Characterization of the Mouse Brain Proteome Using Global Proteomic Analysis Complemented with Cysteinyl-Peptide Enrichment (original) (raw)
. Author manuscript; available in PMC: 2007 Apr 10.
Published in final edited form as: J Proteome Res. 2006 Feb;5(2):361–369. doi: 10.1021/pr0503681
Abstract
Given the growing interest in applying genomic and proteomic approaches for studying the mammalian brain using mouse models, we hereby present a global proteomic approach for analyzing brain tissue and for the first time a comprehensive characterization of the whole mouse brain proteome. Preparation of the whole brain sample incorporated a highly efficient cysteinyl-peptide enrichment (CPE) technique to complement a global enzymatic digestion method. Both the global and the cysteinyl-enriched peptide samples were analyzed by SCX fractionation coupled with reversed phase LC-MS/MS analysis. A total of 48,328 different peptides were confidently identified (>98% confidence level), covering 7792 non-redundant proteins (∼34% of the predicted mouse proteome). 1564 and 1859 proteins were identified exclusively from the cysteinyl-peptide and the global peptide samples, respectively, corresponding to 25% and 31% improvements in proteome coverage compared to analysis of only the global peptide or cysteinyl-peptide samples. The identified proteins provide a broad representation of the mouse proteome with little bias evident due to protein pI, molecular weight, and/or cellular localization. Approximately 26% of the identified proteins with gene ontology (GO) annotations were membrane proteins, with 1447 proteins predicted to have transmembrane domains, and many of the membrane proteins were found to be involved in transport and cell signaling. The MS/MS spectrum count information for the identified proteins was used to provide a measure of relative protein abundances. The mouse brain peptide/protein database generated from this study represents the most comprehensive proteome coverage for the mammalian brain to date, and the basis for future quantitative brain proteomic studies using mouse models. The proteomic approach presented here may have broad applications for rapid proteomic analyses of various mouse models of human brain diseases.
Keywords: cysteinyl-peptide enrichment, mass spectrometry, mouse brain proteome, protein categorization, proteome coverage
Introduction
The immense complexity of the mammalian brain presents one of the most significant challenges in current biomedical science. Recent advances in molecular imaging, genomics, and proteomics technologies offer significant potential not only for gaining a better understanding of brain function and dysfunction, but also for achieving more effective treatments for neurological disorders such as Alzheimer's and Parkinson's diseases that are becoming increasingly prevalent. Mice have been widely used as brain research models due to their genetic accessibility and suitability as models for many aspects of human biology1. Genome-wide mapping of mouse brain gene expression has been recently demonstrated by employing either high throughput in situ hybridization2,3 or voxelation technology combined with cDNA microarray approaches4. Similarly, protein abundance patterns in the brain can also be mapped by using imaging mass spectrometry5 or voxelation coupled with high throughput proteomic technologies. The proteomic profiling of different regions or subcellular compartments of the brain6-8 and brain samples from patients suffering from various brain-related diseases9-15 promises new insights into the molecular basis of brain function and the pathogenesis of brain-related diseases.
The majority of proteomic studies of the mammalian brain reported to date have been based on two-dimensional electrophoresis (2DE) separations coupled with in-gel protein digestion and mass spectrometric (MS) identification. Due to limitations of the 2DE-MS method16, extensive coverage of the mouse brain proteome has not been achieved. By using large format gel 2DE, 8767 protein spots were resolved from the mouse brain, from which 466 proteins were identified by MALDI-MS17, 18. In other studies using similar approaches, 30 proteins were identified from mouse cerebellum7, 437 proteins were identified from the cytosolic, mitochondrial and microsomal compartments of rat brain19, and 180 proteins were identified from the parietal cortex lobe of human brain20.
Alternative strategies using multi-dimensional liquid chromatography separations coupled with tandem mass spectrometry (LC/LC-MS/MS) have recently been applied to profile cultured brain single cell population and brain membrane proteome. In a global analysis of a cultured mouse cortical neuron sample, 4542 proteins were identified by LC/LC-MS/MS21. Another study that targeted mouse brain plasma membrane proteins incorporated a novel membrane protein isolation procedure followed by LC-MS/MS analysis and successfully identified 862 proteins from the mouse brain cortex and 1685 proteins from the mouse hippocampus8. However, the whole proteome of complex brain tissue has not been extensively characterized using LCMS/MS approaches.
In this work, we presented a global proteomic approach for comprehensive profiling of the lipid rich brain tissue (∼12% of lipid content) using LC/LC-MS/MS and an extensive protein database for the whole mouse brain. The high lipid content in complex tissue sample presents additional challenge for sample processing and protein identification. The global analysis of tryptic peptides was complemented by cysteinyl-peptide enrichment (CPE), which resulted in significantly increased proteome coverage22, 23. This extensive analysis provided confident identification of > 48,000 different peptides and 7792 non-redundant proteins that correspond to ∼34% of the predicted mouse proteome. The overall false positive rate for the entire set of unique peptide identifications was estimated to be ∼1.5% on the basis of reversed database searching24. Identified membrane proteins and membrane-associated proteins comprised ∼26% of the entire dataset, which is consistent with previous predictions based on statistical analysis25. Our dataset represents the most comprehensive proteome analysis of the mouse brain reported to date, and provides a basis for future quantitative proteome mapping of different regions within the brain, as well as for comparative proteome analysis of specific brain regions between normal and diseased mouse brains.
MATERIALS AND METHODS
Preparation of Mouse Brain Tissue Digests
A single C57BL/6J inbred strain male mouse purchased from the Jackson Laboratory (Bar Harbor, ME) was used in this study. At the age of 8 weeks, the mouse was sacrificed following deep anesthesia with isoflurane. The whole brain was dissected and then snap-frozen in liquid nitrogen. The frozen brain was sliced into ∼20 small tissue pieces of 10-15 mg each for handling purposes. Tissue pieces were transferred into individual Eppendorf tubes that contained 100 μL of 5 mM phosphate buffer (pH 7.0) and vortexed for 2-3 min. The samples were incubated at room temperature for 1 h with gentle shaking, followed by 5 min of sonication in an ice-water bath. 100 μL of trifluoroethanol (TFE) (Sigma, St. Louis, MO) was added to each sample and the samples were incubated at 60 °C for 2 h and then sonicated for 2 min. Protein disulfide bonds were reduced by 5 mM tributylphosphine (TBP) (Sigma) with 30 min incubation at 60 °C. Samples were diluted five-fold with 50 mM NH4HCO3 (pH 7.8) to reduce the TFE concentration to 10% prior to the addition of sequencing-grade modified trypsin (Promega, Madison, WI) at a ratio of 1:50 (w/w, enzyme : protein). Samples were digested at 37 °C overnight with gentle shaking. The digests were purified by loading the samples onto SPE C18 columns (Supelco, Bellefonte, PA) and eluting the peptides with 1 mL of 80% acetonitrile with 0.1% trifluoroacetic acid (TFA) solution. The eluted peptides were concentrated by lyophilization, and samples were pooled for a final total volume of 1.6 mL. A total of 11.2 mg peptides as determined by bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL) were recovered.
