Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells (original) (raw)
Abstract
IL-12 p40–related cytokines such as IL-12 p35/p40 heterodimer and IL-23 (p19/p40) are potent regulators of adaptive immune responses. Little is known, however, about the transcriptional regulation of the p40 gene in vivo. In an attempt toward this goal, we have generated transgenic mice expressing firefly luciferase under the control of the IL-12 p40 promoter. High constitutive transgene expression was found in the small intestine only, whereas little reporter gene activity was observed in other tissues. Within the small bowel, constitutive promoter activity was restricted to the terminal ileum and associated with high expression of p40 mRNA as well as p40 and IL-23 p19/p40 proteins. The cells constitutively producing IL-12 p40 were identified as CD8α and CD11b double-negative CD11c+ lamina propria dendritic cells (LPDCs) that represent a major cell population in the lamina propria of the small intestine, but not in the colon. FISH directly demonstrated the uptake of bacteria by a subset of LPDCs in the terminal ileum that was associated with p40 expression. Furthermore, little or no p40 protein expression in LPDCs was found in the terminal ileum of germfree mice, indicating a key role of the intestinal flora for constitutive p40 expression. In addition, analysis of transgenic mice with a mutated NF-κB target site in the p40 promoter showed a critical role of NF-κB for constitutive transgene expression. Our data reveal important functional differences between the mucosal immune systems of the small and large bowel in healthy mice and suggest that the high bacterial load in the terminal ileum activates p40 gene transcription in LPDCs through NF-κB. These data suggest a predisposition of the terminal ileum to develop chronic inflammatory responses through IL-23 and thus may provide a molecular explanation for the preferential clinical manifestation of Crohn disease in this part of the gut.
Introduction
The cytokine IL-12 consists of two disulfide-linked subunits, p40 and p35, and is produced mainly by monocytes, macrophages, and DCs (1–4). Whereas the p35 subunit is expressed ubiquitously, the expression of p40 is tightly regulated and restricted to cells producing functionally active IL-12 (5). IL-12 is known to be a key regulator of Th1-type immune responses by binding and signaling through the high-affinity IL-12 receptor, consisting of a constitutively expressed β1 chain and an inducible β2 chain, which is restricted to activated T cells and NK cells. After IL-12 binds to its receptor, it induces activation of specific members of the STAT (signal transducers and activators of transcription) family of transcription factors (STAT-3 and STAT-4), which then translocate to the nucleus and bind to genomic promoter regions, including that governing IFN-γ (6, 7). In this way, IL-12 p35/p40 strongly induces differentiation of naive T lymphocytes into Th1 effector cells.
Oppmann and coworkers (8) recently showed that IL-12 p40 can form a novel cytokine, denoted IL-23 (p19/p40), by binding to a protein called p19. IL-23 exhibits biological activities similar to, as well as distinct from, IL-12 (8, 9). In particular, IL-23 preferentially activates memory rather than naive T lymphocytes and augments Th1 cytokine production by T effector cells. Consistently, the IL-23 receptor is highly expressed on memory T cells and consists of the IL-12 β1 chain and a novel IL-23R chain (10). IL-23 signals via Jak2 and activates STAT-3, STAT-4, and STAT-5, although STAT-4 activation is significantly weaker as compared with IL-12 signaling. The importance of IL-23 for chronic inflammatory diseases in vivo has been underlined by the recent finding that transgenic mice overexpressing p19 showed multiorgan inflammation and premature death (11).
Despite the physiologic and pathophysiologic importance of IL-12 p40–related cytokines, little is known about the regulation of IL-12 p40 gene expression. Recent data, however, suggest that IL-12 p40 is mainly regulated at the transcriptional level by both chromatin remodeling through Toll-like receptor signaling and promoter transactivation through regulatory transcription factors (12–15). In particular, various studies have demonstrated the binding of strong transcriptional activators such as NF-κB, C/EBP-β, PU.1, and AP-1 to the p40 promoter region in monocytes and macrophages (12, 13, 16–19). Furthermore, a repressor element (denoted GA-12) was recently identified within the p40 promoter that modulates promoter activity in response to IL-4 and PGE2, suggesting that both positive and negative response elements tightly control p40 gene transcription (16).
Since overexpression of IL-12 p40–related cytokines appears to be a key step in the pathogenesis of Th1-mediated autoimmune and chronic inflammatory diseases, new insights into the transcriptional regulation of IL-12 p40 expression in vivo are required for our understanding of the pathogenesis of these diseases. In an attempt toward this goal, we generated transgenic mice expressing firefly luciferase under the control of the wild-type IL-12 p40 promoter or the p40 promoter with a mutated NF-κB target site. We observed a constitutive IL-12 p40 promoter activity in the distal ileum associated with increased expression of IL-12 p40 protein, heterodimeric p40/p19 (IL-23), and binding of NF-κB to its respective target site in the p40 promoter. NF-κB binding was essential for constitutive p40 expression, because transgenic mice with the mutated NF-κB site showed no constitutive luciferase activity in the distal ileum. Further studies identified CD8α–CD11b–CD11c+ lamina propria DCs (LPDCs) as the cellular source of constitutive p40 expression in the distal ileum and showed a local uptake of bacteria by these LPDCs. In summary, we provide evidence that DCs in lamina propria of the distal but not the proximal ileum or colon maintain an excessive pool of p40 protein leading to high local levels of IL-23.
Methods
Cell culture conditions.
Cell lines were obtained from the American Type Culture Collection, Rockville, Maryland, USA. Cells were cultured in RPMI-1640, supplemented with 10% FCS (PAA Laboratories, Linz, Austria), 20 mM HEPES buffer (Life Technologies Ltd., Paisley, United Kingdom), 2 mM L-glutamine (Life Technologies Ltd.), and 1,000 U/ml penicillin/streptomycin (Biochrom AG, Berlin, Germany).
Generation of IL-12 p40 promoter/luciferase transgenic mice.
The p40 promoter/luciferase reporter gene vector p40/pXP1 has been published previously (16). Briefly, the IL-12 p40 promoter was amplified as an 813-bp fragment (–747 to +66) by PCR and cloned into the SmaI site of the promoterless pXP1 luciferase reporter gene vector (20). From the parental p40/pXP1 vector the NF-κB mutant reporter gene construct was obtained by using the Quikchange Site-Directed Mutagenesis Kit (200518; Stratagene, Heidelberg, Germany), according to the manufacturer’s instructions. The mutant primer sequence (top strand) was 5′-GAACTTCTTGAAATTAGCC CAGAAGG-3′. Successful mutagenesis was verified by sequencing of the NF-κB mutant reporter gene construct.
The constructs used for microinjection were obtained by modifying the p40/pXP1 vector as follows: restriction sites were inserted 5′ and 3′ of the pBR322 backbone by site-directed mutagenesis. The vector was cut, and the resulting 4.7-kb IL-12 p40 promoter/luciferase expression cassette was microinjected into pronuclei of fertilized eggs. Subsequently, the eggs were transferred to the oviduct of pseudopregnant FVB mice. The screening of potentially transgenic mice was performed by isolation of DNA from tail biopsies and subsequent PCR analysis with a set of construct-specific primers (5′-ATGTCTGGATCCAAGCTCAG-3′ and 5′-TTGTCACGATCAAAGGACTCTGG-3′). Mice were bred in our animal facility under normal housing conditions. Germfree mice were established as published previously (21).
Isolation and culture of primary cells.
Peritoneal macrophages were isolated as follows. Mice were injected intraperitoneally with 2.5 ml of Brewer’s thioglycollate medium (Becton Dickinson and Co., Bethesda, Maryland, USA). After 3 days, mice were sacrificed by cervical dislocation and peritoneal cells were isolated by flushing the peritoneum with 6 ml of cold PBS. The cells were resuspended in culture medium and were allowed to adhere for 2 hours. Subsequently, nonadherent cells were removed and adherent peritoneal macrophages were analyzed as specified below.
Mouse spleen cells were obtained by flushing the spleen through a 0.4-μm filter with cold PBS and lysing the erythrocytes with ACK cell lysis buffer (Cambrex, East Rutherford, New Jersey, USA). Splenocytes were then further purified using the MACS system (Miltenyi Biotech GmbH, Bergisch-Gladbach, Germany) with immunomagnetic beads specific for Mac-1, CD3, or B220 (Miltenyi Biotech GmbH) and the isolation of splenic macrophages, T lymphocytes, and B lymphocytes, respectively. Cells were more than 95% pure, as determined by FACS analysis.
LPMCs were isolated from resected small intestine specimens by a technique described previously (22). Briefly, after removal of Peyer’s patches, the intestine was opened longitudinally, washed several times in PBS to remove feces and debris, and cut into small pieces. Tissues were incubated at 37°C in PBS supplemented with 0.145 mg/ml DTT and 0.37 mg/ml EDTA for 15 minutes. The tissue was then digested in RPMI-1640 containing 0.15 mg/ml type II collagenase (Worthington, Lakewood, New Jersey, USA) and 0.1 mg/ml DNase (Roche Molecular Biochemicals, Mannheim, Germany) for 75–90 minutes at 37°C on a shaking platform. CD11c+ and CD11c–CD11b+–enriched LPMCs were finally isolated subsequently using CD11c and CD11b microbeads and MACS techniques (Miltenyi Biotech GmbH).
