Human TBX1 Missense Mutations Cause Gain of Function Resulting in the Same Phenotype as 22q11.2 Deletions (original) (raw)
Abstract
Deletion 22q11.2 syndrome is the most frequent known microdeletion syndrome and is associated with a highly variable phenotype, including DiGeorge and Shprintzen (velocardiofacial) syndromes. Although haploinsufficiency of the T-box transcription factor gene TBX1 is thought to cause the phenotype, to date, only four different point mutations in TBX1 have been reported in association with six of the major features of 22q11.2 deletion syndrome. Although, for the two truncating mutations, loss of function was previously shown, the pathomechanism of the missense mutations remains unknown. We report a novel heterozygous missense mutation, H194Q, in a familial case of Shprintzen syndrome and show that this and the two previously reported missense mutations result in gain of function, possibly through stabilization of the protein dimer DNA complex. We therefore conclude that TBX1 gain-of-function mutations can result in the same phenotypic spectrum as haploinsufficiency caused by loss-of-function mutations or deletions.
The well-known recurrent ∼3-Mb microdeletion within chromosomal band 22q11.2 is associated with a variety of symptoms and syndromes,1–3 including DiGeorge syndrome (DGS [MIM 188400]) and Shprintzen (velocardiofacial) syndrome (VCFS [MIM 192430]). The 22q11.2 deletion syndrome constitutes a major cause of congenital heart disease (CHD), accounting for ∼5% of all CHD cases among live births.4 It is also the second most common causative diagnosis in mental retardation, accounting for ∼2.4% of patients with developmental delay.5 The majority of patients harbor a deletion of either 3 or 1.5 Mb, both of which include the TBX1 gene.6 Of the ∼30 commonly deleted genes, Tbx1 is the only gene that, after an extensive functional analysis in the mouse, has been found to be haploinsufficient with a convincingly similar phenotype to the human syndrome.7 TBX1 is a member of a phylogenetically conserved family of genes that share a common DNA-binding domain, the T-box. Like the related Xenopus T protein (Xbra), TBX1 was shown to bind a palindromic T oligonucleotide as a dimer.8 TBX1 has three isoforms (A, B, and C) that share exons 1–8 but differ in the terminal exons 9A, 9B/10, and 9C.9,10 The 9C transcript is the only isoform highly conserved in mouse and was shown to be the major transcript in humans.10 However, mutational analysis of a total of 162 patients without deletion initially failed to identify pathogenically significant alterations in the human homolog TBX1.9–11 Only in 2003, by the screening of 10 patients with DGS/VCFS without deletion, Yagi et al. identified missense mutations in two patients with sporadic disease and one truncation mutation segregating in a family with the characteristic 22q11.2 deletion phenotype.12 The truncation mutation was shown to result in loss of function due to the deletion of a C-terminal nuclear localization signal of TBX1.13 This mutation and a novel C-terminal truncating mutation identified in another family with the DGS/VCFS phenotype were also shown to result in lack of activation in a transcriptional chloramphenicol acetyltransferase–reporter assay.14 The two missense mutations reported by Yagi et al., however, did not show any significant effect in a reporter assay by use of a GAL4-Tbx1 fusion protein.13 We now report a third missense mutation in TBX1 and provide, for the first time, experimental evidence that the proteins encoded by these three missense mutations are functionally distinguishable from the wild-type protein.