Cysteinyl Peptide Enrichment (CPE)
Approximately 5 mg of the peptide sample was used for cysteinyl-peptide enrichment23. Briefly, the peptides were lyophilized and then re-dissolved in 120 μL of coupling buffer that consisted of 50 mM Tris-HCl, 1 mM EDTA, pH 7.5. After reducing with 5 mM DTT at 37 °C for 1 h, the sample was diluted to 600 μL with coupling buffer, divided into four equal aliquots, and applied to four Handee Mini-spin columns (Pierce), each containing ∼ 100 μL of pre- equilibrated thiopropyl Sepharose 6B resins (Amersham Biosciences, Uppsala, Sweden). The cysteinylpeptides were captured by the beads during 2 h incubation at room temperature with gentle shaking. The unbound portion of the peptide sample was removed from the beads by stringent washing first with the coupling buffer, followed by 2 M NaCl, 80% acetonitrile/0.1% TFA solution, 100% methanol, and again with the coupling buffer. The captured cysteinyl peptides were released by incubating the beads with 120 μL of 20 mM freshly prepared DTT solution at room temperature for 30 min. The released peptides were further alkylated with 40 mM of iodoacetamide for 1 h at room temperature in the dark; alkylated peptide samples were then desalted using SPE C18 columns. After lyophilization, peptides were resuspended in 600 μL of 10 mM ammonium formate in 25% acetonitrile, pH 3.0 for subsequent strong cation exchange (SCX) fractionation.
Strong Cation Exchange (SCX) Fractionation
Both the global tryptic peptide and the enriched cysteinyl-peptide samples were subjected to LC fractionation by SCX chromatography on a 200 mm × 2.1 mm Polysulfoethyl A column (PolyLC, Columbia, MD) preceded by a 10 mm × 2.1 mm guard column, using a flow rate of 0.2 mL/min. LC separations were performed using an Agilent 1100 series HPLC system (Agilent, Palo Alto, CA). Mobile phase solvents consisted of (A) 10 mM ammonium formate, 25% acetonitrile, pH 3.0 and (B) 500 mM ammonium formate, 25% acetonitrile, pH 6.8. Once loaded, isocratic conditions at 100% A were maintained for 10 min. Peptides were separated by using a gradient from 0-50% B over 40 min, followed by a gradient of 50-100% B over 10 min. The gradient was then held at 100% solvent B for another 10 min. Thirty fractions were collected for each of the global and cysteinyl-peptide enriched samples. Following lyophilization, the fractions were dissolved in 25 mM NH4HCO3 and stored at −80 °C.
Capillary LC-MS/MS analysis
Each SCX fraction from both the global and cysteinyl-peptide samples was analyzed with an automated custom-built capillary HPLC system coupled online to an LTQ ion trap mass spectrometer (ThermoElectron, San Jose, CA) by using an electrospray ionization interface. The reversed phase capillary column was prepared by slurry packing 3-μm Jupiter C18 particles (Phenomenex, Torrence, CA) into a 150 μm i.d. × 65 cm fused silica capillary (Polymicro Technologies, Phoenix, AZ). The mobile phase solvents consisted of (A) 0.2% acetic acid and 0.05% TFA in water and (B) 0.1% TFA in 90% acetonitrile. An exponential gradient was used for the separation, which started with 100% A and gradually increased to 60% B over 100 min. The instrument was operated in a data-dependent mode with an m/z range of 400-2000. The five most abundant ions from each MS scan were selected for further MS/MS analysis by using a normalized collision energy setting of 35%. Dynamic exclusion was applied to avoid repeating analyses of the same abundant precursor ion.
Data Analysis
The SEQUEST algorithm (ThermoElectron) was used to search LC-MS/MS data against the mouse International Protein Index (IPI) database (version 1.25, September, 2005, available online at http://www.ebi.ac.uk/IPI/IPIhelp.html) and a sequence-reversed IPI database. For enriched cysteinyl-peptide samples, a static mass modification for cysteinyl residues that corresponded to alkylation with iodoacetamide (57.0215 Da) was applied during the SEQUEST analysis. The following criteria were used to filter raw SEQUEST results: 1) Xcorr ≥ 1.6 for charge state +1 full tryptic peptides; 2) Xcorr ≥ 2.4 for charge state +2 full tryptic peptides and Xcorr ≥ 4.3 for +2 partial tryptic peptides; and 3) Xcorr ≥ 3.2 for charge state +3 full tryptic peptides and Xcorr ≥ 4.7 for +3 partial tryptic peptides. The delta correlation value (ΔCn) > 0.1 was used in all cases. These criteria were established based on probability-based evaluation using sequence-reversed database searching as previously described to provide >95% overall confidence level for the entire set of unique peptide identifications (<5% false positive rate)24. The minimum confidence level is 75% for individual peptide identification even if the peptide has Xcorr at the cutoff value24. In an attempt to remove redundant protein entries, the software tool ProteinProphet was applied as a clustering tool to group similar or related protein entries into a “Protein Group”26. All identified peptides that passed the filtering criteria were assigned an identical probability score of 1, and then entered the ProteinProphet program solely for clustering analysis to generate the final non-redundant list of proteins or protein groups.
Theoretical molecular weight (Mr) and pI values of identified proteins were obtained from the IPI mouse protein database. The identified proteins were categorized based on their cellular locations and biological processes according to Gene Ontology (GO) information obtained from the European Bioinformatics Institute at http://www.ebi.ac.uk/pub/databases/GO/goa/mouse. The TMHMM (www.cbs.dtu.dk/services/TMHMM/) algorithm was used to predict transmembrane domains (TMDs)27 from the identified proteins.
