Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing (original) (raw)
Abstract
Nuclear pre-messenger RNA (pre-mRNA) splicing requires multiple spliceosomal small nuclear RNA (snRNA) and pre-mRNA rearrangements. Here we reveal a new snRNA conformational switch in which successive roles for two competing U2 helices, stem IIa and stem IIc, promote distinct splicing steps. When stem IIa is stabilized by loss of stem IIc, rapid ATP-independent and Cus2p-insensitive prespliceosome formation occurs. In contrast, hyperstabilized stem IIc improves the first splicing step on aberrant branchpoint pre-mRNAs and rescues temperature-sensitive U6–U57C, a U6 mutation that also suppresses first-step splicing defects of branchpoint mutations. A second, later role for stem IIa is revealed by its suppression of a cold-sensitive allele of the second-step splicing factor PRP16. Our data expose a spliceosomal progression cycle of U2 stem IIa formation, disruption by stem IIc, and then reformation of stem IIa before the second catalytic step. We propose that the competing stem IIa and stem IIc helices are key spliceosomal RNA elements that optimize juxtaposition of the proper reactive sites during splicing.
Keywords: Intron, RNA dynamics, helicase, DExD-box protein, Prp5p
Ribonucleoprotein particles (RNPs) present special challenges with respect to their assembly and function. One reason is that structural elements made from folded RNA may need to be rearranged during assembly and function. For example, the RNA that forms the core elements of the ribosome (Noller 2006), telomerase (Collins 2006), and signal recognition particle (Hainzl et al. 2005) must be properly folded to function. The same is true for the spliceosome, which catalyzes pre-messenger RNA (pre-mRNA) splicing in eukaryotes, with the added complexity that assembly of this RNP occurs each functional cycle and requires dramatic changes in RNA structure during its function (Ares and Weiser 1995; Staley and Guthrie 1998).
A number of structural changes must occur in both the RNA and protein components as the spliceosome progresses through the splicing cycle. Important landmarks in the splicing cycle are the two cleavage-ligation steps that result in splicing and several structural changes that coincide with the need for ATP and one or another essential DExD/H family protein (Kramer 1996; Brow 2002). The sequential presentation of the pre-mRNA branchpoint to the 5′ splice site, followed by presentation of the free exon product of the first reaction to the second reactive phosphate center at the 3′ splice site is expected to require significant substrate rearrangement between the first and second catalytic steps. To position the pre-mRNA reactive sites, the spliceosome undergoes changes in composition and structure, many of which are mediated by intra- and intermolecular small nuclear RNA (snRNA) rearrangements (Staley and Guthrie 1998; Collins and Guthrie 2001; Brow 2002; Konarska et al. 2006). Several landmark changes during the first catalytic step include the disruption of extensive base-pairing between U4 and U6 snRNA, the disruption of intra-U2 and U6 snRNA helices to create a new U6–U2 snRNA interaction, and the exchange of U1 snRNA for U6 snRNA at the 5′ splice site. These changes exemplify a set of a highly coordinated and regulated events (Ares and Weiser 1995; Staley and Guthrie 1998; Brow 2002), but the molecular logistics of these RNA rearrangements have been only loosely determined at best, and coordinated events that support these changes remain largely unknown.
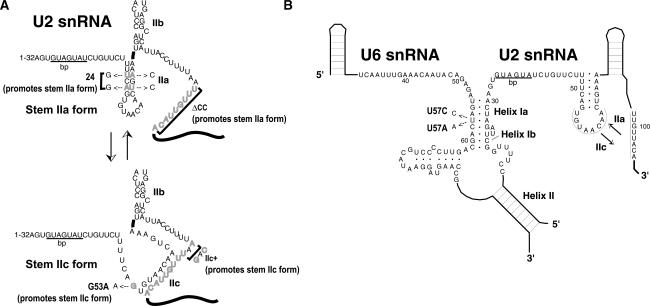
One predicted rearrangement encompasses the U2 snRNA structures stem–loop IIa and the competing structure stem–loop IIc (Zavanelli and Ares 1991; Zavanelli et al. 1994). Numerous studies have demonstrated that stem–loop IIa plays a crucial early role in spliceosome assembly (see Fig. 1A). Stem IIa is required for prespliceosome formation, although its mechanistic function both during and beyond prespliceosomes is unknown (Ares and Igel 1990; Zavanelli and Ares 1991; Wells and Ares 1994; Zavanelli et al. 1994; Ares and Weiser 1995; Wells et al. 1996; Yan and Ares 1996; Yan et al. 1998). A U2 snRNA point mutation, U2–G53A, destabilizes stem IIa and hyperstabilizes the competing stem IIc, causing a cold-sensitive phenotype in vivo and inhibition of prespliceosome formation in vitro (Ares and Igel 1990; Zavanelli and Ares 1991; Zavanelli et al. 1994). Second site intragenic suppressors of U2–G53 act to destabilize structures (including stem IIc) that compete with stem IIa (Zavanelli et al. 1994). Two extragenic suppressor mutations of U2–G53 were identified in U2 snRNP proteins, Cus1p (human SAP145) (Champion-Arnaud and Reed 1994; Wells et al. 1996) and Cus2p (human Tat-SF1) (Yan et al. 1998; Fong and Zhou 2001), but their mechanisms of suppression remain unknown. The early and important role of stem IIa and the conserved but nonessential role of stem IIc has hampered efforts to uncover a functional role for U2 stem IIc during spliceosome assembly and splicing.
Figure 1.
(A) A 5′ portion of yeast U2 snRNA showing stem IIa or stem IIc forms and various mutations used in this study that promote either form. (B) Secondary structure of interactions between U2 and U6 snRNAs required for first transesterification: Mutation at U6–U57 to A or C is indicated.
Here we reveal a new RNA conformational switch in the spliceosome by defining distinct roles for the mutually exclusive formation of U2 stem IIa or stem IIc at several points in spliceosome assembly and splicing. We find that stabilized stem IIa mutants allow rapid formation of prespliceosomes that are both ATP-independent and insensitive to Cus2p regulation in vitro, suggesting that stem IIa formation controls both the rate and ATP dependence of prespliceosome formation. U2 stem IIc is not required for these initial steps, but its formation (or the disruption of stem IIa) contributes to splicing efficiency during the first catalytic step, because the splicing of introns with mutant branchpoint sequences is significantly improved when stem IIc is hyperstabilized. In addition, mutations that hyperstabilize stem IIc suppress the temperatue-sensitive phenotype of U6–U57C, a U6 mutation that inhibits the second step of splicing but also increases the first catalytic step of splicing of mutant branchpoint sequences (McPheeters 1996; Query and Konarska 2004). Finally, we observed genetic suppression by stabilized and hyperstabilized stem IIa mutants of a cold-sensitive allele of Prp16p, a DExD/H box protein that promotes crucial structural transitions between the first and second step of splicing. These data evoke a model in which U2 stem IIa is required to unwind at least once and form at least twice during spliceosome assembly and splicing to support the cyclic exposure and protection of pre-mRNA substrate active sites.