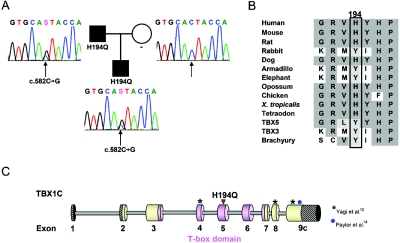
After informed consent for genetic analyses had been obtained, we had tested a series of patients with DGS/VCFS-like phenotype with a set of 10 FISH clones, to exclude typical and atypical 22q11.2 deletions.6 Ten patients without deletion and with a typical 22q11.2 deletion phenotype were then selected for mutational analysis of the TBX1 gene by direct sequencing of all nine exons with exon-intron boundaries, including the three isoforms of exon 9 (GenBank). Direct sequencing of both strands by use of BigDye Terminator Cycle Sequencing Kit v3.1 (Applied Biosystems) was analyzed with an ABI 3730 automated sequencer (Applied Biosystems). Primer sequences and PCR conditions are available on request. In one of these patients, we detected a heterozygous missense mutation, c.582C→G, resulting in the substitution of an evolutionary conserved histidine at position 194 by a glutamine within the T-box domain (fig. 1_A_–1C). This patient of Turkish origin was referred to us at age 2 years because of short stature and speech delay. He was born after 39 wk of gestation, with a birth weight of 2,660 g (−1.9 SD) and a heel-to-crown length of 46 cm (−2.62 SD). At the age of 3 years and 10 mo, he had short stature (height 92.4 cm, −3.01 SD; weight 12.1 kg; BMI 14.17) and a head circumference just below the 3rd percentile (47.5 cm). Apart from an operated club foot, operated undescended testicles, and an innocent cardiac murmur, he had no organic problems or anomalies. Although he reached motor milestones within normal limits (sitting at age 6–7 mo; walking at age 13–14 mo), his speech development was delayed, with only five single words at age 2 years. He therefore received logopedic therapy, and, at age 4 years, he spoke in full sentences. His overall functioning was estimated at the level of low-normal to borderline, but formal testing was not possible so far. His facial features, with small, upslanting palpebral fissures, tubulous nose, short mouth, micrognathia, low-set ears, and flat malar regions with broad cheeks, represented the typical facial gestalt of DGS/VCFS; therefore, he had been considered for TBX1 mutational analysis after exclusion of a 22q11.2 deletion. The patient’s father, who had the same facial features and short stature (160 cm) but normal psychomotor development, was found to have the same heterozygous H194Q mutation (fig. 1A). This mutation was not found in 384 control chromosomes from healthy European individuals. Since the H194Q mutation is located within the T-box domain, it was considered to have a high probability of affecting the function of the protein. To test this hypothesis, we established a novel transcriptional reporter assay. In a cytomegalovirus (CMV) expression vector containing the complete cDNA of human TBX1 isoform C, we introduced the three mutations published by Yagi et al.12 and our novel mutation, by use of site-directed mutagenesis done according to the protocol provided by the Quik Change Site Directed Mutagenesis Kit (Stratagene), with minor modifications.
Figure 1. .
Identification of a mutation in the TBX1 gene in a family with the 22q11 deletion phenotype. A, Pedigree and electropherograms of the mutated genomic sequence of exon 5 of the TBX1 gene from the affected family members and the unaffected mother. The position of the mutated nucleotide is indicated by a black arrow; the identified mutation c.582C→G leads to the amino acid substitution H194Q. B, Alignment of a part of the T-box domain of TBX1 with homologues from various species (UCSC Genome Bioinformatics) and other members of the T-box family. The conserved amino acids are shown with a gray background, and similar amino acids with a light gray background. Residue 194 is indicated with a black box. C, Scheme of TBX1C indicating the T-box (pink), the novel H194Q mutation (red arrowhead), and the mutations published by Yagi et al.12 and Paylor et al.14
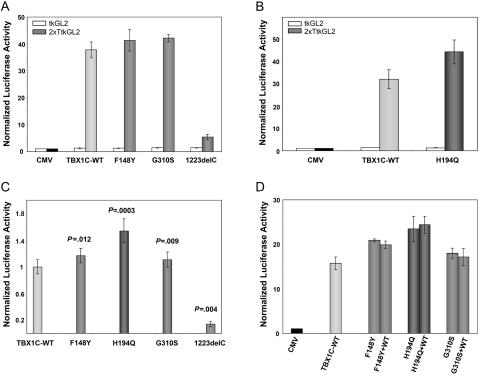
While initial attempts to establish the assay in HEK293 cells demonstrated a very weak and inconsistent transcriptional activity of TBX1C, transcriptional activity was much higher in JEG3 cells, which are derived from human choriocarcinoma. Because JEG3 cells, in contrast to HEK293 cells, constituently express TBX1, this difference suggests that the presence of cofactors may be important for proper TBX1 function. JEG3 cells (American type culture collection cell line HTB-36) cultivated in RPMI medium with 10% fetal calf serum were cotransfected with a CMV expression vector containing a complete wild-type or mutant TBX1C cDNA and a luciferase reporter construct with a herpes simplex thymidine kinase promoter either with two palindromic T-box binding sites inserted upstream of the tk promoter (2xTtkGL2) or without (tkGL2), as well as a renilla luciferase vector to normalize for transfection efficiency. The luciferase reporter construct is similar to that reported by Sinha et al.,8 except that 2xTtkGL2 has two palindromic T-sites, whereas 4xT2tkGL2 had four T-half sites oriented head to tail. None of the TBX1 constructs had any effect on transcription of the tkGL2 reporter construct, which has no T-box binding sites (fig. 2A and 2B). In comparison with the empty CMV vector, the TBX1C wild type showed a 14–46-fold activation of 2xTtkGL2 in three different experiments with three independent transfections each (fig. 2). As expected, the truncating mutation reported by Yagi et al.12 (1223delC) (fig. 2_A_–2C) showed virtually no activation. As a novel finding, the two missense mutations F148Y and G310S, which had behaved similarly to wild type in the assay done by Stoller et al.,13 exhibited a slight but reproducible and statistically significant increase in activation relative to wild-type TBX1C in our assay (fig. 2A and 2C). The novel mutation H194Q showed an even larger increase in activation (fig. 2B and 2C). To better simulate the heterozygous effect of the mutations, cells were also transfected simultaneously with equal amounts of TBX1C wild-type and mutant vectors. No obvious difference was observed in comparison with cells transfected with mutants only (fig. 2D).
