Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint (original) (raw)
Abstract
Inappropriate attachment/tension between chromosomal kinetochores and the kinetochore microtubules activates the spindle assembly checkpoint, which delays anaphase by blocking the ubiquitin-mediated degradation of securin/Pds1p by APCCdc20. The checkpoint proteins Mad2 and Mad3/BubR1 bind to Cdc20, although how they inhibit APCCdc20 is unclear. We investigated the roles of two evolutionarily conserved KEN boxes and a D box within Mad3/BubR1. Although such motifs usually mediate APC-substrate recognition and ubiquitination, they have no apparent role in Mad3p turnover in Saccharomyces cerevisiae. Instead, these motifs are important for Mad3p function in the checkpoint and for binding to Cdc20p. We show that the Mad3p D box and KEN boxes function together to mediate Cdc20p–Mad3p interaction and that Mad3p and an anaphase-promoting complex (APC) substrate, Hsl1p, compete for Cdc20p binding in a D-box- and KEN-box-dependent manner. In vivo, we observed an increased binding of Cdc20p to Mad3p and decreased binding to Hsl1p upon checkpoint activation. Furthermore, we demonstrate that Mad2p stimulates the association between Mad3p and Cdc20p and that this stimulated binding requires KEN box 1 within Mad3p. These findings implicate Mad3p as a pseudosubstrate inhibitor of APCCdc20, competing with APC substrates for Cdc20p binding. We present a model aimed at unifying previous analyses of checkpoint function by focusing on the Mad3–Cdc20 interaction.
Keywords: Anaphase-promoting complex, budding yeast, Cdc20, cell cycle
To maintain genomic integrity, chromosomes must be replicated and segregated with high fidelity. Chromosome duplication results in the pairing of sister chromatids that are held together by the cohesin protein complex (for review, see Nasmyth 2002). Once the kinetochores of the sister chromatids achieve bipolar attachment to the mitotic spindle, the cohesin complex is cleaved and chromosomes segregate to opposite poles (Uhlmann et al. 2000). The dissolution of the cohesin complex requires ubiquitin-mediated proteolysis of the anaphase inhibitor, securin (Pds1p in Saccharomyces cerevisiae), which binds and inhibits separase, a protease whose target is the Scc1 subunit of the cohesin complex (Cohen-Fix et al. 1996; Funabiki et al. 1996a, b; Ciosk et al. 1998; Uhlmann et al. 1999, 2000).
Securin ubiquitination and degradation are mediated by a multisubunit ubiquitin ligase known as the anaphase-promoting complex (APC) or cyclosome (for review, see Harper et al. 2002; Peters 2002). During mitotic cell cycles, the APC requires the binding of either of two WD40-containing proteins, Cdc20 or Cdh1, for its activity. APCCdc20 promotes the degradation of securin at the metaphase-to-anaphase transition, whereas APCCdh1 activity in late mitosis and G1 is required for the degradation of the mitotic cyclins. Cdc20 and Cdh1 bind directly to APC substrates through degradation motifs within the substrate (Burton and Solomon 2001; Hilioti et al. 2001; Pfleger et al. 2001a; Burton et al. 2005). The best characterized of these degradation signals are the D box (Glotzer et al. 1991; King et al. 1996) and the KEN box (Pfleger and Kirschner 2000). In addition, the D box is required for substrate association with the APC (Yamano et al. 2004; Burton et al. 2005; Carroll et al. 2005; Kraft et al. 2005).
During mitosis, errors in chromosome attachment to the mitotic spindle activate the spindle assembly checkpoint at metaphase and delay anaphase onset by inhibiting APCCdc20-mediated securin degradation. The molecular players in the checkpoint include Mad1, Mad2, Mad3 (BubR1 in metazoans), Bub1, Bub3, and Mps1 (for review, see Musacchio and Hardwick 2002; Yu 2002). Both Mad2 and Mad3 bind directly to Cdc20 and can inhibit APCCdc20 ubiquitination activity in vitro (Fang et al. 1998; Hwang et al. 1998; Kim et al. 1998; Sudakin et al. 2001; Tang et al. 2001). In addition, Mad3 associates with Bub3 throughout the cell cycle (Hardwick et al. 2000). Under checkpoint activation conditions Cdc20, Mad2, Mad3/BubR1, and Bub3 form a complex known as the mitotic checkpoint complex (MCC) (Hardwick et al. 2000; Fraschini et al. 2001; Sudakin et al. 2001), which can bind to and inhibit the APC (Sudakin et al. 2001; Tang et al. 2001). Although numerous mechanisms have been proposed by which the binding of checkpoint proteins to Cdc20 could block APC-mediated ubiquitination of securin, a unified molecular understanding has not been achieved. Proposed mechanisms include inhibition of the release of substrate from Mad2–Cdc20 complexes (Pfleger et al. 2001b) and blocking Cdc20–APC association (Tang et al. 2001). In addition, the checkpoint has been proposed to act in part by promoting Cdc20p turnover, since it has been shown that the checkpoint proteins, in particular Mad3p, are important for the rapid APC-mediated degradation of Cdc20p (Pan and Chen 2004). Precisely how Cdc20p is targeted for this degradation is currently unknown.
We have investigated the checkpoint functions of two evolutionarily conserved KEN boxes and a D-box motif within Mad3p. We find that although these motifs have no obvious role in Mad3p turnover, they are required for Mad3p function in the spindle checkpoint. The N-terminal KEN box (KB1) is required for the checkpoint, for interaction with Cdc20p in vivo, and for Mad2p-stimulated binding of Mad3p to Cdc20p in vitro. All three motifs participate in binding Cdc20p in vitro and facilitate a competition between Mad3p and APC substrates for binding Cdc20p. In vivo, we found that checkpoint activation resulted in an increase in Cdc20p binding to Mad3p and a decrease in Cdc20p binding to the APC substrate Hsl1p. These findings suggest that APC substrates and Mad3p share overlapping binding surfaces on Cdc20p and raise the possibility that the spindle checkpoint may act by blocking Cdc20p–substrate interactions. We discuss our results in the context of previous models of spindle checkpoint function and suggest a unified model for how the spindle checkpoint inhibits APCCdc20.
Results
The Mad3p KEN boxes are required for spindle checkpoint function
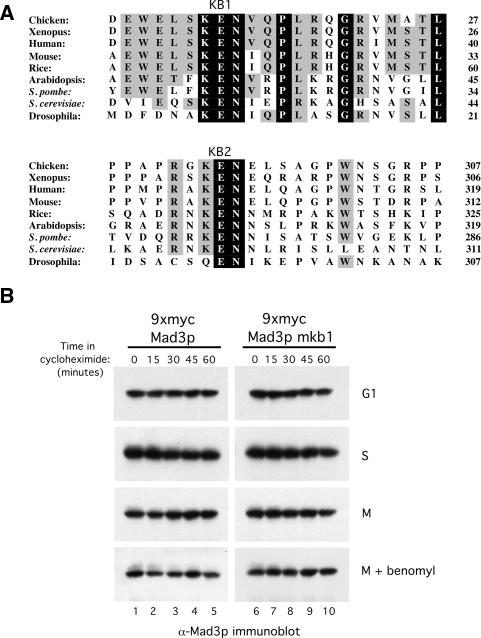
It has been noted previously that Mad3/BubR1 proteins contain a highly conserved KEN-box motif located near their N termini (Fig. 1A, KB1; Murray and Marks 2001). In addition, we noticed a previously unreported conserved KEN box located toward the middle of the Mad3/BubR1 proteins (Fig. 1A, KB2). Since KEN boxes are important for recognition and ubiquitination of substrates by the APC (Harper et al. 2002; Peters 2002), it was suggested that the N-terminal Mad3/BubR1 KEN box might function as a degradation signal and could potentially serve as a mechanism for turning off the mitotic checkpoint signal (Murray and Marks 2001). We therefore mutated this KEN box in budding yeast Mad3p (30KEN32 → AAA, “Mad3p-mkb1”) to examine whether or not it influences the stability of Mad3p. Wild-type and Mad3p-mkb1-expressing strains were arrested at different cell cycle stages followed by the addition of cycloheximide to inhibit protein synthesis. Mad3p levels were monitored by immunoblotting (Fig. 1B). We found that both the wild-type and mkb1 forms of Mad3p were stable through 60 min in all cell cycle stages tested (Fig. 1B). We conclude that KB1 does not have a major role in Mad3p turnover and, as reported previously, that Mad3p is a relatively stable protein (Hardwick et al. 2000). Mutation of KB2 (296KEN298 → AAA, “Mad3p-mkb2”) also had no effect on Mad3p stability (data not shown).
Figure 1.
The conserved KB1 has no apparent role in Mad3p turnover. (A) Alignment of the Mad3/BubR1 proteins from various species is shown with invariant residues shaded in black and highly conserved residues shaded in gray. KB1 and KB2 motifs are labeled. Sequences were aligned using the Clustal W program. Numbers at the right indicate the last residue shown on each line. (B) Both Mad3p WT and mkb1 are stable proteins. Strains containing Mad3p WT (YJB800) and Mad3p-mkb1 (YJB801) were arrested in G1 with α-factor (100 ng/mL), in S phase with hydroxyurea (100 mM), in M phase by expression of GAL-PDS1mdb, or in M phase with an activated spindle checkpoint by expression of GAL-PDS1mdb and the addition of benomyl (50 μg/mL). Samples were taken at the indicated times after cycloheximide addition to inhibit protein synthesis. Mad3p was detected by immunoblotting with anti-Mad3p antibodies.
