The macrophage-stimulating protein pathway promotes metastasis in a mouse model for breast cancer and predicts poor prognosis in humans (original) (raw)
Abstract
A better understanding of tumor metastasis requires development of animal models that authentically reproduce the metastatic process. By modifying an existing mouse model of breast cancer, we discovered that macrophage-stimulating protein promoted breast tumor growth and metastasis to several organs. A special feature of our findings was the occurrence of osteolytic bone metastases, which are prominent in human breast cancer. To explore the clinical relevance of our model, we examined expression levels of three genes involved in activation of the MSP signaling pathway (MSP, MT-SP1, and MST1R) in human breast tumors. We found that overexpression of MSP, MT-SP1, and MST1R was a strong independent indicator of both metastasis and death in human breast cancer patients and significantly increased the accuracy of an existing gene expression signature for poor prognosis. These data suggest that signaling initiated by MSP is an important contributor to metastasis of breast cancer and introduce an independent biomarker for assessing the prognosis of humans with breast cancer.
Keywords: bone metastasis, Ron, tumor, inflammation, prognostic factor
Breast cancer often results in metastasis to many organs, including lymph nodes, bone, lungs, brain, and liver. The most frequent site of breast cancer metastasis is the bone, which occurs in ≈80% of patients with advanced disease (1). To better understand metastasis of breast cancer, models are needed in which metastases spontaneously occur from tumors arising in the orthotopic site. Such models have been described, although they use immortalized cell lines and usually require immunodeficient hosts. We describe here the modification of a mouse model of breast cancer in which tumors originate from primary breast epithelial cells, and metastasis occurs from an orthotopic tumor in immunocompetent animals. This experimental system allows us to efficiently examine the effect of individual genes or combinations of genes on tumor behavior. In this study, we examined the role of macrophage-stimulating protein (MSP) in breast tumor growth and metastasis.
MSP was originally identified as a serum protein that elicited macrophage chemotaxis and activation (2, 3). MSP is secreted as an inactive single-chain precursor (pro-MSP), which becomes active after proteolytic cleavage to yield a disufide-linked heterodimer. The protease that activates pro-MSP was isolated from the extracellular membranes of macrophages (4) and has recently been identified as membrane-type serine protease-1 (MT-SP1, also known as matripase), which is expressed on macrophages and several types of epithelial cells, including breast cells (5).
The biological effects of MSP are not restricted to macrophages. In particular, MSP can promote migration of various epithelial cell lines (6, 7), and the receptor for MSP, macrophage-stimulating-1 receptor (MST1R, also known as Ron), can induce an epithelial-to-mesenchymal transition in immortalized canine kidney cells in vitro (8). Evidence is accumulating that the MSP/MST1R pathway is involved in cancer. For example, overexpression of MST1R caused transformation of cultured epithelial cell lines (9, 10) and led to tumor development in lungs of transgenic mice (11).
MT-SP1 and MST1R are overexpressed in ≈45% and 50% of human infiltrating breast carcinomas, respectively (12, 13). A recent study showed that overexpression of MST1R, together with its homolog Met, correlated with reduced disease-free survival of breast cancer patients (14). These data suggest that the MSP/MT-SP1/MST1R pathway may play a role in breast cancer. We used a mouse model of breast cancer to demonstrate that MSP facilitates tumor growth and metastasis, particularly to bones. We also found that concomitant expression of components of the MSP pathway represented a highly accurate independent prognostic indicator for metastasis and death and significantly increased the accuracy of an existing poor prognosis gene expression signature for breast cancer (15, 16).
Results
Development of a Versatile Orthotopic Mouse Model of Breast Tumorigenesis and Metastasis.
To rigorously address the role of MSP in tumor progression and metastasis in vivo, we required a mouse model system in which metastasis might occur directly from the primary site, rather than as a result of cells having been injected into the bloodstream. We began with mammary tumors from transgenic mice that expressed the Polyomavirus Middle T antigen under control of the mouse mammary tumor virus promoter (17) (MMTV-PyMT). We chose the PyMT model, because detailed analysis has shown that PyMT tumors resemble human breast cancers. Specifically, PyMT tumors develop through a series of distinct histological stages that resemble progression of human breast cancer to malignancy (18). In addition, PyMT tumors display biomarkers that are relevant to progression of the human disease, such as loss of estrogen and progesterone receptors and overexpression of ErbB2/Neu and cyclin D1 (18). The PyMT oncogene also elicits a well characterized signaling cascade (19–21), leading to activation of pathways known to be involved in human breast cancers (22).