Figure 2. .
Transcriptional activation of a luciferase reporter constructs by wild-type (WT) and mutant TBX1C. A–D, JEG-3 cells were transiently transfected with reporter constructs containing a promoter without the T-Box binding sites (tkGL2 [_white bars_]) or with T-Box binding sites (2xTtkGL2, bars with different shades of gray). Cotransfection was performed with either a CMV control vector, the TBX1C wild-type construct, or the mutant constructs. Results are normalized for transfection efficiency to a cotransfected renilla luciferase vector and expressed as average values ± SEM of three independent transfections. The results were confirmed by repeating the triple transfections and measurements for two to three times respectively. A and B, Representative examples of results of single experiments with three independent transfections for each combination. In comparison to the CMV vector TBX1C shows a 30–40-fold increased activation on a reporter containing T-Box binding elements (2xTtkGL2 [_gray bars_]), whereas there is virtually no activation of the reporter construct without T-Box binding sites (tkGL2 [_white bars_]). The truncating mutation 1223delC shows a clear lack of activation, whereas the missense mutations indicate an increase in activation of the luciferase reporter construct. Due to the small number of measurements, none of these results from a single experiment reaches statistical significance with the Mann-Whitney-Wilcoxon test. Similar results were obtained with two further experiments with three independent transfections each (data not shown). C, To compare results from independent experiments, the activity of mutant TBX1C was normalized to that of wild-type TBX1C, and P values were obtained using the Mann-Whitney-Wilcoxon test for unpaired samples of totally nine independent transfections in three independent experiments (six transfections in two experiments for 1223delC). D, To simulate heterozygosity of the mutations, cells were simultaneously transfected with equal amounts of wild-type and mutant TBX1C construct DNA. No obvious difference was observed in comparison with cells transfected with mutant TBX1C only.
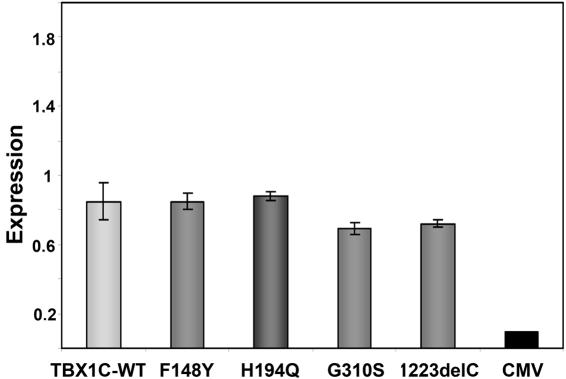
To exclude differences in TBX1 expression as the cause of the activity enhancement, we established a real-time PCR assay to analyze mRNA levels in the transfected cells. Cells were cotransfected in duplicate with the TBX1 wild-type or mutant vector and a renilla luciferase vector and were harvested after 48 h. Quantitative multiplex PCR was performed with an exogenous _TBX1_-specific TaqMan probe and a control probe detecting renilla luciferase, to normalize for transfection efficiency. Data from quadruplicate measurements were analyzed using the ΔΔCt method as described elsewhere.15 Results showed no increase in expression levels of the mutants compared with that of the wild type (fig. 3). These findings support a gain of function rather than a dosage effect as the mechanism responsible for the differential behavior of the wild type and the TBX1 missense mutations in our assay. Nevertheless, the slightly lower mRNA level of the G310S construct could at least partially account for its less pronounced increase in transcriptional activity.
Figure 3. .
Expression of wild-type (WT) and mutant TBX1C in JEG3 cells. JEG3 cells were transiently transfected with CMV, wild-type and mutant TBX1C and TBX1C expression measured with a Taqman assay. No increased expression was observed for the mutants in comparison to the wild type. Results are normalized for transfection efficiency against the cotransfected renilla vector and expressed as average value ± SEM of quadruplicate measurements of two independent transfections, respectively. By use of the Mann-Whitney-Wilcoxon test for unpaired samples, P values for comparison with the wild-type levels are 1 each for F148Y and H194Q, 0.24 for G310S, and 0.44 for 1223delC.