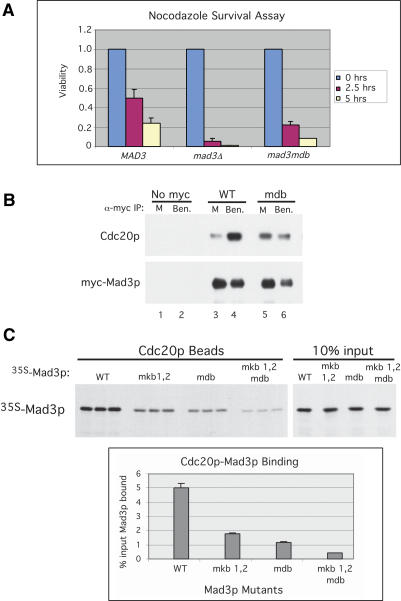
Given the high conservation of KB1 and KB2, we examined whether either is required for Mad3p function in the spindle checkpoint using a nocodazole survival assay. In this assay (Straight and Murray 1997), cells are exposed to nocodazole to disrupt the mitotic spindle and activate the checkpoint for short periods of time prior to plating in the absence of nocodazole. Cells with a functional checkpoint arrest in metaphase and resume dividing when the drug is removed, thus retaining a high level of viability. In contrast, cells that lack an intact checkpoint undergo anaphase in the presence of nocodazole and missegregate their DNA, resulting in low cell viability. We exposed MAD3, _mad3_Δ, mad3mkb1, and mad3mkb2 cells to nocodazole for 0, 2.5, and 5 h. Approximately 200 cells were plated onto medium lacking nocodazole for each time point, and the number of resulting colonies was counted. Cell viability was calculated by dividing the number of colonies formed at the 2.5- and 5-h time points by the number of colonies formed in the absence of nocodazole (0 h). Figure 2A shows the results from three independent experiments and demonstrates that both of the mad3mkb strains lost viability when exposed to nocodazole as rapidly as _mad3_Δ cells, whereas MAD3 cells maintained a much higher level of viability.
Figure 2.
The KEN boxes of Mad3p are required for spindle checkpoint function. (A) Nocodazole survival assay. Wild-type (YJB15), _mad3_Δ (YJB522), mad3mkb1 (YJB804), and mad3mkb2 (YJB932) strains were exposed to nocodazole (15 μg/mL) for 0, 2.5, and 5 h and then plated on YPD plates lacking the drug. Viability was calculated by dividing the number of colonies formed by cells treated with nocodazole for 2.5 and 5 h by the number formed by untreated cells. Error bars depict the standard deviations from three independent experiments. (B) Mps1p overexpression assay. Wild-type, mad3mkb1, and mad3mkb2 strains either with (YJB900, YJB901, and YJB923) or without (YJB894, YJB895, and YJB919) a GAL-myc-MPS1 plasmid (E591) were plated on glucose- and galactose-containing plates. Unlike mad3mkb1 and mad3mkb2 strains, wild-type cells overexpressing Mps1p underwent checkpoint arrest as shown by their inability to form colonies in the presence of galactose. Schematic indicates the positions of the different strains on the plates. (C) mad3mkb1 and mad3mkb2 cells fail to arrest in metaphase in response to benomyl. Wild-type (YJB894), mad3mkb1 (YJB895), and mad3mkb2 (YJB919) strains were arrested in G1 with α-factor (100 ng/mL) and then released synchronously into medium either lacking or containing benomyl (50 μg/mL) to induce the spindle checkpoint. (Top right panel) Pds1p is stabilized in wild-type cells in the presence of benomyl, reflecting a mitotic arrest and indicating the presence of a functional spindle checkpoint. In contrast, Pds1p levels fluctuated in mad3mkb1 and _mad3mkb_2 cells, which entered and then exited mitosis (bottom panels), similar to wild-type cells in the absence of benomyl (top left panel).
We next assessed checkpoint function following activation of the spindle assembly checkpoint via a mechanism other than spindle disruption. Overexpression of the mitotic checkpoint kinase Mps1p activates the spindle assembly checkpoint without any obvious perturbation of the mitotic spindle (Hardwick et al. 1996). Wild-type, mad3mkb1, and mad3mkb2 strains with or without an integrated GAL-MPS1 plasmid were plated onto either glucose- or galactose-containing medium (Fig. 2B). As expected, the strain containing wild-type MAD3 and the GAL-MPS1 plasmid was unable to grow on galactose due to activation of the checkpoint. However, MAD3 cells could grow in the absence of the GAL-MPS1 plasmid or in the presence of the plasmid but on glucose-containing medium, where Mps1p was not overexpressed. In contrast, both mad3mkb1 and mad3mkb2 strains grew on galactose whether or not they contained the GAL-MPS1 plasmid (Fig. 2B), indicating the absence of a functional checkpoint.
A third way to determine the effects of the Mad3p mutations on checkpoint function is to examine the stability of Pds1p under checkpoint activation conditions, which cause stabilization of Pds1p in wild-type cells. Cells expressing Mad3p, Mad3p-mkb1, or Mad3p-mkb2 were arrested in G1 with the mating pheromone α-factor and then released synchronously into medium containing the spindle disrupting agent benomyl to “trap” cells in the subsequent metaphase by activation of the spindle assembly checkpoint. Cells with an intact checkpoint should arrest in metaphase with stabilized Pds1p, whereas cells with a defective checkpoint will be unable to inhibit APCCdc20 and will continue to cycle, which will be reflected in the degradation of Pds1p. In wild-type cells, Pds1p levels oscillated in the absence of benomyl (Fig. 2C, top left), whereas benomyl-induced activation of the spindle checkpoint promoted the stabilization of Pds1p in these cells (Fig. 2C, top right). In contrast, Pds1p continued to cycle in cells expressing Mad3p-mkb1 or Mad3p-mkb2 in the presence of benomyl (Fig. 2C, bottom panels), indicating that these cells lack a functional spindle checkpoint.
KB1 is required for Cdc20p binding in vivo
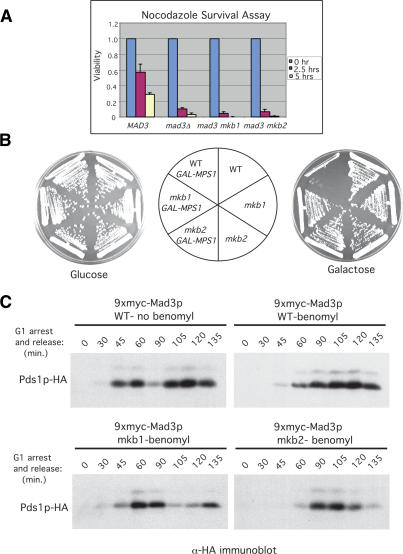
Since KEN boxes mediate APC-substrate binding to Cdc20p and Cdh1p (Burton and Solomon 2001; Pfleger et al. 2001a), it seemed plausible that a KEN box might also mediate the binding of a checkpoint protein to Cdc20p. We therefore examined if Mad3p-mkb1 and Mad3p-mkb2 could bind to Cdc20p using a coimmunoprecipitation assay. Extracts were prepared from cells arrested in G1, when Cdc20p levels are low, and in mitosis and checkpoint-activated cells, when Cdc20p levels are high (Prinz et al. 1998; Pan and Chen 2004). The strains for this experiment contained an integrated copy of GAL-PDS1mdb, which arrests cells in metaphase in galactose-containing medium due to the inability of the APCCdc20 to bind to and promote the degradation of Pds1p-mdb, thereby preventing separase activation (Cohen-Fix et al. 1996). Cells were arrested either in G1 using the mating pheromone α-factor, in M phase by expression of Pds1p-mdb, or in M phase with checkpoint activation by Pds1p-mdb expression and nocodazole addition.
Mad3p was immunoprecipitated from cell extracts, and the amounts of coimmunoprecipitating Cdc20p were examined by immunoblotting (Fig. 3A). Wild-type Mad3p associated with a low level of Cdc20p in metaphase and with a higher level of Cdc20p following checkpoint activation (Fig. 3A, lanes 2,3). The same pattern of association was observed when Cdc20p was immunoprecipitated followed by immunoblotting for Mad3p (Fig. 3B, lanes 1–3). No detectable association between Cdc20p and Mad3p was observed in G1 cells (Fig. 3A,B, lane 1), likely due to the low levels of Cdc20p in these cells (Fig. 3B, lane 1, bottom panel). In contrast, Mad3p-mkb1 was unable to associate with Cdc20p (Fig. 3A,B, lanes 5,6). Similar results were obtained when the checkpoint was activated by overexpression of Mps1p (Supplementary Fig. 1). Despite its inability to bind Cdc20p, Mad3p-mkb1 retained wild-type binding to Bub3p (Supplementary Fig. 2). Mad3p-mkb2 displayed wild-type association with Cdc20p in mitosis but a smaller increase in binding upon checkpoint activation (Fig. 3C, lanes 5,6). Mutation of the KEN boxes had no effect on Mad3p levels (Figs. 3A,C [bottom panels], 1B). As previously observed, the association between wild-type Mad3p and Cdc20p was dependent on Mad2p (Fig. 3D, cf. lanes 3–6; Hwang et al. 1998; Hardwick et al. 2000).