Cells were treated with the following reagents as specified in Results: 100 U/ml mouse IFN-γ (Genzyme Pharmaceuticals, Cambridge, Massachusetts, USA), 1 μg/ml bacterial LPS (Sigma-Aldrich, St. Louis, Missouri, USA), 0.001% fixed Staphylococcus aureus cells (SACs) (Pansorbin cells; Calbiochem, La Jolla, California, USA), 5 ng/ml mouse IL-4 (PharMingen, San Diego, California, USA), 50 ng/ml PMA (Sigma-Aldrich), and 1 μg/ml ionomycin (Sigma-Aldrich).
Isolation of proteins.
Extraction of nuclear proteins was carried out by the method of Schreiber et al. (23). Gut proteins were isolated by homogenization of gut samples in mPER protein extraction reagent (Pierce Biotechnology, Rockford, Illinois, USA) supplemented with Complete proteinase inhibitor (Roche Molecular Biochemicals). Protein concentrations were measured with a protein assay kit (Bio-Rad, Munich, Germany).
Electrophoretic mobility-shift assay.
Oligonucleotides for electrophoretic mobility-shift assay (EMSA) were synthesized, annealed, gel purified, and end labeled with [32P] γ-ATP (>5,000 Ci/mmol; Amersham Life Sciences Inc., Arlington Heights, Illinois, USA) using T4 polynucleotide kinase (New England Biolabs Inc., Beverly, Massachusetts, USA). Twenty-five thousand counts per minute radiolabeled p40 NF-κB-probe (5′-GAACTTCTTGAAATTCCCCCAGAAGG-3′) was added to the binding reaction that also contained 1 μg synthetic DNA duplex of poly(dIdC) (Pharmacia Biotech Inc., Piscataway, New Jersey, USA), 3 μg nuclear proteins, and binding buffer (25 mM HEPES, pH 7.5, 150 mM KCl, 5 mM DTT, 10% glycerol). For supershift assays, 2 μg of Ab’s specific for NFATc or NF-κB p50, p65, c-Rel, and RelB (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) were used. Complex formation was allowed to proceed for 30 minutes at room temperature. Finally, the complexes were separated from unbound DNA by native PAGE on 5% gels. The gels were exposed to Kodak MS films on intensifying screens at –80°C.
Isolation of mRNA and RT-PCR.
Total RNA was isolated using TRIzol (Sigma-Aldrich) according to the manufacturer’s recommendations. Reverse transcription into complementary DNA was performed using the MMLV reverse transcriptase (Life Technologies Inc.). PCR was performed using the RedTaq PCR reagents (Sigma-Aldrich) and the following primers derived from previously published sequence data: IL-12 p40, 5′-GGAGACCCTGCCCATTGAACT-3′ and 5′-CAACGTTGCATCCTAGGATCG-3′; IL-23 p19, 5′-TGCTGGATTGCAGAGCAGTAA-3′ and 5′-CTGGAGGAGTTGGCTGAGTC-3′; IL-17, 5′-TGGCGGCTACAGTGAAGGCA-3′ and 5′-ACAATCGAGGCCACGCAGGT-3′; and β-actin, 5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′ and 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′. PCR products were analyzed on 1% agarose gels.
Western blot analysis.
For IL-12 p40 Western blot analysis, intestinal specimens were snap-frozen in liquid nitrogen, and proteins were isolated as described above. Proteins (50 μg) were separated on Laemmli SDS-PAGE gels and transferred to nitrocellulose membranes (Schleicher & Schuell GmbH, Dassel, Germany) by semidry blotting. A nonreducing sample buffer (Carl Roth GmbH, Karlsruhe, Germany) was chosen to detect p40, p40/p19, and p40/p35 at the same time. Nonspecific binding sites were blocked with PBS, 5% milk powder, 0.1% Tween-20, followed by sequential incubation in 0.2 μg/ml rabbit anti–mouse IL-12 p40 (Santa Cruz Biotechnology Inc.) and 1:2,000 horseradish preoxidase–labeled anti–rabbit IgG (Santa Cruz Biotechnology Inc.). Detection of IL-12 p40–specific complexes was performed with the ECL system (Amersham Life Sciences Inc.) and Biomax MR films (Eastman Kodak Co., Rochester, New York, USA). Densitometry of Western blots was performed using the ChemiImager 5500 software (Alpha Innotech, San Leandro, California, USA). To detect IL-23 p19/p40, a recently described specific IL-23 Ab (24) was used that was generated by immunization of rabbits with a purified glutathione S-transferase fusion protein. Detection of p19/p40 complexes was performed analogous to the p40 Western blotting procedure described above.
ELISA for IL-12 p40 and p70.
To measure IL-12 protein production, 106 primary monocytes per well were seeded out in 1-ml culture medium in triplicate in 48-well tissue-culture plates and incubated at 37°C in humidified 5% CO2 atmosphere in the presence or absence of different stimuli, as indicated above. After 48 hours, cell-free culture supernatants were removed and assayed for p70 and p40 concentration using ELISA (22).
Immunohistochemistry.
Cryosections or cytospins of CD11c-enriched LPMCs (100,000 cells per slide) were analyzed by immunofluorescence or diaminobenzidine (DAB) staining. For cytospins, CD11c-positive cells were isolated from the terminal ileum as described above. Immunofluorescence was performed using the tyramide signal amplification Cy3 system (PerkinElmer Life Sciences, Heidelberg, Germany) and a fluorescence microscope (Olympus fluorescence microscope; Olympus America Inc., Melville, New York, USA). In brief, cryosections were fixed in ice-cold acetone for 10 minutes followed by sequential incubation with methanol, avidin/biotin (Vector Laboratories, Burlingame, California, USA), and protein-blocking reagent (DAKO Corp., Wiesbaden, Germany) to eliminate unspecific background staining. Slides were then incubated overnight with primary Ab’s specific for IL-12 p40, CD11c, CD11b (Santa Cruz Biotechnology Inc.), or firefly luciferase (Europa Bioproducts Ltd., Wicken, United Kingdom). Subsequently, the slides were incubated for 30 minutes at room temperature with biotinylated secondary Ab’s (Dianova, Darmstadt, Germany). All samples were finally treated with streptavidin-HRP and stained with tyramide (Cy3 or FITC), according to the manufacturer’s instructions (PerkinElmer Life Sciences). Before examination, the nuclei were counterstained with Hoechst 3342 (Molecular Probes Inc., Eugene, Oregon, USA).
For luciferase staining with DAB, cryosections were fixed in ice-cold acetone for 10 minutes followed by sequential incubation with methanol, 3% H2O2, avidin/biotin (Vector Laboratories), and protein-blocking reagent (DAKO Corp.) to eliminate unspecific background staining. Slides were then incubated overnight with an Ab specific for firefly luciferase (Europa Bioproducts Ltd.). Subsequently, the samples were incubated with biotinylated secondary Ab’s (Sigma-Aldrich) for 1 hour at room temperature. All samples were finally treated with streptavidin and stained with the DAB chromogen according to the manufacturer’s instructions (DAKO Corp.). Before examination, the nuclei were counterstained with hematoxylin.
FISH of bacterial rRNA.
FISH hybridization of bacterial rRNA on glass slides was performed as described previously (25, 26). The universal eubacterial oligonucleotide probe EUB-338 (GCT GCC TCC CGT AGG AGT) and the control probe NONEUB-338 (CGA CGG AGG GCA TCC TCA) complementary to EUB-338 to exclude nonspecific binding of the probes were synthesized and 5-prime labeled (Metabion, Planegg-Martinsried, Germany) with the fluorochrome Cy3 or FITC. Slides with cryosections were fixed in PFA, washed in PBS, and incubated with 25 ng of each oligonucleotide added in 50 μl of hybridization buffer containing 20% formamide for 90 minutes at 46°C before washing with the same stringency. Signal specificity was demonstrated by using Escherichia coli as positive control with the EUB-338 probe and by comparing with the nonrelated Cy3-labeled control NONEUB-338 oligonucleotide. For some experiments, immunofluorescence staining was performed additionally.
Transient transfections and reporter gene analysis.
Ten micrograms of the p40/pXP1 reporter gene vector along with 2 μg of a β-galactosidase expression vector were transfected into 107 cells using the DEAE transfection method. After 18 hours, the cells were stimulated as described above. The stimulation was allowed to proceed for 8 hours before the cells were harvested, washed in PBS, and lysed in cell lysis buffer (Promega Corp., Madison, Wisconsin, USA). For the analysis of reporter gene transgenic mice, tissue samples were directly homogenized in cell lysis buffer (Promega Corp.). Luciferase activity was measured as light emission over a period of 10 seconds after addition of luciferase assay buffer (Promega Corp.) with a standard luminometer (Sirius; Berthold Detection Systems GmbH, Pforzheim, Germany). Luciferase activity was normalized to the β-galactosidase expression level of the lysate or where applicable to the protein content of the solution.
Statistical analysis.
Data from transfection experiments and Western blot densitometry were analyzed by the Student t test using the software program Excel.
