Characterization of Gene Expression Profiles in Intraductal Papillary-Mucinous Tumors of the Pancreas (original) (raw)
Abstract
The molecular pathology of precursor lesions leading to invasive pancreatic ductal adenocarcinomas remains relatively unknown. We have applied cDNA microarray analysis to characterize gene expression profiles in a series of intraductal papillary-mucinous tumors (IPMTs) of the pancreas, which represents one of the alternative routes of intraepithelial progression to full malignancy in the pancreatic duct system. Using a cDNA microarray containing 4992 human genes, we screened a total of 13 IPMTs including nine noninvasive and four invasive cases. Expression change in more than half of the tumors was observed for 120 genes, ie, 62 up-regulated and 58 down-regulated genes. Some of the up-regulated genes in this study have been previously described in classical pancreatic carcinomas such as lipocalin 2, galectin 3, claudin 4, and cathepsin E. The most highly up-regulated genes in IPMTs corresponded to three members of the trefoil factor family (TFF1, TFF2, and TFF3). Immunohistochemistry performed on five genes found to be differentially expressed at the RNA level (TFF1, TFF2, TFF3, lipocalin 2, and galectin 3) showed a good concordance between transcript level and protein abundance, except for TFF2. Hierarchical clustering organized the cases according to the dysplastic and invasive phenotype of theIPMTs. This analysis has permitted us to implicate several genes (caveolin 1, glypican 1, growth arrest-specific 6 protein, cysteine-rich angiogenic inducer 61) in tumor progression. The observation that several genes are differentially expressed both in IPMTs and pancreatic carcinomas suggests that they may be involved at an early stage of pancreatic carcinogenesis.
Intraductal papillary-mucinous tumors (IPMTs) are a distinct form of exocrine pancreatic neoplasm characterized by dilated ducts that are lined by a proliferation of papillary mucinous epithelium. 1-3 Although IPMTs usually show a favorable outcome compared with classical ductal pancreatic adenocarcinomas, all gradations from low- and high-grade dysplasia through to invasive carcinoma may be encountered. This uncommon type of pancreatic tumor represents a clinically detectable model of intraepithelial neoplasia.
Whereas considerable insights into the genetic basis of classical ductal pancreatic adenocarcinoma have been generated, less is known about the genetic alterations in progenitor lesions. 4-6 Since a progressive accumulation of genetic alterations is now widely accepted for the development of tumors, the identification of the molecular events involved in each step of tumor progression is essential in understanding pancreatic carcinogenesis. Various factors account for our knowledge in this field being less advanced than for tumors in other organs such as colorectal adenoma/carcinoma. In contrast to colonic adenomas, the pancreatic pre-neoplastic lesions are almost always discovered microscopically only after fixation, are relatively inaccessible to biopsy and below the resolution of current imaging modalities. Such lesions are heterogeneous in their topography and degree of dysplasia, are often situated in an abundant stroma, and the possibility of ductal colonization from the invasive component is not always easy to exclude. Furthermore, until recently, 7 a standard nomenclature of pancreatic intraepithelial neoplasia (PanIN) was not established making the comparison of studies between different investigators difficult. Even though laser capture microdissection enables the procurement of pure cell populations, this technique is extremely laborious and limited by the difficulties in grading the dysplasia on frozen sections.
To avoid these problems, we have decided to analyze the gene expression profile in IPMTs, which could potentially represent an early lesion in pancreatic carcinogenesis. As with PanIN, IPMTs can progress from hyperplasia to dysplastic lesions to an invasive carcinoma. However, despite this similar morphological progression, some histological features differentiate these two types of lesions, with a macropapillary proliferation and marked mucosecretion being the characteristics of IPMTs. 8 Furthermore, the prevalence of Ki-ras, p53, CDKN2/p16, and MADH4 mutations appear lower in IPMTs than in PanIN suggesting that the genetic pathway leading to ductal cancer in these two types of preinvasive lesions could be different 9,10 Depending on their mucin expression profiles, two types of IPMTs have been recently characterized. 11,12 Whereas the majority of IPMTs exhibit high expression of MUC2 and usually a good prognosis, some of them reveal a pattern similar to classical ductal adenocarcinoma with MUC1 but no MUC2 expression.
Gene expression patterns derived from cDNA microarray data have been used increasingly to identify genes associated with various human malignancies and are starting to allow tumor classification and prediction of clinical behavior on the basis of molecular information. 13-15 We carried out global analysis of the expression profiles of approximately 5000 gene elements in a series of IPMTs by using custom-built cDNA microarrays.
Materials and Methods
Tissues and Cell Line
IPMT specimens were obtained from 13 patients undergoing pancreaticoduodenectomy, in accordance with institutional guidelines on the use of human tissue. Fresh surgical resection specimens were dissected macroscopically along the dilated main pancreatic duct and exhibited intraductal growth patterns forming polypoid intraluminal masses. Samples of these tumors were snap-frozen in liquid nitrogen within 20 to 30 minutes of harvesting and thereafter stored at −80°C. IPMTs were classified as either non invasive (NInv) lesions (nine cases labeled NInv-626, -628, -630, -632, -633, -635, -636, -638, -s11) or invasive (Inv) carcinoma (four cases labeled Inv-640, -641, -733, -s6) when the intraductal proliferation was associated with an infiltrative component. These latter corresponded in three cases to tubular adenocarcinoma and the remaining case to a colloid carcinoma. A pool of normal pancreas was prepared from two donor tissues and two normal adjacent pancreatic parenchymas from ampullary tumors. A human pancreatic duct epithelial cell line (HPDE) immortalized by transduction with the E6/E7 genes of human papillomavirus (HPV) was kindly provided by Dr Ming-Sound Tsao (University of Toronto, Ontario, Canada) and was grown in keratinocyte serum-free medium as described. 16
RNA Preparation
Histological evaluation of frozen sections after H&E staining demonstrated that all selected IPMT samples contained at least 70% tumor cells. Trimmed pancreatic blocks were cut into 8-μm sections using a cryostat. A total of 100 to 200 frozen sections per specimen were cut and maintained on dry ice for RNA extraction. Every 20 sections, an H&E staining was performed to evaluate the cellular composition. A macrodissection was performed in some cases to enrich the epithelial composition of the samples, particularly in the invasive cases. Total RNA was isolated from both tissues and HPDE cell line using TRIZOL reagent (Gibco BRL, Life Technologies Inc., Frederick, MA) according to the manufacturer’s protocol. The quality of RNA from each sample was verified by agarose gel electrophoresis.