The T-box domain, which is highly conserved in all T-box proteins,16 is responsible for DNA binding and is likely important for dimerization of TBX1, whose three-dimensional structure has not been determined so far. The three-dimensional structure has been solved for the DNA-binding (T-box) domain of the TBX1 homologues TBX317 and the Brachyury transcription factor, Xbra.18 Within the T-box domain, TBX3 and Xbra share sequence identities of 59% and 48% with TBX1, respectively, which is sufficient to obtain a reliable model of the TBX1 tertiary structure. Although TBX3 and Xbra share a highly similar three-dimensional structure for the monomeric proteins, the dimer interface—and thus the quaternary structure—differs significantly.17 A detailed comparison of those residues forming the dimer interface suggests that TBX1 is more similar to Xbra than to TBX3. In particular, TBX1 contains a 3-residue insertion (P248–K250) within the β-sheet that forms the dimer interface in TBX3, leading to steric clashes during TBX1 model building on the TBX3 template (data not shown). This higher similarity of the interface between TBX1 and Xbra is also consistent with biochemical data showing that Xbra and TBX1 bind 40 times more strongly to a palindromic DNA binding site than to a single site, whereas TBX2 (which shares 95% sequence identity with TBX3 in the T-box domain) binds only 4 times stronger.17 This has led to the suggestion that T-box domain proteins fall into different groups that differ in their dimerization properties and their ability to interact with single sites of the palindromic binding sequence.8 Accordingly, TBX1 and Xbra were proposed to belong to the same class, whereas TBX2 and TBX3 belong to a different class.17 Consistent with this observation, we found that, in contrast to TBX2, TBX1 was not active on the 4xT2tkGL2 reporter construct established by Sinha et al.,8 in which four T-half sites are oriented head to tail (unpublished data). In this construct, the orientation of the binding sites would perturb or prohibit interaction of the putative dimer interfaces of T-box proteins bound to adjacent binding sites.
On the basis of these considerations, the structure of the TBX1 DNA binding domain was modeled on the basis of the Xbra DNA crystal structure (Protein Data Bank [PDB] code 1xbr),18 resulting in a dimeric protein with a good overall geometry and no steric clashes. The model of TBX1 was generated using SWISS-MODEL and Swiss-PdbViewer,19 for sequence alignment and for building the three-dimensional structure. The quality of the model was assessed using PROCHECK,20 and WHAT_CHECK,21 and this assessment did not reveal any steric clashes or unfavorable geometries. Mutations were inserted using the Sybyl 6.9 program package (Tripos) and were subsequently refined by 100 steps of energy minimization. Hydrogen bonds were identified using Insight II (Accelrys) with the standard cutoff criteria, and protein-protein as well as protein-DNA contacts were analyzed using LIGPLOT.22
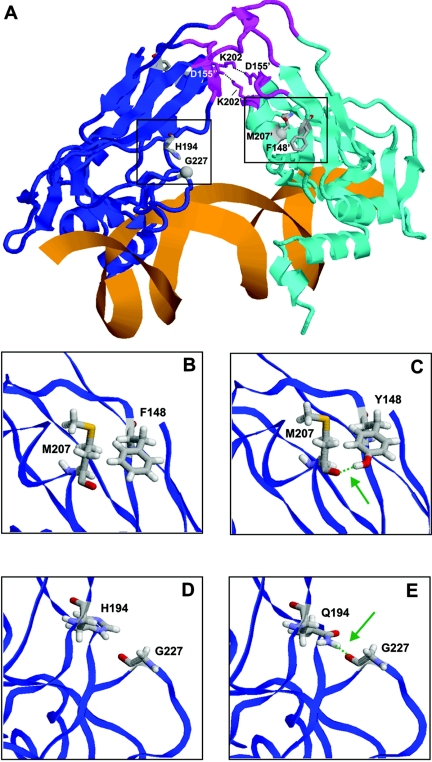
The dimer interface in TBX1 is formed by residues D151–D155 and P200–K202, which are conserved overall between TBX1 and Xbra. Interestingly, the two nonconserved residues (D155 and K202) form an additional salt bridge across the dimer interface of TBX1 (fig. 4A), thus supporting the correctness of the modeled quaternary structure. In the first step, the local effect of the mutations was examined by modeling the mutant proteins and by analyzing the changes of the interactions at the site of the mutation. F148 is a nonconserved residue within the family of T-box transcription factors, and a serine is present at the equivalent sequence position in TBX3 and Xbra. Apart from hydrophobic packing, F148 does not form any specific side-chain interactions (fig. 4B). In contrast, the hydroxyl group present in the side chain of the Y148 mutant allows formation of a hydrogen bond to the backbone carbonyl oxygen of M207 (fig. 4C).
