uPAR and HER-2 gene status in individual breast cancer cells from blood and tissues (original) (raw)
Abstract
Overexpression of urokinase plasminogen activator system or HER-2 (erbB-2) in breast cancer is associated with a poor prognosis. HER-2 overexpression is caused by HER-2 gene amplification. The anti-HER-2 antibody trastuzumab significantly improves clinical outcome for HER2-positive breast cancer. Drugs that target the uPA system are in early clinical trials. The aims of this study were to determine whether urokinase plasminogen activator receptor (uPAR) gene amplification occurs and whether analysis of individual tumor cells (TCs) in the blood or tissue can add information to conventional pathological analysis that could help in diagnosis and treatment. Analysis of individual TCs indicates that uPAR amplification occurs in a significant portion of primary breast cancers and also circulating tumor cells (CTCs) from patients with advanced disease. There was complete concordance between touch preps (TPs) and conventional pathological examination of HER-2 and uPAR gene status in primary tumors. There was also excellent concordance of HER-2 gene status between primary tumors and CTCs provided that acquisition of HER-2 gene amplification in CTCs was taken into account. Unexpectedly, gene amplification of HER-2 and uPAR occurred most frequently in the same TC and patient, suggesting a biological bias and potential advantage for coamplification. Expression of HER-2 and uPAR in primary tumors predicted gene status in 100 and 92% of patients, respectively.
Keywords: oncogene coamplification, expression, amplification
The HER-2/neu proto-oncogene is amplified in ≈25% of invasive breast cancers. HER-2 overexpression has been associated with a decreased time of disease-free survival and other markers of poor prognosis. Trastuzumab (Herceptin, Genentech, South San Francisco, CA) is a HER-2-directed humanized antibody that is effective for the treatment of patients with metastatic or early stage breast cancer (1–3). The methods used for selecting patients for trastuzumab therapy are immunohistochemical (IHC) and gene-based FISH analysis of primary tumor. Retrospective studies have shown that measurement of gene amplification is the best predictive marker of response to trastuzumab-based therapy (4–6).
Activation of the urokinase plasminogen activator (uPA) system is also associated with a poor prognosis in breast cancer, with the greatest amount of evidence for uPA overexpression. However, it is well documented that uPA receptor (uPAR) overexpression is also associated with increased tumor aggressiveness and worse disease-free survival and overall survival in breast and other cancers (7–15). Very little is known about the regulation of uPAR expression. Its gene resides on the long arm of chromosome 19, encodes a protease of molecular mass 35 kDa, and is linked to the outer surface of the plasma membrane by a glycosyl-phosphatidylinositol anchor. When uPAR interacts with one of its ligands, uPA, plasminogen is cleaved to active plasmin, which degrades several extracellular matrix (ECM) components and also activates many promatrix metallo-proteases. As ECM is the major physical obstacle for cancer cells to penetrate and invade the surrounding tissue, proteolytic degradation of ECM allows cellular migration and penetration through tissue boundaries, and facilitates metastasis. In addition, other signaling pathways are activated to induce cellular proliferation, motility, and additional remodeling of the ECM. As a result, overexpression in breast cancer and many other cancers (7–15) is associated with a poor prognosis, and therapies targeting the uPA system are under development.
Here, we have analyzed uPAR and HER-2 gene status in individual tumor cells (TCs) from TPs of primary breast carcinomas and also from circulating TCs (CTCs) in patients with advanced recurrent breast carcinoma. One objective was to determine whether the uPAR gene, like HER-2, can be amplified. A second objective was to demonstrate the advantages of individual TC-analysis over conventional pathological analysis, which determines the average gene and expression status of a population of TCs.
Results
Characteristics of Individual TCs.
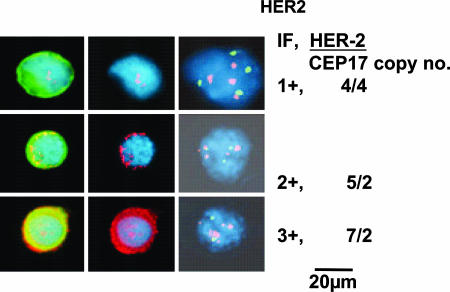
Isolated epithelial cells in our slide-based assay can be unambiguously characterized as TCs because of their distinctive immunophenotype, cytomorphology, and, most importantly, the presence of aneusomy (16–18). Fig. 1Left shows a typical single CTC. As can be seen, the nuclear-to-cytoplasmic ratio is high, the nucleus frequently has irregularities, and cytokeratin stains the cytoplasm at the periphery of the cell causing a ring-like appearance, all being accepted morphological criteria for malignant cells. Fig. 1 Center shows HER-2 expression by immunofluorescence (IF) in the same CTC. Fig. 1 Right shows the results of FISH analysis. The signals can be precisely read on individual CTCs. Hence, it is relatively straight-forward to subjectively quantify individual TCs or CTCs into 1–3+ expression (this classification is unrelated to IHC classification of HER-2 expression in a pathological tissue section), and the copy numbers of HER-2 and CEP17 are shown to the right of Fig. 1.