Figure 3.
Mad3p-mkb1 is defective in its association with Cdc20p. (A) Mad3p was immunoprecipitated from extracts of strains expressing wild-type myc-Mad3p (YJB800) or myc-Mad3p-mkb1 (YJB801) that were arrested in G1, M, or M plus nocodazole (see Materials and Methods). Mad3p and coimmunoprecipitating Cdc20p were detected by immunoblotting. (B) Cdc20p was immunoprecipitated from extracts of strains (YJB800 and YJB801) arrested as described in A. Cdc20p and coimmunoprecipitated Mad3p were detected by immunoblotting as in A. (C) As in A using strains expressing myc-Mad3p wild type (YJB800) or mkb2 (YJB924). (D) Efficient Mad3p association with Cdc20p requires Mad2p. myc-Mad3p was immunoprecipitated using anti-myc antibodies from extracts of M-phase or M-phase plus benomyl arrested cells in wild-type (YJB800) or _mad2_Δ (YJB905) strains. Mad3p and coimmunoprecipitating Cdc20p were detected by immunoblotting. No Cdc20p was detected from a yeast strain lacking myc-Mad3p (YJB793).
During these experiments we noticed that Cdc20p was partially stabilized in the mad3mkb1 and _mad3_Δ strains relative to the MAD3 strain, in which Cdc20 is highly unstable (Pan and Chen 2004). In contrast, mad3mkb2, _mad1_Δ, _mad2_Δ, and _bub1_Δ strains did not stabilize Cdc20 in M-phase-arrested cells (Supplementary Fig. 3). How Cdc20p degradation affects spindle checkpoint function is presently unclear.
Mad3p and APC substrates share binding sites on Cdc20p
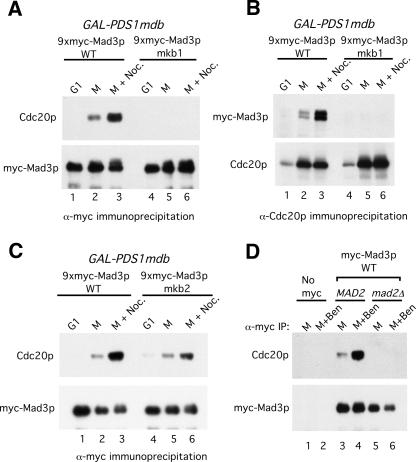
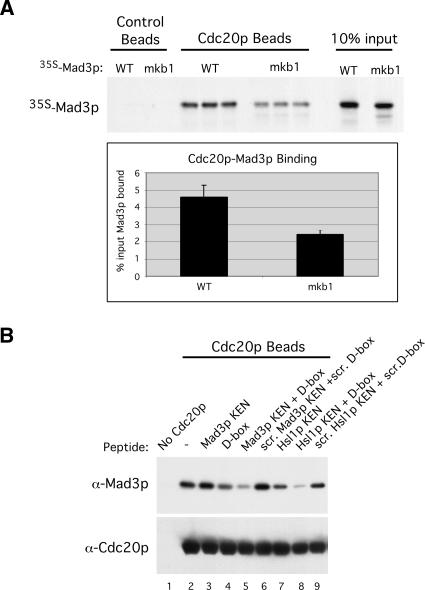
We used in vitro binding assays with recombinant Cdc20p to further explore the binding of Mad3p to Cdc20p. Because of its stronger effect on Cdc20p binding, we analyzed Mad3p-mkb1 more extensively than Mad3p-mkb2 in the following experiments. GST-Cdc20p produced from baculovirus-infected cells and purified on glutathione beads (Burton and Solomon 2001) was incubated with 35S-labeled Mad3p or Mad3p-mkb1 (Fig. 4A). Quantitation from the three independent binding events showed that mutation of KB1 reduced binding to Cdc20p by 48% (Fig. 4A, bottom panel). A somewhat greater effect of the KB1 mutation was observed when using recombinant Mad3p and Mad3p-mkb1 produced and purified from baculovirus-infected cells (Supplementary Fig. 4). Mad3p did not bind to control beads lacking Cdc20p (Fig. 4A; Supplementary Fig. 4).
Figure 4.
KEN-box- and D-box-dependent binding of Mad3p to Cdc20p in vitro. (A) Wild-type Mad3p and Mad3p-mkb1 were translated in vitro in the presence of 35S-methionine and then mixed with control or GST-Cdc20p glutathione beads. The beads were washed, and bound 35S-Mad3p was detected by autoradiography. Ten percent of the input Mad3p is shown in the lanes at the right. The percentages of input Mad3p and Mad3p-mkb1 bound to GST-Cdc20p were quantitated by PhosphorImage analysis using the three independent binding events for each sample. (B) KEN-box and D-box peptides dramatically reduce Mad3p binding to Cdc20p. Wild-type Mad3p was purified from baculovirus-infected cells and incubated with Cdc20p beads that had been preincubated with 500 μM KB1 peptide (“Mad3p KEN”), Hsl1p D-box peptide (“D-box”), Hsl1p KEN-box peptide (“Hsl1p KEN”), combinations of these peptides, or scrambled versions of these peptides (“scr.”), as indicated. The beads were washed, and bound Mad3p and Cdc20p were detected by immunoblotting.
The residual binding of Mad3p-mkb1 to Cdc20p suggested the presence of additional binding motifs within Mad3p. To examine this possibility, we used KEN-box and D-box peptides in a competition assay to determine whether additional degradation motifs might facilitate the binding of Mad3p to Cdc20p. GST-Cdc20p beads were preincubated with peptides corresponding to Mad3p KB1, the Hsl1p KEN box, or the Hsl1p D box (Burton et al. 2005) prior to the addition of purified wild-type Mad3p. Scrambled versions of these peptides were used as negative controls. Bound Mad3p was detected by immunoblotting (Fig. 4B). The Mad3p KB1 peptide on its own had no detectable effect on Mad3p binding to Cdc20p (Fig. 4B, cf. lanes 2 and 3). In contrast, the Hsl1p KEN-box peptide reduced Mad3p binding to Cdc20p (Fig. 4B, lane 7). Surprisingly, the Hsl1p D-box peptide also reduced Mad3p binding to Cdc20p (Fig. 4B, lane 4) and greatly reduced binding when combined with the KEN-box peptides from either Mad3p or Hsl1p (Fig. 4B, lanes 5,8). No inhibition of Mad3p binding was observed when scrambled versions of these peptides were used (Fig. 4B, lanes 6,9). These results suggest that KEN-box and D-box motifs work cooperatively in binding to Cdc20p and confirm the in vivo and in vitro findings using Mad3p-mkb1 that Cdc20p can directly recognize a KEN box in Mad3p. These findings also suggest that Cdc20p may recognize a D box in Mad3p. We also found that the Mad3p KB1 peptide could inhibit the binding of Clb2p and Hsl1p to Cdh1p. As with Cdc20p, the Mad3p KB1 peptide blocked substrate binding to Cdh1p less effectively than the Hsl1p KEN-box peptide, either alone or in combination with the D-box peptide (Supplementary Fig. 5). In all of these experiments, the level of Mad3p binding to Cdc20p was modest, presumably because Mad2p (and possibly other factors) is required for high-affinity binding of Mad3p to Cdc20p (see below and Fig. 3D).
A Mad3p D box is important for Mad3p function and Cdc20p binding
The ability of the Hsl1p D-box peptide to inhibit Mad3p binding to Cdc20p led us to examine whether Mad3p contains a D box. We individually mutated each potential D-box motif within Mad3p, changing “RXXL” to “AXXA.” While mutation of 241RLEL244 and 301RISL304 had no effect on Mad3p function as assessed by the nocodazole survival assay (data not shown), mutation of 429RKAL432 (“Mad3p-mdb”) partially compromised checkpoint function (Fig. 5A). Mad3p-mdb bound to Cdc20p in mitotic cells but showed less enhancement of binding relative to wild-type Mad3p upon checkpoint activation (Fig. 5B, cf. lanes 5,6 and 3,4), similar to what was observed with Mad3p-mkb2 (see Fig. 3C). Also similar to Mad3p-mkb2 cells, Cdc20p was unstable in Mad3p-mdb cells (data not shown).
Figure 5.
Mad3p has a D-box motif important for the spindle checkpoint and Cdc20p binding. (A) Nocodazole survival assay of MAD3 (YJB697), _mad3_Δ (YJB522), and mad3mdb (YJB926) strains. Viability was calculated as in Figure 2A. (B) Mad3p-mdb binds Cdc20p with reduced efficiency. Mad3p (YJB800) and Mad3p-mdb (YJB928) were immunoprecipitated from extracts of cells arrested in M or M plus benomyl (see Materials and Methods). Mad3p and coimmunoprecipitating Cdc20p were detected by immunoblotting. No Mad3p or Cdc20p was immunoprecipitated from a “no myc” control strain extract (YJB793). (C) The Mad3p D box and KEN boxes function together to bind Cdc20p. 35S-methionine labeled wild type (WT), Mad3p-mkb1mkb2, Mad3p-mdb, and Mad3p-mkb1mkb2mdb were produced by in vitro translation and incubated in triplicate with GST-Cdc20p glutathione beads. The beads were washed, and bound proteins were detected by autoradiography. (Top panels) Ten percent of the input Mad3p is shown in the lanes at right. (Bottom panel) The percentage of input Mad3p bound to GST-Cdc20p for each sample was quantitated by PhosphorImage analysis for the three independent binding events.