Tumors initiated by overexpression of transgenic PyMT develop with 100% penetrance and relatively short latency. Whereas many existing transgenic mouse models of breast cancer do not exhibit metastasis, MMTV-PyMT tumors do metastasize, but only to the lungs (17). For our studies, tumors were harvested from either of two sources: MMTV-PyMT transgenic mice or mice bearing PyMT tumors resulting from transplantation of normal mammary epithelial cells that had been transduced in vitro (23) with the PyMT oncogene. To determine whether overexpression of MSP could promote tumor invasion and metastasis, we introduced MSP into tumors elicited by PyMT using either of two methods. In the case of MMTV-PyMT tumors, we introduced a replication-defective mouse stem cell virus (pMIG) that expressed MSP and GFP into primary mammary tumor cells. The tumor cells were infected with either the pMIG (GFP alone) retroviral vector or pMIG-MSP, and infected (GFP+) cells were sorted by flow cytometry and transplanted into cleared mammary fat pads of recipient mice. The transplants gave rise to tumors that expressed either GFP or MSP and GFP. In the case of PyMT tumors generated by retroviral transduction (pMIG-PyMT-IRES-GFP tumors), we introduced pMIG-MSP or pMIG into primary mammary tumor cells along with a retrovirus that contained a puromycin-resistance gene. The cells were selected for 2 days in puromycin before transplantation into cleared mammary fat pads. In both types of experiments, the transplanted tumor cells expressed the PyMT transgene and/or transduced genes, as expected. The results were similar using either method (data not shown), but we found generation of PyMT tumors by retroviral transduction to be advantageous, because the cell-sorting step was eliminated. Expression of GFP provided a reliable sensitive identifier of tumor cells that had metastasized to distant organs.
MSP Facilitated the Growth and Dissemination of Mammary Tumors in Mice.
Tumors that expressed MSP (PyMT+MSP tumors) initially grew twice as fast as control tumors (PyMT+pMIG), although tumor growth reached a plateau, and the control tumors eventually achieved sizes comparable to PyMT+MSP tumors (Fig. 1A). Tumors that expressed MSP were more locally invasive than were control tumors [supporting information (SI) Fig. 4 A and B]. MSP was detected only in PyMT+MSP tumors (SI Fig. 4_C_). Both groups of tumors expressed MT-SP1 (SI Fig. 4_D_), but phosphorylated MSP receptor was detected only in tumors expressing MSP (SI Fig. 4_E_). Thus, overexpression of MSP in MMTV-PyMT tumors resulted in activation of the MSP receptor.
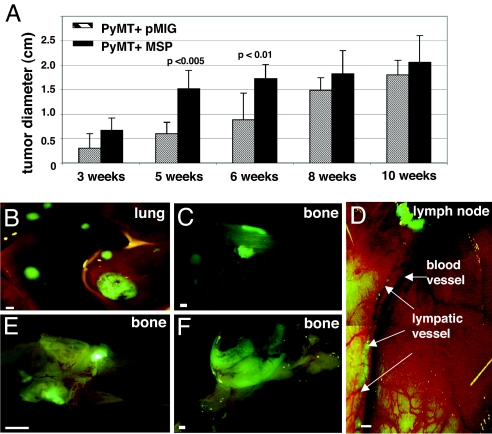
Fig. 1.
Overexpression of MSP in tumors initiated by PyMT results in faster tumor growth and increased metastasis. (A) Diameter of PyMT+pMIG or PyMT+MSP tumors as a function of time after transplantation (measured upon necropsy). The data are represented as a mean ± standard deviation. (B–F) Fluorescent whole-mount images of PyMT+MSP metastases in lung, bone, and lymphatics. (E and F) Fluorescent whole-mount images of larger bone metastases in mice from which the primary tumors (PyMT+MSP) were surgically removed 6 months before analysis. (Scale bars: A, B, D–F, 1 mm; C, 100 μm.)