Figure 4. .
A, Three-dimensional model of human Tbx1 in complex with DNA (orange). The subunits of the dimeric protein are shown in backbone presentation (blue and cyan) and the α-helices and β-sheets are depicted schematically. The three loops predicted to form the dimer interface are shown (magenta), and residues D155 and K202 that form salt bridges (black dotted lines) across the dimer interface are shown in stick presentation. A prime distinguishes residues belonging to different subunits. Residues F148 and H194, for which mutations were observed, are shown in stick presentation, and sequence positions that were predicted to form novel interactions with these residues in the mutant proteins are shown as balls. The spatial vicinity (black boxes) of these residues is shown as an enlargement for the wild-type and mutant proteins in panels B–E. B–E, Detailed analysis of the structural effects of F148Y and H194Q mutants. Models of the wild-type and mutant proteins are shown in the left and right panels, respectively. Structurally important residues are shown in stick presentation and are colored according to the atom types. Green dotted lines indicate important hydrogen bonds, which are apparently affected by the mutations, and the site of the hydrogen bond is marked (green arrow). B and C, Effect of the F148Y mutation. The presence of a tyrosine at position 148 allows the formation of a novel side-chain hydrogen bond to the carbonyl oxygen of M207 (E), which cannot be formed by the phenylalanine in the wild type (D). D and E, Effect of the H194Q mutation. The presence of a glutamine at position 194 allows the formation of a novel side-chain hydrogen bond to the carbonyl oxygen of G227 (E), which cannot be formed by the histidine in the wild type (D).
The second site of mutation located at position 194 contains a histidine in TBX1, whereas other T-box transcription factors generally possess a tyrosine at the respective sequence position. The ring of H194 points toward the loop containing G227 (fig. 1D), but even if a protonation of the histidine ring at Nδ is assumed, the distance to the backbone of G227 would be too large for the formation of a hydrogen bond. This situation is predicted to be different in the H194Q mutant, since, because of the more extended structure of the side chain, a hydrogen bond is predicted to be formed between the side chain amide group of Q194 and the backbone carbonyl oxygen of G227 (fig. 4E). Thus, both mutations are expected to increase the protein stability by additional hydrogen bonds.
To assess the global effect of this stabilization on protein function, the localization of the mutations in the structure was examined in more detail. Both mutations are directly linked via loops to key residues of the predicted dimer interface. F148 belongs to the same peptide stretch as D155, and H194 to the same stretch as K202 (fig. 4A). As described above, D155 and K202 form salt bridges across the predicted dimer interface and are thus expected to be crucial for dimer formation and stabilization in TBX1. The additional interactions found in the F148Y and H194Q mutants are expected to stabilize the respective loops, thus facilitating dimer formation. In addition, the mutations might also increase the rigidity of the entire structure by stabilizing the relative orientation of the predicted dimer interface and the DNA-binding region of TBX1. For example, G227, which is stabilized by an additional hydrogen bond in the H194Q mutant, is located proximal to the residues N220 and S233, which are involved in DNA binding. Thus, there might also exist an increased cooperativity between dimer formation and DNA binding in the mutants. This would explain why Stoller et al.13 did not detect any effect of the F148Y missense mutation by use of GAL4 DNA-binding domain fusions that did not involve T-box–DNA interactions.
In summary, molecular modeling suggests that—consistent with their similar effects seen in our transcriptional activity assay—the F148Y and H194Q mutations may have similar effects on the TBX1 structure, probably resulting in an increased dimer stability and/or DNA-binding affinity. The G310S mutation reported by Yagi et al.12 also showed an increased transcriptional activation. Residue 310 lies outside of but adjacent to the T-box and, therefore, may also affect DNA binding and/or protein stabilization. Alternatively, this mutation may alter protein function through another mechanism.