Results
IL-12 p40 promoter luciferase transgenic mice display a cell type and activation-specific expression of the transgene.
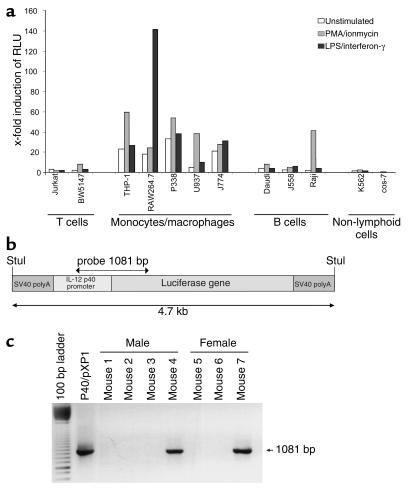
To analyze functional aspects of the IL-12 p40 promoter, we performed transient transfection assays using reporter gene vectors. In these studies, we cloned an 800-bp fragment of the human IL-12 p40 promoter upstream of the luciferase reporter gene of the promoterless pXP1 vector. The resulting construct, denoted p40/pXP1, was then transfected into cell lines of various origin to analyze activity and inducibility of the promoter in response to LPS and IFN-γ or PMA and ionomycin. The IL-12 p40 promoter was found to be active and inducible only in cell lines of myeloid origin, but not in B or T cell lines or other nonlymphoid cell lines (Figure 1a), except for the Raji cell line, an EBV-transformed B cell line capable of producing IL-12 p40 (2). Thus, the 800-bp promoter fragment confers cell type and stimulation-specific activation of the IL-12 p40 promoter in transient transfection assays.
Figure 1.
Generation of IL-12 p40 promoter/luciferase transgenic mice. (a) Reporter gene analysis of the p40/pXP1 reporter gene construct in various cell lines. The IL-12 p40 promoter was cloned as a Stul restriction enzyme fragment upstream of the luciferase reporter gene into the pXP1 vector yielding the p40/pXP1 vector. P40/pXP1 was transfected in various cell lines using the DEAE transfection method. Cells were left untreated or were stimulated for 8 hours with PMA/ionomycin or LPS/IFN-γ, as indicated. Data represent average values of two independent experiments and are presented as fold induction of relative light units (RLU) as compared with the transfection of the empty pXP1 reporter gene vector. (b and c) Map of the luciferase expression cassette and generation of IL-12 p40 promoter/luciferase transgenic mice. A 4.7-kb IL-12 p40 promoter/luciferase expression cassette was used for the generation of transgenic animals. The screening of transgenic mice was performed by isolation of tail DNA and subsequent PCR analysis with a set of construct-specific primers, giving rise to a single band of 1,081 bp. Several founder mice were identified by PCR that were used for subsequent analysis.
Although transient transfections are a very powerful tool for the functional analysis of regulatory sequences, the reporter gene vector remains episomal, and they thus do not necessarily reflect the complex biochemical processes involved in endogenous gene regulation in the nucleus. To analyze the activity of the IL-12 p40 promoter in cells and whole organs in vivo, bacterial sequences were removed from the p40/pXP1 reporter gene vector and the resulting construct (Figure 1b) was injected into the pronuclei of fertilized eggs. Twelve founder FVB mice proved to be transgenic by PCR analysis (Figure 1c). These founder mice and their littermates were phenotypically normal and showed no organ pathology (data not shown).
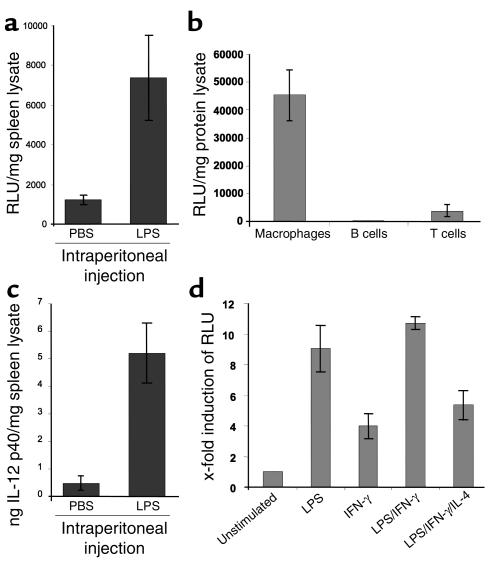
To analyze reporter gene transgenic mice, animals were injected intraperitoneally with a sublethal dose of LPS and subsequently screened for luciferase expression. We found a strong upregulation of reporter gene activity and IL-12 p40 protein expression in the spleen compared with the control PBS-injected animals (Figure 2a), demonstrating the capacity of spleen cells to activate the IL-12 p40 promoter upon LPS stimulation. Next, we isolated T cells, B cells, and macrophages from spleens of IL-12 p40 promoter/luciferase transgenic mice. The cells were simulated with anti-CD3/anti-CD28 or LPS/SACs, respectively, and subjected to reporter gene analysis. High luciferase expression was detected in splenic macrophages, but not in T or B lymphocytes (Figure 2b), suggesting macrophage-specific expression of the transgene in the spleen. Furthermore, peritoneal macrophages showed a tenfold induction of reporter gene expression upon stimulation with LPS and LPS/IFN-γ, but only moderately with IFN-γ alone (Figure 2c). Such induction was suppressed by the addition of IL-4, a known inhibitor of IL-12 expression (27, 28), to the culture medium of stimulated peritoneal macrophages. Taken together, these findings demonstrate that the 800-bp promoter fragment is sufficient to drive inducible, cell type, and activation-specific expression in a transgenic mouse model in vivo.
Figure 2.
Cell-specific and stimulation-dependent expression of the transgene. (a) Upper panel: Luciferase expression in the spleen of control and LPS-injected transgenic mice. Mice were injected intraperitoneally with 200 μg of LPS. After 4 hours, splenic luciferase expression was assessed. Data are shown as mean values of three mice per group ± SD. LPS injection led to increased luciferase activity indicating LPS-dependent p40 promoter activity in spleen cells. Lower panel: IL-12 p40 protein levels in the spleen of the same animals as above were determined by ELISA analysis of splenic cell lysates. Mean values ± SD are shown. (b) Luciferase expression in splenic macrophages, T cells, and B cells of transgenic mice upon stimulation with LPS/SAC (macrophages, B cells) or anti-CD3 plus anti-CD28 Ab’s (T cells) for 24 hours. Date represent mean values ± SD of three mice per group. (c) Luciferase activity in peritoneal macrophages of transgenic mice. Cells were isolated as described in Methods and stimulated for 24 hours as indicated. The data represent mean values ± SD of three independent experiments.
The IL-12 p40 promoter is constitutively activated in the terminal ileum of healthy mice.
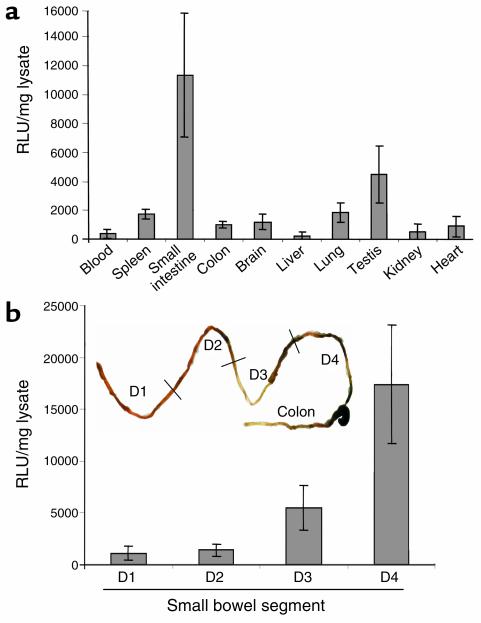
In subsequent studies, we analyzed constitutive luciferase activity in a variety of organs from healthy transgenic mice. Whereas most tissues such as spleen and lung (Figure 3a) of the transgenic mice showed only little reporter gene activity, moderate expression was found in testis consistent with the known IL-12 production and STAT-4 expression in this organ (29, 30). Surprisingly, all founder lines displayed high luciferase activity in the small intestine but not in the colon, thus providing evidence for constitutive IL-12 p40 promoter activity in the former organ. Furthermore, Peyer’s patch–free small bowel preparations showed high luciferase activity, excluding the possibility that constitutive promoter activity was mainly localized in Peyer’s patches. To verify whether the constitutive IL-12 p40 promoter activity was distributed equally within the small intestine, the organ was removed, and Peyer’s patches were excised. The small intestine was then divided into four parts of equal length (proximal D1 to distal D4), and luciferase activity was determined in the gut homogenates. Interestingly, constitutive luciferase activity was restricted to the distal small intestine (D3 and D4), whereas the proximal samples (D1, D2) showed only basal levels of activity (Figure 3b). Thus, the distal but not the proximal small intestine shows high constitutive IL-12 p40 promoter activity in vivo.
Figure 3.