In our experiments, lung metastasis occurred at a frequency consistent with that previously reported for MMTV-PyMT mice (17) (see Table 1). Tumors that expressed MSP, however, metastasized earlier than control tumors (SI Fig. 5) and displayed a broadened spectrum of metastasis to a variety of other organs including lymph nodes, spleen, and bone (Table 1 and Fig. 1 B–F). Control tumors, even at the maximal tumor size, were never as metastatic as tumors expressing MSP, and never metastasized to bone (Table 1). Liver metastases were observed infrequently from both tumors expressing MSP and control tumors (Table 1). Brain metastases were not apparent, although brains were examined only in animals that displayed severe metastasis to other organs (data not shown).
Table 1.
MSP increases metastatic frequency of PyMT tumors to lung, lymph nodes, spleen, and bones
| Site of metastasis | PyMT+pMIG, % | PyMT+MSP, % | P value* |
|---|---|---|---|
| Lung | 42/82 (51) | 68/99 (68) | <0.025 |
| Lymph node | 15/82 (18) | 54/99 (54) | <0.001 |
| Spleen | 3/65 (4.6) | 25/80 (31) | <0.001 |
| Bone | 0/75 (0) | 20/95 (21) | <0.001 |
| Liver | 3/65 (4.6) | 5/65 (7.6) | Not significant† |
MSP Promoted Osteolytic Bone Metastasis.
A prominent effect of MSP expression in tumors was metastasis to bone, which occurred in ≈20% of the mice (Table 1). The most common site for bone metastasis was the distal femur, but metastases were also observed on the ribs, sternum, and tibia (data not shown). Computed tomography (CT) scans revealed loss of bone density due to osteolysis, which was apparent on CT cross-section images (Fig. 2A) and in 3D rendering of the CT data (Fig. 2B). Metastasis was confirmed after CT scanning by the presence of GFP-positive tumor cells (Fig. 2C). We observed GFP-positive tumor cells in the bone marrow both with fluorescent microscopy and by flow cytometry (data not shown), but tumors that expressed MSP were also able to invade through the leg muscle and directly into the bone (data not shown). Histologically, osteolysis was manifested by the appearance of pits in the bone matrix (Fig. 2E) and by the presence of tartrate-resistant acid phosphatase (TRAP)-positive osteoclasts at the site of resorption (Fig. 2F). Using an in vitro bone pit assay, we found that tumor cells that expressed MSP, but not control tumor cells, were able to stimulate activation of osteoclast-like cells, which resulted in erosion of bone matrix (Fig. 2 H and I). These data are consistent with our observation that bone osteolysis resulting from metastasis of tumors that expressed MSP was associated with an increase in osteoclasts at sites of bone metastases in vivo.
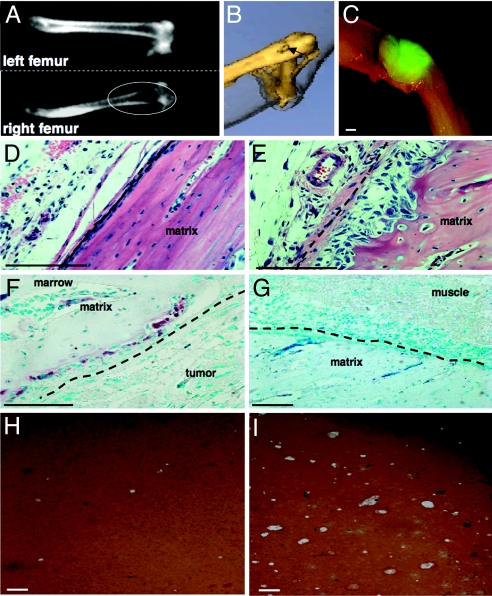
Fig. 2.
Tumor cells overexpressing MSP metastasized to bone, resulting in osteolytic lesions. (A) CT cross-section images of the normal left femur and the affected right femur from a mouse bearing a PyMT+MSP tumor. The images were taken at the same time, and only the right femur was affected by metastasis. (B) 3D rendering of CT data from the same mouse as in A. The arrow shows loss of bone density in the femur just above the knee. (C) Fluorescent whole-mount microscopy of the femur after the CT scan, showing a GFP+ PyMT+MSP tumor near the right knee. (D) Hematoxylin/eosin stain of a section of a normal femur. (E) Hematoxylin/eosin stain of a femur near a metastatic tumor, showing pitting of the bone beneath the periosteum (dashed line). (F) TRAP stain of the bone, showing osteoclasts in the bone pits (purple stain). The periosteum is indicated by the dashed line. (G) TRAP stain of the same bone in the region distal to the tumor. (Scale bar: C, 1 mm; D–G, 100 μm.) (H and I) In vitro bone pit assays using osteoclast-like cells derived from mouse bone marrow and cocultured with PyMT+pMIG primary tumor cells (H) or PyMT+MSP bone metastasis tumor cells (I). The bone matrix stains brown, and the pits induced by osteoclasts appear as white patches. (Scale bars, 100 μm.)