RESULTS
Global Analysis coupled with Cysteinyl-Peptide Enrichment
We applied multidimensional LC-MS/MS (offline SCX fractionation coupled with capillary reversed phase LC-MS/MS) to analyze the global tissue lysate of a whole mouse brain. To further improve the overall dynamic range of detection, we coupled the global analysis method with a recently developed high efficiency CPE method22, 23. Figure 1 shows the overall experimental workflow and peptide/protein identification results. A total of 60 LC-MS/MS analyses were performed for these samples and ∼751,000 MS/MS spectra were obtained. Following database searching and data filtering, a total of 34,241 peptides that corresponded to 6228 proteins were identified from the global tryptic peptide sample and 17,562 peptides that correspond to 5933 proteins were identified from the cysteinyl-enriched peptide sample. 96.5% of the 17,562 peptides identified from the cysteinyl-enriched peptide sample are cysteinyl-peptides, demonstrating the high specificity of the CPE approach. A total of 48,328 different peptides were identified from the combined mouse brain proteome dataset, covering 7792 non-redundant proteins after analysis by ProteinProphet to remove possible redundant proteins. All LC-MS/MS datasets were further analyzed to estimate the false positive rate of peptide identifications, using SEQUEST to search against a “sequence reversed” mouse IPI protein database in a similar fashion to that previously described24. The false positive rate of identification for the 48,328 peptides was estimated to be ∼1.5% on the basis of the reversed database searching results, which demonstrates a very high confidence level for this set of peptide identifications. 5636 proteins (72%) were identified based on using two or more different peptides. For those proteins identified with a single peptide, the Xcorr cutoff values were established to provide a minimum confidence level of 75% for these identifications. In addition, no significant difference in the distributions of peptide Xcorr scores was observed between multiple peptide proteins and proteins identified with only one or two peptide identifications.
Figure 1.
A flowchart showing the experimental strategy: preparation of the whole mouse brain using a combination of global tryptic digestion and cysteinyl-peptide enrichment (CPE) methodology, followed by SCX fractionation and LC-MS/MS analysis of each fraction, and the peptide/protein identification results from both preparation methods.
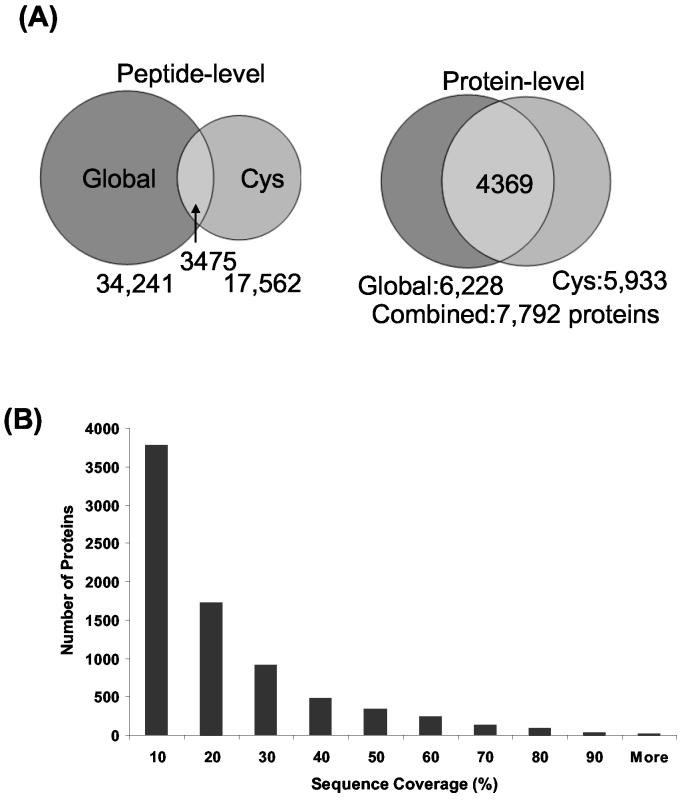
Figure 2A shows the numbers of peptide/protein identifications from the global peptide and cysteinyl-peptide samples and illustrates the complementary nature of the two samples. At the peptide level, the analysis of these two samples resulted in nearly two completely different subsets of peptides with only 3475 peptides (7% of the 48,328 unique peptides) common to both samples. The coupling of CPE leads to a ∼41% increase in total unique peptide identifications compared with the global analysis alone. Despite the very low overlap at the peptide level, 4369 proteins (56% of the 7792 non-redundant proteins) were identified from both the global and cysteine-enriched digests. Increased sequence coverage for these common proteins was observed as a result of the contributions from different sets of peptides identified from the global and the enriched peptide samples. In addition, by reducing sample complexity, CPE allowed significantly more low abundance proteins to be identified. As shown, 1564 proteins were identified exclusively from the cysteinyl-enriched samples, providing an improvement of ∼25% in overall proteome coverage. Figure 2B shows the distribution of percent sequence coverage for the total number of identified proteins. Note that although nearly half of the proteins were identified with <10% sequence coverage, more than 500 proteins were identified with >50% sequence coverage.
Figure 2.
(A) Comparisons of peptide and protein identifications resulting from the global tryptic peptide and cysteinyl-enriched peptide samples. The comparisons were plotted as Venn diagrams at both the peptide level (left) and the protein level (right). (B) Distribution of total sequence coverage in identified proteins. The number of identified proteins is plotted against the percent sequence coverage.