The observation of an increased transcriptional activation induced by the TBX1 missense mutations was initially surprising, because usually the DGS/VCFS phenotype results from haploinsufficiency due to a deletion in the 22q11.2 region or, as shown recently, to truncating mutations within the TBX1C gene.12–14 Nevertheless, our observations are in accordance with findings in mice, in which both over- and underexpression of Tbx1 resulted in a phenotype similar to that of DGS/VCFS in humans.23,24 However, the human phenotype associated with duplication 22q11.2, although overlapping with the clinical spectrum of the corresponding deletion, is distinct, with a different facial gestalt (table 1).25–31 Because our novel and the previously published12 TBX1 missense mutations with increased transcriptional activity are associated with the typical facial gestalt of 22q11.2 deletion, the effect of these specific gain-of-function mutations that probably enhance protein stabilization seems to be a better phenocopy of 22q11.2 deletion than that of increased 22q11.2 dosage. Apart from the characteristic facial gestalt, patients with TBX1 point mutations, like patients with 22q11.2 deletion, show a large phenotypical variability even within the same family, including congenital heart defects, abnormal thymus and parathyreoid, velopharyngeal dysfunction, and psychiatric disorders.12,14 Our observation adds short stature and, possibly, developmental delay to the phenotypic spectrum of TBX1 mutations. In 22q11.2 deletion, this variability is thought to be caused by modifying genes inside or outside the deletion.32–36 Although both features are part of the 22q11.2 deletion spectrum,2 our index patient is the only patient with TBX1 point mutation yet reported to show developmental delay.12,14 Therefore, an additional factor causing the boy’s developmental problems cannot be excluded. Analyses concerning chromosomal abnormalities, subtelomeric rearrangements, fragile-X syndrome, and skewed X inactivation in the mother revealed normal results. Nevertheless, further studies are necessary to confirm an association between developmental delay and a TBX1 point mutation.
Table 1. .
Overview of Phenotype in 22q11.2 Copy-Number Aberrations and TBX1 Mutations[Note]
| H194Q(_n_=2) | |||||||
|---|---|---|---|---|---|---|---|
| Trait | TBX1 Stop Mutation12,14(_n_=7) | F148Y12(_n_=1) | G310S12(_n_=1) | Patient | Father | Deletion2(_n_=558)(%) | Duplication27(_n_=35)a(%) |
| Congenital heart defect | 3 | + | + | − | − | 57 | 19 |
| Hypocalcemia | 1 | − | + | − | − | 60 | … |
| Cleft palate | − | − | − | − | − | 9 | 33 |
| Velopharyngeal insufficiency | 3 | + | + | − | − | 32 | 59 |
| Short stature | − | − | − | + | + | 36 | 50 |
| Mental retardation and/or developmental delay | − | − | − | (+) | − | 38 | 91 |
| Psychiatric disorders | 2 | − | − | − | − | 3029 | … |
| VCFS facial gestaltb | 7 | + | + | + | + | 10012 | … |
In summary, we report the third missense mutation and the fifth overall mutation in TBX1 expanding the associated phenotype to include short stature and, possibly, developmental delay. We provide the first evidence that TBX1 missense mutations may alter the transcriptional activity of the TBX1 protein, possibly through stabilization of the protein-protein or protein-DNA interaction. We also conclude that TBX1 gain-of-function mutations can result in the same phenotypic spectrum as haploinsufficiency caused by loss-of-function mutations or deletions.
Acknowledgments
We thank the family, for participation in this study, and Brigitte Dintenfelder, Daniela Schweitzer, and Michaela Kirsch, for expert technical assistance. C.E.C. was supported by National Institutes of Health grant DK48796.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for TBX1C [accession numbers NT_011519.10 and NM_080647.1], TBX3 [accession numbers NT_009775 and NM_016569], TBX5 [accession numbers NT_009775 and NM_000192.3], and Brachyury [accession numbers NT_007422 and NM_003181.2])
- LIGPLOT, http://www.ebi.ac.uk/Thornton/software.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DGS and VCFS)
- PROCHECK, http://www.ebi.ac.uk/Thornton/software.html
- SWISS-MODEL, http://swissmodel.expasy.org/