Analysis of various organs from IL-12 p40 promoter/luciferase transgenic mice. (a) Luciferase activity in different tissues of untreated, healthy transgenic mice. Luciferase expression was measured in a standard luminometer after homogenization of whole organs from transgenic mice. Results were normalized to the protein content of the homogenates and are presented as relative light units (RLU) per milligram of protein extract ± SD of five independent experiments. All founder lines of IL-12 p40 promoter/luciferase transgenic mice showed the highest constitutive activity in the small intestine, whereas little activity was seen in the spleen, liver, kidney, heart, and colon. (b) Luciferase activity in different segments of the small intestine from untreated, healthy transgenic mice upon removal of the Peyer’s patches. The small intestine was divided into four segments of equal length (from the proximal segment D1 to the distal segment D4), and luciferase expression was measured in a standard luminometer after homogenization of gut samples from transgenic mice. Results were normalized to the protein content of the homogenates and are presented as relative light units per milligram of protein extract ± SD of three independent experiments. All founder lines of IL-12 p40 promoter/luciferase transgenic mice showed the highest constitutive activity in the distal segment of the small intestine, whereas little activity was seen in proximal segments.
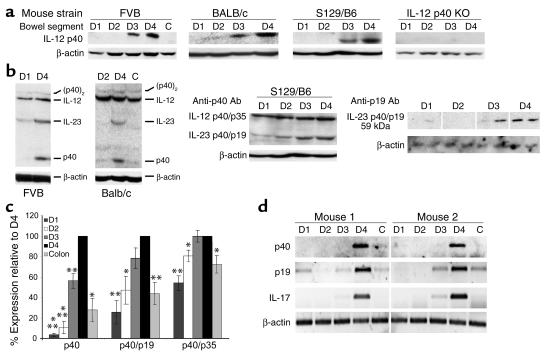
To verify whether this constitutive promoter activity in the distal small bowel gives rise to increased local IL-12 p40 protein levels, we performed Western blot analysis of D1–D4 protein extracts as well as from colon samples of wild-type and transgenic FVB mice. Accordingly, tissue proteins were isolated and subjected to Western blot analysis using an Ab specific for murine IL-12 p40. Consistent with the reporter gene data, significantly higher levels of monomeric p40 protein were detected in distal samples of the small intestine than in proximal samples or colon samples (Figure 4a). To rule out the possibility that this could be a strain-specific observation in FVB mice, we performed Western blot analysis of gut samples from BALB/c and S129/B6 mice. Consistent with the data in FVB mice, we observed high levels of monomeric IL-12 p40 protein only in the distal samples D3 and D4 in both strains (Figure 4a).
Figure 4.
Nonreducing Western blot analysis of gut samples from the small intestine (D1–D4) and colon (C). Samples were analyzed for p40, p40/p19 (IL-23), p40/p35 (IL-12), and p40 homodimer (p40)2 levels. β-Actin staining served as loading control. (a) Western blot for monomeric p40 protein using extracts from different mouse strains. Extracts from IL-12 p40 S129/B6 KO mice served as negative control (far right panel). (b) Analysis of higher molecular weight p40 complexes in FVB and BALB/c mice (left panels) and S129/B6 mice (middle panel, same blot as in a). A marked increase of IL-23 p40/p19, but not IL-12 p40/p19 levels was noted in the distal small bowel as compared with the proximal segments of the small intestine. Right panel: These data were confirmed by Western blot analysis using an IL-23–specific AB (four mice per group, two shown). (c) Densitometry of above p40 Western blots. Data are reported as percentage of expression as compared with D4 (100%). Data represent average values ± SD of six to eight mice per group. Statistically significant differences (*P < 0.05, **P < 0.01, ***P < 0.001) are indicated. (d) RT-PCR analysis of RNA isolated from the small (D1–D4) and large (C) intestine of healthy FVB mice. A marked upregulation in D4 as compared to the proximal small intestine and colon was seen for the mRNAs of p19 and p40, but, importantly, also for the mRNA of IL-17, a recently identified target gene of IL-23 in memory T cells (56).
Interestingly, signals corresponding to the p40/p19 heterodimer (IL-23) were also strongly increased in samples from the terminal ileum as compared with the proximal samples and colonic samples, as determined by Western blot analysis using a p40-specific Ab in three different strains of mice (Figure 4b; left panels and middle panel). This finding was confirmed by additional Western blot experiments with an IL-23–specific Ab demonstrating high constitutive expression of IL-23 in the terminal ileum as compared with the proximal small intestine (Figure 4b; right panel). In contrast, signals corresponding to the IL-12 p40/p35 heterodimer as well as levels of p40 homodimers were almost comparable between proximal and distal samples and colonic samples (Figure 4, b and c). Furthermore, both p19 and p40 mRNA levels were strongly increased in the terminal ileum as compared with the proximal small bowel and colon, as determined by RT-PCR (Figure 4d). These findings indicate a constitutive activity of the IL-12 p40 promoter in the distal ileum of healthy mice giving rise to an excess of p40 mRNA, monomeric p40 protein, and IL-23.
Intestinal IL-12 p40 is mainly produced by CD11c+ LPDCs.
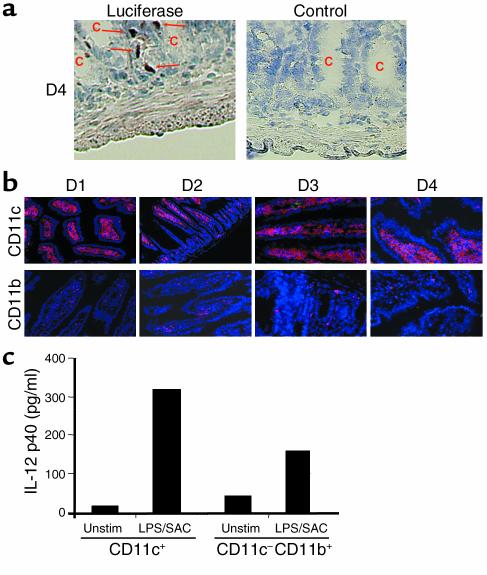
To identify the cellular source of intestinal IL-12 p40 promoter activity, immunohistochemistry studies for luciferase expression were performed. Interestingly, luciferase positive cells in transgenic animals were seen mainly in the lamina propria below the crypts, suggesting that the cells producing IL-12 p40 constitutively reside in the lamina propria (Figure 5a). Accordingly, we attempted to detect CD11b+ macrophages and CD11c+ DCs as known producers of IL-12 p40 in the lamina propria by immunohistochemistry. Whereas only a limited number of lamina propria cells stained positive for CD11b, a major cell population in the lamina propria stained positive for the CD11c, suggesting the existence of LPDCs (Figure 5b). To assess the capacity of both cell populations to produce IL-12 p40, LPMCs were isolated from the small intestine of healthy mice and enriched for CD11c+ DCs and CD11c–CD11b+ macrophages using immunomagnetic beads. Isolated cells were left untreated or were stimulated with LPS/SACs for 24 hours followed by analysis of supernatants for IL-12 p40. As shown in Figure 5c, both cell populations produced IL-12 p40 in response to LPS/SAC stimulation, although CD11+ DCs produced higher amounts than CD11b+ macrophages. These data are consistent with a model in which CD11c+ cells of the small intestine produce large amounts of IL-12 p40 in vivo. No statistically significant differences could be observed in the overall number of CD11c+ cells when samples within the small intestine (D1–D2 versus D3–D4) were compared (not shown), excluding the possibility that the differences in p40 levels between the proximal and distal small bowel are simply due to changes in the local number of producer cells. A comparison of the small intestine with the colon, however, demonstrated a strongly reduced number of CD11c+ cells in colon cross sections (Figure 6a). Moreover, whereas DCs in the small intestine were located in large numbers in the lamina propria, colonic DCs were found mainly in lymphoid follicles and subepithelial regions.
Figure 5.
IL-12 p40 is produced largely by CD11c+ DCs located below the crypts in the lamina propria. (a) Cryosections of transgenic mice were analyzed by immunohistochemistry for luciferase expression. One representative staining for luciferase in the small intestine (D4) of a transgenic animal out of four is shown (left panel). Luciferase-expressing cells were mainly seen in the lamina propria below the crypts (arrows). No staining was seen in sections from transgenic mice treated with an isotype control Ab right panel), the proximal segments of the small intestine (D1, D2), and in healthy nontransgenic control mice (not shown). (b) Detection of CD11c+ and CD11b+ cells in the lamina propria of transgenic mice. More CD11c+ than CD11b+ cells were detected in the lamina propria, suggesting that many cells in the lamina propria of healthy mice carry surface markers of DCs. No differences in the staining patterns of CD11b+ and CD11c+ cells were noted between the proximal and the distal segments of the small bowel. (c) IL-12 p40 cytokine levels in supernatants from CD11c+ enriched DCs and CD11c–CD11b+ enriched macrophages isolated from the lamina propria of healthy mice. Lamina propria cells were isolated as described in Methods and purified using the MACS system. To measure IL-12 p40 protein production, 500,000 cells/well were seeded out in 1 ml culture medium in triplicate and incubated in the presence or absence of LPS/SAC. After 24 hours, supernatants were removed and assayed for p40 concentration by ELISA. Unstim, unstimulated.