Mouse Brain Proteome Coverage
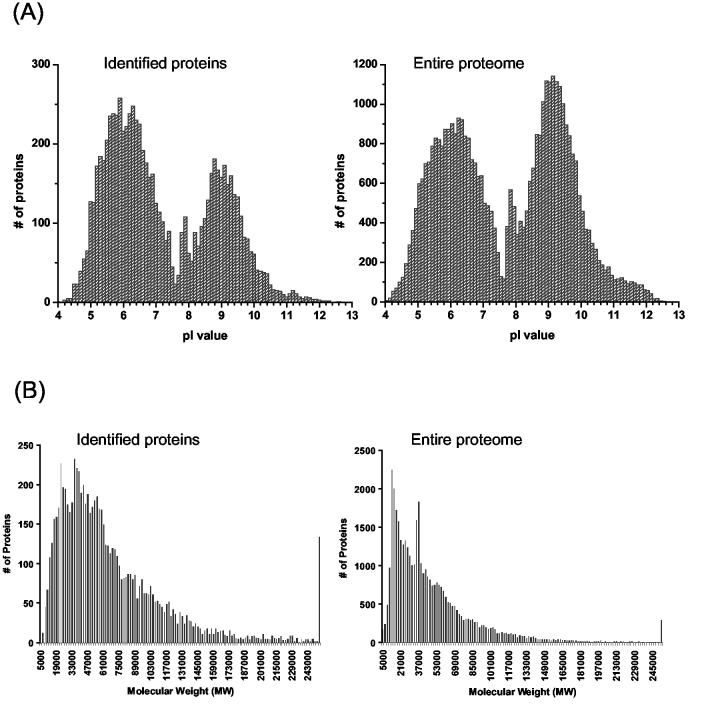
The 48,328 different peptides identified from the mouse brain dataset cover 13,828 of the total 40,745 protein entries (with at least one peptide per protein entry) in the mouse IPI protein database, representing ∼34% coverage of the total proteins in the database. Because multiple protein entries can share the same identified peptide, a set of 7792 non-redundant proteins/protein groups was generated using ProteinProphet analysis to remove possible redundant protein entries. Figure 3 shows the histograms of the pI value distribution and molecular weight (MW) distribution patterns for the identified proteins, which were compared with distribution patterns for the 40,745 proteins from the IPI mouse protein database. Figure 3A shows the similarity between the distribution of the estimated pI values for the identified proteins and the distribution of the estimated pI values for the entire proteome. These tri-modal pI distributions are consistent with previous predictions for eukaryotes28. The region predicted to be rich in membrane associated proteins (pI>8)28 appears slightly underrepresented compared to the whole proteome. Similar patterns were also observed between the MW distributions of the identified proteins and the whole proteome (Figure 3B). The identified proteins were detected with masses between 5 kDa and 840 kDa; 48.4% of these proteins were in the 20-60 kDa range, which is in agreement with the 50.1% calculated for the predicted mouse proteome. The proteins with the lowest and highest mass were annotated as thymosin β10 (5026 Da) and similar to mucin 16 (839,304 Da), respectively. Only a slight bias against lower MW proteins was observed that can be attributed to the smaller numbers of possible/detectable peptides.
Figure 3.
Comparisons of protein distributions, based on their physiochemical characteristics: (A) pI, and (B) molecular weight (MW), between the identified proteins in the dataset and the entire mouse proteome. Predicted pI values were obtained from the IPI mouse protein database and the pI distributions were plotted with 0.2 pH unit increment. MW values were also obtained from the IPI mouse protein database and the MW distributions were plotted with 2 kDa increments.
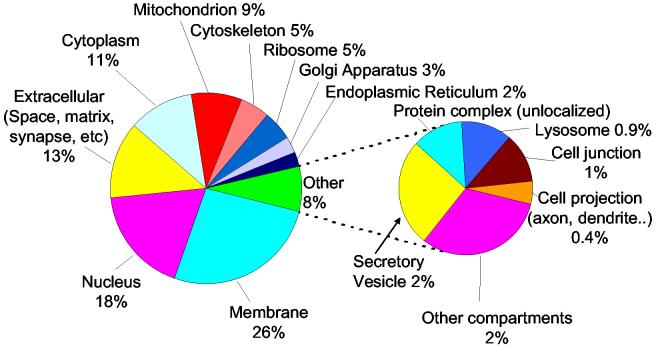
The identified proteins were categorized based on their cellular localizations by using GO information. Figure 4 graphically depicts the GO cellular locations assigned to 3472 (44%) of the 7792 non-redundant proteins. Proteins from most subcellular compartments were identified, with the majority of these proteins being from the plasma membrane, nucleus, extracellular regions, cytoplasm, and mitochondria. 420 proteins were annotated as mitochondrial proteins without prior organelle purification. For comparison, a total of 615 proteins were reported for an extensive characterization of the human heart mitochondrial proteome29. Since only 44% of identified proteins in our study are presently annotated, the actual number of mitochondrial proteins in the present work may well be significantly higher than 420. Overall, these results indicate that the set of mouse brain proteins identified using the present approach provides a substantially unbiased representation of the mouse brain proteome with regard to protein pI, MW, and subcellular localization.
Figure 4.
Protein categorization using GO identification numbers based on their cellular location, comprising 44% of the total 7792 identified proteins in the dataset.
Identification of Brain Membrane Proteins
Identification of membrane proteins is particularly challenging due to their hydrophobic nature. Several 2DE-MS-based studies specifically aimed at identifying brain membrane proteins have reported up to 500 protein identifications from either the brain microsomal fraction19, the synaptic plasma membrane fraction30, or the postsynaptic density fraction31, 32. More recently, Nielsen, et al. reported the identification of 862 proteins from the mouse brain cortex and 1685 proteins from the mouse hippocampus as a result of incorporating a novel membrane isolation procedure prior to LC-MS/MS analysis8.
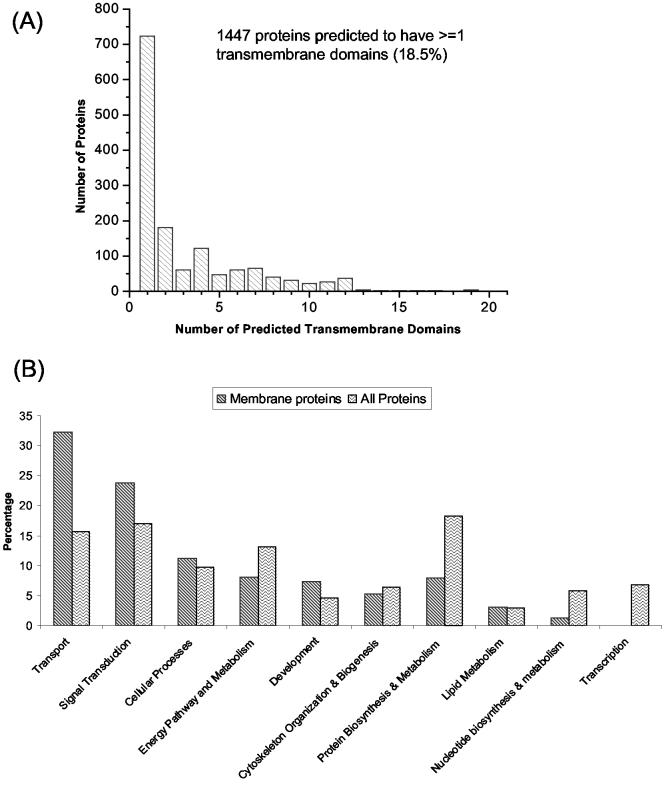
In this work, ∼26% of the annotated protein identifications were membrane proteins (Figure 4), which is consistent with predictions from topology studies that speculated that 20-30% of all open reading frames (ORFs) encode for membrane proteins25, 33, 34. Our results are alsoconsistent with the 28% annotated membrane proteins in a previous study that employed high pH and proteinase K for mouse brain homogenate digestion35. Moreover, Figure 5A shows 1447 proteins (18.6%) are predicted by the TMHMM 2.0 algorithm33 to have transmembrane domains (TMDs) and >700 proteins to contain 2-19 TMDs. These results suggest our global proteomic approach is not significantly biased against membrane proteins. A comparison of the list of non-redundant proteins with the previously published list of ∼2000 proteins from the mouse brain cortex and hippocampus8 reveals an ∼75% overlap between the two datasets, further supporting the overall good membrane proteome coverage of the current dataset.