Fig. 1.
Correlation of HER-2 expression and gene amplification in CTCs. (Left) Representative CTCs: nucleus (blue); cytokeratin IF (green); HER-2 IF (red) (overlap of red/green is yellow). (Center) The same cells: nucleus (blue); HER-2 (red). (Right) FISH analysis: HER-2 copies (spectrum orange); CEP 17 copies (spectrum green). HER-2 expression, 1–3+, and HER-2 copy number/CEP17 copy number are shown at right.
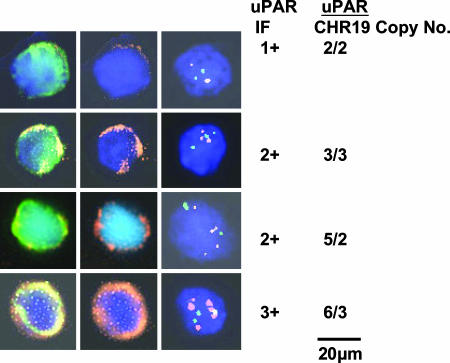
Fig. 2 shows uPAR gene amplification and expression in representative CTCs, similar to Fig. 1.
Fig. 2.
Correlation of uPAR expression and gene amplification in CTCs. (Left) Representative CTCs. (Center) The same cells: nucleus (blue); uPAR (orange). (Right) FISH analysis: uPAR copies (spectrum orange); chr 19 copies (spectrum green); uPAR expression, 1–3+, and uPAR copy number/chr 19 copy number are shown at right.
Correlation of HER-2 and uPAR Gene Status Between Touch Preps and Primary Tumors.
For individual TC–analysis of tumor tissue, TPs from the breast tumor's cut surface was used. For interpretation of the tumor's gene status, it is necessary to determine whether the gene status from TPs is concordant with conventional pathological analysis.
The results of HER-2 gene status using TPs (100 TCs per patient) compared with the pathologist's analysis showed concordance in all 21 patients [100% concordance, with 95% confidence interval (CI) of 84–100%]. Moreover, the ratios using TP and conventional pathological analysis were very close in each patient, HER-2+ patients: average 3.67 (SD 1.35) and 4.57 (SD 1.81), respectively; HER-2− patients: average 1.43 (SD 0.18) and 1.25 (SD 0.25), respectively. The results of uPAR gene status using TPs compared with the pathologist's analysis were also concordant in all seven patients (100% concordance with 95% CI of 59–100%). Moreover, the ratios using TP and conventional pathological analysis were very close in each patient, uPAR+ patients: average 2.40 (SD 0.17) and 2.17 (SD 0.06), respectively; uPAR− patients: average 1.55 (SD 0.10) and 1.35 (SD 0.18), respectively. However, we have observed discordances in the past of HER-2 gene status between CTCs and pathological analysis that were caused by heterogeneity in the tumor. Hence, it is essential to extend the uPAR correlations to a much larger number of tumors. Therefore, TPs can be used to determine HER-2 and, potentially, uPAR gene status in patients.
In HER-2-amplified cases using TPs and CTCs, 92% (23 of 25) were also uPAR gene amplified, whereas in HER-2 nonamplified patients, only 3% (1 of 39) were uPAR gene amplified. Therefore, HER-2 and uPAR gene status are highly correlated in patients (P < 0.01). Comparison of the extent of HER-2 to uPAR gene amplification in HER-2+ patients showed 3.4 (SD1.4) to 2.3 (SD 0.28) respectively. Thus, HER-2 amplification is significantly higher than uPAR amplification if the latter is calculated by HER-2 criteria. However, expression of uPAR to be described below indicated that the most effective method to obtain concordance with uPAR gene amplification was to subtract copy numbers of CHR19 from those of uPAR rather than use the ratio. Using this method of calculation, the average gene amplification value of uPAR was 3.2 (SD 0.9), and the concordance of uPAR expression and amplification was 96% vs. 93% using HER-2 criteria. Clearly, more studies are needed to determine how to calculate uPAR gene status. The clinical significance of the above differences is not known.