Coordinate Overexpression of MSP, MT-SP1, and MST1R Correlated with Metastasis and Death in Human Breast Cancer.
To explore the relevance of our findings to human breast cancer, we examined microarray gene expression data from 295 breast cancer patients from the Netherlands Cancer Institute studies (16). We found that MSP gene expression alone was not prognostic for outcome in human breast cancer (data not shown); however, because the biological effects of MSP depend on its proteolytic activation and the ability to bind to its receptor (2), expression of all three genes (MSP, MT-SP1, and MST1R) would be required for activation of the MSP pathway. Thus, we determined whether MSP, MT-SP1, and MST1R were overexpressed coordinately in breast cancer.
The arrays contained one oligonucleotide each corresponding to MT-SP1 and MST1R but happened to contain two different oligonucleotides corresponding to MSP. Some patients appeared to express MSP differently according to the two MSP oligonucleotides. This was purely an empirical observation, although there are two different MSP transcripts (24), which have different affinities for the two MSP oligonucleotides (data not shown). Faced with uncertainties about which MSP oligonucleotide might provide the more decisive conclusion, we analyzed the results with all four possible permutations: using oligonucleotide 1 only, using oligonucleotide 2 only, combining results obtained with each oligonucleotide (identifying patients positive for either oligonucleotide), and using both oligonucleotides (identifying patients positive for both oligonucleotides). Every permutation revealed significant prognostic value when the three components of the MSP signaling pathway were expressed concomitantly, although the strength and scope of the prognoses varied. In the presentation that follows, we use the results with oligonucleotide 2 for illustrative purposes, with secondary reference to the other permutations presented in SI Text.
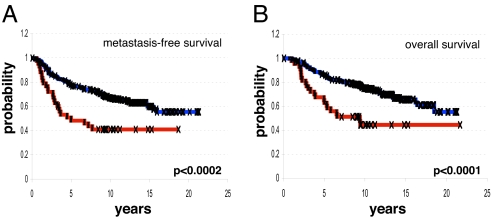
Using MSP oligonucleotide 2, we found that 43/295 (14.6%) of primary breast tumors in the Netherlands Cancer Institute (NKI) data set coordinately expressed MSP, MT-SP1, and MST1R at above-average levels. The number of patients defined as overexpressing MSP, MT-SP1, and MST1R ranged from 7.5% to 19.6%, depending on the MSP oligonucleotide used (SI Table 4). Kaplan–Meier analysis showed that patients whose tumors overexpressed MSP/MT-SP1/MST1R had significantly shorter metastasis-free survival (P = 0.0002, Fig. 3A) and overall survival (P = 0.0001, Fig. 3B) compared with patients whose tumors did not overexpress all three genes. The correlation with poor outcome was significant, no matter which MSP oligonucleotide(s) was used (SI Fig. 6). Patients whose tumors coordinately expressed MSP/MST1R/MT-SP1 below the mean did not benefit from significantly longer metastasis-free survival or overall survival, and overexpression of MSP, MT-SP1, or MST1R alone, or any combination of two of these three genes, did not reveal any significant association with clinical outcome in this data set (data not shown).
Fig. 3.
Overexpression of the MSP/MST1R pathway in human breast cancer is associated with metastasis and death. (A) Kaplan–Meier analysis showing development of metastases over time for two groups of patients: those whose tumors expressed MSP/MST1R/MT-SP1 (according to MSP oligonucleotide 2) at levels above the mean (red line), and those whose tumors did not express all three genes above the mean (blue line). (B) Kaplan–Meier analysis showing time to death for the same groups of patients as in A. P values were calculated by using the log-rank test.