Results
A mutation that stabilizes U2 stem IIa bypasses ATP- and Cus2p-dependent regulation of prespliceosome formation
Normally, ATP and the DExD/H protein Prp5p are required for prespliceosome formation in vitro (Ruby et al. 1993). However we have previously observed that prespliceosomes can slowly form in the absence of ATP in _cus2_Δ extracts (Perriman and Ares 2000) and that yeast expressing ATP-binding-deficient forms of Prp5p mutants are rescued by _cus2_Δ or stabilizing and hyperstabilizing U2–stem IIa mutations in vivo (Perriman et al. 2003). We were therefore interested in determining whether U2 snRNPs with stabilized stem IIa could bypass both the ATP requirement and the Cus2p regulation for prespliceosome assembly in vitro. To test this, we measured the rate of prespliceosome formation in vitro in extracts from cells expressing U2-ΔCC, a U2 snRNA mutant predicted to strongly stabilize stem IIa of U2 snRNA. U2-ΔCC is deleted for the conserved complementarity (CC), which pairs with the loop of stem IIa to form competing stem IIc (see Fig. 1). When the CC is deleted, stem IIa is favored due to the inability of the competing stem IIc to form (Fig. 1A; Zavanelli and Ares 1991; Zavanelli et al. 1994). In vivo structure probing of RNA from cells expressing U2-ΔCC demonstrates that the bulk of U2 snRNPs contain U2 snRNA in the stem IIa form (data not shown). We prepared splicing extracts from U2-ΔCC strains with and without Cus2p and tested them for their ability to form prespliceosomes (Fig. 2A).
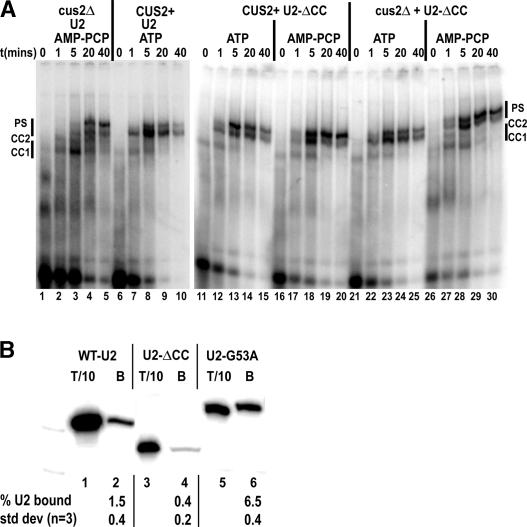
Figure 2.
U2 stem IIa formation regulates the rate, Cus2p, and ATP dependence of prespliceosome formation. (A) U2-ΔCC rapidly forms Cus2p and ATP-independent prespliceosomes at identical rates in vitro. Shown is native gel analysis of a 0- to 40-min time course of spliceosome assembly on pre-RP51A in U6-depleted _cus2_Δ (lanes 1–5), CUS2+ (lanes 6–10), CUS2+ U2-ΔCC (lanes 11–20), or _cus2_Δ U2-ΔCC (lanes 21–30) splicing extracts in the presence of AMP-PCP (lanes 1–5,16–20,26–30) or ATP (lanes 6–10,11–15,22–25). Samples were taken at 0 min (lanes 1,6,11,16,21,26), 1 min (lanes 2,7,12,17,22,27), 5 min (lanes 3,8,13,18,23,28), 20 min (lanes 4,9,14,19,24,29), and 40 min (lanes 5,10,15,20,25,30) after pre-RP51A addition. (CC1) Commitment complex 1; (CC2) commitment complex 2; (PS) prespliceosomes. (B) HA-Cus2p preferentially coimmunoprecipitates U2 snRNA that favors U2–stem IIc. Extracts prepared from strains containing HA-Cus2p and U2 (lanes 1,2), U2-ΔCC (lanes 3,4), or U2–G53A (lanes 5,6) were incubated with anti-HA antibody 12CA5 prebound to Protein A Sepharose and washed with 50 mM NET buffer. Bound RNA was extracted and used as a template for primer extension with U2 snRNA-specific primer. Lanes are 1/10 total (1,3,5) and coimmunopreciptated fractions (2,4,6). Below is average percentage of U2 bound and standard deviations from three independent coimmunopreciptations.
Remarkably, U2-ΔCC snRNPs support rapid prespliceosome formation whether or not ATP or Cus2p is present. Prespliceosome formation in CUS2+ (Fig. 2A, lanes 11–20) or _cus2_Δ (Fig. 2A, lanes 21–30) U2-ΔCC splicing extracts with ATP (Fig. 2A, lanes 11–15,21–25) or AMP-PCP (Fig. 2A, lanes 16–20,26–30) assemble at wild-type rates (Fig. 2A, lanes 6–10) and are splicing competent (data not shown). The rates are significantly faster than those in _cus2_Δ extracts in the presence of AMP-PCP (Fig. 2A, lanes 1–5). This suggests that unlike the wild-type U2 snRNP, the U2-ΔCC snRNP no longer responds to the rate enhancement provided by ATP and is insensitive to the regulatory influence of Cus2p in the absence of ATP (Perriman and Ares 2000; Perriman et al. 2003). From this, we conclude that removing the downstream conserved complementary sequence from U2 snRNA, (favoring U2–stem IIa and preventing the formation of competing stem IIc) bypasses the regulation exerted by ATP and Cus2p on the rate of prespliceosome formation. This differs significantly from wild-type U2 snRNA, because ATP and Cus2p no longer affect the ability or rate of formation of prespliceosomes. These data strongly implicate U2 stem IIa as the rate-determining product of an ATP- and Cus2p-regulated reaction during prespliceosome formation in vitro. We suggest that Prp5p mediates this action (Ruby et al. 1993; Perriman and Ares 2000; Perriman et al. 2003).
The ability of stem IIc to form in the U2 snRNP correlates with increased Cus2p binding
Since U2-ΔCC allows rapid formation of prespliceosomes in a fashion that is insensitive to Cus2p (or ATP) imposed regulation, we postulated that Cus2p may preferentially bind the stem IIc-containing form of the U2 snRNP. If true, we might detect more efficient coimmunoprecipitation between Cus2p and U2 snRNA mutants that predominantly adopt the stem IIc form, since that form is completely absent in the Cus2p-insensitive ΔCC mutant. To test this, we immunoprecipitated Cus2p from splicing extracts of either wild-type U2 (mostly stem IIa, small fraction stem IIc) (Ares and Igel 1990), U2-ΔCC (only stem IIa, stem IIc deleted) (Fig. 1A) or U2–G53A (mostly stem IIc) (Fig. 1A; Zavanelli and Ares 1991) and measured the amount of U2 RNA coprecipitated with Cus2p by primer extension (Fig. 2B).