Array Fabrication
The custom cDNA microarrays (5K1) used in this study were obtained through the Cancer Research UK/Ludwig/Welcome Trust consortium at the Sanger Center, Cambridge, UK, where the glass arrays were manufactured and quality controlled. The 5K1 slides used contained 5184 elements comprising 4992 human I.M.A.G.E clones together with positive and negative controls. The spotting patterns and the complete annotated list of these cDNA are available at the ICRF Microarray web site.
cDNA Preparation, Hybridization, and Scanning
Labeling of 23 μg of total RNA was achieved by direct incorporation of Cy5-dCTP or Cy3-dCTP (Amersham Pharmacia Biotech, Amersham, UK) in a reverse transcription reaction using anchored oligodeoxythymidylate primer (Oligonucleotide Service, Clare Hall Laboratories, Potters Bar, UK) and Superscript II reverse transcriptase (Gibco BRL, Frederick, MD). The details of the hybridization and washing protocols are available online. Complementary DNA from the HPDE cell line, which constituted the reference sample, was used in all hybridizations. For each experiment Cy5-dCTP-tagged cDNAs were mixed with Cy3-dCTP-tagged common reference cDNA (HPDE) and subsequently co-hybridized to a microarray. Two of the tumor samples were hybridized in duplicate. Fluorescence intensities of hybridized microarray slides after washing were scanned by dual-laser Affymetrix 418 Scanner (Affymetrix, High Wycombe, UK).
Image and Data Analysis
Signal intensity values of each element were extracted using the ImaGene software program, version 4.1 (BioDiscovery, Los Angeles, CA) and then exported to Microsoft Excel. Spots with intensity values less than twofold that of the local background were flagged and discarded.
Background correction was performed in Excel by subtracting the local background intensity value around each spot from the reported signal intensities from the Cy5 and Cy3 channels. A global intensity correction factor was calculated using the sum of all Cy5 median probe signals divided by the sum of all Cy3 median probe signals. This factor was used for data normalization. The ratio of the normalized values from both the Cy5 and Cy3 channels were calculated and used to determine differential gene expression levels. Only genes with expression changes of at least fourfold in more than half of tumors were selected for this analysis. Using such stringent criteria, the possibility of false positives was remote.
Before clustering the normalized expression ratio values were log2 transformed and were filtered for genes with a detectable signal in at least 50% of the experiments, thus yielding 2906 genes from the data set. Subsequently the data were imported to the software program Cluster 17 and genes were ordered using a self-organizing map algorithm with the number of nodes set to √n. The genes and tumor samples were clustered by average-linkage hierarchical clustering of an uncentered Pearson correlation similarity matrix. The results were visualized with the software program TreeView. 17
SAGE Analysis
We have searched the SAGEmap database (available online) using the Gene-to-Tag online analysis tools. This program allows one to view the expression levels of selected SAGE tags in all available SAGE libraries. We have focused our analysis on SAGE libraries of four pancreatic cell lines (CAPAN1, CAPAN2, HS766T, and Panc1), two primary pancreatic adenocarcinomas (Panc 91–16113 and Panc 96–6252), and two short-term cultures of normal pancreatic ductal epithelium (HX and H126). The expression level was presented as Tags per million, which reflects the levels of the corresponding transcript (“virtual Northern”).
Immunohistochemistry
Immunohistochemical analysis was performed on 30 formalin-fixed, paraffin-embedded IPMTs including the 13 cases analyzed by cDNA microarrays, as well as on a tissue microarray (TMA) block containing 100 classical ductal pancreatic adenocarcinomas. The construction of this TMA was performed using a tissue arrayer (Beecher Instruments, Silver Spring, MD) as described previously by Kallioniemi et al. 18 All of the tumors were reviewed by one pathologist (B.T.).
Paraffin-embedded IPMTs were deparaffinized and rehydrated by standard procedures. The tissue sections were treated by boiling with 10 mmol/L sodium citrate (pH 6.0) for 20 minutes and cooling to room temperature for antigen retrieval. They were treated with 1% hydrogen peroxide to inactivate endogenous peroxidase. Sections were blocked in 10% normal serum for 10 minutes and incubated for one hour with the appropriate primary antibody. The monoclonal antibodies used were against trefoil factor 1 (TFF1, pS2, diluted 1/800), trefoil factor 2 (TFF2, hSP, diluted 1/25) (kindly provided by G. Elia and the Research Monoclonal Antibody Service, Cancer Research UK, London, UK), 19,20 and galectin 3, diluted 1:100 (Research Diagnostics, Flanders, NJ). The polyclonal antibodies used were against trefoil factor 3 (TFF3) (kindly provided by A. Giraud, Footscray, Australia) 21 and lipocalin 2 (kindly provided by M. Gould, Madison, WI). 22 The sections were incubated for one hour with biotinylated secondary antibody and then stained using Vectastain Elite ABC (Vector Laboratories, Inc.) avidin-biotin-peroxidase complex for 30 minutes. Positive controls were carried out in parallel using paraffin-embedded material from human gastric mucosa (for TFF1 and TFF2), colonic mucosa (for TFF3), and normal pancreas containing large ducts (for lipocalin 2 and galectin 3).
Results
Gene Expression Profile of IPMTs and Normal Pancreas
Tables 1 and 2 ▶ ▶ contain lists of the genes that were found over- and underexpressed more than fourfold relative to the HPDE cells in more than half of the IPMTs analyzed (62 and 58 genes, respectively). Three of the genes showing the highest level of expression belong to the trefoil factor family. Several up-regulated genes (Table 1) ▶ found in this study have previously been described in classical pancreatic adenocarcinoma such as lipocalin 2, claudin 4, cathepsin E, galectin 3, and anti-elastase. In addition, we have identified other genes such as CD55 and galectin 9 which have not yet been reported in tumors of this organ. The group of down-regulated genes (Table 2) ▶ in our series of IPMTs included various keratins 1,6,15 and adhesive molecules (laminin, cadherin) as well as cyclin D2, growth arrest-specific 6 protein (Gas6), glypican 1, and caveolin 1.
Table 1.