- Swiss-PdbViewer, http://www.expasy.ch/spdbv/
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/
- WHAT_CHECK, http://swift.cmbi.ru.nl/gv/whatcheck/
References
- 1.McDonald-McGinn DM, Tonnesen MK, Laufer-Cahana A, Finucane B, Driscoll DA, Emanuel BS, Zackai EH (2001) Phenotype of the 22q11.2 deletion in individuals identified through an affected relative: cast a wide FISHing net! Genet Med 3:23–29 [DOI] [PubMed] [Google Scholar]
- 2.Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, Seidel H, Schuffenhauer S, Oechsler H, Belohradsky B, Prieur M, et al (1997) Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet 34:798–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scambler PJ (2000) The 22q11 deletion syndromes. Hum Mol Genet 9:2421–2426 10.1093/hmg/9.16.2421 [DOI] [PubMed] [Google Scholar]
- 4.Wilson DI, Cross IE, Wren C (1994) Minimum prevalence of chromosome 22q11 deletions. Am J Hum Genet Suppl 55:A169 [Google Scholar]
- 5.Rauch A, Hoyer J, Guth S, Zweier C, Kraus C, Becker C, Zenker M, Huffmeier U, Thiel C, Ruschendorf F, et al (2006) Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A 140:2063–2074 [DOI] [PubMed] [Google Scholar]
- 6.Rauch A, Zink S, Zweier C, Thiel CT, Koch A, Rauch R, Lascorz J, Huffmeier U, Weyand M, Singer H, et al (2005) Systematic assessment of atypical deletions reveals genotype-phenotype correlation in 22q11.2. J Med Genet 42:871–876 10.1136/jmg.2004.030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldini A (2005) Dissecting contiguous gene defects: TBX1. Curr Opin Genet Dev 15:279–284 10.1016/j.gde.2005.03.001 [DOI] [PubMed] [Google Scholar]
- 8.Sinha S, Abraham S, Gronostajski RM, Campbell CE (2000) Differential DNA binding and transcription modulation by three T-box proteins, T, TBX1 and TBX2. Gene 258:15–29 10.1016/S0378-1119(00)00417-0 [DOI] [PubMed] [Google Scholar]
- 9.Chieffo C, Garvey N, Gong W, Roe B, Zhang G, Silver L, Emanuel BS, Budarf ML (1997) Isolation and characterization of a gene from the DiGeorge chromosomal region homologous to the mouse Tbx1 gene. Genomics 43:267–277 10.1006/geno.1997.4829 [DOI] [PubMed] [Google Scholar]
- 10.Gong W, Gottlieb S, Collins J, Blescia A, Dietz H, Goldmuntz E, McDonald-McGinn DM, Zackai EH, Emanuel BS, Driscoll DA, et al (2001) Mutation analysis of TBX1 in non-deleted patients with features of DGS/VCFS or isolated cardiovascular defects. J Med Genet 38:E45 10.1136/jmg.38.12.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conti E, Grifone N, Sarkozy A, Tandoi C, Marino B, Digilio MC, Mingarelli R, Pizzuti A, Dallapiccola B (2003) DiGeorge subtypes of nonsyndromic conotruncal defects: evidence against a major role of TBX1 gene. Eur J Hum Genet 11:349–351 10.1038/sj.ejhg.5200956 [DOI] [PubMed] [Google Scholar]
- 12.Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S, et al (2003) Role of TBX1 in human del22q11.2 syndrome. Lancet 362:1366–1373 10.1016/S0140-6736(03)14632-6 [DOI] [PubMed] [Google Scholar]
- 13.Stoller JZ, Epstein JA (2005) Identification of a novel nuclear localization signal in Tbx1 that is deleted in DiGeorge syndrome patients harboring the 1223delC mutation. Hum Mol Genet 14:885–892 10.1093/hmg/ddi081 [DOI] [PubMed] [Google Scholar]
- 14.Paylor R, Glaser B, Mupo A, Ataliotis P, Spencer C, Sobotka A, Sparks C, Choi CH, Oghalai J, Curran S, et al (2006) Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc Natl Acad Sci USA 103:7729–7734 10.1073/pnas.0600206103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thiel CT, Kraus C, Rauch A, Ekici AB, Rautenstrauss B, Reis A (2003) A new quantitative PCR multiplex assay for rapid analysis of chromosome 17p11.2-12 duplications and deletions leading to HMSN/HNPP. Eur J Hum Genet 11:170–178 10.1038/sj.ejhg.5200920 [DOI] [PubMed] [Google Scholar]
- 16.Plageman TF Jr, Yutzey KE (2005) T-box genes and heart development: putting the “T” in heart. Dev Dyn 232:11–20 10.1002/dvdy.20201 [DOI] [PubMed] [Google Scholar]
- 17.Coll M, Seidman JG, Muller CW (2002) Structure of the DNA-bound T-box domain of human TBX3, a transcription factor responsible for ulnar-mammary syndrome. Structure 10:343–356 10.1016/S0969-2126(02)00722-0 [DOI] [PubMed] [Google Scholar]