For wild-type extracts, ∼1.5 ± 0.4% of U2 snRNA can be coimmunopreciptated with Cus2p (Fig. 2B, lanes 1,2; see also Yan et al. 1998). By comparison, fourfold less U2-ΔCC is recovered (0.4 ± 0.2%) (Fig. 2B, lanes 3,4). This lower binding is consistent with the hypothesis that U2-ΔCC has reduced ability to bind Cus2p. In contrast, when compared with wild-type U2 snRNA, fourfold greater U2–G53A is associated with Cus2p (6.5 ± 0.4%) (Fig. 2B, lanes 5,6). Since U2-ΔCC is unable to form stem IIc and U2–G53A forms mostly stem IIc (Zavanelli et al. 1994), these data argue that Cus2p preferentially interacts with the U2 snRNP when the U2 snRNA is folded in the stem IIc form. We do not know whether Cus2p binds directly to U2 snRNA or whether it binds to other proteins whose binding or conformation is influenced by stem IIa and stem IIc. Nonetheless, these findings support models in which Cus2p enables the formation of stem IIa by binding snRNPs containing U2 snRNA in other conformations, and dissociates from the complex after stem IIa is formed.
A test for contribution of U2 stems IIa and IIc to the fidelity of branchpoint recognition
The loss of Cus2p-mediated regulation of, or any dependence on ATP for prespliceosome assembly suggests the possibility of a concomitant loss of a fidelity check that normally ensures correct prespliceosome assembly. To test whether the bypass of regulation observed in U2-ΔCC might also reduce the fidelity of branchpoint selection, we measured the effects of U2 snRNA mutations on splicing of reporter pre-mRNAs with mutant branchpoints (Lesser and Guthrie 1993), expecting that the bypass condition might allow splicing of messages with branchpoint mutations. We tested three U2 alleles with differing abilities to form stem IIa or stem IIc (see Fig. 1A) and a series of pre-mRNA mutations created in the ACT1-CUP1 reporter plasmids (see Fig. 3A; Lesser and Guthrie 1993). If the same alleles that accelerate and relieve the ATP dependence of prespliceosome formation also allow mutant branchpoints to be more efficiently spliced, then U2-ΔCC allele should suppress the reporter mutations and cause an increase in copper resistance.
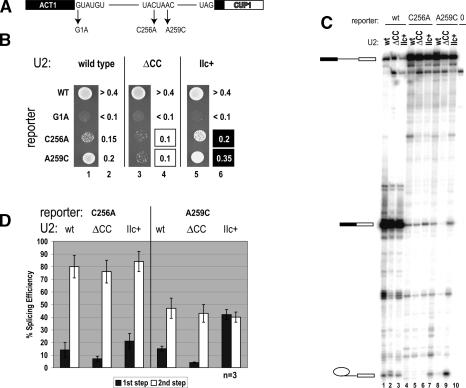
Figure 3.
Stabilized U2 stem IIa decreases, but hyperstabilized U2 stem IIc increases, the first step of splicing on C256A and A259C branch site mutant ACT1-CUP1 reporter in vivo. (A) ACT1-CUP1 reporter pre-mRNA indicating 5′ splice site and branch site mutants used in this study. (B) Copper growth of strains expressing ACT1-CUP1 reporters (indicated) and either wild-type (lanes 1,2), U2-ΔCC (lanes 3,4), or U2–IIc+ (lanes 5,6). Lanes 1, 3, and 5 show growth on 0.15 mM Cu++, while lanes 2, 4, and 6 indicate the highest copper concentration allowing growth. Boxes indicate strains where growth was observed on higher (black) or lower (white) Cu++ when compared with wild-type U2 snRNA. (C) Primer extension analysis of RNAs from strain DS4D containing wild type (lanes 1–3), C256A (lanes 4–7), A259C (lanes 8–10), A259G, and one of U2 (lanes 1,4,7), U2-ΔCC (lanes 2,5,8), or U2–IIC+ (lanes 3,6,9). Primer complementary to the 3′ exon was used to visualize pre-mRNA, mRNA, and lariat intermediate indicated from top to bottom of gel, respectively. Lane 0 (10) are RNA from isogenic yeast lacking the ACT1-CUP1 reporter. (D) Quantitation of results from three independent experiments, an example of which is presented in B. Dark bars show first-step efficiency calculated as M + L/(P + M + L) and light bars show second-step efficiency calculated as M/(M + L) as per Query and Konarska (2004). Percent splicing efficiency is normalized to the first-step efficiency of wild-type pre-ACT1-CUP1 in wild-type strain set at 100.
Surprisingly, the reverse occurs, and U2-ΔCC reduces copper resistance of branchpoint mutants C256A and A259C (Fig. 3B, lanes 3,4). Equally unexpected, but consistent with this result, the U2–IIc+ allele, in which stem IIc is hyperstabilized, increases the limited copper resistance provided by the mutant reporters C256A and A259C (Fig. 3B, lanes 5,6). These findings suggest that stabilizing stem IIa might increase the fidelity of branchpoint selection, whereas hyperstabilizing stem IIc might relax the fidelity of branchpoint selection. This is the first hint that despite its nonessential nature, stem IIc may function before or during the first catalytic step of splicing.
Influence of U2 stems IIa and IIc on the catalytic steps of splicing
To test whether the above reporter results are due to changes in mutant branchpoint use and to investigate the possible role of these U2 snRNA stems on the catalytic steps of splicing, we performed primer extension on the reporter-derived RNAs (Fig. 3C). Following Query and Konarska (2004), we measured first- and second-step splicing efficiencies by quantitating pre-mRNA (P), lariat intermediate (L), and mature mRNA (M) and calculate the first step efficiency as M + L/P + M + L, while the second-step efficiency is calculated as M/M + L (Fig. 3D). All three U2 snRNA alleles efficiently splice wild-type pre-mRNA (Fig. 3C, lanes 1–3). Mutation of the intron branchpoint in the presence of wild-type U2 snRNA results in severe first-step splicing defects (Fig. 3C, C256A, lane 4, or A259C, lane 7; Fouser and Friesen 1986; Burgess and Guthrie 1993; McPheeters 1996; Query and Konarska 2004). Consistent with the in vivo copper resistance assay, splicing of C256A (Fig. 3C, lane 5), and A259C (Fig. 3C, lane 8) substrates is reduced when U2-ΔCC is expressed. In contrast, expression of U2–IIc+ with hyperstable stem IIc results in slight suppression of the splicing defects of C256A (Fig. 3C, lane 6) and significant suppression for A259C (Fig. 3C, lane 9).
Quantitation of these results demonstrates that the first step of splicing of A259C pre-mRNAs is significantly reduced in strains expressing U2-ΔCC (increased first-step fidelity) but significantly increased (reduced first-step fidelity) in strains expressing U2–IIc+ (Fig. 3D). Similarly, the first step of splicing of C256A pre-mRNAs is also reduced with U2-ΔCC and increased with U2–IIc+, but we have not determined that this change is statistically different from wild-type U2 (Fig. 3D), although the copper resistance phenotype is clear (Fig. 3B). In contrast, all three U2 snRNAs demonstrate equal levels of efficiency of the second step of splicing for both C256A and A259C pre-mRNAs (Fig. 3D). From these data we conclude that eliminating U2–stem IIc inhibits, whereas hyperstabilizing U2–stem IIc promotes the first step of splicing. Together with the fact that stem IIa is a rate-determining requirement for prespliceosome formation (Fig. 2), these data suggest that optimal progression from prespliceosomes through the first step of splicing requires unwinding of U2 stem IIa to form U2 stem IIc.