Distribution of HER-2 and uPAR Gene Status in Individual TCs of HER-2+ Cases.
We classified the gene status of individual cells (TPs of primary tumors and CTCs) into four subsets: both genes amplified, only one of the genes amplified, or neither amplified. The results of gene amplifications for TCs from 12 HER-2+ primary tumors (100 TCs per patient) and CTCs from 2 HER-2+ patients (50 and 100 CTCs per patient) are shown in Table 1.
Table 1.
Distribution of HER-2 and uPAR gene amplification in individual TCs of HER-2+ patients
| Subset | % of TCs amplified in individual patients | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TPs of HER-2+ primary tumors | CTCs | |||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
| Both genes amplified | 86 | 58 | 81 | 45 | 50 | 75 | 80 | 67 | 73 | 73 | 90 | 88 | 55 | 63 |
| Only Her-2 gene amplified | 3 | 2 | 16 | 24 | 26 | 11 | 19 | 17 | 10 | 10 | 7 | 5 | 16 | 37 |
| Only uPAR gene amplified | 2 | 27 | 0 | 9 | 5 | 3 | 1 | 0 | 2 | 0 | 0 | 2 | 7 | 0 |
| Neither gene amplified | 9 | 13 | 3 | 22 | 19 | 11 | 0 | 16 | 15 | 17 | 3 | 5 | 22 | 0 |
As can be seen, there is a marked preference for amplification of both HER-2 and uPAR genes to occur in the same TC rather than in separate TCs in HER-2-amplified primary tumors, as well as CTCs from two patients with advanced disease. Using linear regression models, both the HER-2 copy number ability to predict uPAR copy number and the HER-2 ratio predicting uPAR ratio also gave highly significant P values for each patient. The patients described in Table 1 all had a sufficient number of HER-2-amplified TCs to allow statistical analysis of gene status. Patients with HER-2-negative primary tumors (16 patients) or CTCs (24 patients) did not have a sufficient number of HER-2- or uPAR-amplified TCs to perform meaningful analysis. Thirteen HER-2-amplified cases with CTCs did not have sufficient numbers of CTCs for statistical analysis except for the two patients in Table 1. However, there was the same preference for coamplification as shown in Table 1 in virtually every patient.
Correlation of HER-2 Gene Status Between CTCs and Primary Tumor.
The HER-2 gene status of CTCs in 52 patients with recurrent or progressive advanced breast cancer was compared with the pathologist's analysis of the HER-2 gene status of their primary tumor. Consistent with our prior results (19), five formerly HER-2− cases acquired HER-2+ CTCs upon recurrence or progression. Their patterns of HER-2 distribution were similar to those whose primary tumor was HER-2+. We did not consider these patients as discordant (i.e., if HER-2 status had been determined at the same point in time for CTCs and tumor tissue, there would have been concordance). However, in two patients whose primary tumor was HER-2+ and trastuzumab treatment had been stopped 3–4 months earlier, the HER-2 analysis of CTCs yielded ratios of 1.6 and 1.3, respectively. We consider these two patients as discordant, although we cannot exclude an effect of past treatment with trastuzumab on our results. This finding implies a 96% concordance for HER-2 gene status between CTCs and primary tumor analysis by pathologists (95% CI of 87–100%).
Correlation of Expression of HER-2 and uPAR with Gene Status in Primary Tumors and CTCs.
We studied the correlation between expression and gene status in 72 patients using TPs of the primary tumor or CTCs from patients who had at least 10 CTCs. We used 10 consecutive CTCs (a bin) and called the entire bin 1+, 2+, or 3+ (see Fig. 1) if five or more of the cells were classified with the same expression. If there was discordance among bins within a patient, we used the classification of the majority of the bins to determine the classification of the patient. Patients in which the majority of bins were evaluated as 3+ were considered to be HER2 or uPAR gene amplified; otherwise, the patient's tumor was considered nonamplified.