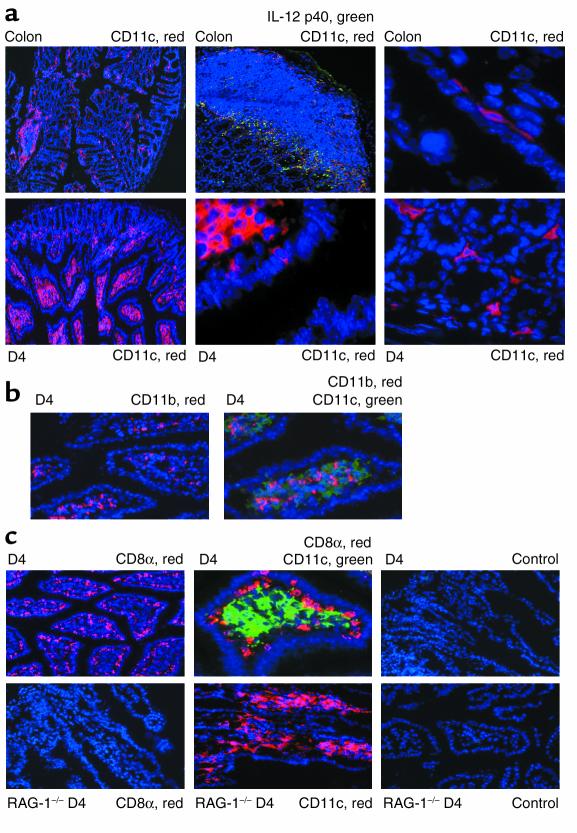
Figure 6.
LPDCs are differentially located in the small intestine as compared with the colon and are largely CD11b–CD8α–. Location of LPDCs in the colon and small bowel. Cryosections were analyzed by immunofluorescence using the tyramide signal amplification Cy3 or FITC system and fluorescence microscopy. Cryosections were stained for CD11c, IL-12 p40, CD11b, and CD8a, as indicated. Nuclei were counterstained in blue. (a) Differential localization of LPDCs in the colon (upper panels) and small intestine (lower panels). The lamina propria of the small bowel contained many CD11c+ LPDCs as well as some CD11c+ subepithelial cells, whereas few LPDCs were seen in the colon. (b) Staining for CD11b+ (red), CD11c+ (green), and CD11b+ plus CD11c+ (yellow) cells in the distal small bowel of healthy mice, showing that most CD11c+ LPDCs do not coexpress CD11b. (c) Staining for CD11c (green), CD8α (red), and CD11c plus CD8α (yellow) cells in the distal small bowel (D4) of healthy FVB mice (upper panels), showing that most CD11c+ LPDCs do not coexpress CD8a. CD11c+ LPDCs did express MHC class II molecules, as determined by immunohistochemistry, however (not shown). No staining for CD8α was noted in T cell–deficient RAG knockout mice (RAG–/–; lower panels), although many CD11c+; LPDCs were detected in these animals.
Recently, three functionally different subpopulations of CD11c+ DCs have been identified in murine Peyer’s patches: CD11b+CD8α–, CD11b–CD8α+, and CD11b–CD8α– cells (31, 32). To investigate whether the LPDCs in the small bowel might carry these markers, too, immunofluorescence staining of cryosections was performed. Accordingly, consecutive cryosections were stained for CD11c and CD11b. As shown in Figure 6b, few CD11c+ cells stained positive for CD11b. Furthermore, as shown in Figure 6c (upper panels), Ab’s to CD8α stained a cell population located in the subepithelium rather than in the lamina propria. In contrast, this cell population was not detectable in cryosections from the small intestine of RAG-1 knockout mice lacking T cells and B cells (Figure 6c, lower left panel), suggesting that these cells represent intraepithelial lymphocytes rather than LPDCs. Thus, IL-12 producing LPDCs are largely negative for CD11b and CD8α and therefore resemble a double-negative DC population similar to that recently described in Peyer’s patches.
Binding of NF-κB p50/p65 to the IL-12 p40 promoter is upregulated in the terminal ileum.
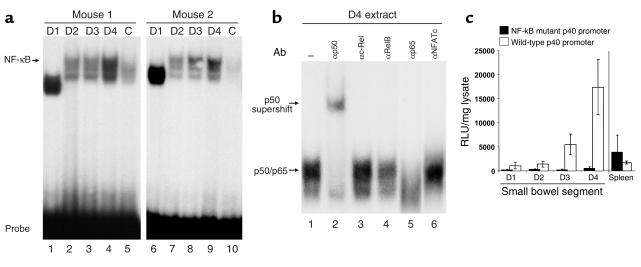
The transcription factor NF-κB is a key regulator of inducible IL-12 p40 promoter activity and is known to be activated by bacterial products by Toll-like receptor signaling (33, 34). To analyze whether binding of this transcription factor to its site in the IL-12 p40 promoter is enhanced in the distal versus the proximal small bowel, we performed EMSA experiments. Therefore, protein extracts of D1–D4 were incubated with an oligonucleotide corresponding to the IL-12 p40 promoter NF-κB site, and protein/DNA complexes were analyzed by native PAGE. As shown in Figure 7a, complex binding to the p40 NF-κB site was strongly increased in extracts of the distal versus the proximal small intestine. Supershift analysis demonstrated that complexes were composed mainly of the NF-κB subunits p50 and p65 (Figure 7b), since the addition of Ab’s specific for the these proteins led to a retarded migration (p50) or complete abrogation of the complex (p65).
Figure 7.
Binding of NF-κB to the IL-12 p40 promoter is upregulated in the distal small intestine. (a) EMSA analysis: 40 μg of tissue lysate from the small intestine (D1–D4) of FVB mice was incubated with a radiolabeled probe corresponding to the IL-12 p40 promoter NF-κB site. Protein/DNA complexes were analyzed on a 5% native polyacrylamide gel. Two representative experiments out of four are shown. (b) For supershift analysis, D4 protein lysate was preincubated with 2 μg of Ab’s specific for the indicated transcription factors prior to the addition of radiolabeled probe. The locations of the p50/p65 complex and the p50 supershift are indicated. (c) Constitutive luciferase activity in samples D1 (proximal) to D4 (distal) of the small bowel and spleen of IL-12 p40 wild-type promoter/transgenic mice and NF-κB mutant promoter/luciferase transgenic mice. Luciferase expression was measured in a standard luminometer after homogenization of organ samples of transgenic mice. Results were normalized to the protein content of the homogenate and are presented as relative light units per milligram of protein extract ± SD of three independent experiments with independent founder mice. A striking reduction of luciferase activity was noted in the distal small intestine of mice carrying a loss-of-function mutation in the NF-κB–binding site of the IL-12 p40 promoter as compared with transgenic mice carrying the wild-type p40 promoter upstream of the luciferase gene. In contrast, no reduction of luciferase activity in spleen cell lysates was noted.
To functionally analyze the role of NF-κB for constitutive p40 promoter activation in the distal small intestine, reporter gene transgenic mice were generated carrying a loss-of-function mutation in the NF-κB target site of the IL-12 p40 promoter. This mutation has been shown previously to prevent binding of NF-κB to its target site in the p40 promoter (16). Healthy transgenic mice carrying a mutant p40 promoter construct upstream of the luciferase gene were sacrificed, and the gut and spleen were analyzed for luciferase activity. No constitutive promoter activity was measured in the samples D1–D4 from the small intestine (Figure 7c), providing evidence for a key role of NF-κB for constitutive p40 promoter activity in the distal ileum in vivo.
Bacteria are necessary for p40 expression in the distal ileum and are actively taken up from the intestinal lumen by LPDCs.
NF-κB is known to be activated via Toll-like receptor-signaling by bacterial antigens. To further analyze whether bacteria play a role for high p40 gene expression in the distal ileum, Western blot analysis for p40 was performed using extracts from the proximal and distal ileum of healthy mice bred under germfree conditions. Interestingly, no increased p40 signals in the distal ileal samples were noted in germfree mice as compared with mice housed under conventional conditions (Figure 8a; left panels). Since the number of CD11c+ LPDCs was not significantly reduced in germfree mice (Figure 8a; right panel), these data suggested that bacterial antigens or products induce p40 promoter activation and p40 protein production in LPDCs in vivo.
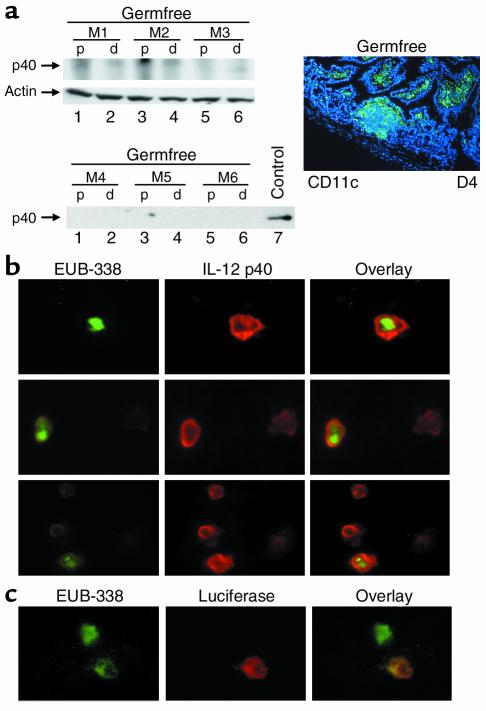
Figure 8.