Figure 5.
(A) Histogram illustrating the distributions of the identified proteins that have transmembrane domains (TMD), as predicted by TMHMM. The two axes represent the number of TMDs (1-19) and the corresponding total number of proteins having the same amount of TMDs. (B) Comparison of the distributions of functional categories between membrane proteins and all proteins according to GO biological processes. A total of 4428 proteins were annotated by GO biological processes from the entire dataset, among which 1332 annotated proteins were membrane proteins.
Figure 5B compares the distributions of functional categories between annotated membrane proteins (1332 proteins) and all annotated proteins (4428 proteins) according to GO biological processes. As shown, the percentages of membrane proteins involved in the transport and signal transduction categories are significantly higher than the percentages of the global proteins involved in these two categories. More than 400 membrane proteins (33%) that include ion channels, receptors, intracellular protein transporters, synaptic transmitters, etc. were annotated as being involved in the transport category. For example, seven different syntaxin proteins (Syntaxin 1A, 1B, 4, 7, 8, 12, 16) and ten different synaptotagmin proteins (synaptotagmin I, II, III, V, VI, IX, X, XI, XII, XIII) were identified. Syntaxin and synaptotagmin are SNARE (soluble _N_-ethylmaleimide-sensitive-factor attachment protein receptor) associated proteins that play important roles in synaptic vesicle exocytosis36. Additionally, 83 ion channel proteins that included calcium, sodium, potassium, and chloride channels, as well as 240 different kinds of intracellular or plasma membrane receptor proteins were identified. In addition to proteins involved in transport, >300 membrane proteins (24%) were annotated as being involved in the signal transduction category.
Relative Abundances of Identified Proteins within the Mouse Brain
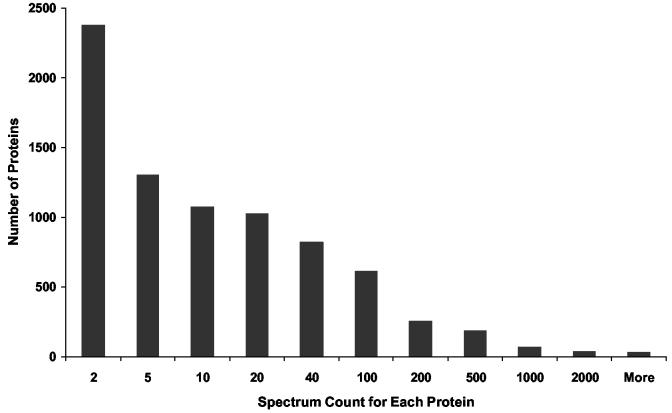
In a recent human blood plasma proteome profiling study37, we observed good correlation between the protein concentration (μg/uL) and the number of LC-MS/MS spectra (spectrum count) used to identify each protein. A similar observation was also reported for spectrum counts and the abundances of yeast proteins38. These studies laid the foundation for using spectrum count information as a qualitative or semi-quantitative measure of relative protein abundances within the mouse brain sample. It should be noted that the spectrum count information provides an estimation of the relative abundances of different proteins within the same sample in terms of mass rather than mole quantities. Figure 6 shows the distribution of spectrum counts for the identified proteins. Notably, the majority of proteins were identified with fewer than 10 total spectra, presumably due to their relatively low abundances. A total of 73 proteins were identified in >1000 spectra, and this list of proteins includes many expected high abundance structural proteins and metabolic enzymes, such as tubulin, actin, spectrin, clathrin, dynamin, triosephosphate isomerase, pyruvate kinase, enolase, creatine kinase, adolase, etc. Other examples of proteins included on this list are elongation factors, histone H2B, sodium-dependent glutamate/aspartate transporter 2, synapsin I, Na+/K+ transporting ATPase, and ATP synthase. On the other hand, all transcription factors, which are generally expressed in cells at relatively low levels, were identified with spectrum counts of <50. The spectrum count data for different protein isoforms of a specific protein class also reflect the relative abundance levels for these proteins. For example, the brain expresses relatively high levels of syntaxin 1 and SNAP-25, which are involved in synaptic vesicle exocytosis39. In our dataset, syntaxin 1A, 1B, and SNAP-25 were all identified with >300 spectrum counts, while other detected syntaxin isoforms including syntaxin 4, 7, 8, 12, and 16 were identified by <50 spectra. Synaptotagmin I and II were observed to be the two relatively abundant proteins among all identified isoforms with 559 and 109 spectrum counts, respectively, while the rest of the isoforms had <50 spectrum counts. These relative abundance results agree well with the expression data from immunoblotting experiments40. A complete list of the 7792 identified proteins along with the total number of unique peptide and spectrum count information for each protein is provided as a Supplementary Table.
Figure 6.
The distribution of spectrum counts for identified proteins. The number of identified proteins was plotted against the spectrum count for each protein.
Our database provides some qualitative or semi-quantitative insights into the relative abundances of certain neuronal receptors and their subunits and isoforms. In general, these receptors are expected to be expressed with relatively low abundances. Table 1 represents the list of all detected major small molecule neurotransmitter (glutamate, GABA, glycine, acetylcholine, catecholamines, purine, and histamine) receptors. The spectrum count information was used in conjunction with the number of peptides per protein to provide a semi-quantitative estimation of abundances of different receptors and their subunits. On the basis of these estimations, glutamate receptors, especially ionotropic AMPA, NMDA types, and metabotropic mGlu I and mGlu II, are the most abundant receptors. Some of the receptor subunits were observed with significantly higher abundance than others. For example, NMDA type is mainly represented by Grin1 and Grin2b subunits and to a lesser extent, by Grin2d subunit (16 and 76 spectrum counts vs. 2). Kainate type receptors are represented only by one subunit Grik5 (KA2) and appear in much lower abundance than AMPA and NMDA types, presumably due to lower expression levels and/or limited distribution of kainite receptors. Indeed, KA2 is the only receptor subunit reported to be ubiquitously expressed throughout the nervous system41. Surprisingly, the subunits of the least studied orphan type of glutamate receptors Grid1 and Grid2 are well represented and comparable in number to the well-studied AMPA and NMDA types. The other abundant family of receptors is the GABA receptors, represented by ionotropic A and metabotropic B types. Because type C is localized predominantly in the retina42, the overall abundance of this receptor is too low to be detected. The other families of small molecule receptors detected in relatively low abundance are glycine, acetylcholine, catecholamine, purine, and histamine. Similar semi-quantitative analyses can be performed for other protein types.