Because overexpression of MSP/MT-SP1/MST1R significantly correlated with overall metastasis (Fig. 3A), we investigated whether overexpression of these genes correlated with metastasis to specific organs. We found that patients with tumors that overexpressed MSP/MT-SP1/MST1R experienced a significantly increased incidence of metastasis to bone (P < 0.025), lung (P < 0.001), liver (P < 0.05), and brain (P < 0.05). Bone was the most common site of metastasis for all patients (Table 2).
Table 2.
Percentage of patients that experienced metastasis to various sites
| Site of metastasis | _MSP/MT-SPT/MST1R_+ tumors (n = 13), % | Other tumors (n = 252), % | P value |
|---|---|---|---|
| Bone | 44.2 | 27.4 | <0.025 |
| Lung | 25.6 | 6.7 | <0.001 |
| Brain | 14.0 | 4.8 | <0.05 |
| Liver | 27.9 | 15.9 | <0.05 |
| Lymph | 11.6 | 4.8 | <0.1 |
| Meninges | 7.0 | 1.6 | <0.1 |
| Pleura | 14.0 | 7.9 | 0.2 |
| Skin | 7.0 | 2.4 | 1 |
| Any lymph node | 14.0 | 8.3 | 1 |
| Peritoneum | 4.7 | 2.8 | 1 |
Overexpression of MSP, MT-SP1, and MST1R Was an Independent Prognostic Factor for Metastasis and Death in Human Breast Cancer Patients.
We analyzed potential associations of MSP/MST1R/MT-SP1 overexpression with other clinical parameters, including patient age, number of positive lymph nodes, tumor size, histological grade, and estrogen receptor status. We found no statistically significant association with factors other than metastasis and death (data not shown). We also found no significant association between MSP/MST1R/MT-SP1 overexpression and previously described gene expression signatures (15, 16, 25, 26). To more rigorously address the independence of MSP/MST1R/MT-SP1 overexpression in prognosticating patient outcome, we carried out multivariate proportional-hazards analysis (27) using distant metastasis as the first event and overall survival as endpoints. We used multivariate analysis to compare the prognostic value of our results to the value of parameters currently used in the clinic and of recently identified gene expression signatures (15, 16, 25, 26). We found that coordinate overexpression of MSP/MST1R/MT-SP1 was an independent prognostic factor for risk of both metastasis and death, with a hazard ratio of 2.87 and 3.22, respectively (P ≤ 0.001; Table 3 and SI Table 5). The only other independent prognostic factors for survival in this analysis were tumor diameter (P = 0.05) and a “poor-prognosis” gene expression signature (P ≤ 0.001), which consists of 70 genes whose coordinate expression is strongly prognostic for a short interval to metastasis (15). The signature was identified by supervised classification, trained on a subset of the patient samples included in our analysis (15, 16). Overexpression of MSP/MST1R/MT-SP1 was a strong independent prognostic factor for risk of both metastasis and death, no matter which MSP oligonucleotide(s) was used (SI Table 4).
Table 3.
Multivariate Cox proportional-hazards analysis of the risk of death in breast cancer patients
| Variable (endpoint: survival) | Hazard ratio (95% CI)* | P value |
|---|---|---|
| MSP/MT-SP1/MST1R expression (above mean vs. below mean) | 3.22 (1.98–5.26) | <0.001 |
| Poor-prognosis signature (vs. good-prognosis signature) | 3.49 (1.74–7.00) | <0.001 |
| Tumor diameter (>2 vs. <2 cm) | 1.55 (1.00–2.37) | 0.05 |
| Patient age (>40 vs. <40 years) | 0.69 (0.44–1.07) | 0.1 |
| Wound signature (positive vs. negative) | 1.41 (0.86–2.33) | 0.18 |
| Sorlie subtype (basal or luminal B vs. normal or luminal A) | 1.39 (0.76–2.54) | 0.29 |
| Tumor grade (grade III vs. grades I or II) | 1.10 (0.67–1.79) | 0.71 |
| Estrogen receptor expression (positive vs. negative) | 0.95 (0.59–1.54) | 0.84 |
| Lymph node status (positive vs. negative) | 1.00 (0.66–1.53) | 1 |
Assessment of MSP/MST1R/MT-SP1 Expression Improved the Prognostic Accuracy of the 70-Gene Poor Prognosis Signature.