Up-Regulated Genes in IPMT
| GenBank | IPMT/normal ratio* | Gene name |
|---|---|---|
| L15203 | 84.05342119 | Trefoil factor 3 |
| R47791 | 74.44313188 | Fc fragment of IgG binding protein |
| X00474 | 73.71753476 | Trefoil factor 1 |
| K01396 | 72.52858397 | SERPINA1, protease inhibitor 1 (anti-elastase) |
| NM_005423 | 65.87522866 | Trefoil factor 2 |
| X65614 | 34.55119433 | S100 calcium-binding protein P |
| M63438 | 34.41986467 | Immunoglobulin kappa constant |
| X72964 | 32.06439961 | Centrin, EF-hand protein, 2 |
| AB014458 | 28.24171627 | Ubiquitin specific protease 1 |
| D37931 | 25.4556301 | RNase 4 |
| K01228 | 24.88210491 | Proalpha 1 (I) chain of type I procollagen |
| Y09788 | 21.91818393 | Mucin 5 (MUC5B) |
| X99133 | 18.6584518 | Lipocalin 2 |
| J00220 | 18.59502296 | Human Ig germline H-chain G-E-A region A: gamma-3 5′ flank |
| X16468 | 17.9974938 | Collagen alpha-1 type II |
| T97774 | 16.86863153 | V-fos FBJ murine osteosarcoma viral oncogene homolog |
| X51405 | 15.97881199 | Carboxypeptidase E |
| AL034553 | 15.36310833 | Human DNA sequence from clone 914P20 on chromosome 20q13.13-13.2 |
| D26129 | 15.36298093 | Ribonuclease A (RNase A) |
| AL021786 | 13.99414013 | Human DNA sequence from PAC 696H22 on chromosome Xq21.1-21.2 |
| M62628 | 12.90351069 | Alpha-1 Ig germline C-region membrane-coding region, 3′ end |
| NM_001688 | 12.16965341 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit b, isoform 1 |
| W19215 | 11.92086509 | Decay accelerating factor for complement (CD55, Cromer blood group system) |
| 50999_A | 11.20921769 | Aldehyde dehydrogenase 1, soluble |
| U24163 | 11.19879063 | Frizzled-related protein |
| AB000712 | 10.05233687 | Claudin 4 |
| NM_000177 | 10.0001507 | Gelsolin (amyloidosis, Finnish type) |
| NM_012105 | 9.029424784 | Beta-site APP-cleaving enzyme 2 |
| AF053630 | 8.779707909 | SERPINB1, Protease inhibitor 2 (anti-elastase) |
| J05036 | 8.404941313 | Cathepsin E |
| AF014398 | 8.313108875 | Inositol(myo)-1(or 4)-monophosphatase 2 |
| NM_001311 | 8.22755819 | Cysteine-rich protein 1 (intestinal) |
| U6431 | 8.128671636 | Human Crk-associated substrate related protein Cas-L mRNA |
| AB000712 | 8.113322204 | Claudin 4 |
| U67963 | 7.415174703 | Lysophospholipase-like |
| T74333 | 6.970991547 | Potassium channel subunit (HOHO1) |
| X02761 | 6.808717044 | Fibronectin precursor |
| AC002477 | 6.752661909 | Zinc finger protein 183 (RING finger, C3HC4 type) |
| AA292185 | 6.390520701 | galectin 9 |
| M97796 | 6.351884437 | Inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| NM_00673 | 6.233377328 | Lymphoid blast crisis oncogene |
| AF047439 | 6.226569876 | Chromosome 1 open reading frame 8 |
| M19384 | 6.208406619 | annexin A5 |
| AF011794 | 6.126962289 | Cell cycle progression 8 protein |
| W47253 | 6.070612413 | Homo sapiens, clone IMAGE:4183312, mRNA, partial cds |
| D8729 | 6.037013846 | Thiosulfate sulfurtransferase (rhodanese) |
| S68287 | 5.754454821 | Aldo-keto reductase family 1, member C4 (chlordecone reductase) |
| AF042081 | 5.597599936 | SH3 domain binding glutamic acid-rich protein like |
| D50312 | 5.574152091 | Potassium inwardly-rectifying channel, subfamily J, member 8 |
| R11913 | 5.514858025 | ATPase, Na+/K+ transporting, beta 1 polypeptide |
| AI381734 | 5.415021469 | glutaminyl-peptide cyclotransferase |
| M59807 | 5.139030307 | Natural killer cell transcript 4 |
| AB018335 | 5.081112454 | KIAA0792 gene product |
| NM_004417 | 5.000361416 | Dual specificity phosphatase 1 |
| L20826 | 4.921863064 | Plastin 1 (I isoform) |
| N93058 | 4.881253571 | galectin 3 |
| J04164 | 4.832592455 | Interferon induced transmembrane protein 1 (9–27) |
| D87742 | 4.79820912 | KIAA0268 protein |
| AA680040 | 4.711578012 | clone IMAGE:1126607 |
| U04810 | 4.707383209 | Trophinin associated protein (tastin) |
| M10036 | 4.39852969 | Triosephosphate isomerase |
| X70326 | 3.903669545 | Macrophage myristoylated alanine-rich C kinase substrate |
| M75884 | 3.566595357 | Sterol carrier protein 2 |
Table 2.