Hyperstabilized stem IIc can suppress the temperature-sensitive phenotype of U6–U57C
U6–U57C, a temperature-sensitive allele of U6 snRNA, increases the first catalytic step but inhibits the second catalytic step of splicing of pre-mRNAs with A259C branchpoint mutations (McPheeters 1996; Query and Konarska 2004). Because both U2–IIc+ and U6–U57C can suppress the first-step splicing defects of aberrant branchpoint pre-mRNAs, we tested the combination of U2–IIc+, or U2-ΔCC with U6–U57C (Fig. 4A). To our surprise we found that U2–IIc+, but not U2-ΔCC, can efficiently suppress U6–U57C at 37°C. As U6–U57C has demonstrated effects on both the first and second steps in splicing (McPheeters 1996; Query and Konarska 2004), our genetic suppression may indicate one of two possible effects: either that U2–stem IIc acts together with U6–U57 to promote the first step of splicing, or that U2–stem IIc somehow helps U6–U57 during its second-step function.
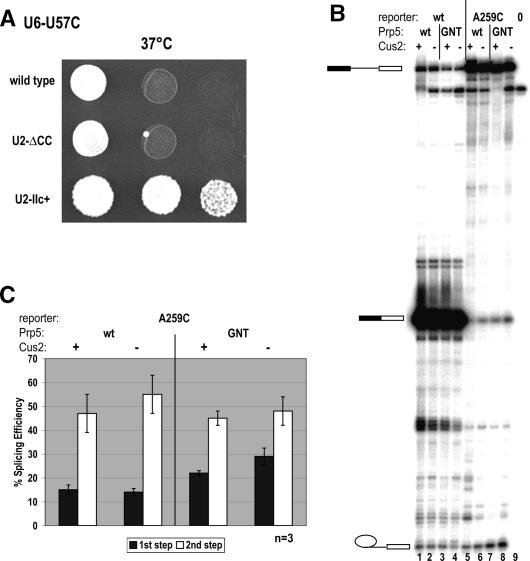
Figure 4.
Stem IIc interacts functionally with U6–U57C, while _cus2_Δ in combination with a Prp5p ATPase mutant increases the first step of splicing on A259C pre-mRNA. (A) Hyperstabilized stem IIc can rescue temperature-sensitive U6–U57C. Growth of fourfold serial dilutions of strain YHM118 carrying temperature-sensitive U6–U57C at restrictive temperature (37°C) is shown with one of U2, U2-ΔCC, or U2–IIc+ snRNAs. (B) A Prp5p ATPase mutant can increase the first step of splicing on A259C ACT1-CUP1 pre-mRNA, and _cus2_Δ can enhance this. Primer extension analysis of RNAs from DS4D containing wild type (lanes 1–4) or A259C (lanes 5–8) and Prp5p (lanes 1,2,5,6) or Prp5-GNTp (lanes 3,4,7,8) and CUS2+ (lanes 1,3,5,7) or _cus2_Δ (lanes 2,4,6,8). Lane 0 is control RNA as in Figure 3. Product designations are as in Figure 3. (C) Quantitation of results from three independent experiments, an example of which is presented in A. Calculations of percent efficiency is as in Figure 3.
The loss of Cus2p and ATPase-deficient Prp5p can also suppress first-step splicing defects
The behavior of hyperstabilized stem IIa or stem IIc mutants suggests that this part of U2 snRNA structure is rearranged between the time of prespliceosome formation and the first catalytic step of splicing. Since our previous data implicate Cus2p and the ATPase activity of Prp5p in regulating U2 snRNA structure before and during prespliceosome formation, we questioned whether these two proteins might also play a role in later U2 snRNA rearrangements. We therefore used primer extension analysis to analyze splicing efficiency of the wild-type and A259C ACT1-CUP1 reporters in strains expressing wild-type Prp5p, or the ATP-binding-deficient Prp5p allele Prp5-GNT, with and without Cus2p (Fig. 4B,C; Perriman et al. 2003).
Strikingly, there is an increase in first-step efficiency on A259C branch site mutants in _cus2_Δ Prp5-GNTp strains (Fig. 4B, lane 8). CUS2+ Prp5-GNTp strains also display a moderate increase in first-step efficiency when compared with _CUS2+_-containing wild-type Prp5p (Fig. 4B, cf. lanes 7 and 5). In contrast, we observed no differences in splicing efficiency of wild-type (Fig. 4B, lanes 1,2) or A259C (Fig. 4B, lanes 5,6) pre-mRNAs when wild-type Prp5p is expressed with or without Cus2p. Quantitation of these results demonstrates that expression of Prp5-GNTp in combination with _cus2_Δ can increase the first-step splicing efficiency of A259C pre-mRNA twofold when compared with splicing of the same pre-mRNA in the presence of wild-type Prp5p (Fig. 4C). These findings suggest that while Prp5p’s ATP-binding activity and its regulator, Cus2p, are not required for the first catalytic step, they do impact fidelity during this step, because removing them increases the efficiency of splicing on A259C mutant branchpoints. Thus, fidelity is relaxed by removal of Prp5p ATP-binding activity and Cus2p.
Genetic interactions between U2 and Prp16p or U6–U57A suggest a role for U2 stem IIa between the first and second catalytic steps
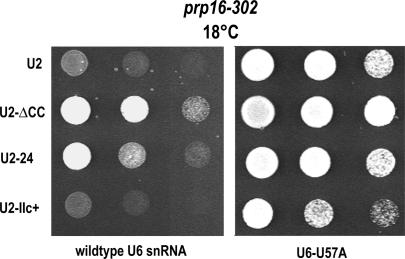
U2–IIc+ suppresses first-step splicing defects on aberrant branchpoint pre-mRNAs and genetically suppresses U6–U57C, a U6 allele that is blocked at the second step, but favors first-step catalysis on A259C branchpoint mutations (Fig. 4). This finding suggests that stem IIc and U6–U57 impact similar steps in the splicing cycle. In addition, U6–U57C is synthetic lethal with the cold-sensitive prp16-302 allele, a Prp16p mutation also known to block the second step and increase the first catalytic step of splicing of A259C branchpoint mutations. In contrast to U6–U57C, a U6–U57A mutation favors second-step catalysis on branchpoint mutations and can suppress the prp16-302 cold-sensitive phenotype (McPheeters 1996; Query and Konarska 2004; Villa and Guthrie 2005). Because of these genetic links between U6–U57, PRP16, and hyperstabilized U2–stem IIc, we wondered whether U2 snRNA mutations that stabilize U2–stem IIa or U2–stem IIc might affect the cold-sensitive phenotype of the prp16-302 allele and its rescue by U6–U57A.