MSP, MT-SP1, and MST1R are not members of the 70-gene prognostic signature (15), yet we found that coordinate overexpression of these three genes was a strong independent prognostic factor for poor outcome. Despite its high specificity, however, coordinate overexpression of MSP/MT-SP1/MST1R as a single prognostic factor was not sensitive enough to identify all patients that experienced metastasis. This prompted us to investigate whether predictions based on overexpression of MSP/MT-SP1/MST1R could provide additional prognostic benefit when used together with the 70-gene signature.
Using MSP oligonucleotide 2, we found that detection of MSP/MT-SP1/MST1R overexpression in tumors can improve the accuracy of prognoses for poor outcome made by the 70-gene “poor prognosis” signature. The 70-gene signature predicted that 61% of the 295 patients would have a poor outcome, but only half of these patients actually developed metastases or died by 2005 (51% accuracy over a 10-year minimum followup time). We found that 27/295 patients (9.1%) displayed both the 70-gene “poor prognosis” signature and overexpression of MSP/MT-SP1/MST1R. Over the same 10-year minimum followup period, 82% of these patients developed metastasis or died. Therefore, a combination of the 70-gene “poor prognosis” signature and overexpression of MSP, MT-SP1, and MST1R provided a significant improvement in prognostic accuracy over the 70-gene “poor prognosis” signature alone (P < 0.01), and this applied to 9.1% of breast cancer patients in this study. Overexpression of MSP/MST1R/MT-SP1 always significantly improved the accuracy of the 70-gene “poor prognosis” signature, no matter which MSP oligonucleotide(s) was used (SI Table 6). The number of breast cancer patients in this study that displayed both the 70-gene “poor prognosis” signature and overexpression of MSP/MT-SP1/MST1R, and would thus benefit from the improved accuracy of the prognosis, ranged from 4% to 12%, depending on which MSP oligonucleotide(s) was used (SI Table 6).
MSP/MT-SP1/MST1R Was Validated as a Marker of Poor Prognosis in an Independent Set of Patients.
To validate our finding that MSP/MT-SP1/MST1R was a prognostic factor for poor outcome in breast cancer, we analyzed data from an independent study. The second data set comprised microarray gene expression data from 162 primary tumors collected from a diverse cohort of patients from four institutions (28, 29). We refer to this collection as the University of North Carolina Utah (UNC/Utah)/Utah data set. Patients in the UNC/Utah collection differed from the NKI cohort with respect to both age and stage of disease and are likely more representative of all women with breast cancer (see SI Text).
We found that 10/162 (6.2%) of primary breast tumors in the UNC/Utah collection coordinately expressed MSP, MT-SP1, and MST1R at above-average levels. Kaplan–Meier analysis showed that patients whose tumors overexpressed MSP/MT-SP1/MST1R had significantly shorter relapse-free survival (P = 0.03; SI Fig. 7) compared with the rest of the patients. We did not see a significant association with MSP/MT-SP1/MST1R status and overall survival to date (data not shown), although the patients in the UNC/Utah collection have been followed for a much shorter time than those in the NKI collection (median time, 21.5 months vs. 10.2 years).
We also investigated whether MSP/MT-SP1/MST1R status could improve the accuracy of the 70-gene “poor” prognosis in the UNC/Utah dataset. We found that, in these patients to date, the accuracy of the 70-gene “poor prognosis” signature was 28%. When MSP/MT-SP1/MST1R expression and the 70-gene “poor prognosis” signature were combined, the accuracy was 56% (P = 0.1). Although not statistically significant, these data show a trend toward improvement of accuracy of the 70-gene “poor prognosis” signature in the UNC/Utah patients. Significance will likely be improved upon analysis of more patients and/or a longer clinical followup time.
Discussion
We have used a mouse model of breast cancer to demonstrate that MSP promotes tumor growth and metastasis, including osteolytic metastasis to bone, from the site of the primary tumor. These findings provide the first indication that MSP facilitates metastasis. Our results complement a recent finding that targeted deletion of the MST1R kinase domain in the MMTV-PyMT model of breast cancer caused a reduction in tumor growth and metastasis to lungs (30). The MST1R study, however, could not investigate a role for the MSP pathway in metastasis to other organs, because the mice used in that study do not exhibit metastasis to sites other than lung (17, 30).