Down-Regulated Genes in IPMT
| GenBank | IPMT/normal ratio* | Gene name |
|---|---|---|
| NM_005063 | 0.05018865 | Stearoyl-CoA desaturase (delta-9-desaturase) |
| Y16788 | 0.080235625 | keratin, hair, acidic, 3A |
| AF016903 | 0.095060176 | Agrin precursor |
| AF016903 | 0.098051946 | Heparin-binding growth factor binding protein |
| M17183 | 0.102191105 | Parathyroid hormone-related protein precursor |
| NM_000402 | 0.103378948 | Glucose-6-phosphate dehydrogenase |
| AB006867 | 0.123618379 | SRY (sex determining region Y)-box 20 |
| X63629 | 0.129128659 | Cadherin 3, P-cadherin (placental) |
| X63629 | 0.134671471 | Glypican 1 |
| Z18951 | 0.152828573 | Caveolin-1 |
| NM_003561 | 0.154060474 | Phospholipase A2, group X |
| M59911 | 0.156069179 | Integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| L12468 | 0.156751867 | Glutamyl aminopeptidase (aminopeptidase A) |
| U78773 | 0.160828547 | KRAB-associated protein 1 |
| 74621 | 0.168671808 | Prion protein (p27–30) |
| U78773 | 0.170308631 | Chromosome segregation 1 (yeast homolog)-like |
| AB011105 | 0.185293743 | Laminin, alpha 5 |
| NM_000623 | 0.19194852 | Bradykinin receptor B2 |
| X16662 | 0.192238268 | Annexin A8 |
| R61450 | 0.192509605 | KIAA0586 gene product |
| AA058860 | 0.195476748 | Fascin (Strongylocentrotus purpuratus) homolog 2 (actin-bundling protein, retinal) |
| AF104913 | 0.19666058 | Eukaryotic translation initiation factor 4 gamma, 1 |
| AF104913 | 0.19917747 | Thymidine kinase 1, soluble |
| L34155 | 0.200643346 | Laminin, alpha 3 |
| J00269 | 0.205955309 | Keratin 6A |
| AF025437 | 0.206367669 | Thyroid hormone receptor interactor 6 |
| T48883 | 0.22936451 | Ubiquitin-conjugating enzyme E2M |
| AF070561 | 0.231003177 | Chromosome 19 open reading frame 3 |
| AF201934 | 0.233927594 | DC12 protein |
| K00557 | 0.23893984 | TRAF interacting protein |
| W57890 | 0.240813641 | Cystatin A (stefin A) |
| J03075 | 0.243279271 | Protein kinase C substrate 80K-H |
| AF000421 | 0.248910632 | Polymerase I and transcript release factor |
| M21551 | 0.254327104 | Neuromedin B |
| U19796 | 0.261800808 | Melanoma-associated antigen recognised by cytotoxic T lymphocytes |
| J04173 | 0.269622449 | Phosphoglycerate mutase 1 (brain) |
| AB014589 | 0.26975328 | Likely ortholog of mouse variant polyadenylation protein CSTF-64 |
| M14505 | 0.271017542 | Cyclin-dependent kinase 4 |
| D00510 | 0.27795818 | Annexin A6 |
| AF025439 | 0.279642427 | Pyruvate kinase, muscle |
| L10678 | 0.280981633 | Profilin 2 |
| AB015983 | 0.282894774 | Pyruvate kinase, liver and RBC |
| NM_013442 | 0.289765016 | Stomatin-like 2 |
| S70154 | 0.290724406 | Acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) |
| AF053470 | 0.296710961 | Bladder cancer associated protein |
| D13643 | 0.320990498 | Seladin-1 |
| AF069984 | 0.324228613 | Nitrilase 1 |
| AF025437 | 0.328151563 | Thyroid hormone receptor interactor 6 |
| X00351 | 0.339905385 | Actin, beta |
| M98776 | 0.346805982 | Keratin 1 (epidermolytic hyperkeratosis) |
| X53778 | 0.349484869 | Glyceraldehyde-3-phosphate dehydrogenase |
| X07696 | 0.350902035 | Keratin 15 |
| M16342 | 0.365604705 | Heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| L13720 | 0.381172162 | Growth arrest-specific 6 |
| L12350 | 0.388118649 | Thrombospondin 2 |
| X05908 | 0.390394458 | Annexin A1 |
| D13639 | 0.461065789 | Cyclin D2 |
| R38335 | 0.659152931 | Ribonuclease HI, large subunit |
One of our experiments was performed to compare the expression profiles between HPDE cells and bulk normal pancreas. The majority of genes were commonly expressed in both, with only 60 genes over-represented and 16 genes under-represented in the RNA extracted from whole pancreas compared to the ductal epithelial cells. As expected, a large proportion of the over-represented genes identified in normal pancreatic tissue corresponded to genes expressed in acinar cells (pancreatitis-associated protein, phospholipase), endocrine cells (pancreatic polypeptide, secretory granule, glucagon), and inflammatory and fibroblast cells, which were not present in the ductal cell line used as control in our series.
Identification of Tumor Groups by Hierarchical Clustering
The expression profiles of IPMTs were monitored by means of cDNA microarrays containing 4992 genes. In total, 13 different samples derived from IPMTs, which included nine noninvasive cases (NInv-626, -628, -630, -632, -633, -635, -636, -637, -s11) and four invasive cases (Inv-640, -641, -733, -s6) were used. Hybridizations for tumors NInv-635 and NInv-s11 were performed in replicates.
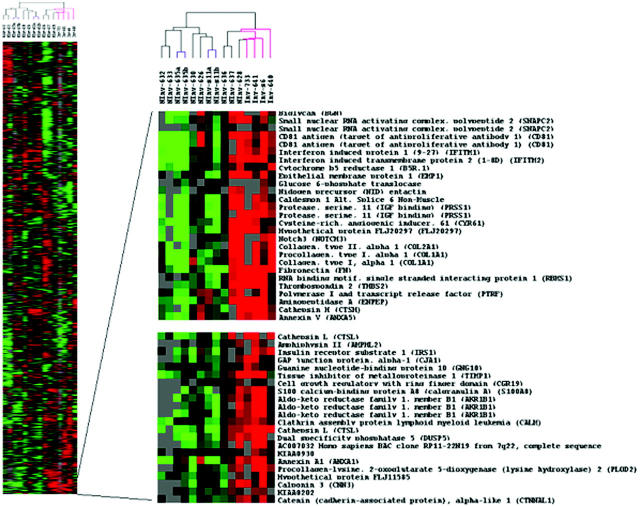
To reveal possible distinctions between the invasive and noninvasive tumors, a subset of 2906 genes (which was obtained after filtering for positive signals in at least 50% of experiments) was selected. Hierarchical clustering was performed to group both the selected genes and the 13 IPMTs, including the two replicates (a total of 15 experiments) according to their overall similarities in the levels of expression. The tumors were arranged on a dendrogram, which contained two main branches with all of the invasive cases occupying the same branch (Figure 1) ▶ . Three of the four invasive IPMTs (InvT-641, -733 and -s6) clustered tightly next to each other forming a distinct subgroup of one of the main branches together with one noninvasive case (NInv-628) which on further macroscopic examination corresponded to an IPMT with a particular micropapillary oncocytic dysplasia. The remaining invasive case (Inv-640), which did not cluster with the others, showed a ductal invasion pattern.
Figure 1.