Table 1.
List of detected major small molecule neurotransmitter receptors.
| Reference IDa | Genea | Receptor Family | Class | Type | Peptidesb | Unique Peptidesb | Spectrum Count |
|---|---|---|---|---|---|---|---|
| IPI00407939.1 | Gria1 | Glutamate | Ionotropic | AMPA | 12 | 7 | 21 |
| IPI00228621.1 | Gria2 | Glutamate | Ionotropic | AMPA | 23 | 16 | 54 |
| IPI00407594.1 | Gria4 | Glutamate | Ionotropic | AMPA | 13 | 7 | 18 |
| IPI00118385.1 | Grin1 | Glutamate | Ionotropic | NMDA | 11 | 11 | 16 |
| IPI00321320.3 | Grin2b | Glutamate | Ionotropic | NMDA | 28 | 27 | 76 |
| IPI00131279.1 | Grin2d | Glutamate | Ionotropic | NMDA | 2 | 2 | 2 |
| IPI00123538.1 | Grik5 | Glutamate | Ionotropic | Kainate | 3 | 3 | 3 |
| IPI00123542.1 | Grid1 | Glutamate | Ionotropic | Orphan | 6 | 5 | 9 |
| IPI00123534.1 | Grid2 | Glutamate | Ionotropic | Orphan | 8 | 7 | 15 |
| IPI00110402.1 | Grm1 | Glutamate | Metabotropic | mGlu I | 15 | 14 | 21 |
| IPI00136716.1 | Grm3 | Glutamate | Metabotropic | mGlu II | 15 | 14 | 44 |
| IPI00113772.1 | Gabra1 | GABA | Ionotropic | A | 7 | 3 | 11 |
| IPI00110598.1 | Gabra2 | GABA | Ionotropic | A | 4 | 1 | 5 |
| IPI00110601.3 | Gabra3 | GABA | Ionotropic | A | 2 | 1 | 2 |
| IPI00109065.1 | Gabra4 | GABA | Ionotropic | A | 1 | 1 | 1 |
| IPI00221880.1 | Gabra5 | GABA | Ionotropic | A | 2 | 0 | 3 |
| IPI00119283.1 | Gabrb1 | GABA | Ionotropic | A | 4 | 0 | 6 |
| IPI00323554.3 | Gabrb2 | GABA | Ionotropic | A | 6 | 2 | 8 |
| IPI00130546.1 | Gabrb3 | GABA | Ionotropic | A | 7 | 3 | 10 |
| IPI00387194.1 | Gabrg1 | GABA | Ionotropic | A | 1 | 1 | 3 |
| IPI00228358.1 | Gabrg2 | GABA | Ionotropic | A | 4 | 4 | 10 |
| IPI00309158.1 | Gabbr1 | GABA | Metabotropic | B | 14 | 14 | 36 |
| IPI00409336.1 | Gpr51 | GABA | Metabotropic | B | 4 | 4 | 5 |
| IPI00270265.1 | Glra2 | Glycine | Ionotropic | 1 | 1 | 1 | |
| IPI00380761.1 | Glra3 | Glycine | Ionotropic | 2 | 2 | 3 | |
| IPI00108776.1 | Glrb | Glycine | Ionotropic | 1 | 1 | 2 | |
| IPI00115712.2 | Chrna4 | Acetylcholine | Ionotropic | Nicotinic | 3 | 3 | 3 |
| IPI00308091.1 | Chrnb2 | Acetylcholine | Ionotropic | Nicotinic | 1 | 1 | 1 |
| IPI00112942.1 | Chrm2 | Acetylcholine | Metabotropic | Muscarinic | 2 | 2 | 2 |
| IPI00136358.1 | Chrm4 | Acetylcholine | Metabotropic | Muscarinic | 1 | 1 | 1 |
| IPI00314429.2 | Adra1b | Catecholamine | Metabotropic | α-Adrenergic | 2 | 2 | 3 |
| IPI00228756.1 | Adrb3 | Catecholamine | Metabotropic | β-Adrenergic | 1 | 1 | 1 |
| IPI00123490.2 | Drd1a | Catecholamine | Metabotropic | Dopaminergic | 2 | 2 | 2 |
| IPI00133472.1 | Drd3 | Catecholamine | Metabotropic | Dopaminergic | 1 | 1 | 1 |
| IPI00130271.1 | P2rx7 | Purine | Ionotropic | P2x | 4 | 4 | 4 |
| IPI00112417.1 | P2ry6 | Purine | Metabotropic | P2y | 1 | 1 | 1 |
| IPI00136348.2 | Hrh1 | Histamine | Metabotropic | 1 | 1 | 1 |
DISCUSSION
Proteomic characterization of mammalian tissue samples is generally challenged by the extreme complexity and broad range of relative protein abundances. In this work, we demonstrated the direct analysis of lipid rich brain tissue by applying two-dimensional LC-MS/MS to global tryptic and cysteinyl-enriched peptide mixtures to attain extensive coverage of the mouse brain proteome without any subcellular fractionation or enrichment. When combined, the results led to significantly improved overall proteome coverage, as well as protein sequence coverage due to the complementary nature of the global and CPE analyses. The overall complexity of the enriched sample is significantly reduced compared with the global peptide sample, which enables effective detection of significantly more low abundance proteins and provides improved proteome coverage. A limitation of the CPE method is that many of the proteins that contain few cysteinyl-residues are not detectable; however, this limitation is nicely complemented by the global approach. As illustrated in Figure 2, the combined analyses led to 25% and 31% increases in overall proteome coverage compared to the analysis of only the global and the CPE samples, respectively. This approach will be suitable for rapid proteomic analysis of various mouse models of human brain diseases. The ability to achieve significantly improved proteome coverage by an alternative approach to subcellular fractionation for reducing sample complexity offers several advantages. First, it reduces cross-contamination (e.g. variations in partitioning between fractions) in that are problematic for subsequent quantitative analyses. Additionally, many biological studies involve very small clinical samples, which are not amenable to subcellular fractionation. The CPE strategy offers better sensitivity due to the effective handling of relatively small samples.