Although primary tumors expressing MSP grew faster than control tumors, we did not detect an increase in the number of cycling cells or a decrease in apoptotic cells (data not shown), suggesting that MSP does not promote metastasis simply by promoting tumor cell survival or proliferation. Also, the increased incidence of metastasis does not appear to be secondary to a rise in tumor cell numbers: transplantation of 10-fold more control tumor cells than MSP-expressing tumor cells caused the control tumors to grow faster but did not recapitulate the pattern or frequency of metastasis seen in tumors expressing MSP (A.L.W. and J.M.B., unpublished data). MSP may instead promote metastasis by affecting the ability of tumor cells to migrate, invade the extracellular matrix and blood vessels, and/or grow in distant tissues. The direct activation of osteoclasts by tumor cells expressing MSP suggests a mechanism by which MSP could facilitate growth in the bone.
Macrophage infiltration is a feature of invasive cancers and is associated with poor prognosis in breast cancer (31, 32). Macrophages are important for lung metastasis in the MMTV-PyMT mice from which we obtained the tumors used in our studies (33). Because MSP activates macrophages, it is possible that the effects of MSP occur through a mechanism involving inflammation. Our in vitro data showed that recombinant MSP was sufficient to stimulate mammary epithelial cells to invade extracellular matrix, indicating that these cells can directly respond to MSP. The effect of MSP was augmented, however, when macrophages were cocultured with the epithelial cells (A.L.W. and J.M.B., unpublished data). We also attempted to address the requirement for macrophages in our model system in vivo using _csf1_-deficient (op/op) mice (34), but we found large variations in tumor growth and metastasis in the op/op background that made our results impossible to interpret. Therefore, it is still unknown whether macrophages play a role in MSP-induced metastasis in vivo.
The replication of clinically relevant metastasis in our mouse model prompted us to investigate MSP gene expression in human breast cancer. Analysis of a total of 457 breast tumors from two independent studies showed a significant correlation between MSP/MT-SP1/MST1R overexpression and poor outcome. Approximately two-thirds of patients whose tumors overexpressed MSP/MT-SP1/MST1R had poor outcome: they experienced increased metastasis to bone, lung, liver and brain and had shorter survival times. It should be noted, however, that whereas nearly 80% of women with breast cancer experience bone metastasis, a much smaller fraction of breast cancers overexpressed MSP/MT-SP1/MST1R in our analyses (up to 19.6% in the NKI data set and 6.2% in the UNC/Utah set). These data suggest that pathways other than the MSP pathway may contribute to bone metastasis.
Analysis of expression of MSP/MT-SP1/MST1R in concert with the previously described 70-gene “poor prognosis” signature significantly increased the accuracy with which poor outcome was prognosticated for breast cancer patients. These data suggest that a greater prognostic power would result from addition of MSP, MT-SP1, and MST1R oligos to the existing 70-gene array, so that MSP/MT-SP1/MST1R expression status can be considered in parallel with the prognosis signature. Even the least stringent analysis of the NKI data (which defined patients according to a positive signal from either MSP oligonucleotide) significantly increased the accuracy of the 70-gene signature for 12% of patients, which would currently represent >25,000 people per year based on an annual diagnosis rate of >212,000 (see www.komen.org).
The gene expression data used in the first part of this study were gathered from 295 tumors collected from patients with stage I or II breast cancer (16). Our data indicate that coordinate overexpression of MSP, MT-SP1, and MST1R is a prognostic factor for poor outcome in early stage disease, which may aid in treatment decisions. We also validated our findings on an independent collection of 162 breast tumors from individuals with a wider range of disease stages, albeit with shorter followup to date.
Together, our findings implicate MSP as an important contributor to metastasis and highlight how the study of mouse models can inform the analysis of clinical data. Our model should be useful for preclinical studies to determine whether disruption of the MSP pathway is effective in slowing tumor growth and reducing or preventing metastasis.
Experimental Procedures
Animals.
Animals were handled according to protocols approved by the University of California, San Francisco, Institutional Animal Care and Use Committee. MMTV-PyMT mice were obtained from The Jackson Laboratory, (Bar Harbor, ME). Some mice underwent a second surgery to remove the primary tumor, which was 2–2.5 cm in diameter at the time of resection.
Infection of Cells with Retroviruses.