To test this, we introduced mutant U2 alleles into a prp16-302 yeast strain carrying either wild-type U6 snRNA or U6–U57A and analyzed growth at 25°C (data not shown) or 18°C (Fig. 5). In the presence of wild-type U6 snRNA, the stabilized stem IIa mutant U2-ΔCC and the hyperstabilized stem IIa mutant U2-24 (see Fig. 1A) can suppress the prp16-302 defect. Under the same conditions, hyperstabilized stem IIc allele U2–IIc+ does not improve the growth of prp16-302 cells. At 25°C, all five U2 snRNA alleles support growth with either U6 or U6–U57A alleles (data not shown). Thus, favoring U2–stem IIa can suppress the cold-sensitive phenotype of prp16-302.
Figure 5.
Genetic interactions between U2–stem IIa, U6–U57A, and Prp16p, a DExD/H protein involved in first-to-second step transition suggest a role for U2–IIa at this transition. U2-ΔCC and U2-24, but not U2–IIc+, can suppress the cold-sensitive phenotype of prp16-302 at 18°C. This rescue is enhanced when U2-ΔCC and U6–U57A (also a suppressor of prp16-302) (McPheeters 1996) or U2-24 and U6–U57A are coexpressed. Growth of fourfold dilutions of strain YHM187 at restrictive temperature (18°C) is shown with one of U2, U2-ΔCC, or U2-24 as the sole source of U2 snRNA and U6, or U6–U57A as the sole source of U6 snRNA.
We next analyzed growth in the presence of the U6–U57A suppressor and the mutant U2 alleles (Fig. 5). Consistent with previous analysis (McPheeters 1996), U6–U57A can suppress the prp16-302 cold-sensitive phenotype when wild-type U2 snRNA is expressed. Coexpression of U6–U57A plus U2-ΔCC increases the growth rate of the prp16-302 cells, and this growth is better than that observed with U6–U57A or U2-ΔCC allele alone. When U2-24 is coexpressed, suppression is equivalent to that observed with wild-type U2 snRNA, suggesting that direct hyperstabilization of stem IIa has no additional effect on U6–U57A suppression of prp16-302, although alone, U2-24 can suppress. In contrast, U2–IIc+ reduces U6–U57A suppression of prp16-302. Taken together, we conclude that favoring stem IIa by removing the competing stem IIc (U2-ΔCC) or hyperstabilizing stem IIa directly (U2-24) enables suppression of prp16-302, while the combination of U2-ΔCC and U6–U57A can enhance this suppression. This result strongly suggests that U2 stem IIa formation, and loss of stem IIc promotes the same rearrangement between the first and second catalytic step as Prp16p and U6–U57.
Discussion
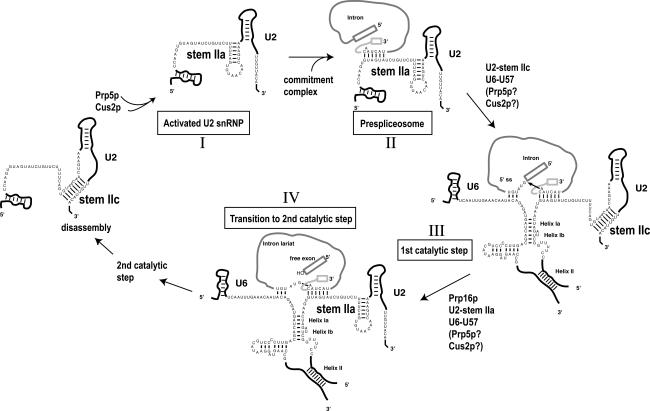
Conformational changes in RNA–RNA interactions within the spliceosome are poorly understood yet essential for accurate pre-mRNA splicing. Here we present data in support of a model in which two competing U2 RNA helices, stem IIa and stem IIc, contribute to structural alterations of U2 snRNA from its initial entry into the spliceosome through multiple steps in the splicing cycle (Fig. 6). We suggest that U2 dynamics promote assembly and rearrangement (stem IIa) or catalysis (stem IIc), allowing the proper pre-mRNA reactive sites to be exposed. Thus, remodeling U2 snRNA between stem IIa and stem IIc forms helps to drive the transition of the spliceosome from one step to another. First stem IIa enhances prespliceosome formation, then stem IIc aids the first catalytic step, and this is followed by a requirement for stem IIa during the Prp16p catalyzed transition from first to second step (Fig. 6). Since mutants that stabilize stem IIc have a cold-sensitive phenotype in vivo, but stem IIa stabilizing mutants have no obvious growth defects (M. Perriman and R.J. Ares, unpubl.), the rate-limiting step in U2 snRNP function in vivo must be its initial recruitment to form prespliceosomes. In addition, at later steps, where hyperstabilized stem IIa interferes with optimal splicing progression, factors must be in place to efficiently unwind even strong U2–stem IIa alleles.
Figure 6.
A model showing the roles of U2–stem IIa, U2–stem IIc in steps leading to the first-to-second step transition in pre-mRNA splicing. Formation of U2–stem IIa as the rate-limiting and ATP-determining product (I) leading to prespliceosomes (II) (Fig. 2). This step is catalyzed by ATPase activity of Prp5p and regulated by Cus2p. (III) A role for formed U2–stem IIc, and not U2–stem IIa, during the first step (Fig. 3). (IV) A role for reformed U2–stem IIa between first and second step either as an important scaffold for PRP16 action at catalytic site or a direct PRP16 substrate in disrupting U2–stem IIc (see also Fig. 5).
U2–stem IIa controls the rate of initial U2 snRNP recruitment to form prespliceosomes
In vitro prespliceosome assembly experiments indicate that the ability of U2 snRNA to form U2–stem IIa dictates the rate of prespliceosome assembly (Figs. 2, 6 [steps I, II]). Furthermore, in splicing extracts expressing U2-ΔCC snRNPs, which lack the ability to form stem IIc, prespliceosomes formation is rapid and insensitive to Cus2p and ATP, indicating that when stem IIa is formed, these factors are no longer rate limiting for prespliceosome formation (Fig. 2A). In support of this, we observed a strong Cus2p-binding preference for U2 snRNPs containing U2 snRNA folded in the stem IIc formation (Fig. 2B). Together with our previous analyses (Yan et al. 1998; Perriman and Ares 2000; Perriman et al. 2003), this indicates that in wild-type cells, U2–stem IIa formation requires the unwinding of U2–stem IIc to form stem IIa in a reaction that is controlled by Cus2p and catalyzed by the ATP-dependent activity of Prp5p (Fig. 6, step I). We propose that Prp5p facilitates the disruption of a Cus2p–U2 interaction in a fashion that culminates in stem IIa formation (Fig. 1; Perriman and Ares 2000; Perriman et al. 2003; M. Perriman and R.J. Ares, unpubl.). In this view, U2 mutations that increase the intrinsic rate of U2–stem IIa formation (such as U2-ΔCC) relieve the ATP requirement for prespliceosome formation. In essence, the rate of U2–stem IIa formation controls the rate of stable U2 snRNP recruitment to the pre-mRNA, forming the prespliceosome. This is the first role for stem IIa.