The effectiveness of the current approach for high dynamic range proteome profiling is evidenced by the confident identification of 7792 non-redundant proteins and >48,000 different peptides from the whole mouse brain homogenate without any prefractionation at the protein level. These non-redundant proteins cover ∼34% of all proteins in the current mouse IPI protein database, which provides significantly higher coverage than any previously reported for the mouse brain proteome. On the basis of protein pI, molecular weight, and cellular localization analyses, this set of identified proteins provides an overall unbiased representation of the mouse proteome with extensive coverage. Of particular interest are the ∼2000 membrane proteins (26%) and ∼1,000 extracellular proteins that were identified without any specific enrichment of membrane fractions. The many ion channel proteins, receptor proteins, and signaling proteins identified from the annotated membrane proteins cover ∼75% of the proteins identified from the most recent extensive mouse brain membrane proteome study8. Additionally, the spectrum count data for each identified protein provide proteome-wide semi-quantitative information on the relative abundances of mouse brain proteins.
We anticipate the comprehensive peptide/protein database developed in this study will have significant implications for neuroscience research in general. Particularly, the significantly improved coverage of membrane proteins will facilitate efforts aimed at a variety of functional studies such as cell signaling, cell-to-cell interactions, and profiling of surface membrane protein in diseases. Perhaps more apparently, this database will be a valuable resource for mouse brain proteomic research by providing a solid foundation for future quantitative proteomic studies of either spatially or temporally controlled protein abundance measurements that employ mouse models of disease. For example, in a recent study, a voxelation technique has been applied for 3D-mapping of gene expressions in the mouse brain4. This same technique is currently being applied to map protein expressions in the mouse brain. In an initial demonstration, we efficiently processed and identified proteins in mouse brain voxel samples using LC-MS/MS and LCFTICR43. In the next step to quantitatively map the mouse brain proteome, the current database will serve as the extensive reference database of mass and elution time tags needed to analyze voxelated samples using stable isotope 16O/18O labeling and the accurate mass and time tag (AMT) approach44, 45.
Acknowledgements
The authors thank the NIH (grant RR018522 to Richard D. Smith and grants R01 DA015802 and R01 NS050148 to Desmond J. Smith) for support of this research and the Environmental Molecular Sciences Laboratory (EMSL) for use of the instrumentation applied in this research. EMSL is a U.S. Department of Energy (DOE) national scientific user facility located at the Pacific Northwest National Laboratory in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL01830.
Footnotes
Supporting Information Available: The complete list of unique proteins and unique peptides are provided as Supplemental Table 1 and 2, respectively.
References
- 1.Peterson AS. Pixelating the brain. Genome Res. 2002;12(2):217–218. doi: 10.1101/gr.227802. [DOI] [PubMed] [Google Scholar]
- 2.Carson JP, Thaller C, Eichele G. A transcriptome atlas of the mouse brain at cellular resolution. Curr Opin Neurobiol. 2002;12(5):562–5. doi: 10.1016/s0959-4388(02)00356-2. [DOI] [PubMed] [Google Scholar]
- 3.Allen Brain Atlas. www.brain-map.org.
- 4.Singh RP, Smith DJ. Genome scale mapping of brain gene expression. Biol Psychiatry. 2003;53(12):1069–74. doi: 10.1016/s0006-3223(03)00238-5. [DOI] [PubMed] [Google Scholar]
- 5.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat. Med. 2001;7493-6(4):493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- 6.Fountoulakis M. Application of proteomics technologies in the investigation of the brain. Mass Spectrom Rev. 2004;23(4):231–58. doi: 10.1002/mas.10075. [DOI] [PubMed] [Google Scholar]
- 7.Beranova-Giorgianni S, Pabst MJ, Russell TM, Giorgianni F, Goldowitz D, Desiderio DM. Preliminary analysis of the mouse cerebellum proteome. Brain Res. Mol. Brain Res. 2002;98:135–140. doi: 10.1016/s0169-328x(01)00333-3. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen PA, Olsen JV, Podtelejnikov AV, Andersen JR, Mann M, Wisniewski JR. Proteomic mapping of brain plasma membrane proteins. Mol. Cell Proteomics. 2005;4:402–408. doi: 10.1074/mcp.T500002-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Tsuji T, Shiozaki A, Kohno R, Yoshizato K, Shimohama S. Proteomic profiling and neurodegeneration in Alzheimer's disease. Neurochem. Res. 2002;27:1245–1253. doi: 10.1023/a:1020941929414. [DOI] [PubMed] [Google Scholar]
- 10.Butterfield DA, Boyd-Kimball D, Castegna A. Proteomics in Alzheimer's disease: insights into potential mechanisms of neurodegeneration. J. Neurochem. 2003;86:1313–1327. doi: 10.1046/j.1471-4159.2003.01948.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Goodlett DR. Proteomic approach to studying Parkinson's disease. Mol. Neurobiol. 2004;29:271–288. doi: 10.1385/MN:29:3:271. [DOI] [PubMed] [Google Scholar]
- 12.Tribl F, Gerlach M, Marcus K, Asan E, Tatschner T, Arzberger T, Meyer HE, Bringmann G, Riederer P. “Subcellular Proteomics” of Neuromelanin Granules Isolated from the Human Brain. Mol. Cell Proteomics. 2005;4:945–957. doi: 10.1074/mcp.M400117-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Davidsson P, Sjogren M. The use of proteomics in biomarker discovery in neurodegenerative diseases. Dis. Markers. 2005;21:81–92. doi: 10.1155/2005/848676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd-Kimball D, Sultana R, Fai Poon H, Lynn BC, Casamenti F, Pepeu G, Klein JB, Butterfield DA. Proteomic identification of proteins specifically oxidized by intracerebral injection of amyloid beta-peptide (1-42) into rat brain: implications for Alzheimer's disease. Neuroscience. 2005;132:313–324. doi: 10.1016/j.neuroscience.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 15.Lovell MA, Xiong S, Markesbery WR, Lynn BC. Quantitative proteomic analysis of mitochondria from primary neuron cultures treated with amyloid beta peptide. Neurochem. Res. 2005;30:113–122. doi: 10.1007/s11064-004-9692-5. [DOI] [PubMed] [Google Scholar]