With regard to primary tumors, 27 TPs (100 TCs per patient; i.e., 10 bins) were evaluated for correlation between expression and gene status. Using one sample binomial proportion, the results of TPs indicate a concordance of 100% (95% CI, 89–100%) for HER-2. HER-2 expression is very significantly associated with HER-2 gene amplification (P < 0.001 using exact binomial test). For uPAR, the concordance was 96% (95% CI, 83–100%) if the copy number of chromosome 19 was subtracted from the copy number of uPAR. uPAR expression is very significantly associated with uPAR gene amplification (P < 0.001 using exact binomial test).
With regard to CTCs, 10 patients (193 CTCs) exhibited HER-2 amplification; 36 patients (670 CTCs) were nonamplified. There was a concordance in 44 of 46 patients between HER-2 expression and gene status in the CTCs (96% concordance, 95% CI, 86–99%). For uPAR, the concordance was 92% (95% CI, 65–100%, 11 of 12 patients). The results suggest a significance of uPAR amplification associated with uPAR expression (P = 0.02 using exact binomial test). Therefore, correlation between protein expression and gene status for HER-2 and uPAR in patients is very high. However, expression of HER-2 and uPAR proteins cannot be quantitatively compared because different fluorochromes were conjugated to the two antibodies.
Subset Analysis.
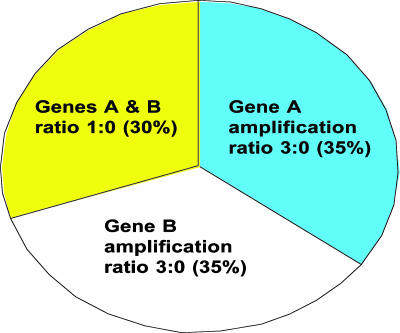
Fig. 3 illustrates a theoretical division of a population of TCs from a patient into three subsets, each with a different level of oncogene status. As can be seen, neither oncogene A nor B make the patient a candidate for targeted therapy for either oncogene; amplification of each gene = 1.7 using HER-2 gene criteria. However, if there were effective targeting drugs for both oncogenes, the patient might qualify as a candidate for simultaneous administration of both drugs (amplification of A and B together = 2.4).
Fig. 3.
Theoretical division of CTCs into subsets with different degrees of amplification for two oncogenes (A and B).
Discussion
The two major conclusions to be derived from the present study of individual TCs are: (i) Individual tumor cell analysis can provide additional information not available from conventional pathological analysis i.e., multigene analysis of single tumor cells, subdividing a population of tumor cells into subsets that each display a particular genetic change and allowing more precise predictability of gene amplification from gene expression. (ii) uPAR gene amplification is described in breast cancer. To our knowledge, this is the first report of such amplification in any cancer.
Individual tumor cell analysis is a relatively recent assay confined primarily to CTCs and bone marrow. In the present study, we have extended such analysis to TPs from primary tumor. Multigene analysis was demonstrated in individual cells and the implications will be discussed below. Conventional pathological analysis and touch prep assays showed complete concordance with HER-2 and uPAR gene status in 21 and 7 consecutive patients, respectively, thereby validating the use of TPs for this purpose (P = 0.0001). Similar results were obtained with HER-2 gene status of CTCs and pathological analysis of primary tumors provided acquisition (i.e., clonal selection/expansion) of HER-2 gene amplification in CTCs associated with cancer progression was taken into consideration (19). Expression of HER-2 protein in 52 cases using either their primary tumors or CTCs showed a minimum of 96% concordance with HER-2 gene amplification determined by the pathologist. As shown in Fig. 1, subjective quantification of intensity of staining of nonoverlapping tumor cells flattened on a slide is readily achievable.
The above results contrast with the limitation of conventional pathological analysis for marker and gene status. For example, to determine HER-2 gene status, the pathologist selects one site at the growing edge of the tumor; paraffin sections are prepared necessitating denaturation and loss of protein antigens by the fixatives; 60 nuclei are counted from overlapping cells cut horizontally; estimates of the intensity of staining by immunohistochemistry are made, and if necessary, FISH for determining HER-2 gene status is performed. After appropriate calculation, the average of the population determines the gene status. The results are associated with significant errors in estimating the extent of IHC-staining to determine gene status (4–6) and continued controversy concerning the method for calculating gene amplification (20–25). Also, it is not possible to provide the additional information concerning individual TCs cited above. These considerations in no way challenge the necessity for conventional pathological analysis but indicate potential benefits if individual tumor cell assay can be incorporated into pathological analysis in the future.