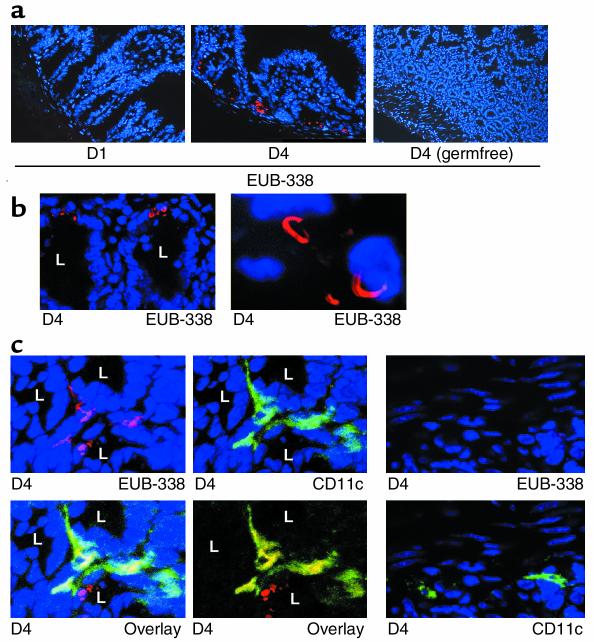
Bacteria in the distal small intestine drive constitutive intestinal IL-12 p40 expression. (a) Western blot for IL-12 p40 of gut samples (D1 plus D2: p; D3 to D4: d) derived from three mice (M1, M2, M3) bred under germfree conditions (control), no constitutive p40 protein expression was seen under germfree conditions. The number of CD11c+ LPDCs in the terminal ileum of germfree mice was comparable to that in the ileum of mice bred under specific pathogen-free conditions, as demonstrated by immunofluorescence analysis (right panel) and quantification of fluorescence-positive cells (not shown). (b) FISH analysis on CD11c-enriched lamina propria cells from the distal small intestine of healthy FVB mice using a universal, FITC-labeled eubacterial oligonucleotide probe (EUB-338) and simultaneous immunocytochemical analysis of IL-12 p40 expression. Three representative high-power fields are shown. Image arithmetic (overlay) demonstrated colocalization of bacteria and p40 protein expression in CD11c-enriched lamina propria cells. The CD11c-enriched lamina propria cells did not express CD8α or CD11b, as shown by double-staining analysis (not shown). (c) FISH analysis of CD11c enriched lamina propria cells of the distal small intestine of healthy p40 promoter transgenic FVB mice using a universal, FITC-labeled eubacterial oligonucleotide probe (EUB-338) and simultaneous immunohistochemical analysis of luciferase expression. Colocalization of bacteria and luciferase protein expression was noted in lamina propria cells. No staining was observed using a control probe (NONEUB-338) complementary to EUB-338 to exclude nonspecific binding (not shown).
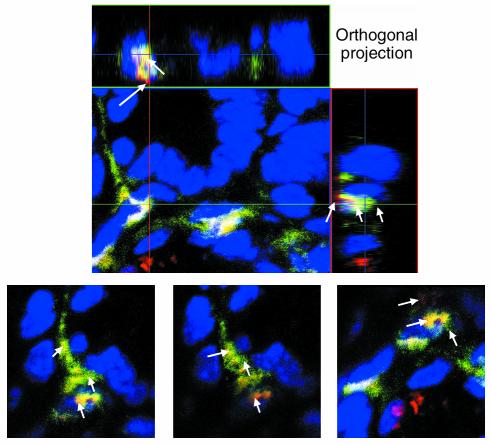
To directly test the concept that bacterial antigens induce p40 production in LPDCs, we performed FISH analysis on CD11c-enriched lamina propria cells with the FITC-labeled oligonucleotide EUB-338 directed to a RNA sequence present in the rRNA of all eubacteria. Interestingly, FISH analysis and immunocytochemical studies showed colocalization of bacteria with p40 protein in CD11c-enriched lamina propria cells from the distal small bowel (Figure 8b). Furthermore, EUB-338–positive cells in the terminal ileum of p40 promoter transgenic mice were found to express luciferase (Figure 8c), suggesting that bacterial antigens drive p40 production in vivo. To further analyze whether p40-producing LPDCs may take up bacterial antigens from the intestinal lumen in healthy mice in vivo, we performed FISH analysis of gut cryosections with the Cy3-labeled oligonucleotide EUB-338. As shown in Figures 9, a and b, the probe for eubacteria stained positive in the crypts of the distal small intestine (D4), but not in the proximal small intestine (D1), providing evidence for a higher concentration of bacteria in the crypts of the terminal ileum. To analyze whether CD11c+ LPDCs stain positive for bacterial rRNA, double and triple fluorescence experiments were performed by staining cryosections from the distal ileum for CD11c (FITC) and EUB-338 (Cy3) (Figure 9 and 10). In addition to hybridizing to bacteria in the gut lumen, EUB-338–stained cells in the lamina propria of D4 that were CD11c positive, suggesting that these LPDCs directly sample bacterial antigens from the intestinal lumen in vivo, resulting in p40 protein production. Further quantification showed that less than 10% of LPDCs stained positive for EUB-338, whereas the majority of CD11c LPDCs showed no staining (Figure 9d).
Figure 9.
Bacteria in the distal small intestine are actively taken up by LPDCs in close proximity to the crypts. (a) FISH analysis using a universal, Cy3-labeled eubacterial oligonucleotide probe (EUB-338). Whereas marked staining was seen in the distal small bowel (D4), no staining was detected in the proximal samples (D1). Furthermore, no staining was observed using a control probe (which was NONEUB-338) complementary to EUB-338 to exclude nonspecific binding (not shown). (b) Analysis of the above FISH experiment by confocal laser microscopy. Higher magnifications showed that the bacteria in the terminal ileum (D4) of healthy mice typically had a curved appearance. L, crypt lumen. (c) Immunofluorescence triple-staining analysis of CD11c (FITC: green), Hoechst 3342 (blue), and FISH (Cy3: red), using confocal laser microscopy in D4. CD11c+ plus FISH double-positive cells (yellow, image arithmetic overlay) were identified in the distal small intestine (D4), suggesting uptake of bacteria by LPDCs in vivo. (d) Immunofluorescence triple-staining analysis of CD11c (FITC: green), Hoechst 3342 (blue), and FISH (Cy3: red) using confocal laser microscopy in D4. The majority of CD11c -positive LPDC cells were negative for EUB-338.
Discussion
The ability to produce IL-12 p40–related cytokines such as IL-12 p35/p40 and IL-23 p19/p40 is a fundamental property of macrophages and DCs in various infectious and autoimmune diseases (22, 35–43). Little is known about the transcriptional regulation of p40 gene expression in vivo, however. The data presented in this manuscript provide strong evidence for an unexpected constitutive transcriptional activity of the IL-12 p40 promoter and IL-23 p19/p40 production in the distal ileum of healthy mice. LPDCs were identified as main producers of p40 in response to the bacterial flora by activation of NF-κB. These results implicate important functional differences between the mucosal immune systems of the colon and the proximal and distal parts of the small intestine.
To gain insights into the transcriptional regulation of the IL-12 p40 promoter in various organs and cell types in vivo, we have generated IL-12 p40 promoter/luciferase transgenic mice. Importantly, all transgenic founder lines were phenotypically normal, and the transgene expression correlated with that of the endogenous IL-12 p40 gene. Consistently, IL-12 p40 promoter activity measured as luciferase expression was highly inducible by LPS and LPS/IFN-γ in isolated peritoneal macrophages. Furthermore, analysis of p40 promoter activity in isolated spleen cells of transgenic mice demonstrated an inducible expression in macrophages but no significant reporter gene activity in B or T lymphocytes. Our data therefore provide strong evidence that an 800-bp fragment of the IL-12 p40 promoter is sufficient to confer cell type–and stimulus-specific promoter activity in vivo. This finding is consistent with data from transient transfection assays performed by different groups, including ours (12, 13, 16, 44).
Surprisingly, we found high constitutive luciferase activity in samples from the small bowel, but only basal levels in the colon and various other organs such as spleen and lung. Within the intestine, high luciferase activity was detected in the distal small bowel of transgenic mice only (even upon removal of the Peyer’s patches). In contrast, only basal levels of activity were found in proximal samples and the colon, suggesting a constitutive transcriptional activity of the IL-12 p40 promoter in the distal ileum of healthy mice. Furthermore, Western blots using extracts from three different mouse strains showed high levels of monomeric IL-12 p40 protein only in samples from the distal small intestine but not in proximal samples. These data suggest that p40 is produced in large excess in the terminal ileum by transcriptional mechanisms. A part of this excess p40 protein may heterodimerize with p19 to form IL-23, which would explain the significantly higher level of IL-23 p19/p40 in the distal ileum when compared with proximal samples and colonic samples, as determined by Western blot analysis. The differential expression of IL-12 p35/p40 and IL-12 antagonizing p40 homodimers versus IL-23 p19/p40, however, implicates the existence of additional regulatory mechanisms. In the case of IL-23 this mechanism is most likely related to local tissue levels of the p40-binding partner p19. Indeed, p19 mRNA expression in the terminal ileum was upregulated as compared with the proximal small bowel, suggesting that high local tissue levels of p19 and p40 in the former region result in IL-23 p19/p40 protein production. In any case, however, these differences in tissue levels of IL-12 p40 and IL-23 may have important functional consequences, since IL-23 (in contrast to IL-12 p35/p40) induces the proliferation of mouse memory (CD4+CD45RBlow) T cells and acts more broadly as an end-stage effector cytokine in chronic inflammation through direct actions on macrophages (45). One may therefore speculate that higher levels of IL-23 in the terminal ileum may serve to expand and activate a pool of memory T cells and macrophages in this part of the gut upon initiation of a specific immune response. This may lead to a predisposition for chronic inflammation with continuous activation of memory cells and macrophages in the terminal ileum.