A role for U2–stem IIc during the first catalytic step
Next, stem IIa must unwind for the first step of splicing, a reaction that is enhanced by the formation of stem IIc (Fig. 6, step III). This finding ends several years of conjecture about a role for the phylogenetically conserved stem IIc helix (Lamond et al. 1989; Ares and Igel 1990; Zavanelli and Ares 1991; Barabino et al. 1992; Datta and Weiner 1992; Zavanelli et al. 1994; Ares and Weiser 1995; Staley and Guthrie 1998). We show that hyperstabilization of stem IIc, at the expense of stem IIa, can suppress first-step splicing defects of mutant branchpoint pre-mRNAs, revealing a positive role for stem IIc during this step (Fig. 3), a finding also supported by Hilliker et al. (2007). This requirement must be mediated through the unwinding of U2–stem IIa, since hyperstable stem IIa alleles have the opposite effect and enhance first-step splicing defects. We also find that hyperstabilized U2–stem IIc (U2–IIc+) can suppress the temperature-sensitive U6–U57C mutation (Fig. 4A). U6–U57 lies two nucleotides upstream of the AGC triad in a critical region of U6 snRNA (Hilliker and Staley 2004). Previous findings have suggested the involvement of U6–U57 in mutually exclusive interactions with U2 snRNA forming helix Ia (Madhani and Guthrie 1992) or the U6 3′ extension (Sun and Manley 1995). U6–U57C inhibits the second step (but relaxes the fidelity of the first step) on mutant branchpoint pre-mRNAs (Fig. 4; McPheeters 1996; Query and Konarska 2004); thus, suppression of its temperature sensitivity by U2–IIc+ may reveal a role for U2–stem IIc during the second catalytic step, a hypothesis supported by Hilliker et al. (2007).
The combined absence of Cus2p and the ATP-binding activity of Prp5p also impacts the first catalytic step by suppressing the ACT1-CUP1 mutant branchpoint first-step splicing defects (Fig. 4). This suppression is consistent with a positive role for stem IIc during the first step because it occurs as a consequence of the loss of factors that ordinarily promote stem IIa formation. Stabilization of stem IIa during the first step prevents efficient splicing progression (see Fig. 3). Thus, a plausible hypothesis for the role for Prp5p and Cus2p in the first step could be to impose a checkpoint for noncanonical pre-mRNAs, ensuring that they are discarded rather than spliced. This function also implies that Cus2p and Prp5p can access spliceosomes that have matured to activated spliceosomes, and hints that the two proteins may remain to carry out a role later in perhaps helping reform stem IIa. Alternatively, the ATP-independent and uncontrolled prespliceosome formation observed in vitro with the U2-ΔCC extracts (Fig. 2) could have some correlation in vivo with wild-type U2 snRNA, when Cus2p and ATP-binding Prp5p are absent. In this scenario, the splicing of the branchpoint mutant pre-mRNA is enhanced through the loss of fidelity at the prespliceosome assembly step. An increase in step one for these mutant pre-mRNAs is then explained not by stabilization of stem IIa during the first step (because factors that enhance this are missing), but rather by a simple increase in the amount of prespliceosomes that form on the mutant pre-mRNA in the first place.
U2–stem IIa reforms for the transition to second step
After stem IIc contributes to the first step, stem IIa forms again between the first and second step (Fig. 6, step IV). Stabilized U2–stem IIa can rescue growth of the ATPase-deficient cold-sensitive Prp16 allele, prp16-302 (Fig. 5; Hilliker et al. 2007). This rescue is improved when combined with U6–U57A, suggesting inter-U2 and U6 snRNA communication that impacts efficiency from the first step through to the transition to second step. Curiously, stable U2–stem IIc does not enhance the prp16-302 mutation (Fig. 5; data not shown), suggesting that if Prp16p unwinds U2–stem IIc, some other activity of prp16-302 is cold sensitive, or that Prp16p does not unwind stem IIc. These data suggest a cooperative role for U2–stem IIa with Prp16p during the first-to-second step transition. If true, and stem IIc is unwound to form stem IIa, Cus2p and Prp5p may once again aid this step. Alternatively, since stabilizing U2–stem IIa aids an ATPase-deficient Prp16p allele, stem IIa formation (although our genetics suggests that this is not via stem IIc disruption) may be a direct or indirect substrate or product of Prp16p ATPase activity.
Placing new pieces in a dynamic puzzle
We suggest that these changes in U2 snRNA structure provide important spliceosomal architecture that alternately protects or displays pre-mRNA reactive sites. When stabilized stem IIa or stem IIc mutants are expressed, the balance between the two structures is tipped, leading to mistimed rearrangements. This then allows otherwise discarded mutant precursors to enter and proceed through steps of splicing and splice with increased frequency (Burgess and Guthrie 1993; Query and Konarska 2004; Villa and Guthrie 2005).
Genetic and biochemical links between alleles of U6 snRNA, PRP16, and PRP8, also implicated in suppression of branchpoint mutations, have led to a simple two-state hypothesis, and we can now place our data in the context of this model (Burgess and Guthrie 1993; McPheeters 1996; Query and Konarska 2004). Here, the spliceosome maintains equilibrium between two conformational states that favor either the first or second catalytic step. Artificially favoring one or other state (i.e., via a suppressor) alters the balance and facilitates progression of mutant substrates through the spliceosome (Query and Konarska 2004; Villa and Guthrie 2005). In essence, we are stabilizing spliceosomal reactants that are normally in flux and this allows aberrant substrates to participate in splicing. Our findings place dynamic U2–stem IIa and stem IIc rearrangements at stages in spliceosome assembly and splicing previously recognized through specific alleles of U6 snRNA and PRP16 (Burgess and Guthrie 1993; McPheeters 1996; Query and Konarska 2004; Villa and Guthrie 2005). Whereas hyperstabilized U2–stem IIc alleles behave similarly to U6–U57C or prp16-302 in aiding aberrant pre-mRNAs at the first catalytic step, stabilized U2–stem IIa alleles behave like U6–U57A, and rescue prp16-302 in aiding transition to the second catalytic step. Thus, stabilizing one or other U2 snRNA structure at the appropriate time in spliceosome assembly or catalysis promotes progression by favoring necessary conformations. While the overlapping phenotypes we observed with previously described first- or second-step suppressors demonstrate a role for U2 stem IIa or stem IIc at similar and distinct stages in spliceosome assembly, further experimentation is required to determine the exact structural basis and placements of these.
Implications of forming, unforming, and reforming U2–stem IIa
What might be the structural advantage to switching between U2–stem IIa and U2–stem IIc? It has been previously noted that U2–stem IIa bears a striking resemblance to the 715 stem–loop in Escherichia coli 23S ribosomal RNA (Culver et al. 1999). The 715 stem–loop lies on the interface between 30S and 50S subunits and forms an intersubunit contact with S15. Although 23S has no potential to form an equivalent to stem IIc, U2–stem IIa may hold a comparable position on the surface of the U2 snRNP, forming similar inter-snRNP contacts, perhaps with U1 or U6 snRNP.