The finding of uPAR gene amplification in 20–25% of breast cancer patients is consistent with published data on overexpression of uPAR as a frequent occurrence in breast cancer. Overexpression of uPAR is associated with increased metastases, heightened malignancy and a shorter survival time (7–15). Indeed, overexpression of uPA, a ligand for uPAR, is the only breast cancer marker with Level of Evidence-1 for a poor prognosis (26–28). Because there are drugs already in early clinical trials (29, 30) and a large number of candidate drugs from animal models and cell culture experiments (7, 8) whose aim is to block the function of uPA–uPAR interaction, it is important to determine the criteria for candidacy of breast cancer patients for these putative drugs. The present results represent a first step toward this goal. The next is a large clinical trial to further test protein expression, gene status, and correlation of these parameters to clinical outcomes. Only then can tentative criteria for treatment with putative targeting drugs for uPAR gene amplification be established.
Analysis of uPAR and HER-2 gene status in TPs of primary breast carcinomas and CTCs of patients with advanced breast carcinomas revealed a marked bias for amplification of both oncogenes in the same TC. The higher the HER-2 ratio, the more likely it was to have coamplification and to have higher levels of uPAR amplification. The concordance of gene amplification patterns between primary tumor and CTCs obtained much later in the course of disease (taking into account acquisition of HER-2 gene amplification) also supports the validity of using CTCs in monitoring the genetic status of tumors (19). The likelihood of HER-2 and uPAR gene overexpression occurring in the same primary breast cancer by conventional pathological analysis has been reported (31) and is consistent with coamplification. The possible clinical implications of coamplification on the responsiveness of HER-2+ patients to trastuzumab require study.
Coamplification suggests crosstalk and cooperativity between the HER-2 and uPAR signaling pathways. A prior report (32) using breast cancer cell lines showed that overexpression of HER-2 led to up-regulation and activation of phosphokinase C (PKC). In breast cancer patients' samples, HER-2 overexpressing tumors showed significantly higher PKC membrane staining compared with HER-2 low expressing tumors. PKC is coimmunoprecipitated with Src and activity can be decreased by a Src inhibitor (PP2) and by the expression of a dominant negative mutant of Src. In addition, HER-2-mediated up-regulation of uPAR is reduced by either PKC or Src inhibitors. Inhibition of PKC or Src in HER-2 overexpressing breast cancer cells by chemical inhibitors, dominant-negative mutants, or si-RNA decreased uPAR expression and cancer cell invasion. Taken together, these results indicate that HER-2-activated Src and PKC kinases play critical roles in the HER-2-mediated up-regulation of uPAR and the enhancement of cancer cell invasiveness.
HER-2 and uPAR gene amplification are the main contributors to overexpression of their respective receptors. Using strict criteria, EGFR is also amplified and overexpressed in 6% of breast cancer patients (33). In contrast, there is considerable evidence that overexpression of ligands depends largely on up-regulation of transcriptional/translational mechanisms (31). The question arises as to whether overexpression of target receptors in breast and other cancers mainly depends on gene amplification (or mutation), whereas overexpression of their ligands depends mainly on regulation by nongenomic mechanisms as observed in most normal cells. Thus, from the evolutionary viewpoint of the cancer cell, amplification of receptor genes permanently marks the TC as a prime candidate for increased malignancy. In contrast, amplification of the ligand gene, whose product is secreted, is an inefficient way to gain increased malignancy because “receptive” TCs are needed to accomplish increased malignancy. For gene-amplified TCs, only the widely available transcriptional/translational mechanisms for up-regulation of ligands in malignant or normal cells (stromal, etc.) (34, 35) are needed for increased malignancy. Analysis of additional ligand and receptor genes by FISH are needed to answer this question.
Materials and Methods
Patient Selection and Data Recording.
Women with a documented histological diagnosis of primary breast carcinoma or those with advanced metastatic breast carcinoma and normal age-matched controls were recruited. A few patients with primary breast cancer had received brief preoperative chemotherapy, and most patients with advanced breast cancer were experiencing progression of cancer on chemotherapy, along with trastuzumab in HER-2+ cases. Pertinent demographic and clinical data were recorded for every patient. All specimens were obtained with informed consent and collected using protocols approved by the Institutional Review Board at the University of Texas Southwestern Medical Center.
Tissue Acquisition.