Recent data suggest that DCs in the intestine and elsewhere in the body control the critical balance between self-tolerance and the development of chronic inflammatory and autoimmune diseases (31, 46–50). This function probably involves both endocytosis of apoptotic epithelial cells and direct sampling of luminal bacteria, followed by antigen presentation to intestinal T cells (46, 51, 52). Here, we have identified a subset of LPDCs as cellular source of constitutive IL-12 p40 promoter activity and IL-12 p40 gene expression in the terminal ileum. In fact, isolated CD11c+ LPDC produced high amounts of IL-12 p40 protein upon activation with LPS and SACs. In addition, immunofluorescence studies demonstrated luciferase expression by lamina propria cells in the terminal ileum as well as colocalization of bacteria and luciferase expression in transgenic mice. Finally, further characterization of LPDCs showed that they are largely CD8α and CD11b double negative and thus reminiscent of a DC subset recently characterized in murine Peyer’s patches that produces high levels of IL-12 in response to S. aureus antigen and IFN-γ stimulation (31). Importantly, LPDCs represented a large population of LPMCs in the small bowel, whereas only very few CD11c+ DCs were seen in lymphoid follicles and subepithelial areas of the colon. This finding suggests that LPDCs represent a prominent population of professional APCs in the small but not the large bowel.
The above data raise the question about the cause for a difference in IL-12 p40 and IL-23 production between LPDCs from the proximal and distal small intestine of healthy mice. First, there could be a difference in the number of producer cells between the distal and proximal small intestine. Quantification, however, revealed no significant differences in the absolute number of CD11c+ LPDCs or CD11b+CD11c– macrophages between the proximal and distal small bowel (Becker et al., unpublished data). Second, producer cells of IL-12 p40 could be differentially activated through the local microenvironment. In this context, bacterial antigens and products such as LPS or CpG-DNA have been shown to be strong transcriptional activators of the IL-12 p40 promoter (12, 13, 16, 19, 44). Interestingly, FISH experiments using a specific probe for eubacteria demonstrated localization of bacteria in p40-expressing cells and endocytosis of bacteria by a subset (10%) of CD11c+ LPDCs in the crypts of the terminal ileum but not the proximal parts of the small bowel. These differences might be due to an increased bacterial load of the terminal ileum as compared with the proximal small bowel or to local changes in the composition of the microflora. Consistent with this idea, no increase in constitutive p40 expression was observed in the terminal ileum of mice raised under germfree conditions. Thus, these data indicate that the local bacterial flora in the terminal ileum but not in the proximal small bowel induces increased expression of the p40 gene by activating the p40 promoter. Furthermore, gel-shift experiments using a p40 promoter NF-κB probe demonstrated differences in complex formation between proximal and distal samples, suggesting a constitutive NF-κB p50/p65 activation in the terminal ileum that drives IL-12 p40 promoter activation and p40 protein production. Consistently, reporter gene transgenic mice carrying a loss-of-function mutation in the NF-κB motif of the p40 promoter showed no constitutive p40 promoter activity in the terminal ileum. Taken together, these data provide strong evidence that the intestinal microflora drives constitutive NF-κB activation in CD11c+ LPDCs from the terminal ileum in healthy mice, thereby causing high IL-12 p40 promoter activation in vivo.
In summary, this manuscript demonstrates constitutive transcriptional activity of the IL-12 p40 promoter in LPDCs of the distal ileum in healthy mice and implicates important functional differences between the mucosal immune systems of the colon and the proximal and distal small intestine. This idea is underlined by the recent observation that antigen processing by 20S proteasomes is more effective in the small bowel as compared with the colon and other organs (53). Our data, however, indicate that the bacterial flora in the terminal ileum activates p40 gene transcription in local LPDCs by NF-κB. These findings suggest a predisposition of the terminal ileum to develop chronic inflammatory responses by LPDC-derived IL-23 and thus may provide a molecular explanation for the preferential clinical manifestation of Crohn disease (a Th1-mediated chronic inflammatory bowel disease; refs. 54, 55) in this part of the gut. Consistent with this idea, an uptake of bacteria by LPDCs of the terminal ileum was only seen in the crypts where the earliest pathological manifestations of ileitis in Crohn disease are known to occur.
Figure 10.
Detailed analysis of double staining. Immunofluorescence triple-staining analysis of CD11c (FITC: green), Hoechst 3342 (blue), and FISH (Cy3: red) in D4 using confocal laser microscopy. Orthogonal projections of the sample in three dimensions are shown. Imaging was performed using a multitracking program (Carl Zeiss, Oberkochen, Germany), which eliminates bleed through of other channels. Control images with a single laser activated were collected, and these control images demonstrated no bleed through (not shown). Horizontal and vertical sections from the same images on LPDCs are presented in the upper panels followed by consecutive sections in the lower panels, showing distinct positive red and green areas (arrows) as well as double positivity for CD11c plus FISH in yellow (arrows).
Acknowledgments
The authors thank Kurt Reiffenberg (Animal Research Facility, University of Mainz) for the generation and breeding of the reporter gene transgenic mice. M.F. Neurath was supported by grants from the Deutsche Forschungsgemeinschaft (Ne490/4-1), the Sonderforschungsbereich 548, and the Innovationsstiftung Rheinland-Pfalz.
Footnotes
See the related Commentary beginning on page 648.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: signal transducers and activators of transcription (STAT); lamina propria DC (LPDC); Staphylococcus aureus cell (SAC); electrophoretic mobility-shift assay (EMSA); diaminobenzidine (DAB).
References
- 1.Gubler U, et al. Coexpression of two distinct genes is required to generate secreted bioactive cytotoxic lymphocyte maturation factor. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4143–4147. doi: 10.1073/pnas.88.10.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi M, et al. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J. Exp. Med. 1989;170:827–845. doi: 10.1084/jem.170.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern AS, et al. Purification to homogeneity and partial characterization of cytotoxic lymphocyte maturation factor from human B-lymphoblastoid cells. Proc. Natl. Acad. Sci. U. S. A. 1990;87:6808–6812. doi: 10.1073/pnas.87.17.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolf SF, et al. Cloning of cDNA for natural killer cell stimulatory factor, a heterodimeric cytokine with multiple biologic effects on T and natural killer cells. J. Immunol. 1991;146:3074–3081. [PubMed] [Google Scholar]
- 5.D’Andrea A, et al. Production of natural killer cell stimulatory factor (interleukin 12) by peripheral blood mononuclear cells. J. Exp. Med. 1992;176:1387–1398. doi: 10.1084/jem.176.5.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbulescu K, et al. IL-12 and IL-18 differentially regulate the transcriptional activity of the human IFN-gamma promoter in primary CD4+ T lymphocytes. J. Immunol. 1998;160:3642–3647. [PubMed] [Google Scholar]
- 7.Jacobson NG, et al. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J. Exp. Med. 1995;181:1755–1762. doi: 10.1084/jem.181.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oppmann B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 9.Belladonna ML, et al. IL-23 and IL-12 have overlapping, but distinct, effects on murine dendritic cells. J. Immunol. 2002;168:5448–5454. doi: 10.4049/jimmunol.168.11.5448. [DOI] [PubMed] [Google Scholar]
- 10.Parham C, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J. Immunol. 2002;168:5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- 11.Wiekowski MT, et al. Ubiquitous transgenic expression of the IL-23 subunit p19 induces multiorgan inflammation, runting, infertility, and premature death. J. Immunol. 2001;166:7563–7570. doi: 10.4049/jimmunol.166.12.7563. [DOI] [PubMed] [Google Scholar]
- 12.Ma X, et al. The interleukin 12 p40 gene promoter is primed by interferon gamma in monocytic cells. J. Exp. Med. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy TL, Cleveland MG, Kulesza P, Magram J, Murphy KM. Regulation of interleukin 12 p40 expression through an NF-kappa B half-site. Mol. Cell Biol. 1995;15:5258–5267. doi: 10.1128/mcb.15.10.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- 15.Weinmann AS, et al. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat. Immunol. 2001;2:51–57. doi: 10.1038/83168. [DOI] [PubMed] [Google Scholar]