- 16.Lubec G, Krapfenbauer K, Fountoulakis M. Proteomics in brain research: potentials and limitations. Prog. Neurobiol. 2003;69:193–211. doi: 10.1016/s0301-0082(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 17.Gauss C, Kalkum M, Lowe M, Lehrach H, Klose J. Analysis of the mouse proteome. (I) Brain proteins: separation by two-dimensional electrophoresis and identification by mass spectrometry and genetic variation. Electrophoresis. 1999;20(3):575–600. doi: 10.1002/(SICI)1522-2683(19990301)20:3<575::AID-ELPS575>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Klose J, Nock C, Herrmann M, Stuhler K, Marcus K, Bluggel M, Krause E, Schalkwyk LC, Rastan S, Brown SDM, Bussow K, Himmelbauer H, Lehrach H. Genetic analysis of the mouse brain proteome. Nat. Genet. 2002;30:385–393. doi: 10.1038/ng861. [DOI] [PubMed] [Google Scholar]
- 19.Krapfenbauer K, Fountoulakis M, Lubec G. A rat brain protein expression map including cytosolic and enriched mitochondrial and microsomal fractions. Electrophoresis. 2003;24(11):1847–70. doi: 10.1002/elps.200305401. [DOI] [PubMed] [Google Scholar]
- 20.Langen H, Berndt P, Roder D, Cairns N, Lubec G, Fountoulakis M. Two-dimensional map of human brain proteins. Electrophoresis. 1999;20:907–916. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<907::AID-ELPS907>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 21.Yu LR, Conrads TP, Uo T, Kinoshita Y, Morrison RS, Lucas DA, Chan KC, Blonder J, Issaq HJ, Veenstra TD. Global analysis of the cortical neuron proteome. Mol. Cell Proteomics. 2004;3:896–907. doi: 10.1074/mcp.M400034-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Liu T, Qian WJ, Strittmatter EF, Camp DG, Anderson GA, Thrall BD, Smith RD. High throughput comparative proteome analysis using a quantitative cysteinyl-peptide enrichment technology. Anal. Chem. 2004;76:5345–5353. doi: 10.1021/ac049485q. [DOI] [PubMed] [Google Scholar]
- 23.Liu T, Qian WJ, Chen WU, Jacobs JM, Moore RJ, Anderson DJ, Gritsenko MA, Monroe ME, Thrall BD, Camp DG, Smith RD. Improved Proteome Coverage by Using High Efficiency Cysteinyl Peptide Enrichment: The Human Mammary Epithelial Cell Proteome. Proteomics. 2005;5(5):1263–1273. doi: 10.1002/pmic.200401055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, Smith RD. Probability-Based Evaluation of Peptide and Protein Identifications from Tandem Mass Spectrometry and SEQUEST Analysis: The Human Proteome. J. of Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 25.Wallin E, von Heijne G. Genome-wide Analysis of Integral Membrane Proteins from Eubacterial, Archaean, and Eukaryotic Organisms. Protein Sci. 1998;7(4):1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 27.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 28.Schwartz R, Ting CS, King J. Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome. Res. 2001;11:703–709. doi: 10.1101/gr.gr-1587r. [DOI] [PubMed] [Google Scholar]
- 29.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21(3):281–6. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 30.Stevens SM, Jr., Zharikova AD, Prokai L. Proteomic analysis of the synaptic plasma membrane fraction isolated from rat forebrain. Brain Res Mol Brain Res. 2003;117(2):116–28. doi: 10.1016/s0169-328x(03)00282-1. [DOI] [PubMed] [Google Scholar]
- 31.Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T, Yamauchi T. Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J Neurochem. 2004;88(3):759–68. doi: 10.1046/j.1471-4159.2003.02136.x. [DOI] [PubMed] [Google Scholar]
- 32.Li KW, Hornshaw MP, Van Der Schors RC, Watson R, Tate S, Casetta B, Jimenez CR, Gouwenberg Y, Gundelfinger ED, Smalla KH, Smit AB. Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J Biol Chem. 2004;279(2):987–1002. doi: 10.1074/jbc.M303116200. [DOI] [PubMed] [Google Scholar]
- 33.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 34.Ahram M, Springer DL. Large-scale proteomic analysis of membrane proteins. Expert Rev. Proteomics. 2004;1:293–302. doi: 10.1586/14789450.1.3.293. [DOI] [PubMed] [Google Scholar]
- 35.Wu CC, MacCoss MJ, Howell KE, Yates JR. A method for the comprehensive proteomic analysis of membrane proteins. Nat. Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 36.Tucker WC, Weber T, Chapman ER. Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science. 2004;304(5669):435–8. doi: 10.1126/science.1097196. [DOI] [PubMed] [Google Scholar]
- 37.Qian WJ, Jacobs JM, Camp DG, II, Monroe ME, Moore RJ, Gritsenko MA, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Smith RD. Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics. 2005;5(2):572–584. doi: 10.1002/pmic.200400942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Low SH, Miura M, Weimbs T. SNARE expression and localization in renal epithelial cells suggest mechanism for variability of trafficking phenotypes. Am J Physiol Renal Physiol. 2002;283(5):F1111–22. doi: 10.1152/ajprenal.00185.2002. [DOI] [PubMed] [Google Scholar]
- 40.Osborne SL, Herreros J, Bastiaens PI, Schiavo G. Calcium-dependent oligomerization of synaptotagmins I and II. Synaptotagmins I and II are localized on the same synaptic vesicle and heterodimerize in the presence of calcium. J Biol Chem. 1999;274(1):59–66. doi: 10.1074/jbc.274.1.59. [DOI] [PubMed] [Google Scholar]
- 41.Wisden W, Seeburg PH. A complex mosaic of high-affinity kainate receptors in rat brain. J Neurosci. 1993;13(8):3582–98. doi: 10.1523/JNEUROSCI.13-08-03582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feigenspan A, Wassle H, Bormann J. Pharmacology of GABA receptor Cl- channels in rat retinal bipolar cells. Nature. 1993;361(6408):159–62. doi: 10.1038/361159a0. [DOI] [PubMed] [Google Scholar]
- 43.Wang H, Qian WJ, Mottaz HM, Clauss TRW, Anderson DJ, Moore RJ, Camp DG, Khan AH, Sforza DM, Pallavicini M, Smith DJ, Smith RD. Development and Evaluation of a Micro- and Nano-Scale Proteomic Sample Preparation Method. J. Proteome Res. doi: 10.1021/pr050160f. In press, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qian WJ, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Moore RJ, Anderson DJ, Zhang R, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Camp DG, Smith RD. Quantitative Proteome Analysis of Human Plasma following in Vivo Lipopolysaccharide Administration Using 16O/18O Labeling and the Accurate Mass and Time Tag Approach. Mol. Cell. Proteomics. 2005;4(5):700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qian WJ, Camp DG, Smith RD. High Throughput Proteomics Using Fourier Transform Ion Cyclotron Resonance (FTICR) Mass Spectrometry. Expert Review of Proteomics. 2004;1(1):89–97. doi: 10.1586/14789450.1.1.87. [DOI] [PubMed] [Google Scholar]