The University of Texas Southwestern Tissue Repository is a core facility that provided frozen breast cancer tissue from which we made touch preps of primary cancers. Tumor tissue was washed for 2 min two times in 1× PBS and was cut into four sections. Each section was then touched gently onto a poly(l-lysine)-coated slide. Slides were air dried and fixed with 95% ethanol for 10 min. The clinical pathology department has paraffin block specimens corresponding to all of the frozen specimens.
Collection of Blood Samples.
Thirty milliliters of blood were drawn from the antecubital vein of patients into 10-ml Vacutainer tubes (BD Biosciences, San Diego, CA) containing EDTA, or into CellSave tubes from Immunicon (Huntingdon, PA). The EDTA samples were processed within 4 h of collection, and the CellSave samples were processed within 24 h.
Cell Lines.
Carcinoma cell lines SKBr3 (breast), Colo205 (colon), and PC3 (prostate) were used for testing of antibodies and ferrofluids. Cells were grown in RPMI medium 1640 containing 10% FCS.
Ferrofluids.
CTCs were immunomagnetically enriched by using ferrofluid (Immunicon) that is conjugated to a monoclonal mouse antibody against a human pan-epithelial cell adhesion molecule (GA73.3, provided by D.H.)
Isolation of CTCs.
Briefly, 5-ml aliquots of blood were incubated with ferrofluid and placed in a magnetic field to draw the tumor cell–ferrofluid complexes to the side of the tube, allowing the unwanted portion containing normal blood cells and plasma to be aspirated away. Buffer was added, and the sample was centrifuged to remove unbound ferrofluid. The pellet was resuspended in PBS and placed on a slide to air dry. For details, see ref. 13.
Antibodies.
We used the following antibodies for direct staining: (i) pan-anti-cytokeratin clone C11-FITC (Sigma, St. Louis, MO), (ii) anti-CD45 (clone 9.4 from American Type Culture Collection) conjugated in our laboratory to Alexa Fluor 546 (Molecular Probes, Eugene, OR), (iii) anti-HER81 mouse monoclonal antibody (E. Vitetta, University of Texas Southwestern Medical Center) conjugated to Alexa Fluor 594, and (iv) anti-uPAR mouse monoclonal antibody, clone 62022, (R & D Systems, Minneapolis, MN) conjugated in our laboratory to Alexa Fluor 546.
IF.
Staining.
The detection of epithelial cells is accomplished by IF using anti-CK and anti-CD45. Briefly, nonspecific sites were blocked with PBS plus 5% BSA for 30 min. Then, the diluted CK, CD45, and HER-2 or uPAR antibodies were added and the slides incubated for 45 min at room temperature, then washed three times in PBS-1% Tween. After air-drying, the slides were mounted and the nucleus counterstained with mounting media containing DAPI (Vectashield, Vector Laboratories, Burlingame, CA). The slides were stored at 4°C.
Detection of TCs.
Slides were examined manually for the presence of epithelial cells using a fluorescent microscope (Axiophot, Zeiss, Jena, Germany). Scanning for CK+ and CD45− cells was performed with a single bandpass filter for FITC and Alexa Fluor 546. To assure that the FITC staining was associated with the correct cell, the same area was evaluated with a dual band-pass filter for FITC/DAPI. As the microscope is equipped with an automated stage, the location of each tumor cell on the slide was recorded and stored. An image from each positive cell was acquired to confirm that the same cell was relocated when performing multicolor FISH.
Multicolor FISH.
Hybridization of touch preps and CTCs.
Hybridization and posthybridization procedures were performed according to manufacturer's instructions (Abbott Molecular, Des Plaines, IL). HER-2 FISH was analyzed first, and then the TC was reprobed for uPAR.
Evaluation.
After hybridization, the cell image from automated relocation and the previously recorded image were visually compared to confirm relocation accuracy. The FISH signals of TCs were evaluated by using specific filters for each fluorochrome. One probe set contained a FISH probe hybridizing to a region including the HER-2 gene at 17q12 and a probe controlling for chromosome 17 aneusomy hybridizing to the peri-centromeric alpha satellite region of chromosome 17 (PathVysion, Abbott Molecular). The second probe set contained a FISH probe hybridizing to a region including the uPAR gene at 19q13.31, labeled with SpectrumOrange, and a control probe targeting the 19p13 locus on the opposite arm of chromosome 19 (target region includes ZNF44 and MAN2B1 genes), labeled with SpectrumGreen. For each relocated cell, an image was recorded to document the results of FISH. The leukocytes on the patients' slides were used as control cells for the hybridization quality. Gene amplification within a cell was defined as either ≥2 HER-2 signals per chromosome 17 peri-centromeric signals, or ≥2 uPAR signals per chromosome 19 p13 signals.
