarity of the terminal sequences at both ends is reduced to two inverted repeats which share a central motif, GCTXCGC. This motif is characteristic of linear rhodococcal plasmids and is suggested to be involved in extending the 5′ lagging strand after each replication round (3, 7). In addition to this conserved central motif, the linear plasmids pBD2 and pHG201 from R. opacus MR11 and pRHL2 from Rhodococcus sp. strain RHA1 (16) show significant similarities over a range of 1,000 bp at the left termini and of 130 bp of the right termini. The short rhodococcal TIRs are in contrast to the long TIRs of the Streptomyces linear plasmids, which comprise many palindromes with the potential to form very stable complex secondary structures at the 3′ ends of linear replicons. Although the significance is not clear, the conservation of these structures suggests an important biological role, which obviously is not conserved with respect to linear rhodococcal plasmids.
Nucleotide sequence accession number.
The sequence of the linear plasmid pBD2 from R. erythropolis BD2 is available in GenBank under the accession number AY223810. The graphical representation and a detailed annotation are available at the Laboratorium für Genomanalyse, Göttingen, Germany, at http://www.g2l.bio.uni-goettingen.de.
Acknowledgments
The work carried out by the Laboratorium für Genomanalyse was supported by grants from the Ministry of Science and Culture of Lower Saxony (Germany) and the Academy of Sciences of Göttingen.
We thank Maria Kesseler and Jutta Kalkus for providing the ends of the linear plasmid. We are grateful to Arnim Wiezer and Heiko Liesegang for assistance in analyzing the sequence information and for submission of the sequence information to the databanks.
REFERENCES
- 1.Anderson, M. L. M., and B. D. Young. 1985. Quantitative filter hybridization, p. 73-110. In B. D. Hames and S. J. Higgins (ed.), Nucleic acid hybridization. IRL Press, Ltd., Oxford, United Kingdom.
- 2.Bao, K., and S. N. Cohen. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 15**:**1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. W. 1996. Complications and implications of linear bacterial chromosomes. Trends Genet. 12**:**192-196. [DOI] [PubMed] [Google Scholar]
- 4.Dabrock, B., M. Kesseler, B. Averhoff, and G. Gottschalk. 1994. Identification and characterization of a transmissible linear plasmid from Rhodococcus erythropolis BD2 that encodes isopropylbenzene and trichloroethene catabolism. Appl. Environ. Microbiol. 60**:**853-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dabrock, B., J. Riedel, J. Bertram, and G. Gottschalk. 1992. Isopropylbenzene (cumene)—a new substrate for the isolation of trichloroethene-degrading bacteria. Arch. Microbiol. 158**:**9-13. [DOI] [PubMed] [Google Scholar]
- 6.Kalkus, J., C. Dorrie, D. Fischer, M. Reh, and H. G. Schlegel. 1993. The giant linear plasmid pHG207 from Rhodococcus sp. encoding hydrogen autotrophy: characterization of the plasmid and its termini. J. Gen. Microbiol. 139**:**2055-2065. [DOI] [PubMed] [Google Scholar]
- 7.Kalkus, J., R. Menne, M. Reh, and H. G. Schlegel. 1998. The terminal structures of linear plasmids from Rhodococcus opacus. Microbiology 144**:**1271-1279. [DOI] [PubMed] [Google Scholar]
- 8.Kesseler, M., E. R. Dabbs, B. Averhoff, and G. Gottschalk. 1996. Studies on the isopropylbenzene 2,3-dioxygenase and the 3-isopropylcatechol 2,3-dioxygenase genes encoded by the linear plasmid of Rhodococcus erythropolis BD2. Microbiology 142**:**3241-3251. [DOI] [PubMed] [Google Scholar]
- 9.Labbe, D., J. Garnon, and P. C. Lau. 1997. Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl-degrading bacterium, Rhodococcus sp. strain M5. J. Bacteriol. 179**:**2772-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Dantec, C., N. Winter, B. Gicquel, V. Vincent, and M. Picardeau. 2001. Genomic sequence and transcriptional analysis of a 23-kilobase mycobacterial linear plasmid: evidence for horizontal transfer and identification of plasmid maintenance systems. J. Bacteriol. 183**:**2157-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maes, T., D. Vereecke, T. Ritsema, K. Cornelis, H. N. Thu, M. Van Montagu, M. Holsters, and K. Goethals. 2001. The att locus of Rhodococcus fascians strain D188 is essential for full virulence on tobacco through the production of an autoregulatory compound. Mol. Microbiol. 42**:**13-28. [DOI] [PubMed] [Google Scholar]
- 12.Mead, D. A., E. Szczesna-Skorupa, and B. Kemper. 1986. Single-stranded DNA ‘blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1**:**67-74. [DOI] [PubMed] [Google Scholar]
- 13.Meinhardt, F., R. Schaffrath, and M. Larsen. 1997. Microbial linear plasmids. Appl. Microbiol. Biotechnol. 47**:**329-336. [DOI] [PubMed] [Google Scholar]
- 14.Polo, S., O. Guerini, M. Sosio, and G. Deho. 1998. Identification of two linear plasmids in the actinomycete Planobispora rosea. Microbiology 144**:**2819-2825. [DOI] [PubMed] [Google Scholar]
- 15.Redenbach, M., K. Ikeda, M. Yamasaki, and H. Kinashi. 1998. Cloning and physical mapping of the _Eco_RI fragments of the giant linear plasmid SCP1. J. Bacteriol. 180**:**2796-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu, S., H. Kobayashi, E. Masai, and M. Fukuda. 2001. Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 67**:**2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahl, G. M., K. A. Lewis, J. C. Ruiz, B. Rothenberg, J. Zhao, and G. A. Evans. 1987. Cosmid vectors for rapid genomic walking, restriction mapping, and gene transfer. Proc. Natl. Acad. Sci. USA 84**:**2160-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu, X., and K. L. Roy. 1993. Complete nucleotide sequence of a linear plasmid from Streptomyces clavuligerus and characterization of its RNA transcripts. J. Bacteriol. 175**:**37-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamada, A., H. Kishi, K. Sugiyama, T. Hatta, K. Nakamura, E. Masai, and M. Fukuda. 1998. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 64**:**2006-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamasaki, M., M. Redenbach, and H. Kinashi. 2001. Integrated structures of the linear plasmid SCP1 in two bidirectional donor strains of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 264**:**634-642. [DOI] [PubMed] [Google Scholar]