Variation in expression of 2906 genes in 13 IPMTs. Each row represents individual genes and each column an experimental sample. The ratio of the abundance of transcripts of each gene to its median abundance across all tumors is represented by the color: green (transcript levels below median), black (equal to the median), red (greater than the median), and gray (technically inadequate or missing data). Color intensity reflects the magnitude of the ratio relative to the median for each set of samples. A: Overall groupings of genes and samples. B: The dendrogram of samples showed two main branches with all of the invasive cases (colored in pink) occupying the same branch. Two separate clusters were identified containing genes differentially expressed between invasive and noninvasive tumors.
As expected, replicate samples of IPMTs (Ninv-635 and NInv-s11) clustered together, demonstrating the overall reproducibility of the technique, and were highly correlated (P = 0.945 and P = 0.946, respectively).
Gene clusters containing genes specifically up- or down-regulated in both invasive and noninvasive IPMTs were identified. The two most representative of these clusters comprised groups of genes that were highly expressed in invasive tumors but not in the noninvasive tumors (Figure 1) ▶ . Both clusters contain a spectrum of genes in which notable groups include those involved in matrix remodeling (such as cathepsin L, PLOD 2, TIMP 1) and angiogenic factors (thrombospondin 2, CYR61) which have been previously related to the invasive process. Several other genes down-regulated in invasive tumors were identified (cysteine-rich protein 1 (CRIP1), galectin 9, Frizzled-related protein, and CD55), but do not belong to any particular cluster.
Results of SAGE Analysis
Using the Gene-to-Tag analysis tool (available online), we were able to investigate further expression patterns of several of the overexpressed genes identified in our analysis. More specifically, TFF1, TFF2, TFF3, galectin 3, SH3 domain-binding glutamic acid-rich protein, plastin 1, lipocalin 2, mucin 5B, claudin 4, fibronectin 1, cathepsin E, cysteine-rich protein 1, and protease inhibitor 2 were found to have high levels of expression in pancreatic adenocarcinoma material.
Immunohistochemical Analysis
To validate the data obtained by cDNA array analysis, the expression of four up-regulated genes (TFF1, TFF2, TFF3, and galectin 3) was investigated at the protein level by immunohistochemistry in 30 IPMTs, as well as on a tissue array block comprising 100 pancreatic adenocarcinomas. In the normal pancreas, no immunostaining was observed for TFF2, whereas a weak signal for TFF1 was present at the apical pole of rare epithelial cells lining interlobular ducts. Weak expression of TFF3 was observed in ductal mucinous metaplasia and in endocrine islets. Moderate galectin 3 immunoreactivity was observed in normal ductal cells and in pancreatic nerves. Lipocalin 2 was highly expressed by normal ductal cells. Increased membranous and cytoplasmic expression for TFF1 (Figure 2A) ▶ , TFF3 (Figure 2C) ▶ , and galectin 3 (Figure 2D) ▶ was observed in 90%, 80%, and 100% of IPMTs and in 59%, 46%, and 85% of classical pancreatic adenocarcinomas respectively. In contrast, expression of TFF2 (Figure 2B) ▶ was present in only 20% of IPMTs and in 5% of classical pancreatic adenocarcinomas, whereas the normal antral glands exhibited strong immunoreactivity. High expression of lipocalin 2 was observed in 97% of IPMTs and 95% of classical pancreatic adenocarcinomas, respectively.
Figure 2.
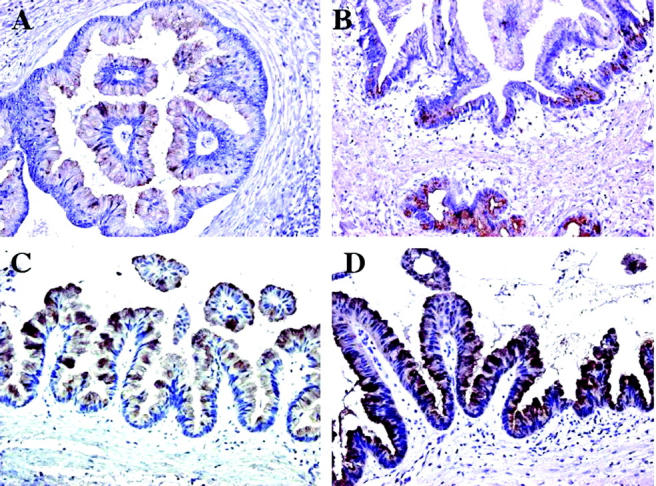
Immunohistochemical staining of IPMT. Intense homogeneous positive staining was seen in neoplastic epithelial cells with TFF1 (A), TFF3 (C), and galectin 3 (D). Immunoreactivity for TFF2 was typically heterogeneous and only moderately intense (B).
Discussion
cDNA microarray technology provides the means to analyze the expression level of thousands of genes simultaneously and to obtain a more complete understanding of the many events that characterize different stages of a tumor process. Whereas abnormalities in Ki-ras, p53, CDKN2/p16, MADH4 and allelic losses (9p, 17p, 18q) have been reported in different grades of PanIN, 4-6 global gene expression profiling has not yet been carried out. To clarify the mechanisms of pancreatic carcinogenesis, we have analyzed a series of IPMTs which to date constitute the best clinically detectable model of pancreatic preinvasive lesion despite some morphological and genetic differences compared with the PanIN. 8,9 Our findings indicate that many genes differentially expressed in classical ductal adenocarcinomas show a similar pattern in IPMTs, suggesting that they are involved in the early stages of pancreatic carcinogenesis.