Acknowledgments
We thank Keren Elias and Linda Flores for their work as our Research Coordinators, Erica Garza and Linda Berry for administrative assistance, Jennifer Sayne and Dr. Richard Scheuermann for providing tissue specimens from the University of Texas Southwestern Medical Center repository, and D. Hayes for critical review of this manuscript. This work was supported by National Cancer Institute Grant CA 101790-01A3 and Susan B. Komen Foundation Grant BCTR0504014.
Abbreviations
uPA
urokinase plasminogen activator
uPAR
uPA receptor
TC
tumor cell
CTC
circulating TC
IF
immunofluorescence
CI
confidence interval
TP
touch prep.
Note Added in Proofs.
The findings of Urban et al. (36) are highly pertinent to our findings. For amplification of HER-2 to make a breast cancer more aggresive, the tumor must also have uPA (a ligand for uPAR) overexpression. Therefore, we speculate that coamplification of uPAR and HER-2 may contribute to the unexpected and provocative findings of Urban et al.
Footnotes
Conflict of interest statement: J.U. holds stock in Immunicon Corporation.
See Commentary on page 17073.
References
- 1.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 2.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, et al. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 3.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, et al. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 4.Yeon CH, Pegram MD. Invest New Drugs. 2005;23:391–409. doi: 10.1007/s10637-005-2899-8. [DOI] [PubMed] [Google Scholar]
- 5.Ross JS, Fletcher JA, Bloom KJ, Linette GP, Stec J, Summans WF, Pusztai L, Hortobagyi GN. Mol Cell Proteomics. 2004;3:379–398. doi: 10.1074/mcp.R400001-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 7.Sidenius N, Blasi F. Cancer Metastasis. 2003;22:205–222. doi: 10.1023/a:1023099415940. [DOI] [PubMed] [Google Scholar]
- 8.Romer J, Nielsen BS, Ploug M. Curr Pharm Des. 2004;10:2359–2376. doi: 10.2174/1381612043383962. [DOI] [PubMed] [Google Scholar]
- 9.Grondahl-Hansen J, Peters HA, van Putten WL, Pappot H, Ronne E, Dano K, Klijn JG, Foekens JA. Clin Cancer Res. 1995;1:1079–1087. [PubMed] [Google Scholar]
- 10.de Witte JH, Foekens JA, Brunner N, Heuvel JJ, van Tienoven T. Br J Cancer. 2001;85:85–92. doi: 10.1054/bjoc.2001.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemsen A, Riethdorf L, Brunner N, Berger J, Ebel S, Thomssen C, Janicke F, Pantel K. Int J Cancer. 2003;107:903–909. doi: 10.1002/ijc.11488. [DOI] [PubMed] [Google Scholar]
- 12.Riisbro R, Christensen IJ, Piironen T, Greenall M, Larsen B, Stephens RW, Han C. Clin Cancer Res. 2002;8:1132–1141. [PubMed] [Google Scholar]
- 13.Duggan C, Maquire T, McDermott E, O'Higgins N, Fennelly JJ, Duffy MJ. Int J Cancer. 1995;61:597–600. doi: 10.1002/ijc.2910610502. [DOI] [PubMed] [Google Scholar]
- 14.Kotzsch M, Farthmann J, Meye A, Fuessel S, Baretton G, Tjan-Heijnen VC. Eur J Cancer. 2005;41:2760–2768. doi: 10.1016/j.ejca.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Werle B, Kotzsch M, Lah TT, Kos J, Gabrijelcic-Geiger D. Anticancer Res. 2004;24:4147–4161. [PubMed] [Google Scholar]