We assessed the roles of the two KEN boxes and the D box in Mad3p binding to Cdc20p in vitro. Mutations of both KEN boxes reduced Cdc20p binding by 67% compared with wild-type Mad3p, which was somewhat greater than the effect of mutating KB1 alone (48%) (cf. Figs. 4A and 5C). The D-box mutation on its own reduced Mad3p binding to Cdc20p by 76%, and the combination of all three mutations reduced binding by 92% (Fig. 5C). Thus, as with APC substrates, Mad3p binds to Cdc20p via both D-box and (two) KEN-box motifs.
Mad3p and Hsl1p compete for binding to Cdc20p
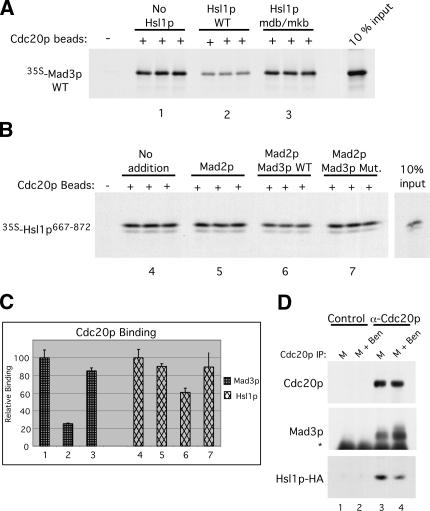
Since Cdc20p recognizes Mad3p via its KEN boxes and a D box, we were interested in whether Mad3p and an APC substrate might compete with one another for binding to Cdc20p. We used a competition assay in which GST-Cdc20p beads were preincubated with a large excess of the APC-substrate Hsl1p (Burton and Solomon 2000, 2001) prior to the addition of 35S-Mad3p. Control competitions included no Hsl1p or the addition of Hsl1p containing a mutated D box and a KEN box (Hsl1p mdb/mkb) that cannot bind Cdc20p (Burton and Solomon 2001). Preincubation with wild-type Hsl1p reduced the binding of Mad3p to Cdc20p by 70% (Fig. 6A,C, bars 1,2), suggesting that Mad3p and Hsl1p share binding sites on Cdc20p. This inhibition of binding was specific as Hsl1p mdb/mkb had little effect on the Mad3p–Cdc20p interaction (Fig. 6A,C, cf. bars 1 and 3).
Figure 6.
Mad3p and Hsl1p compete for Cdc20p binding. (A) Hsl1p wild type but not mdb/mkb can compete with Mad3p for binding to Cdc20p. GST-Cdc20p beads were preincubated in the absence or the presence of wild-type or mdb/mkb MBP-Hsl1p followed by incubation with wild-type 35S-Mad3p as in Figure 4A. Bound 35S-Mad3p was detected by autoradiography. (B) Mad3p WT but not Mad3p-Mut. can compete with Hsl1p667–872 for binding to Cdc20p. Cdc20p beads were preincubated with Mad2p alone or with Mad2p and wild-type or triply mutated Mad3p followed by incubation with 35S-Hsl1p667–872. Bound 35S-Hsl1p667–872 was detected by autoradiography. (C) The relative levels of wild-type 35S-Mad3p (left bars 1–3) or 35S-Hsl1p667–872 (right bars 4–7) bound to Cdc20p were quantitated by PhosphorImage analysis of the samples shown in A and B. The bindings of 35S-Mad3p and 35S-Hsl1p667–872 to Cdc20p in the absence of competitor proteins were given relative values of 100. Error bars represent the standard deviations from the three independent binding events shown for each sample. (D) Cell extracts were prepared from a strain (YJB864) constitutively expressing Hsl1p667–872-HA that was arrested in M phase or M phase plus benomyl to induce the spindle checkpoint. Cdc20p was immunoprecipitated, and coimmunoprecipitating Mad3p and Hsl1p-HA levels were assessed with anti-Mad3p and anti-HA antibodies, respectively. (Lanes 3,4) Checkpoint activation increased the amount of Mad3p associated with Cdc20p and decreased the amount of Hsl1p-HA associated with Cdc20p. (Lanes 1,2) None of these proteins was detected using a control preimmune serum. Asterisk denotes IgG heavy chain.
We next performed the converse experiment and examined whether Mad3p could inhibit the binding of Hsl1p to Cdc20p. This experiment was performed in the presence of Mad2p since Mad2p is required for the efficient binding of Mad3p to Cdc20p in vivo (Fig. 3D; Hwang et al. 1998; Hardwick et al. 2000) and since Mad2p stimulates the binding of Mad3p to Cdc20p in vitro (see below). Cdc20p beads were preincubated with Mad2p alone or with Mad2p and either a wild-type Mad3p–Bub3p complex (“Mad3p WT”) or a Mad3p-mkb1mkb2mdb–Bub3p complex (“Mad3p Mut.”) that were produced and purified from Escherichia coli (see Materials and Methods). 35S-Hsl1p667–872 was added, and the bound Hsl1p was visualized by autoradiography and quantitated by PhosphorImage analysis (Fig. 6B,C). The combination of Mad2p and the Mad3 WT complex reduced Hsl1p binding by ∼40% relative to Hsl1p binding in the absence of Mad proteins. Mad2p alone or in combination with the Mad3p Mut. complex had little effect on the Cdc20p–Hsl1p interaction. Thus, Mad3p and Hsl1p compete with each other for binding to Cdc20p in vitro. Hsl1p appeared to be the more effective competitor in these experiments. This observation may reflect the better quality and the greater quantity of recombinant Hsl1p that was added compared with Mad3p, which has been more difficult to produce in large quantities. Also, Hsl1p is an extremely tight-binding APC substrate originally identified as an APC substrate in a two-hybrid screen using Cdc20p as the bait (Burton and Solomon 2000, 2001). We have not performed similar competition experiments using Pds1p, the canonical Cdc20p substrate, since we have been unable to detect Pds1p binding to Cdc20p in vitro (data not shown).
These competition experiments in vitro predict that Mad3p binding to Cdc20p will reduce the amount of an APC substrate bound to Cdc20p in vivo. To test this prediction, Cdc20p was immunoprecipitated from extracts of cells arrested in M phase or M phase with benomyl to induce the mitotic checkpoint. Coimmunoprecipitation of Mad3p and of a constitutively expressed fragment of Hsl1p-HA (Hsl1p667–872-HA) was then examined (Fig. 6D). Spindle checkpoint activation induced an increase in the amount of Mad3p associated with Cdc20p but a significant decrease in the amount of bound Hsl1p-HA (Fig. 6D, middle and bottom panels, lanes 3,4). These findings are in agreement with the competition experiments and indicate that checkpoint activation inhibits substrate association with Cdc20p. The residual association of Hsl1p-HA with Cdc20p may reflect the fact that Hsl1p was significantly overexpressed whereas Mad3p was expressed at normal levels. It may have little practical significance, particularly if the MCC (which contains Mad3p) sequesters APCCdc20 in an inactive complex (see Discussion) such that the non-APC-bound Cdc20p–Hsl1p complexes would not be able to associate with the APC.
Mad2p stimulates Cdc20p–Mad3p interaction in a KB1-dependent manner
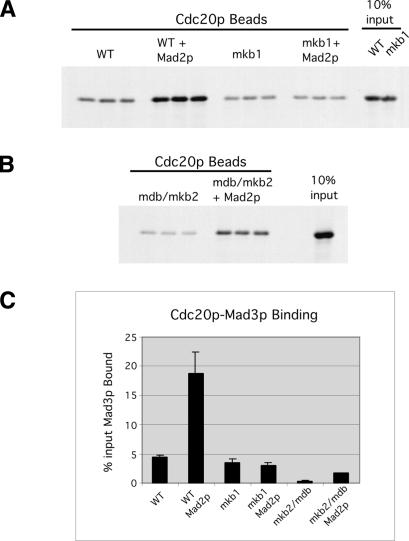
Since Mad2p is required for Mad3p association with Cdc20p in vivo (Fig. 3D; Hwang et al. 1998; Hardwick et al. 2000), we investigated whether Mad2p might also affect Mad3p binding to Cdc20p in vitro. The binding assays were carried out as before except that the Cdc20p beads were preincubated with Mad2p prior to the addition of 35S-Mad3p. Mad2p enhanced Mad3p binding to Cdc20p fourfold relative to Mad3p–Cdc20p binding in the absence of Mad2p (Fig. 7A,C). Interestingly, this Mad2p-stimulated binding depended on KB1 within Mad3p as Mad2p had no effect on the binding of 35S-Mad3p-mkb1 to Cdc20p (Fig. 7A,C). This KB1-dependent effect of Mad2p explains why mutation of KB1 had a dramatic effect on Mad3p binding to Cdc20p in vivo (Fig. 3A) but a muted effect in vitro in the absence of Mad2p (Fig. 4A). In contrast to KB1, KB2 and the D box were not required for the Mad2-stimulated association between Mad3p and Cdc20p. Although the overall level of 35S-Mad3p-mkb2mdb binding to Cdc20p was low, Mad2p still stimulated binding approximately fourfold relative to the binding observed in the absence of Mad2p (Fig. 7B,C), a similar stimulation to what was observed for wild-type Mad3p with Mad2p. These findings demonstrate that KB1 functions specifically in the high-affinity Mad2p-stimulated Mad3p–Cdc20p interaction and explains why Mad3p-mkb1 does not interact with Cdc20p in vivo. To our knowledge, this is the first demonstration that Mad2p has a direct effect on the binding of Mad3p to Cdc20p.