Our data extends the cross-talk between the U2 and U6 snRNPs by revealing new genetic dependencies between U6–U57 and stem IIa and stem IIc. The link between these parts of U6 and U2 snRNA, in combination with the close proximity of stem IIa and stem IIc to the branchpoint interaction sequence, places these structures and their dynamic switching at the very heart of the catalytic core. This arrangement could represent a critical region that adopts “open” and “closed” conformations, depending on active site accessibility requirements during splicing. Perhaps formed stem IIa helps present the U2 snRNA branchpoint interaction region to the pre-mRNA branchpoint for initial prespliceosome assembly. Following this, released stem IIa and formed stem IIc provides enough torsion within the assembling spliceosome to enable the 2′ hydroxyl of the U2 snRNP-bound branchpoint adenosine, to attack at the 5′ splice site, thus allowing critical contacts for the first catalytic step. Precedence for this comes from documented changes in the accessibility of the U2 branchpoint interaction region from early U2 snRNP assembly through to active spliceosomes (O’Day et al. 1996; Wiest et al. 1996; Abu Dayyeh et al. 2002; Dybkov et al. 2006; Rhode et al. 2006), and raises the intriguing possibility that these changes require concomitant rearrangement from stem IIa to stem IIc. Finally, reformed stem IIa helps the assembling spliceosome to prepare for second-step chemistry. Previous data have implicated this Prp16p_-_mediated transition step as an important checkpoint to ensure fidelity of splice site choice (Burgess et al. 1990; Burgess and Guthrie 1993; Villa and Guthrie 2005). Our data demonstrating decreased splicing (increased fidelity) of aberrant pre-mRNAs and prp16-302 suppression by stabilized stem IIa alleles supports a role for stem IIa in also ensuring fidelity at this step.
Materials and methods
Strains and reporter plasmids
The construction of strain DS4D; MATa, prp5∷Kanr, cus2∷Kanr, snr20∷HIS3, trp1, ura3, leu2, lys2, pIP45 (PRP5 + SNR20 on URA; note that snr20 encodes U2 snRNA) is described in Perriman et al. (2003). YHM187 and YHM118 were obtained from David McPheeters (Case Western Reserve University, Cleveland, OH) (Madhani and Guthrie 1992). pGAC reporter constructs containing ACT1-CUP1 fusion with site-specific mutations at 5′ splice site or branchpoint sequence were obtained from Christine Guthrie (University of California at San Francisco, San Francisco, CA) and are described in detail in Lesser and Guthrie (1993).
Splicing extracts and spliceosome assembly assays
Splicing extracts were isolated as described and derive from yeast strain RP01 (Yan et al. 1998) cotransformed with pRS314CUS2 (CUS2+) or pRS314 (_cus2_Δ) and various U2 genes on a LEU2 plasmid. Splicing and spliceosome assembly were as described (Perriman and Ares 2000; Perriman et al. 2003). U6 snRNA depletion (Fig. 2) was done by adding 45 nM U6-specific oligonucleotide (see Perriman and Ares 2000) and incubating for 15 min at 25°C.
Immunoprecipitation
Protein A-Sepharose beads were swollen overnight in NET-50 (50 mM Tris-CL at pH 7.5, 0.01% Nonidet P-40, 50 mM NaCl) at 100 mg/4 mL at 4°C with slow rotation. A 20-uL packed volume was suspended in 0.4 mL of NET-50 and incubated with 5 μL of 12CA5 antibody for 1 h at 4°C. Twenty microliters of appropriate splicing extract were incubated with 12CA5-bound beads in 0.4 mL of NET-50 for 1 h at 4°C. Immune complexes were collected and washed, and RNA was isolated by addition of 100 uL of 0.3 mM sodium acetate (pH 5.2), 0.2% SDS, 1 mM EDTA, and 10 ug of proteinase K, then incubated for 10 min at 65°C. RNAs were phenol-extracted and ethanol-precipitated, and recovered RNAs were subjected to primer extension using U2-specific oligonucleotide.
Copper sensitivity assays
Yeast cultures were grown to mid-log phase in SCD-Leu medium to maintain the ACT1-CUP1 reporter plasmid. Cultures were diluted to OD600 0.002 and 10 uL of drops were plated on a series of plates containing increasing Cu++ from 0, 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, and 0.4 mM. Plates were assayed for growth after 4 d at 30°C.
Primer extensions
Three micrograms of total yeast RNA from various DS4D yeast strains were annealed to 0.2 ng of primer 3 in 5 uL (5′-ATT AATTCGCTGAACCCG-3′) by incubation for 5 min at 65°C, followed by incubation for 20 min at 45°C. A 5-uL cocktail containing 5 mM DTT, 0.125 mM dNTPs, 100 mM Tris-CL (pH 8.3), 150 mM KCl, 6 mM MgCl2, and 50 U SuperScript III was added and the mix was incubated for 35 min at 48°C. Five microliters of a solution of 10 μg/mL RNaseA, 30 mM EDTA, and 0.6 mM sodium acetate (pH 5.2) were added and incubated for 5 min at 48°C, followed by addition of 5 uL of a solution of 0.2% SDS, 20 ug/mL Proteinase K, 0.6 mM sodium acetate (pH 5.2), and a further 5-min incubation. Samples were ethanol-precipitated and resuspended in 1 uL of 20 ug/mL proteinase K, 25 mM EDTA, and 1 uL formamide loading dye. Products were resolved in 6% denaturing PAGE and visualized on a Typhoon imaging system.
Acknowledgments
We thank Melissa Jurica, Grant Hartzog, and Jon Staley for comments and advice on this manuscript; Jon Staley and Angela Hilliker for sharing unpublished results; John Abelson for plasmid U2-ΔCC; David McPheeters for U6–U57 plasmids and YHM118 and YHM187 yeast strains; and Christine Guthrie for ACT1-CUP1 reporter constructs. This work was supported by GM 040478 from the National Institute of Health to M.A.