- 16.Becker C, et al. Regulation of IL-12 p40 promoter activity in primary human monocytes: roles of NF-kappaB, CCAAT/enhancer-binding protein beta, and PU.1 and identification of a novel repressor element (GA-12) that responds to IL-4 and prostaglandin E(2) J. Immunol. 2001;167:2608–2618. doi: 10.4049/jimmunol.167.5.2608. [DOI] [PubMed] [Google Scholar]
- 17.Ma X, Gri G, Trinchieri G. A novel Ets-2-related nuclear factor is involved in transcriptional activation of the human interleukin-12 p40 gene promoter in response to interferon-gamma and LPS stimulation of monocytic cells. Ann. N. Y. Acad. Sci. 1996;795:357–360. doi: 10.1111/j.1749-6632.1996.tb52692.x. [DOI] [PubMed] [Google Scholar]
- 18.Ma X, Neurath M, Gri G, Trinchieri G. Identification and characterization of a novel Ets-2-related nuclear complex implicated in the activation of the human interleukin-12 p40 gene promoter. J. Biol. Chem. 1997;272:10389–10395. doi: 10.1074/jbc.272.16.10389. [DOI] [PubMed] [Google Scholar]
- 19.Plevy SE, Gemberling JH, Hsu S, Dorner AJ, Smale ST. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol. Cell Biol. 1997;17:4572–4588. doi: 10.1128/mcb.17.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nordeen SK. Luciferase reporter gene vectors for analysis of promoters and enhancers. Biotechniques. 1988;6:454–458. [PubMed] [Google Scholar]
- 21.Waidmann M, et al. Microflora reactive IL-10 producing regulatory T cells are present in the colon of IL-2 deficient mice but lack efficacious inhibition of IFN-gamma and TNF-alpha production. Gut. 2002;50:170–179. doi: 10.1136/gut.50.2.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neurath MF, Fuss I, Kelsall BL, Stuber E, Strober W. Antibodies to interleukin 12 abrogate established experimental colitis in mice. J. Exp. Med. 1995;182:1281–1290. doi: 10.1084/jem.182.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pirhonen J, Matikainen S, Julkunen I. Regulation of virus-induced IL-12 and IL-23 expression in human macrophages. J. Immunol. 2002;169:5673–5678. doi: 10.4049/jimmunol.169.10.5673. [DOI] [PubMed] [Google Scholar]
- 25.Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kempf VA, Trebesius K, Autenrieth IB. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 2000;38:830–838. doi: 10.1128/jcm.38.2.830-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Andrea A, Ma X, Aste-Amezaga M, Paganin C, Trinchieri G. Stimulatory and inhibitory effects of interleukin (IL)-4 and IL-13 on the production of cytokines by human peripheral blood mononuclear cells: priming for IL-12 and tumor necrosis factor alpha production. J. Exp. Med. 1995;181:537–546. doi: 10.1084/jem.181.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch F, et al. High level IL-12 production by murine dendritic cells: upregulation via MHC class II and CD40 molecules and downregulation by IL-4 and IL-10. J. Exp. Med. 1996;184:741–746. doi: 10.1084/jem.184.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naz RK, Evans L. Presence and modulation of interleukin-12 in seminal plasma of fertile and infertile men. J. Androl. 1998;19:302–307. [PubMed] [Google Scholar]
- 30.Yamamoto K, et al. cDNA cloning, expression and chromosome mapping of the human STAT4 gene: both STAT4 and STAT1 genes are mapped to 2q32.2 ? q32.3. Cytogenet. Cell Genet. 1997;77:207–210. doi: 10.1159/000134578. [DOI] [PubMed] [Google Scholar]
- 31.Iwasaki A, Kelsall BL. Unique functions of CD11b+, CD8 alpha+, and double-negative Peyer’s patch dendritic cells. J. Immunol. 2001;166:4884–4890. doi: 10.4049/jimmunol.166.8.4884. [DOI] [PubMed] [Google Scholar]
- 32.Iwasaki A, Kelsall BL. Localization of distinct Peyer’s patch dendritic cell subsets and their recruitment by chemokines macrophage inflammatory protein (MIP)-3alpha, MIP-3beta, and secondary lymphoid organ chemokine. J. Exp. Med. 2000;191:1381–1394. doi: 10.1084/jem.191.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 34.Zhang FX, et al. Bacterial lipopolysaccharide activates nuclear factor-kappaB through interleukin-1 signaling mediators in cultured human dermal endothelial cells and mononuclear phagocytes. J. Biol. Chem. 1999;274:7611–7614. doi: 10.1074/jbc.274.12.7611. [DOI] [PubMed] [Google Scholar]
- 35.Bucht A, et al. Expression of interferon-gamma (IFN-gamma), IL-10, IL-12 and transforming growth factor-beta (TGF-beta) mRNA in synovial fluid cells from patients in the early and late phases of rheumatoid arthritis (RA) Clin. Exp. Immunol. 1996;103:357–367. doi: 10.1111/j.1365-2249.1996.tb08288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuss IJ, et al. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. Crohn’s disease LP cells manifest increased secretion of IFN-gamma, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J. Immunol. 1996;157:1261–1270. [PubMed] [Google Scholar]
- 37.Gately MK, et al. Interleukin-12 antagonist activity of mouse interleukin-12 p40 homodimer in vitro and in vivo. Ann. N. Y. Acad. Sci. 1996;795:1–12. doi: 10.1111/j.1749-6632.1996.tb52650.x. [DOI] [PubMed] [Google Scholar]
- 38.McIntyre KW, et al. Reduced incidence and severity of collagen-induced arthritis in interleukin-12-deficient mice. Eur. J. Immunol. 1996;26:2933–2938. doi: 10.1002/eji.1830261219. [DOI] [PubMed] [Google Scholar]
- 39.Ozmen L, et al. Interleukin 12, interferon gamma, and tumor necrosis factor alpha are the key cytokines of the generalized Shwartzman reaction. J. Exp. Med. 1994;180:907–915. doi: 10.1084/jem.180.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trembleau S, Penna G, Gregori S, Gately MK, Adorini L. Deviation of pancreas-infiltrating cells to Th2 by interleukin-12 antagonist administration inhibits autoimmune diabetes. Eur. J. Immunol. 1997;27:2330–2339. doi: 10.1002/eji.1830270930. [DOI] [PubMed] [Google Scholar]
- 41.Wysocka M, et al. Interleukin-12 is required for interferon-gamma production and lethality in lipopolysaccharide-induced shock in mice. Eur. J. Immunol. 1995;25:672–676. doi: 10.1002/eji.1830250307. [DOI] [PubMed] [Google Scholar]
- 42.Zipris D, et al. Cytokine gene expression in islets and thyroids of BB rats. IFN-gamma and IL-12p40 mRNA increase with age in both diabetic and insulin-treated nondiabetic BB rats. J. Immunol. 1996;156:1315–1321. [PubMed] [Google Scholar]
- 43.Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- 44.Cowdery JS, Boerth NJ, Norian LA, Myung PS, Koretzky GA. Differential regulation of the IL-12 p40 promoter and of p40 secretion by CpG DNA and lipopolysaccharide. J. Immunol. 1999;162:6770–6775. [PubMed] [Google Scholar]
- 45.Cua DJ, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 46.Huang FP, et al. A discrete subpopulation of dendritic cells transports apoptotic intestinal epithelial cells to T cell areas of mesenteric lymph nodes. J. Exp. Med. 2000;191:435–444. doi: 10.1084/jem.191.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwasaki A, Kelsall BL. Mucosal immunity and inflammation. I. Mucosal dendritic cells: their specialized role in initiating T cell responses. Am. J. Physiol. 1999;276:G1074–G1078. doi: 10.1152/ajpgi.1999.276.5.G1074. [DOI] [PubMed] [Google Scholar]
- 48.Iwasaki A, Kelsall BL. Freshly isolated Peyer’s patch, but not spleen, dendritic cells produce interleukin 10 and induce the differentiation of T helper type 2 cells. J. Exp. Med. 1999;190:229–239. doi: 10.1084/jem.190.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinman RM, Turley S, Mellman I, Inaba K. The induction of tolerance by dendritic cells that have captured apoptotic cells. J. Exp. Med. 2000;191:411–416. doi: 10.1084/jem.191.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu K, et al. Immune tolerance after delivery of dying cells to dendritic cells in situ. J. Exp. Med. 2002;196:1091–1097. doi: 10.1084/jem.20021215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rescigno M, Rotta G, Valzasina B, Ricciardi-Castagnoli P. Dendritic cells shuttle microbes across gut epithelial monolayers. Immunobiology. 2001;204:572–581. doi: 10.1078/0171-2985-00094. [DOI] [PubMed] [Google Scholar]
- 52.Rescigno M, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 53.Kuckelkorn U, et al. Link between organ-specific antigen processing by 20S proteasomes and CD8(+) T cell-mediated autoimmunity. J. Exp. Med. 2002;195:983–990. doi: 10.1084/jem.20011199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atreya R, et al. Blockade of IL-6 trans-signaling suppresses T cell resistance against apoptosis in chronic intestinal inflammation: Evidence in Crohn’s disease and experimental colitis in vivo. Nat. Med. 2000;6:583–588. doi: 10.1038/75068. [DOI] [PubMed] [Google Scholar]
- 55.Friedman, S., and Blumberg, R.S. 1999. Inflammatory bowel disease. In Harrison’s principles of internal medicine. 15th edition. McGraw-Hill. New York, New York, USA. 1679–1691.
- 56.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]