- 18.Muller CW, Herrmann BG (1997) Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature 389:884–888 10.1038/39929 [DOI] [PubMed] [Google Scholar]
- 19.Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- 20.Laskowski RA, Moss DS, Thornton JM (1993) Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231:1049–1067 10.1006/jmbi.1993.1351 [DOI] [PubMed] [Google Scholar]
- 21.Hooft RW, Vriend G, Sander C, Abola EE (1996) Errors in protein structures. Nature 381:272 10.1038/381272a0 [DOI] [PubMed] [Google Scholar]
- 22.Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8:127–134 10.1093/protein/8.2.127 [DOI] [PubMed] [Google Scholar]
- 23.Funke B, Epstein JA, Kochilas LK, Lu MM, Pandita RK, Liao J, Bauerndistel R, Schuler T, Schorle H, Brown MC, et al (2001) Mice overexpressing genes from the 22q11 region deleted in velo-cardio-facial syndrome/DiGeorge syndrome have middle and inner ear defects. Hum Mol Genet 10:2549–2556 10.1093/hmg/10.22.2549 [DOI] [PubMed] [Google Scholar]
- 24.Liao J, Kochilas L, Nowotschin S, Arnold JS, Aggarwal VS, Epstein JA, Brown MC, Adams J, Morrow BE (2004) Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum Mol Genet 13:1577–1585 10.1093/hmg/ddh176 [DOI] [PubMed] [Google Scholar]
- 25.Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE (1999) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8:1157–1167 10.1093/hmg/8.7.1157 [DOI] [PubMed] [Google Scholar]
- 26.Ensenauer RE, Adeyinka A, Flynn HC, Michels VV, Lindor NM, Dawson DB, Thorland EC, Lorentz CP, Goldstein JL, McDonald MT, et al (2003) Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet 73:1027–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Portnoi MF, Lebas F, Gruchy N, Ardalan A, Biran-Mucignat V, Malan V, Finkel L, Roger G, Ducrocq S, Gold F, et al (2005) 22q11.2 Duplication syndrome: two new familial cases with some overlapping features with DiGeorge/velocardiofacial syndromes. Am J Med Genet A 137:47–51 [DOI] [PubMed] [Google Scholar]
- 28.Yobb TM, Somerville MJ, Willatt L, Firth HV, Harrison K, MacKenzie J, Gallo N, Morrow BE, Shaffer LG, Babcock M, et al (2005) Microduplication and triplication of 22q11.2: a highly variable syndrome. Am J Hum Genet 76:865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy KC, Jones LA, Owen MJ (1999) High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry 56:940–945 10.1001/archpsyc.56.10.940 [DOI] [PubMed] [Google Scholar]
- 30.Shprintzen RJ, Goldberg RB, Young D, Wolford L (1981) The velo-cardio-facial syndrome: a clinical and genetic analysis. Pediatrics 67:167–172 [PubMed] [Google Scholar]
- 31.Wilson DI, Burn J, Scambler P, Goodship J (1993) DiGeorge syndrome: part of CATCH 22. J Med Genet 30:852–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gothelf D, Eliez S, Thompson T, Hinard C, Penniman L, Feinstein C, Kwon H, Jin S, Jo B, Antonarakis SE, et al (2005) COMT genotype predicts longitudinal cognitive decline and psychosis in 22q11.2 deletion syndrome. Nat Neurosci 8:1500–1502 10.1038/nn1572 [DOI] [PubMed] [Google Scholar]
- 33.Gothelf D, Michaelovsky E, Frisch A, Zohar AH, Presburger G, Burg M, Aviram-Goldring A, Frydman M, Yeshaya J, Shohat M, et al (2006) Association of the low-activity _COMT_158 Met allele with ADHD and OCD in subjects with velocardiofacial syndrome. Int J Neuropsychopharmacol 31:1–8 10.1038/sj.npp.1301265 [DOI] [PubMed] [Google Scholar]
- 34.Rauch A, Devriendt K, Koch A, Rauch R, Gewillig M, Kraus C, Weyand M, Singer H, Reis A, Hofbeck M (2004) Assessment of association between variants and haplotypes of the remaining TBX1 gene and manifestations of congenital heart defects in 22q11.2 deletion patients. J Med Genet 41:e40 10.1136/jmg.2003.010975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shashi V, Keshavan MS, Howard TD, Berry MN, Basehore MJ, Lewandowski E, Kwapil TR (2006) Cognitive correlates of a functional COMT polymorphism in children with 22q11.2 deletion syndrome. Clin Genet 69:234–238 10.1111/j.1399-0004.2006.00569.x [DOI] [PubMed] [Google Scholar]
- 36.Stalmans I, Lambrechts D, De Smet F, Jansen S, Wang J, Maity S, Kneer P, von der Ohe M, Swillen A, Maes C, et al (2003) VEGF: a modifier of the del22q11 (DiGeorge) syndrome? Nat Med 9:173–182 10.1038/nm819 [DOI] [PubMed] [Google Scholar]