Figure 7.
Mad2p stimulates Mad3p binding to Cdc20p in vitro in a KB1-dependent manner. GST-Cdc20p beads were preincubated with or without Mad2p followed by the addition of either wild-type or mkb1 35S-labeled Mad3p (A) or 35S-labeled Mad3p-mkb2mdb (B). (C) The amount of bound Mad3p was detected by PhosphorImage analysis. Error bars represent the standard deviations for the three independent binding events shown for each sample.
Discussion
We have explored the roles of three degradation motifs in Mad3/BubR1 proteins, two invariant KEN boxes and one Destruction box. Although such motifs are usually associated with APC substrates, Mad3p is highly stable. Nevertheless, the presence of each motif is important for Mad3p function in the spindle assembly checkpoint, and all three motifs are involved in docking Mad3p to Cdc20p in vitro. KB1 plays a particularly important role in Mad3p function, as its presence is required for Mad3p binding to Cdc20p in cell extracts and for the Mad2p-enhanced binding of Mad3p to Cdc20p in vitro. Peptide competition experiments suggest that all three Mad3p motifs are recognized by the same surfaces on Cdc20p that Cdc20p uses to bind APC substrates, indicating that Mad3p binds Cdc20p as a pseudosubstrate. This mode of interaction was reflected in competition between Mad3p and Hsl1p for binding to Cdc20p both in vitro and in vivo.
Although both KB2 and the D box are required for checkpoint function and play roles in Mad3p binding to Cdc20p in vitro, neither is absolutely required for Mad3p binding to Cdc20p in vivo, and their precise roles remain unclear. Interestingly, mutations of these motifs have similar effects on Mad3p binding to Cdc20p, with wild-type levels of binding during mitosis but attenuated binding upon checkpoint activation. Likewise, mutation of KB1 and deletion of MAD2 have similar effects on Mad3p binding to Cdc20p in vivo, in both cases reducing binding to undetectable levels. In vitro, Mad2p enhanced the binding of Mad3p to Cdc20p, and this enhancement required KB1 but not KB2 or the D box. It is conceivable that KB2 and the D box function together, and that KB1 plays a distinct role in concert with Mad2p. Being at the N terminus of Mad3p, KB1 may be more exposed, and the initial Mad2p-dependent binding to Cdc20p in cells may act through KB1, perhaps exposing KB2 and the D box for subsequent interactions.
It is not clear why Mad3p contains so many Cdc20p-binding motifs, particularly two KEN boxes. It is possible that these motifs function redundantly, just as multiple functional D boxes exist in some APC substrates. More complex and speculative scenarios can also be envisioned. For instance, one KEN box might interact directly with Mad2p or the APC, while the other two motifs interact with Cdc20p. (Along these lines, we cannot fully exclude the possibility that Mad2p promotes Mad3p binding to Cdc20p via a weak direct interaction with KB1, although it is also clear that Cdc20p can recognize KB1 in vitro.) It is also possible that Mad3p might use the three binding motifs to bind two molecules of Cdc20p at the same time, one serving the usual function of directing the complex to the APC and the second acting as a substrate, thereby accounting for the known requirement for Mad3p in promoting rapid Cdc20p degradation (Pan and Chen 2004).
Our working model is that Mad3/BubR1 proteins are the primary biochemical effectors of the spindle assembly checkpoint. The checkpoint begins with the catalytic conformational activation of Mad2 at improperly attached kinetochores (Luo et al. 2002; Sironi et al. 2002; De Antoni et al. 2005; Nezi et al. 2006). Activated Mad2 then binds to Cdc20 and recruits Mad3/BubR1. Mad2p serves two important roles in this process. First, it couples information from kinetochores to the effector, Mad3/BubR1. Second, the specificity of Mad2p for Cdc20p over Cdh1p may also give specificity to Mad3p, which might otherwise be expected to be able to bind to and inhibit Cdh1p. Mad3p binding to Cdc20p appears to have two consequences, blocking of substrate binding to Cdc20p and, at least in budding yeast, the promotion of Cdc20p degradation. Both actions will tend to inhibit APC-mediated degradation of substrates such as Pds1p/securin. The high conservation of the KEN boxes in Mad3/BubR1 proteins suggests that this mechanism of spindle checkpoint function is conserved in eukaryotes from yeast to man. We have confirmed a previous report (Pan and Chen 2004) indicating that Mad3p helps to promote rapid Cdc20p turnover and found that Cdc20p binding via KB1 is required for this effect (Supplementary Fig. 3). The relative roles of substrate competition and Cdc20p degradation in spindle checkpoint function are not yet clear. Although Cdc20p degradation may have an important role in the checkpoint, it is not absolutely required since mad3mkb2 and mad3mdb cells are defective in the checkpoint despite their rapid degradation of Cdc20p. Recent mathematical modeling studies predict that sequestration of Cdc20p may be more important for checkpoint function than Cdc20p turnover (Doncic et al. 2006).
Pseudosubstrate inhibition may provide a general mechanism for inhibiting the APC. Emi1 is a metazoan cell cycle regulator that inhibits the APC during S phase to allow the accumulation of cyclins (Reimann et al. 2001a, b). It was recently found that Emi1 is a pseudosubstrate inhibitor of APCCdh1 (Miller et al. 2006). Emi1 possesses both a D box and a zinc-binding region (ZBR). The D box is necessary for APC-binding, whereas the ZBR is required for APC inhibition; mutation of the ZBR converts Emi1 from an inhibitor into an APCCdh1 substrate (Miller et al. 2006). Our findings indicate that both APCCdh1 and APCCdc20 can be inhibited by pseudosubstrates, Emi1 and Mad3p, respectively. It is presently unclear why Mad3p does not function as an APC substrate. Unlike Emi1, Mad3p does not contain a ZBR. Perhaps Mad3p, Mad2p, or even Bub3p possesses an additional domain that, like the ZBR, serves to inhibit the enzymatic activity of the APC. Alternatively, Mad3p may simply be a very poor APC substrate, perhaps having been selected through evolution to lack lysines in readily ubiquitinatable positions. However, in cell extracts, we observe a concomitant loss of Hsl1p binding and an increase of Mad3p binding to Cdc20p upon activation of the mitotic checkpoint consistent with Mad3p functioning as a pseudosubstrate.
Toward a unified model of spindle checkpoint biochemistry
Several seemingly distinct models have been proposed for the inhibition of APCCdc20 by the spindle assembly checkpoint. The following review of these models suggests that most are consistent with our proposal that Mad3/BubR1 proteins are the primary effectors of the checkpoint.
BubR1 itself has been reported to inhibit APCCdc20 activity in vitro (Tang et al. 2001; Fang 2002), which would make sense if BubR1 acts as a competitive inhibitor. Although BubR1 has also been reported to block the binding of Cdc20p to the APC in vitro (Tang et al. 2001), it is clear that BubR1, Cdc20p, and the APC can associate simultaneously in cell extracts (Sudakin et al. 2001). Similarly, we have found that wild-type Mad3p (but not Mad3p-mkb1) coimmunoprecipitates both Cdc20p and the APC in M-phase-arrested or checkpoint-induced cell extracts (Supplementary Fig. 6), confirming that Cdc20p, Mad3p, and the APC can form a ternary complex in yeast cell extracts.
Mad2 can inhibit APCCdc20 in vitro in the absence of any other checkpoint protein (Fang et al. 1998; Tang et al. 2001; Fang 2002). However, very high (superphysiological) concentrations of Mad2 are required for this inhibition. In addition, the combination of Mad2 and BubR1 yields a far more potent inhibitor of APCCdc20 than Mad2 alone (Tang et al. 2001; Fang 2002). Moreover, overexpression of mad2+ in fission yeast can induce the spindle checkpoint, but only in strains expressing Mad3p (Millband and Hardwick 2002). These observations are consistent with a primary role for Mad3/BubR1 and suggest that Mad2 functions in concert with Mad3/BubR1 to inhibit APCCdc20 in vivo.
An interesting observation concerning the biochemistry of Mad2 function may help explain both how it inhibits APCCdc20 in vitro and how it recruits Mad3/BubR1 proteins to Cdc20. It has been reported that Mad2 slows the dissociation of substrates from Cdc20 in vitro (Pfleger et al. 2001b). The tighter binding of substrates reflected in this slow substrate release could inhibit ubiquitination by preventing a structural rearrangement necessary for a step in the ubiquitination cycle. Alternatively, slow substrate release could be paralleled by a slowed release of ubiquitinated product and a consequent reduction in the rate of bulk ubiquitination. In addition to enhancing substrate binding to Cdc20, Mad2p enhances the binding of Mad3p to Cdc20p via KB1 of Mad3p. Although the key function of Mad2p may be to enhance Mad3p binding to Cdc20p, a biochemical consequence of this action might be to enhance substrate binding to Cdc20p, resulting in the ability of Mad2 to inhibit APCCdc20 when examined on its own in vitro.