Footnotes
References
- Abu Dayyeh B.K., Quan T.K., Castro M., Ruby S.W., Quan T.K., Castro M., Ruby S.W., Castro M., Ruby S.W., Ruby S.W. Probing interactions between the U2 small nuclear ribonucleoprotein and the DEAD-box protein, Prp5. J. Biol. Chem. 2002;277:20221–20233. doi: 10.1074/jbc.M109553200. [DOI] [PubMed] [Google Scholar]
- Ares M., Jr., Igel A.H., Igel A.H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes & Dev. 1990;4:2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- Ares M., Jr., Weiser B., Weiser B. Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog. Nucleic Acid Res. Mol. Biol. 1995;50:131–159. doi: 10.1016/s0079-6603(08)60813-2. [DOI] [PubMed] [Google Scholar]
- Barabino S.M., Sproat B.S., Lamond A.I., Sproat B.S., Lamond A.I., Lamond A.I. Antisense probes targeted to an internal domain in U2 snRNP specifically inhibit the second step of pre-mRNA splicing. Nucleic Acids Res. 1992;20:4457–4464. doi: 10.1093/nar/20.17.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Burgess S.M., Guthrie C., Guthrie C. A mechanism to enhance mRNA splicing fidelity: The RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- Burgess S., Couto J.R., Guthrie C., Couto J.R., Guthrie C., Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990;60:705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- Champion-Arnaud P., Reed R., Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes & Dev. 1994;8:1974–1983. doi: 10.1101/gad.8.16.1974. [DOI] [PubMed] [Google Scholar]
- Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C.A., Guthrie C., Guthrie C. Genetic interactions between the 5′ and 3′ splice site consensus sequences and U6 snRNA during the second catalytic step of pre-mRNA splicing. RNA. 2001;7:1845–1854. [PMC free article] [PubMed] [Google Scholar]
- Culver G.M., Cate J.H., Yusupova G.Z., Yusupov M.M., Noller H.F., Cate J.H., Yusupova G.Z., Yusupov M.M., Noller H.F., Yusupova G.Z., Yusupov M.M., Noller H.F., Yusupov M.M., Noller H.F., Noller H.F. Identification of an RNA-protein bridge spanning the ribosomal subunit interface. Science. 1999;285:2133–2136. doi: 10.1126/science.285.5436.2133. [DOI] [PubMed] [Google Scholar]
- Datta B., Weiner A.M., Weiner A.M. Cross-linking of U2 snRNA using nitrogen mustard. Evidence for higher order structure. J. Biol. Chem. 1992;267:4497–4502. [PubMed] [Google Scholar]
- Dybkov O., Will C.L., Deckert J., Behzadnia N., Hartmuth K., Luhrmann R., Will C.L., Deckert J., Behzadnia N., Hartmuth K., Luhrmann R., Deckert J., Behzadnia N., Hartmuth K., Luhrmann R., Behzadnia N., Hartmuth K., Luhrmann R., Hartmuth K., Luhrmann R., Luhrmann R. U2 snRNA–protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol. Cell. Biol. 2006;26:2803–2816. doi: 10.1128/MCB.26.7.2803-2816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y.W., Zhou Q., Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- Fouser L.A., Friesen J.D., Friesen J.D. Mutations in a yeast intron demonstrate the importance of specific conserved nucleotides for the two stages of nuclear mRNA splicing. Cell. 1986;45:81–93. doi: 10.1016/0092-8674(86)90540-4. [DOI] [PubMed] [Google Scholar]
- Hainzl T., Huang S., Sauer-Eriksson A.E., Huang S., Sauer-Eriksson A.E., Sauer-Eriksson A.E. Structural insights into SRP RNA: An induced fit mechanism for SRP assembly. RNA. 2005;11:1043–1050. doi: 10.1261/rna.2080205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A.K., Staley J.P., Staley J.P. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004;10:921–928. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A.K., Mefford M.A., Staley J.P., Mefford M.A., Staley J.P., Staley J.P. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes & Dev. 2007 doi: 10.1101/gad.1536107. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M.M., Vilardell J., Query C.C., Vilardell J., Query C.C., Query C.C. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol. Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Lamond A.I., Sproat B., Ryder U., Hamm J., Sproat B., Ryder U., Hamm J., Ryder U., Hamm J., Hamm J. Probing the structure and function of U2 snRNP with antisense oligonucleotides made of 2′-OMe RNA. Cell. 1989;58:383–390. doi: 10.1016/0092-8674(89)90852-0. [DOI] [PubMed] [Google Scholar]
- Lesser C.F., Guthrie C., Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H.D., Guthrie C., Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- McPheeters D.S. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA. 1996;2:1110–1123. [PMC free article] [PubMed] [Google Scholar]
- Noller H.F. Biochemical characterization of the ribosomal decoding site. Biochimie. 2006;88:935–941. doi: 10.1016/j.biochi.2006.04.006. [DOI] [PubMed] [Google Scholar]
- O’Day C.L., Dalbadie-McFarland G., Abelson J., Dalbadie-McFarland G., Abelson J., Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J. Biol. Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- Perriman R., Ares M., Jr., Ares M., Jr. ATP can be dispensable for prespliceosome formation in yeast. Genes & Dev. 2000;14:97–107. [PMC free article] [PubMed] [Google Scholar]
- Perriman R., Barta I., Voeltz G.K., Abelson J., Ares M., Jr., Barta I., Voeltz G.K., Abelson J., Ares M., Jr., Voeltz G.K., Abelson J., Ares M., Jr., Abelson J., Ares M., Jr., Ares M., Jr. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl. Acad. Sci. 2003;100:13857–13862. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query C.C., Konarska M.M., Konarska M.M. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- Rhode B.M., Hartmuth K., Westhof E., Luhrmann R., Hartmuth K., Westhof E., Luhrmann R., Westhof E., Luhrmann R., Luhrmann R. Proximity of conserved U6 and U2 snRNA elements to the 5′ splice site region in activated spliceosomes. EMBO J. 2006;25:2475–2486. doi: 10.1038/sj.emboj.7601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S.W., Chang T.H., Abelson J., Chang T.H., Abelson J., Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes & Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Staley J.P., Guthrie C., Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Sun J.S., Manley J.L., Manley J.L. A novel U2–U6 snRNA structure is necessary for mammalian mRNA splicing. Genes & Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- Villa T., Guthrie C., Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes & Dev. 2005;19:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S.E., Ares M., Jr., Ares M., Jr. Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:6337–6349. doi: 10.1128/mcb.14.9.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S.E., Neville M., Haynes M., Wang J., Igel H., Ares M., Jr., Neville M., Haynes M., Wang J., Igel H., Ares M., Jr., Haynes M., Wang J., Igel H., Ares M., Jr., Wang J., Igel H., Ares M., Jr., Igel H., Ares M., Jr., Ares M., Jr. CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes & Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- Wiest D.K., O’Day C.L., Abelson J., O’Day C.L., Abelson J., Abelson J. In vitro studies of the Prp9.Prp11.Prp21 complex indicate a pathway for U2 small nuclear ribonucleoprotein activation. J. Biol. Chem. 1996;271:33268–33276. doi: 10.1074/jbc.271.52.33268. [DOI] [PubMed] [Google Scholar]
- Yan D., Ares M., Jr., Ares M., Jr. Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol. Cell. Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Perriman R., Igel H., Howe K.J., Neville M., Ares M., Jr., Perriman R., Igel H., Howe K.J., Neville M., Ares M., Jr., Igel H., Howe K.J., Neville M., Ares M., Jr., Howe K.J., Neville M., Ares M., Jr., Neville M., Ares M., Jr., Ares M., Jr. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol. Cell. Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavanelli M.I., Ares M., Jr., Ares M., Jr. Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes & Dev. 1991;5:2521–2533. doi: 10.1101/gad.5.12b.2521. [DOI] [PubMed] [Google Scholar]
- Zavanelli M.I., Britton J.S., Igel A.H., Ares M., Jr., Britton J.S., Igel A.H., Ares M., Jr., Igel A.H., Ares M., Jr., Ares M., Jr. Mutations in an essential U2 small nuclear RNA structure cause cold-sensitive U2 small nuclear ribonucleoprotein function by favoring competing alternative U2 RNA structures. Mol. Cell. Biol. 1994;14:1689–1697. doi: 10.1128/mcb.14.3.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]