Due to the low proportion of ductal cells in normal pancreatic parenchyma, and since it is generally agreed that adenocarcinomas arise from these cells, we chose to use as a normal control the immortal epithelial cell line HPDE. This cell line was obtained from a primary culture of normal human pancreatic duct epithelium following transduction of the HPV16-E6/E7 genes. 16 Except for the loss of the p53 functional pathway and a marked reduction of Rb protein level, this cell line demonstrates phenotypic and genotypic resemblance to normal pancreatic ductal cells. 23
Among the 62 genes which were up-regulated in more than half of IPMTs studied (Table 1) ▶ , several (lipocalin 2, galectin 3, claudin 4, cathepsin E, trefoil factors 1 and 2) have previously been described in pancreatic adenocarcinomas either by immunohistochemistry, in situ hybridization, representational difference analysis, or SAGE analysis. 22,24-26
The genes found to be the most highly up-regulated in our cDNA microarray analysis comprise the three members of the trefoil peptide family named TFF1 (pS2), TFF2 (human spasmolytic polypeptide), and TFF3 (intestinal trefoil factor) which all map to the same chromosomal region (21q22.3). These genes show a coordinate overexpression, shared by another gene (SH3 domain-binding glutamic acid-rich protein) situated in close physical proximity. TFFs are principally expressed in the gastrointestinal mucosa and are up-regulated in a variety of ulcerative, pre-neoplastic and neoplastic conditions. 27 Previous studies, analyzing single gene expression, have described up-regulation of TFF1 and TFF2 in 55% and 23% of classical pancreatic adenocarcinomas, respectively. 26,28 It has been recently established that TFFs are frequently found to be co-expressed with mucins (especially MUC2 and MUC5AC, not represented on the 5K1 cDNA array) which interact directly with TFF1 through the binding of two cysteine-rich domains. 29 It is therefore not surprising that we observed an up-regulation of the trefoil factors in IPMTs, as high expression of mucins has been reported in this entity. 11 Unfortunately, the sequences for MUC1, MUC2, and MUC5AC were not present on our array slides. However, in a previous work, 12 we have characterized the mucin expression profile in a large series of IPMT by in situ hybridization and immunohistochemistry. Among the IPMTs analyzed here, 10 cases showed MUC2 and MUC5AC expression, whereas 3 of the 4 invasive cases exhibited MUC1 but no MUC2 expression (data not shown). Cooperation of TFFs with other proteins remains possible, notably with cysteine-rich protein 1 (intestinal), which is present in the set of up-regulated genes in this study. Although we obtained excellent concordance between microarray and immunohistochemical data for TFF1 and TFF3, validation for TFF2 was less clear-cut. This difference between TFF2 RNA and protein expression has been previously reported and could potentially be explained by impaired accessibility of the epitope or a defect of translation. 30
Galectin 3 is a member of the β-galactosidase-binding lectin family. 31 It is involved in diverse physiological and pathological processes, including the regulation of cell growth, cell differentiation and inflammatory responses. Galectin 3 is not only able to bind intracellular ligands such as RNA and Bcl-2, but also a variety of ligands (laminins, lysosome-associated membrane proteins) located in the extracellular matrix. Transfection of galectin 3 into colon cancer cells enhances their potential to form liver metastases in athymic mice. 32 Recently, it has been demonstrated by Northern blot and in situ techniques that galectin 3 is overexpressed in pancreatic adenocarcinoma and, to a lesser extent, in chronic pancreatitis. 33 Our immunohistochemical study confirms that galectin 3 is overexpressed in all IPMTs and in 85% of pancreatic adenocarcinoma analyzed by tissue array. Other genes found to be up-regulated may participate in tumor growth, for example CD55 and the Frizzled-related protein, which we have recently reported to be dysregulated in classical pancreatic adenocarcinoma. 34 The product of the former gene protects cells from attack by complement and has been previously reported to be overexpressed in various tumors. It has been used successfully as a target for both tumor imaging and cancer vaccines. 35,36
Among the down-regulated genes, we note the presence of several genes implicated in epithelial differentiation as well as in cellular adhesion and maintenance of the cytoskeleton. The low levels of expression of genes implicated in cell cycle regulation (cyclin D2, cdk4) in the IPMTs relative to the HPDE cells probably reflects the fact that the cell line was cultured in logarithmic growth phase, whereas only a limited fraction of cells in any solid tumor is actively proliferating. Another explanation for this lower abundance of cyclin D2 in IPMTs is hypermethylation of its promoter, as has been recently reported in breast carcinomas. 37 This hypothesis is supported by the observation that multiple genes in pancreatic carcinomas are hypermethylated. 38 However, we cannot exclude alterations in the cyclin D pathway in the immortalized cell line used as control.
In addition, an important goal of our experiments was to investigate the differences in expression profiles between invasive and noninvasive IPMTs. Encouragingly, we have identified several genes to be highly expressed exclusively in invasive IPMTs such as glypican 1, caveolin 1, growth arrest-specific 6 product, and Notch 3. Overexpression of caveolin 1 (which we have recently reported to be up-regulated in ductal adenocarcinoma) 34 and Notch 3, both of which play important roles in signal transduction, have also been implicated in breast, lung, and prostatic carcinomas. 39-41 The overexpression of glypican 1, which belongs to the heparan sulfate proteoglycan family, could be secondary to the high heparanase activity present in pancreatic carcinoma. 42,43 The Gas6 protein, which interacts with a tyrosine kinase receptor, has a role in the anti-apoptotic pathway by increasing nuclear NF-Kappa B activity. 44 Therefore, escaping from the apoptosis signal may be a crucial step for the invasive process in IPMTs as well. Despite the limited number of invasive samples, hierarchical clustering led to a relatively clear separation between these two types of tumors. It is of interest that the sole noninvasive IPMT which clustered with the invasive ones corresponded to a particular histological form with micropapillary oncocytic dysplasia. 45 A closely related cluster of positively co-varying transcripts in invasive tumors consisted of genes with functions related to matrix remodeling and angiogenesis. Similarly to others’ findings, 25,46 using different techniques such as SAGE analysis and differential display, we have found high expression of several transcripts which are involved in stromal reaction to malignant invasion (fibronectin, proteases, and cathepsin). Such a finding may not be unexpected, as it is well known that invasive pancreatic carcinomas typically exhibit a dense desmoplastic reaction. Thrombospondin 2 and CYR61, two factors implicated in angiogenesis, were found to be up-regulated in invasive IPMTs. The expression of the latter was recently correlated with breast cancer progression. 47
In summary, in this paper we present expression profiles for the uncommon cystic pancreatic tumor called IPMT. The analysis included both noninvasive and invasive cases. Of note, several genes differentially expressed in this study have been previously identified in classical pancreatic carcinoma supporting the proposal that IPMT represents one of the alternative routes of intraepithelial progression to full malignancy in the pancreatic duct system. Among the other genes characterized, some of them were related to the mucosecretion and the invasive pattern of the tumor. Additional studies are required to determine the exact role of these genes and their biochemical pathways in pancreatic carcinogenesis.
Footnotes
Address reprint requests to Prof. N. R. Lemoine, Molecular Oncology Unit, Department of Cancer Medicine, Imperial College School of Medicine, Hammersmith Campus, London W12 0NN, UK. E-mail: nick.lemoine@cancer.org.uk.