The MCC consists of Mad2, BubR1, Bub3, and Cdc20 and is a very potent inhibitor of APCCdc20 (Sudakin et al. 2001). Our results suggest that the MCC contains an already inactive form of Cdc20. It is possible that the MCC’s ability to inhibit APCCdc20 reflects a competition for APC binding between the inactive Cdc20 in the complex and active Cdc20 in the APC assay. Our previous studies indicated that engagement of a D box by Cdh1p directs the Cdh1p–substrate complex to the APC (Burton et al. 2005). If Cdc20p behaves similarly, then Cdc20p bound to the Mad3p D box in a Mad2p–Mad3p–Cdc20p complex might bind preferentially to the APC. Formation of such an MCC–APC complex would simultaneously inhibit the Cdc20p involved in such a com-plex, and, by sequestering the APC, also inhibit any free Cdc20p that might still be able to bind substrates.
Finally, it has been reported that Bub1 phosphorylation of Cdc20 is required for the spindle checkpoint (Tang et al. 2004). It is currently unclear how this finding might be incorporated into our view of Mad3/BubR1 function. It is possible that Bub1 phosphorylation of Cdc20 may help recruit Mad2 or Mad3/BubR1 to Cdc20 or influence the association of the MCC with the APC. Alternatively, this phosphorylation may represent a parallel aspect of checkpoint inactivation of Cdc20.
Although many facets of Mad3/BubR1 function in the spindle checkpoint remain to be elucidated, our results suggest that the essential biochemical function of these proteins is simple: to bind Cdc20 using the same motifs used by APC substrates to bind Cdc20. This pseudosubstrate binding serves to block substrate binding. This observation helps unite previous biochemical studies of the spindle checkpoint’s effects on Cdc20 and provides both a scaffold and specific predictions for further studies.
Materials and methods
Plasmid and yeast strain constructions
The GAL-myc-MPS1 (E591) (Hardwick et al. 1996) and GAL-PDS1mdb (pOC70) (Cohen-Fix et al. 1996) plasmids were kind gifts from Mark Winey (University of Colorado, Boulder, CO) and Orna Cohen-Fix (National Institutes of Health, Bethesda, MD), respectively. _CDC16-HA_-YIplac211 and pVT93 were described previously (Burton et al. 2005). The _mad1_Δ, _mad2_Δ, _mad3_Δ, and _bub1_Δ disruption cassettes were constructed in pRS303 (Sikorski and Hieter 1989). For each gene disruption, the 5′-untranslated region and 3′ region containing the C-terminal coding sequence were subcloned into pRS303 and then cut between these two regions and transformed into yeast to replace the given gene with HIS3 by homologous recombination (Rothstein 1991). The _mad1_Δ, _mad3_Δ, and _bub1_Δ pRS303 disruption cassettes were each linearized with EcoRI, and the _mad2_Δ pRS303 disruption cassette was linearized with EcoRV.
To make myc-tagged versions of Mad3p expressed from the MAD3 promoter, we first cloned the 500 base pairs (bp) upstream of the MAD3 start codon into a c-myc-epitope-tagged YIplac204 vector (Gietz and Sugino 1988) containing either three or nine copies of the myc-epitope tag. The entire coding sequence of MAD3 was placed downstream and in frame of the myc epitope tags using SphI and SalI to generate _Mad3p-3xmyc-MAD3_-YIplac204 and _Mad3p-9xmyc-MAD3_-YIplac204 plasmids. These constructs were cut with AvaI within TRP1 for integration into the TRP1 locus. The myc-tagged forms of wild-type Mad3p were functional in the nocodazole survival assay (Fig. 5A; data not shown).
The mad3mkb1, mad3mkb2, and mad3mdb constructs were generated by QuikChange mutagenesis (Stratagene) using _Mad3p-9xmyc-MAD3_-YIplac204 as a template. The KEN motifs beginning at amino acids 30 and 296 were changed to AAA, and the RKAL motif beginning at amino acid 429 was changed to AKAA.
_BUB3-HA_-YIplac211 was made by ligation of the BUB3 coding sequence into YIplac211 (Gietz and Sugino 1988) containing three copies of the HA-epitope tag at the C terminus of Bub3p. This plasmid was linearized within BUB3 with EcoNI for tagging the chromosomal copy of BUB3 with the HA-epitope tag. Although Bub3p-HA can bind to Mad3p, strains expressing it as the sole form of Bub3p lack a functional spindle checkpoint in the nocodazole survival assay (data not shown).
_PDS1-HA_-YIplac128 was constructed by ligation of the C-terminal half of PDS1 into YIplac211-HA such that PDS1 was in frame with the HA-encoding sequence. The construct was linearized within PDS1 with AvrII for tagging the chromosomal copy of PDS1.
For in vitro transcription/translation of MAD3, the full-length wild-type MAD3, mad3mkb1, mad3mkb2, and mad3mdb coding sequences were cloned into pKB171, using BamHI and XhoI. The double and triple Mad3p mutants, _mad3mkb1mkb2_-pKB171 and _mad3mkb1mkb2mdb_-pKB171, respectively, were generated by QuikChange mutagenesis using mad3mkb1 and mad3mkb1mkb2 as templates, respectively. For in vitro transcription/translation of HSL1, sequences encoding amino acids 667–872 of HSL1 were inserted into pKB171 to generate _HSL1_667–872-pKB171. A wild-type His6-MAD3 baculovirus was a kind gift from Hongtao Yu (Southwestern Medical Center, Dallas, TX). The His6-mad3mkb1 baculovirus was constructed by isolating mad3mkb1 from _mad3mkb1_-pKB171 using BamHI and XhoI and ligating into the pFastBac HTa vector (Invitrogen) cut with BamHI and SalI. The GST-Cdc20p baculovirus was described previously (Burton and Solomon 2001). For Mad3p antibody generation, DNA sequences encoding the last 108 amino acids of Mad3p were ligated to pET28c (Invitrogen) such that the 6xHis tag was in frame at the N terminus of the fusion protein to generate His6-_MAD3C108_-pET28c. For Cdc20p antibody production, DNA encoding the N-terminal 188 amino acids of Cdc20p was placed in frame with the 6xHis tag of pET28c to generate His6-_CDC20N188_-pET28c. The His6-_MAD2_-pET28c contained full-length MAD2 in pET28c. His6-_MAD3-BUB3_-HA-pETDuet-1 (“Mad3p WT”) and His6-_mad3mkb1mkb2mdb-BUB3_-HA-pETDuet-1 (“Mad3p Mut.”) constructs were made by insertion of _BUB3_-HA from YIplac211 into the second multicloning site of the pET-Duet-1 vector (Novagen) followed by insertion of either MAD3 or mad3mkb1mkb2mdb into the first multicloning site of pETDuet-1.
Oligonucleotide sequences used for the QuikChange mutagenesis and for the different plasmid constructions described above are available upon request. All genes amplified by PCR or QuikChange mutagenesis were sequenced in their entirety to verify that only desired sequences were present. Integrations were verified by PCR and, in the case of epitope tagging, by immunoblot analysis.
Yeast culture and cell cycle arrests
Yeast media and protocols were derived from Ausubel et al. (1995). All yeast strains were derivatives of W303-1A (MATa ade2-1 his3-11,15 leu2-3,112 can1-100 ura3-1 trp1-1 ssd1-d) (Rothstein 1991) with the specified alterations (Table 1). For G1, M-phase, and M-phase plus benomyl (or nocodazole) cell cycle arrests, cells were grown to mid-exponential phase in YP medium containing 2% raffinose. For the G1 arrest, the culture was then supplemented with α-factor (100 ng/mL) (Sigma-Aldrich) for 3 h at 30°C until all cells had the “shmoo” morphology. For the M-phase and M-phase plus benomyl or nocodazole arrests, 2% galactose was added to the raffinose-containing culture for 1 h to induce the expression of GAL-PDS1mdb. Cells were then pelleted, resuspended in YP medium with 2% dextrose (M phase), or YP medium with 2% dextrose plus benomyl (50 μg/mL; Dupont) or nocodazole (15 μg/mL; Sigma-Aldrich), and incubated for 2 h at 30°C to allow cells to progress into mitosis and to induce the mitotic checkpoint. For S-phase arrests, cells were grown to mid-exponential phase in YP medium with 2% dextrose. Hydroxyurea (Sigma-Aldrich) was added to a final concentration of 100 mM, and cells were incubated for 2 h at 30°C. For Mad3p and Cdc20p half-life studies, cells were arrested as described above, and cell samples were taken for the zero time points. Cycloheximide (MP Biomedicals, Inc.) was added to a final concentration of 0.5 mg/mL to inhibit protein synthesis. For the G1 arrest and release experiments, cells were grown in YP medium with 2% dextrose to an OD600 = 0.2. α-Factor was added (100 ng/mL) for 2 h at 30°C. A cell sample was taken for the zero time point, and the remaining cells were pelleted, washed, and resuspended in an equal volume of YP medium supplemented with 2% dextrose with or without benomyl (50 μg/mL) and incubated at 30°C. All cell samples were pelleted, washed, and frozen in liquid nitrogen prior to extract preparation.
Table 1.