- 16.Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, Beitsch PD, Leitch M, Hoover S, Euhus D, et al. Clin Cancer Res. 2004;10:8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 17.Fehm T, Morrison L, Saboorian H, Hynan L, Tucker T, Uhr J. Breast Cancer Res Treat. 2002;2379:1–13. doi: 10.1023/a:1019901010758. [DOI] [PubMed] [Google Scholar]
- 18.Fehm T, Sagalowsky AI, Clifford E, Beitsch PD, Saboorian H, Euhus D, Meng S, Morrison L, Tucker T, Lane N, et al. Clin Cancer Res. 2002;8:2073–2084. [PubMed] [Google Scholar]
- 19.Meng S, Tripathy D, Shete S, Ashfaq R, Haley B, Perkins S, Beitsch P, Khan A, Euhus D, Osborne C, et al. Proc Natl Acad Sci USA. 2004;101:9393–9398. doi: 10.1073/pnas.0402993101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Downs-Kelly E, Yoder BJ, Stoler M, Tubbs RR, Skacel M, Grogan T, Roche P, Hicks DG. Am J Surg Pathol. 2005;29:1221–1227. doi: 10.1097/01.pas.0000165528.78945.95. [DOI] [PubMed] [Google Scholar]
- 21.Salido M, Tusquets I, Corominas JM, Suarez M, Espinet B, Corzo C, Bellet M, Fabregat X, Serrano S, Sole F. Breast Cancer Res. 2005;7:R267–R273. doi: 10.1186/bcr996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y, Lespagnard L, Durbecq V, Paesmans M, Desmedt C, Gomex-Galdon M, Veys I, Cardoso F, Sotiriou C, Dileo A, et al. Clin Can Res. 2005;11:4393–4399. doi: 10.1158/1078-0432.CCR-04-2256. [DOI] [PubMed] [Google Scholar]
- 23.Owens MA, Horten BC, Da Silca MM. Clin Breast Cancer. 2004;5:63–69. doi: 10.3816/cbc.2004.n.011. [DOI] [PubMed] [Google Scholar]
- 24.Ross JS, Fletcher JA, Bloom KJ, Linette GP, Stec J, Clark E, Ayers M, Symmans WF, Pusztai L, Hortobagyi GN. Am J Clin Pathol. 2003:S53–S71. doi: 10.1309/949FPQ1AQ3P0RLC0. Dec. [DOI] [PubMed] [Google Scholar]
- 25.Winston JS, Ramanaryanan J, Levine E. Am J Clin Pathol. 2004:S33–S49. doi: 10.1309/9UNL7UXPYO6CPWBQ. Jun. [DOI] [PubMed] [Google Scholar]
- 26.Look MP, Van Putten WLJ, Duffy MJ, Harbeck N, Christensen TJ, Thomssen C, Kates R, Spyratos F, Ferno M, Eppenberger-Castori S, et al. J Natl Cancer Inst. 2002;94:116–128. doi: 10.1093/jnci/94.2.116. [DOI] [PubMed] [Google Scholar]
- 27.Hayes DF. Breast. 2005;14:493–499. doi: 10.1016/j.breast.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 28.Duffy MJ. Clin Chem. 2002;48:1194–1197. [PubMed] [Google Scholar]
- 29.Siafaca K, Dugan J, editors. Future Oncology. Vol 8. Lake Forest, CA: New Medicine; 2005. pp. 1761–1784. [Google Scholar]
- 30.Berlenbilt A, Matulonis UA, Kroener JF, Dezube BJ, Lam GN, Cuasay JC, Brunner N, Jones TR, Silverman MH, Gold MA. Gynecol Oncol. 2005;1:50–57. doi: 10.1016/j.ygyno.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 31.Han B, Nakamura M, Mori I, Nakamura Y, Kakudo K. Oncol Rep. 2005;14:105–112. [PubMed] [Google Scholar]
- 32.Tan M, Sun M, Yin G, Yu D. Oncogene. 2006;25:3286–3295. doi: 10.1038/sj.onc.1209361. [DOI] [PubMed] [Google Scholar]
- 33.Bhargava R, Gerald WL, Li AR, Pan Q, Lal P, Ladanyi M, Chen B. Mod Pathol. 2005;18:1027–1033. doi: 10.1038/modpathol.3800438. [DOI] [PubMed] [Google Scholar]
- 34.Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 35.Helenius MA, Savinainen KJ, Bova GS, Visakorpi T. BJU Int. 2005;97:404–409. doi: 10.1111/j.1464-410X.2005.05912.x. [DOI] [PubMed] [Google Scholar]
- 36.Urban P, Vuaroqueaux V, Labuhn M, Delorenzi M, Wirapati P, Wight E, Senn HJ, Benz C, Eppenberger-Castori S. J Clin Oncol. 2006;24:4245–4252. doi: 10.1200/JCO.2005.05.1912. [DOI] [PubMed] [Google Scholar]