References
- 1.Loftus EV, Jr, Olivares-Pakzad BA, Batts KP, Adkins MC, Stephens DH, Sarr MG, DiMagno EP: Intraductal papillary-mucinous tumors of the pancreas: clinicopathologic features, outcome, and nomenclature. Members of the Pancreas Clinic, and Pancreatic Surgeons of Mayo Clinic. Gastroenterology 1996, 110:1909-1918 [DOI] [PubMed] [Google Scholar]
- 2.Kloppel G: Clinicopathologic view of intraductal papillary-mucinous tumor of the pancreas. Hepatogastroenterology 1998, 45:1981-1985 [PubMed] [Google Scholar]
- 3.Terris B, Ponsot P, Paye F, Hammel P, Sauvanet A, Molas G, Bernades P, Belghiti J, Ruszniewski P, Flejou JF: Intraductal papillary mucinous tumors of the pancreas confined to secondary ducts show less aggressive pathologic features as compared with those involving the main pancreatic duct. Am J Surg Pathol 2000, 24:1372-1377 [DOI] [PubMed] [Google Scholar]
- 4.Luttges J, Schlehe B, Menke MA, Vogel I, Henne-Bruns D, Kloppel G: The K-ras mutation pattern in pancreatic ductal adenocarcinoma usually is identical to that in associated normal, hyperplastic, and metaplastic ductal epithelium. Cancer 1999, 85:1703-1710 [PubMed] [Google Scholar]
- 5.Heinmoller E, Dietmaier W, Zirngibl H, Heinmoller P, Scaringe W, Jauch KW, Hofstadter F, Ruschoff J: Molecular analysis of microdissected tumors and preneoplastic intraductal lesions in pancreatic carcinoma. Am J Pathol 2000, 157:83-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luttges J, Galehdari H, Brocker V, Schwarte-Waldhoff I, Henne-Bruns D, Kloppel G, Schmiegel W, Hahn SA: Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis. Am J Pathol 2001, 158:1677-1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hruban RH, Adsay NV, Albores-Saavedra J, Compton C, Garrett ES, Goodman SN, Kern SE, Klimstra DS, Kloppel G, Longnecker DS, Luttges J, Offerhaus GJ: Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol 2001, 25:579-586 [DOI] [PubMed] [Google Scholar]
- 8.Adsay NV, Pierson C, Sarkar F, Abrams J, Weaver D, Conlon KC, Brennan MF, Klimstra DS: Colloid (mucinous noncystic) carcinoma of the pancreas: Am J Surg Pathol 2001, 25:26-42 [DOI] [PubMed] [Google Scholar]
- 9.Iacobuzio-Donahue CA, Klimstra DS, Adsay NV, Wilentz RE, Argani P, Sohn TA, Yeo CJ, Cameron JL, Kern SE, Hruban RH: Dpc-4 protein is expressed in virtually all human intraductal papillary mucinous neoplasms of the pancreas: comparison with conventional ductal adenocarcinomas. Am J Pathol 2000, 157:755-761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore PS, Orlandini S, Zamboni G, Capelli P, Rigaud G, Falconi M, Bassi C, Lemoine NR, Scarpa A: Pancreatic tumours: molecular pathways implicated in ductal cancer are involved in ampullary but not in exocrine nonductal or endocrine tumorigenesis. Br J Cancer 2001, 84:253-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luttges J, Zamboni G, Longnecker D, Kloppel G: The immunohistochemical mucin expression pattern distinguishes different types of intraductal papillary mucinous neoplasms of the pancreas and determines their relationship to mucinous noncystic carcinoma and ductal adenocarcinoma. Am J Surg Pathol 2001, 25:942-948 [DOI] [PubMed] [Google Scholar]
- 12.Terris B, Dubois S, Buisine MP, Sauvanet A, Ruszniewski P, Aubert JP, Porchet N, Couvelard A, Degott C, Fléjou JF: Mucin gene expression in intraductal papillary-mucinous pancreatic tumors. J Pathol 2002 (in press) [DOI] [PubMed]
- 13.Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM: Expression profiling using cDNA microarrays. Nat Genet 1999, 21:10-14 [DOI] [PubMed] [Google Scholar]
- 14.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES: Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999, 286:531-537 [DOI] [PubMed] [Google Scholar]
- 15.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Nilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM: Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403:503-511 [DOI] [PubMed] [Google Scholar]
- 16.Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao MS: Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol 1996, 148:1763-1770 [PMC free article] [PubMed] [Google Scholar]
- 17.Eisen MB, Spellman PT, Brown PO, Botstein D: Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998, 95:14863-14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallioniemi OP, Wagner U, Kononen J, Sauter G: Tissue microarray technology for high-throughput molecular profiling of cancer. Hum Mol Genet 2001, 10:657-662 [DOI] [PubMed] [Google Scholar]
- 19.Williams R, Elia G, Stamp GW, Oates T, Wright NA, Lalani EN: Characterization of monoclonal antibodies raised to C-terminal peptides of pS2: a major trefoil peptide and motility factor expressed in adenocarcinomas and regions of mucosal injury. Hum Pathol 1996, 27:1259-1266 [DOI] [PubMed] [Google Scholar]
- 20.Elia G, Chinery R, Hanby AM, Poulsom R, Wright NA: The production and characterization of a new monoclonal antibody to the trefoil peptide human spasmolytic polypeptide. Histochem J 1994, 26:644-647 [DOI] [PubMed] [Google Scholar]
- 21.Taupin D, Ooi K, Yeomans N, Giraud A: Conserved expression of intestinal trefoil factor in the human colonic adenoma-carcinoma sequence. Lab Invest 1996, 75:25-32 [PubMed] [Google Scholar]
- 22.Friedl A, Stoesz SP, Buckley P, Gould MN: Neutrophil gelatinase-associated lipocalin in normal and neoplastic human tissues: cell type-specific pattern of expression. Histochem J 1999, 31:433-441 [DOI] [PubMed] [Google Scholar]