Yeast strains used in this study
Nocodazole survival and Mps1p overexpression assays
Cells were grown to OD600 = 0.2 or less in 2 mL YP medium plus 2% dextrose. Cells were counted with a hemocytometer, and ∼200 cells per plate were spread onto two YP dextrose plates for the zero time point. Nocodazole (15 μg/mL) was added to the liquid culture, and cells were incubated for an additional 2.5 and 5 h at 30°C prior to plating as described above. The viability for each strain was determined by dividing the number of colonies formed after treatment with nocodazole for 2.5 and 5 h by the number formed at the 0-h time point. The results in Figures 2A and 5A represent the data obtained from three independent experiments for each strain. To analyze checkpoint competency of yeast strains by Mps1p overexpression, the indicated strains were struck out onto YP dextrose- and YP galactose-containing plates and incubated for 2 and 3 d, respectively, at 30°C.
Protein extract preparation and coimmunoprecipitation analysis
Protein samples for immunoblot analysis of total yeast extracts were prepared in 1× sample buffer as described previously (Burton and Solomon 2001). Approximately 70 μg of protein were loaded per lane for SDS-PAGE, proteins were transferred onto Immobilon-P membranes (Millipore), and the blots were probed for either Cdc20p or Mad3p using affinity-purified anti-Cdc20p or anti-Mad3p polyclonal antibodies, respectively (see below). Where indicated, myc-Mad3p was also detected with 9E10 monoclonal anti-c-myc antibodies (Covance Research Products) or 9E10-HRP antibodies (Santa Cruz Biotechnology). Pds1p-HA, Bub3p-HA, Hsl1p667–872-HA, and Cdc16p-HA were detected with 12CA5 monoclonal anti-HA antibodies (Covance Research Products). For coimmunoprecipitation analysis, yeast extracts were prepared, and anti-myc immunoprecipitations were performed as previously described (Burton et al. 2005). Cdc20p, Mad3p, and Bub3p-HA immunoprecipitations were done similarly, except that 1.0 μL of the appropriate antibody was used rather than 0.25 μL.
Recombinant proteins and in vitro binding assays
His6-Cdc20pN188 and His6-Mad3pC108 (for anti-Cdc20p and anti-Mad3p antibody production, respectively) were produced in BL21 (DE3) cells in 1 L of LB supplemented with kanamycin (30 μg/mL). Cells were induced with 0.3 mM IPTG (Sigma-Aldrich) for 3 h at 37°C and lysed using a French Cell Press at 900 psi. Proteins were purified from a clarified extract on Talon Resin (BD Biosciences) and eluted with 150 mM imidazole (Sigma-Aldrich) in 1× PBS. Purified proteins were injected into rabbits for polyclonal antibody production (Pocono Rabbit Farm). Antibodies were affinity-purified by incubation of crude sera with appropriate antigen that was run on 12% SDS-PAGE and transferred to Immobilon-P. Bound antibodies were eluted with low pH and neutralized (Sharp et al. 1993). Affinity-purified antibodies were used at a 1:500 dilution. His6-Mad2p, His6-Mad3p WT, and His6-Mad3p Mut. proteins were made as above except that cells were induced with 50 μM IPTG overnight at 18°C, and the medium was supplemented with ampicillin (50 μg/mL) for Mad3p expression.
Wild-type and mutant MBP-Hsl1p667–872 and wild-type MBP-Clb2p were produced in E. coli and purified on amylose resin (New England Biolabs) as described previously (Burton and Solomon 2001; Burton et al. 2005). MBP fusion proteins were eluted with 10 mM maltose. GST-Cdc20p was isolated from baculovirus-infected Sf21 cells as previously described (Burton and Solomon 2001). His6-Mad3p and His6-Mad3p-mkb1 were produced in Sf21 insect cells and purified essentially as described for His6-Cdh1p (Burton et al. 2005). 35S-methionine (NEN)-labeled Mad3p and Hsl1p667–872 proteins were expressed from the appropriate pKB171 constructs using the TNT Quick-Coupled Transcription/Translation System (Promega) according to the manufacturer’s instructions. The Hsl1p D-box and KEN-box peptides and scrambled versions of these peptides were described previously (Burton et al. 2005). The Mad3p KB1 and Mad3p scrambled KB1 peptides were synthesized at the W.M. Keck Foundation Biotechnology Resource Laboratory. The sequence of the KB1 peptide is 24E-E-I-E-T-Q-K-E-N-I-L-P-L-K-E-G-R-S-A-A-A-L45-C, and the sequence for the scrambled peptide is A-Q-E-L-I-E-P-G-K-L-A-N-R-I-L-A-E-S-E-T-E-K-C.
For Cdc20p-binding assays with radiolabeled Mad3p, 10 μL of GST-Cdc20p beads were incubated with 15 μL of in vitro translated 35S-Mad3p for 1 h at 4°C in 0.2 mL of immunoprecipitation buffer plus phosphatase inhibitors (50 mM Na+-HEPES at pH 7.6, 100 mM NaCl, 1 mM EGTA, 40 mM EDTA, 20 mM sodium pyrophosphate, 20 mM sodium fluoride, 0.5 mM DTT, 0.1% Tween 20, 10% glycerol) plus 0.1 mg/mL ovalbumin and protease inhibitors (10 mM PMSF, 10 μg each of chymostatin, leupeptin, and pepstatin [Chemicon]). Beads were washed three times with 0.4 mL of the same buffer and transferred to a fresh tube. Proteins were eluted with 2.5× SDS-PAGE sample buffer and run on an 8% SDS-PAGE. Radioactivity was detected by autoradiography and quantitated by PhosphorImage analysis. For Mad2p-stimulated binding of Mad3p to Cdc20p, the binding reaction was carried out as described above except that the Cdc20p beads were incubated with 15 μg of His6-Mad2p for 30 min at 4°C prior to the addition of 35S-Mad3p, 35S-Mad3p-mkb1, or 35S-Mad3p-mkb2mdb.
For competition assays with radiolabeled Mad3p, binding reactions were performed as above except that the GST-Cdc20p beads were preincubated with 50 μg of either MBP-Hsl1p667–872 wild type or mdb/mkb for 10 min on ice prior to addition of 35S-Mad3p. For competition assays with radiolabeled Hsl1p667–872, the GST-Cdc20p beads were preincubated for 30 min with 15 μg of His6-Mad2p and ∼4 μg of the His6-Mad3p WT or His6-Mad3p Mut. complexes prior to the addition of 10 μL of in vitro translated 35S-Hsl1p667–872.
Cdc20p-binding assays with baculovirus-produced Mad3p were essentially the same as above except that ∼0.2 μg of recombinant wild-type or mkb1 Mad3p was added to the binding reaction. Mad3p bound to Cdc20p was detected by immunoblot analysis using anti-Mad3p antibodies. For peptide competition assays, GST-Cdc20p beads were preincubated with 500 μM the indicated peptides in 0.1 mL of immunoprecipitation buffer plus 0.1 mg/mL ovalbumin for 30 min at 4°C prior to addition of 0.2 μg of wild-type Mad3p and incubation for an additional 30 min at 4°C. Beads were washed as indicated above, and Mad3p bound to the beads was detected by immunoblotting with anti-Mad3p antibodies.
Acknowledgments
We thank Orna Cohen-Fix and Mark Winey for plasmids, Hongtao Yu for the Mad3p baculovirus, and the W.M Keck Foundation Biotechnology Resource Laboratory for peptide synthesis. We thank Kevin Hardwick for communicating unpublished results. We thank Jingsheng Si for baculoviral work and Mark Hochstrasser, Aiyang Cheng, and Denis Ostapenko for critical reading of the manuscript. This work was supported by grant GM47830 from the NIH (to M.J.S.) and grant 0455851T from the American Heart Association (to M.J.S.).
Footnotes
References
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., Moore D.D., Seidman J.G., Smith J.A., Struhl K., Seidman J.G., Smith J.A., Struhl K., Smith J.A., Struhl K., Struhl K. Current protocols in molecular biology. Wiley; New York: 1995. [Google Scholar]
- Burton J.L., Solomon M.J., Solomon M.J. Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol. Cell. Biol. 2000;20:4614–4625. doi: 10.1128/mcb.20.13.4614-4625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J.L., Solomon M.J., Solomon M.J. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes & Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J.L., Tsakraklides V., Solomon M.J., Tsakraklides V., Solomon M.J., Solomon M.J. Assembly of an APC–Cdh1–substrate complex is stimulated by engagement of a destruction box. Mol. Cell. 2005;18:533–542. doi: 10.1016/j.molcel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Carroll C.W., Enquist-Newman M., Morgan D.O., Enquist-Newman M., Morgan D.O., Morgan D.O. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr. Biol. 2005;15:11–18. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K., Michaelis C., Shevchenko A., Mann M., Nasmyth K., Shevchenko A., Mann M., Nasmyth K., Mann M., Nasmyth K., Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters J.M., Kirschner M.W., Koshland D., Peters J.M., Kirschner M.W., Koshland D., Kirschner M.W., Koshland D., Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., Mapelli M., Sironi L., Faretta M., Salmon E.D., Sironi L., Faretta M., Salmon E.D., Faretta M., Salmon E.D., Salmon E.D., et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Doncic A., Ben-Jacob E., Barkai N., Ben-Jacob E., Barkai N., Barkai N. Noise resistance in the spindle assembly checkpoint. Mol. Syst. Biol. 2006;2:2006–2027. doi: 10.1038/msb4100070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Yu H., Kirschner M.W., Yu H., Kirschner M.W., Kirschner M.W. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes & Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Beretta A., Sironi L., Musacchio A., Lucchini G., Piatti S., Beretta A., Sironi L., Musacchio A., Lucchini G., Piatti S., Sironi L., Musacchio A., Lucchini G., Piatti S., Musacchio A., Lucchini G., Piatti S., Lucchini G., Piatti S., Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Kumada K., Yanagida M., Kumada K., Yanagida M., Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 1996a;15:6617–6628. [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Yamano H., Kumada K., Nagano K., Hunt T., Yanagida M., Yamano H., Kumada K., Nagano K., Hunt T., Yanagida M., Kumada K., Nagano K., Hunt T., Yanagida M., Nagano K., Hunt T., Yanagida M., Hunt T., Yanagida M., Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996b;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Sugino A., Sugino A. New yeast—Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A.W., Kirschner M.W., Murray A.W., Kirschner M.W., Kirschner M.W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hardwick K.G., Weiss E., Luca F.C., Winey M., Murray A.W., Weiss E., Luca F.C., Winey M., Murray A.W., Luca F.C., Winey M., Murray A.W., Winey M., Murray A.W., Murray A.W. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Hardwick K.G., Johnston R.C., Smith D.L., Murray A.W., Johnston R.C., Smith D.L., Murray A.W., Smith D.L., Murray A.W., Murray A.W. MAD3encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Burton J.L., Solomon M.J., Burton J.L., Solomon M.J., Solomon M.J. The anaphase-promoting complex: It’s not just for mitosis any more. Genes & Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- Hilioti Z., Chung Y.S., Mochizuki Y., Hardy C.F., Cohen-Fix O., Chung Y.S., Mochizuki Y., Hardy C.F., Cohen-Fix O., Mochizuki Y., Hardy C.F., Cohen-Fix O., Hardy C.F., Cohen-Fix O., Cohen-Fix O. The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr. Biol. 2001;11:1347–1352. doi: 10.1016/s0960-9822(01)00399-2. [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Lau L.F., Smith D.L., Mistrot C.A., Hardwick K.G., Hwang E.S., Amon A., Murray A.W., Lau L.F., Smith D.L., Mistrot C.A., Hardwick K.G., Hwang E.S., Amon A., Murray A.W., Smith D.L., Mistrot C.A., Hardwick K.G., Hwang E.S., Amon A., Murray A.W., Mistrot C.A., Hardwick K.G., Hwang E.S., Amon A., Murray A.W., Hardwick K.G., Hwang E.S., Amon A., Murray A.W., Hwang E.S., Amon A., Murray A.W., Amon A., Murray A.W., Murray A.W. Budding yeast Cdc20: A target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Lin D.P., Matsumoto S., Kitazono A., Matsumoto T., Lin D.P., Matsumoto S., Kitazono A., Matsumoto T., Matsumoto S., Kitazono A., Matsumoto T., Kitazono A., Matsumoto T., Matsumoto T. Fission yeast Slp1: An effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- King R.W., Glotzer M., Kirschner M.W., Glotzer M., Kirschner M.W., Kirschner M.W. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol. Biol. Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Vodermaier H.C., Maurer-Stroh S., Eisenhaber F., Peters J.M., Vodermaier H.C., Maurer-Stroh S., Eisenhaber F., Peters J.M., Maurer-Stroh S., Eisenhaber F., Peters J.M., Eisenhaber F., Peters J.M., Peters J.M. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol. Cell. 2005;18:543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Luo X., Tang Z., Rizo J., Yu H., Tang Z., Rizo J., Yu H., Rizo J., Yu H., Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- Millband D.N., Hardwick K.G., Hardwick K.G. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell. Biol. 2002;22:2728–2742. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.J., Summers M.K., Hansen D.V., Nachury M.V., Lehman N.L., Loktev A., Jackson P.K., Summers M.K., Hansen D.V., Nachury M.V., Lehman N.L., Loktev A., Jackson P.K., Hansen D.V., Nachury M.V., Lehman N.L., Loktev A., Jackson P.K., Nachury M.V., Lehman N.L., Loktev A., Jackson P.K., Lehman N.L., Loktev A., Jackson P.K., Loktev A., Jackson P.K., Jackson P.K. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes & Dev. 2006;20:2410–2420. doi: 10.1101/gad.1454006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.W., Marks D., Marks D. Can sequencing shed light on cell cycling? Nature. 2001;409:844–846. doi: 10.1038/35057033. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Hardwick K.G., Hardwick K.G. The spindle checkpoint: Structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: The molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Nezi L., Rancati G., De Antoni A., Pasqualato S., Piatti S., Musacchio A., Rancati G., De Antoni A., Pasqualato S., Piatti S., Musacchio A., De Antoni A., Pasqualato S., Piatti S., Musacchio A., Pasqualato S., Piatti S., Musacchio A., Piatti S., Musacchio A., Musacchio A. Accumulation of Mad2–Cdc20 complex during spindle checkpoint activation requires binding of open and closed conformers of Mad2 in Saccharomyces cerevisiae. J. Cell Biol. 2006;174:39–51. doi: 10.1083/jcb.200602109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Chen R.H., Chen R.H. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes & Dev. 2004;18:1439–1451. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M. The anaphase-promoting complex: Proteolysis in mitosis and beyond. Mol. Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- Pfleger C.M., Kirschner M.W., Kirschner M.W. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes & Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pfleger C.M., Lee E., Kirschner M.W., Lee E., Kirschner M.W., Kirschner M.W. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes & Dev. 2001a;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger C.M., Salic A., Lee E., Kirschner M.W., Salic A., Lee E., Kirschner M.W., Lee E., Kirschner M.W., Kirschner M.W. Inhibition of Cdh1-APC by the MAD2-related protein MAD2L2: A novel mechanism for regulating Cdh1. Genes & Dev. 2001b;15:1759–1764. doi: 10.1101/gad.897901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S., Hwang E.S., Visintin R., Amon A., Hwang E.S., Visintin R., Amon A., Visintin R., Amon A., Amon A. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr. Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Reimann J.D., Freed E., Hsu J.Y., Kramer E.R., Peters J.M., Jackson P.K., Freed E., Hsu J.Y., Kramer E.R., Peters J.M., Jackson P.K., Hsu J.Y., Kramer E.R., Peters J.M., Jackson P.K., Kramer E.R., Peters J.M., Jackson P.K., Peters J.M., Jackson P.K., Jackson P.K. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001a;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- Reimann J.D., Gardner B.E., Margottin-Goguet F., Jackson P.K., Gardner B.E., Margottin-Goguet F., Jackson P.K., Margottin-Goguet F., Jackson P.K., Jackson P.K. Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes & Dev. 2001b;15:3278–3285. doi: 10.1101/gad.945701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sharp A.H., McPherson P.S., Dawson T.M., Aoki C., Campbell K.P., Snyder S.H., McPherson P.S., Dawson T.M., Aoki C., Campbell K.P., Snyder S.H., Dawson T.M., Aoki C., Campbell K.P., Snyder S.H., Aoki C., Campbell K.P., Snyder S.H., Campbell K.P., Snyder S.H., Snyder S.H. Differential immunohistochemical localizaton of inositol 1,4,5-trisphosphate- and ryanodine-sensitive Ca2+ release channels in rat brain. J. Neurosci. 1993;13:3051–3063. doi: 10.1523/JNEUROSCI.13-07-03051.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L., Mapelli M., Knapp S., Antoni A.D., Jeang K.T., Musacchio A., Mapelli M., Knapp S., Antoni A.D., Jeang K.T., Musacchio A., Knapp S., Antoni A.D., Jeang K.T., Musacchio A., Antoni A.D., Jeang K.T., Musacchio A., Jeang K.T., Musacchio A., Musacchio A. Crystal structure of the tetrameric Mad1–Mad2 core complex: Implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Murray A.W., Murray A.W. The spindle assembly checkpoint in budding yeast. Methods Enzymol. 1997;283:425–440. doi: 10.1016/s0076-6879(97)83035-2. [DOI] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., Yen T.J., Chan G.K., Yen T.J., Yen T.J. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Bharadwaj R., Li B., Yu H., Bharadwaj R., Li B., Yu H., Li B., Yu H., Yu H. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- Tang Z., Shu H., Oncel D., Chen S., Yu H., Shu H., Oncel D., Chen S., Yu H., Oncel D., Chen S., Yu H., Chen S., Yu H., Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell. 2004;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Lottspeich F., Nasmyth K., Lottspeich F., Nasmyth K., Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Wernic D., Poupart M.A., Koonin E.V., Nasmyth K., Wernic D., Poupart M.A., Koonin E.V., Nasmyth K., Poupart M.A., Koonin E.V., Nasmyth K., Koonin E.V., Nasmyth K., Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Yamano H., Gannon J., Mahbubani H., Hunt T., Gannon J., Mahbubani H., Hunt T., Mahbubani H., Hunt T., Hunt T. Cell cycle-regulated recognition of the destruction box of cyclin B by the APC/C in Xenopus egg extracts. Mol. Cell. 2004;13:137–147. doi: 10.1016/s1097-2765(03)00480-5. [DOI] [PubMed] [Google Scholar]
- Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 2002;14:706–714. doi: 10.1016/s0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]