- 23.Ouyang H, Mou L, Luk C, Liu N, Karaskova J, Squire J, Tsao MS: Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am J Pathol 2000, 157:1623-1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sessa F, Bonato M, Frigerio B, Capella C, Solcia E, Prat M, Bara J, Samloff IM: Ductal cancers of the pancreas frequently express markers of gastrointestinal epithelial cells. Gastroenterology 1990, 98:1655-1665 [DOI] [PubMed] [Google Scholar]
- 25.Gress TM, Wallrapp C, Frohme M, Muller-Pillasch F, Lacher U, Friess H, Buchler M, Adler G, Hoheisel JD: Identification of genes with specific expression in pancreatic cancer by cDNA representational difference analysis. Genes Chromosomes Cancer 1997, 19:97-103 [DOI] [PubMed] [Google Scholar]
- 26.Collier JD, Bennett MK, Bassendine MF, Lendrum R: Immunolocalization of pS2, a putative growth factor, in pancreatic carcinoma. J Gastroenterol Hepatol 1995, 10:396-400 [DOI] [PubMed] [Google Scholar]
- 27.Wright NA, Hoffmann W, Otto WR, Rio MC, Thim L: Rolling in the clover: trefoil factor family (TFF)-domain peptides, cell migration and cancer. FEBS Lett 1997, 408:121-123 [DOI] [PubMed] [Google Scholar]
- 28.Ohshio G, Suwa H, Kawaguchi Y, Imamura M, Yamaoka Y, Yamabe H, Matsumoto M, Yoshioka H, Hashimoto Y, Takeda H: Differential expression of human spasmolytic polypeptide (trefoil factor family-2) in pancreatic carcinomas, ampullary carcinomas, and mucin-producing tumors of the pancreas. Dig Dis Sci 2000, 45:659-664 [DOI] [PubMed] [Google Scholar]
- 29.Tomasetto C, Masson R, Linares JL, Wendling C, Lefebvre O, Chenard MP, Rio MC: pS2/TFF1 interacts directly with the VWFC cysteine-rich domains of mucins. Gastroenterology 2000, 118:70-80 [DOI] [PubMed] [Google Scholar]
- 30.Gouyer V, Wiede A, Buisine MP, Dekeyser S, Moreau O, Lesuffleur T, Hoffmann W, Huet G: Specific secretion of gel-forming mucins and TFF peptides in HT-29 cells of mucin-secreting phenotype. Biochim Biophys Acta 2001, 1539:71-84 [DOI] [PubMed] [Google Scholar]
- 31.Hughes RC: The galectin family of mammalian carbohydrate-binding molecules. Biochem Soc Trans 1997, 25:1194-1198 [DOI] [PubMed] [Google Scholar]
- 32.Bresalier RS, Mazurek N, Sternberg LR, Byrd JC, Yunker CK, Nangia-Makker P, Raz A: Metastasis of human colon cancer is altered by modifying expression of the β-galactoside-binding protein galectin 3. Gastroenterology 1998, 115:287-296 [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Friess H, Zhu Z, Frigeri L, Zimmermann A, Korc M, Berberat PO, Buchler MW: Galectin-1 and galectin-3 in chronic pancreatitis. Lab Invest 2000, 80:1233-1241 [DOI] [PubMed] [Google Scholar]
- 34.Crnogorac-Jurcevic T, Efthimiou E, Capelli P, Blaveri E, Terris B, Baron A, Jones M, Tyson K, Bassi C, Scarpa A, Lemoine NR: Gene expression profiles of pancreatic cancer and stromal desmoplasia. Oncogene 2001, 20:7437-7446 [DOI] [PubMed] [Google Scholar]
- 35.Denton GW, Durrant LG, Hardcastle JD, Austin EB, Sewell HF, Robins RA: Clinical outcome of colorectal cancer patients treated with human monoclonal anti-idiotypic antibody. Int J Cancer 1994, 57:10-14 [DOI] [PubMed] [Google Scholar]
- 36.Li L, Spendlove I, Morgan J, Durrant LG: CD55 is over-expressed in the tumour environment. Br J Cancer 2001, 84:80-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evron E, Umbricht CB, Korz D, Raman V, Loeb DM, Niranjan B, Buluwela L, Weitzman SA, Marks J, Sukumar S: Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res 2001, 61:2782-2787 [PubMed] [Google Scholar]
- 38.Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH, Goggins M: Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res 2000, 60:1835-1839 [PubMed] [Google Scholar]
- 39.Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, Roberts JR, Carbone DP: Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst 2000, 92:1355-1357 [DOI] [PubMed] [Google Scholar]
- 40.Li L, Yang G, Ebara S, Satoh T, Nasu Y, Timme TL, Ren C, Wang J, Tahir SA, Thompson TC: Caveolin-1 mediates testosterone-stimulated survival/clonal growth and promotes metastatic activities in prostate cancer cells. Cancer Res 2001, 61:4386-4392 [PubMed] [Google Scholar]
- 41.Hayashi K, Matsuda S, Machida K, Yamamoto T, Fukuda Y, Nimura Y, Hayakawa T, Hamaguchi M: Invasion activating caveolin-1 mutation in human scirrhous breast cancers. Cancer Res 2001, 61:2361-2364 [PubMed] [Google Scholar]
- 42.Kleeff J, Ishiwata T, Kumbasar A, Friess H, Buchler MW, Lander AD, Korc M: The cell-surface heparan sulfate proteoglycan glypican-1 regulates growth factor action in pancreatic carcinoma cells and is overexpressed in human pancreatic cancer. J Clin Invest 1998, 102:1662-1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koliopanos A, Friess H, Kleeff J, Shi X, Liao Q, Pecker I, Vlodavsky I, Zimmermann A, Buchler MW: Heparanase expression in primary and metastatic pancreatic cancer. Cancer Res 2001, 61:4655-4659 [PubMed] [Google Scholar]
- 44.Demarchi F, Verardo R, Varnum B, Brancolini C, Schneider C: Gas6 anti-apoptotic signaling requires NF-κB activation. J Biol Chem 2001, 276:31738-31744 [DOI] [PubMed] [Google Scholar]
- 45.Adsay NV, Adair CF, Heffess CS, Klimstra DS: Intraductal oncocytic papillary neoplasms of the pancreas. Am J Surg Pathol 1996, 20:980-994 [DOI] [PubMed] [Google Scholar]
- 46.Ryu B, Jones J, Hollingsworth MA, Hruban RH, Kern SE: Invasion-specific genes in malignancy: serial analysis of gene expression comparisons of primary and passaged cancers. Cancer Res 2001, 61:1833-1838 [PubMed] [Google Scholar]
- 47.Tsai MS, Hornby AE, Lakins J, Lupu R: Expression and function of CYR61, an angiogenic factor, in breast cancer cell lines and tumor biopsies. Cancer Res 2000, 60:5603-5607 [PubMed] [Google Scholar]