Retroviral production and infections were as described (23). Infection of MMTV-PyMT tumor cells was carried out as for normal mammary epithelial cells except that, after collagenase treatment, the tumors were plated on 150-mm tissue culture dishes overnight, then trypsinized and plated at a density of 105 cells/well in six-well dishes for infection the next day. Infected MMTV-PyMT tumor cells were sorted for GFP by using a FACSVantage SE cell sorter, and the cells were transplanted immediately. To generate tumors expressing PyMT by retroviral transduction, normal mammary epithelial cells were infected with pMIG-PyMT. The cells were transplanted into cleared mammary fat pads (106 cells/gland), and tumors developed in ≈3 months. pMIG-PyMT tumor cells were isolated and infected with pMIG-MSP or pMIG, plus a retrovirus containing a puromycin resistance gene at a ratio of 2:1. Puromycin was added at a concentration of 1 μg/ml beginning 1 day after infection. Selection continued until transplantation (4 days after tumor harvest).
CT.
microCT was performed on a combined microSPECT/microCT scanner for small animals (X-SPECT; Gamma Medica Ideas, Northridge, CA). Mice were anesthetized with isoflurane during image acquisition. Tomographic data were acquired at 512 angles over 360° at 50 kilovoltage peak tube potential and 600 μA tube current. Images were reconstructed with the Feldkamp cone-beam algorithm (Gamma Medica Ideas) and imported into Amira (Mercury Computing Systems, Chelmsford, MA) for processing and analysis. Transaxial images were oriented to obtain cross-sectional views through individual vertebrae and along the axes of both femurs. The entire skeleton was surface-rendered as a 3D volumetric image as an aid to visualization.
Osteoclast Assays.
TRAP staining was performed as described (35). For in vitro assays, osteoclast-like cells were prepared from mouse bone marrow as described (36). The cells were plated on osteologic discs (BD Biosciences, San Jose, CA) for bone pit assays. After 5 days, PyMT+pMIG or PyMT+MSP tumor cells were added to the osteoclast cultures at a density of 105 tumor cells per well. Five days later, the cells were removed with bleach, and the discs were stained according to the manufacturer.
Gene Expression Analysis.
Gene expression data for MSP1, MT-SP1, and MST1R were culled for each tumor sample from the NKI data set by mapping Unigene identifiers from build 158, release date January 18, 2003. Two clones were mapped for MSP1, corresponding to Unigene ID 349110. One clone each was mapped for MT-SP1 (Unigene ID 56937) and MST1R (Unigene ID 2942). Gene expression data for all four clones were extracted for each sample and mean-centered across all samples for each clone. Samples were segregated into groups for each analysis. Group A comprised tumors coordinately expressing MT-SP1, MST1R, and MSP (according to oligonucleotide 2) at levels higher than the mean (43 tumors), and group B comprised the remaining samples (252 tumors). Detailed information on analysis of the different MSP/MT-SP1/MST1R groups using all possible permutations can be found in SI Text. Information on metastasis and survival was obtained by using followup data as of January 1, 2005, which represents an update of the 2001 data published previously (16). The same method was used for the UNC/Utah data set (see SI Text for more information).
Statistical Analysis.
Kaplan–Meier survival curves were generated by using the software package WINSTAT FOR EXCEL (R. Fitch Software, Staufen, Germany). Multivariate analysis was performed by using the Cox proportional-hazard method and SPSS 13 software (SPSS, Chicago, IL). Covariates analyzed in the multivariate analysis included the following binary variables: tumor diameter (up to and including 2 vs. >2 cm), positive lymph nodes (zero vs. one or more positive nodes), tumor grade (I or II vs. III), patient age (≤40 vs. >40 year), estrogen receptor status (negative vs. positive), wound signature (“quiescent” vs. “activated”), poor prognosis signature (“good” vs. “poor”), Sorlie subtype (normal-like or luminal A vs. luminal B, basal, or ErbB2-like), and MSP pathway status (group B vs. A). For each sample, assignment according to the wound signature as “quiescent” or “activated” was made on the basis of unsupervised clustering of the entire NKI data set with the wound signature genes, as reported (25, 37). Assignment to the five-class “intrinsic gene signature” or Sorlie subtype was made by matching the expression of the intrinsic genes in each tumor sample to the most similar expression centroid for the five classes, as described (25, 26, 38). Classification of good or poor prognosis according to the Poor Prognosis Signature has been described (15, 16).
Supplementary Material
Supporting Information
Acknowledgments
We thank Luda Urisman for technical assistance, as well as members of the Bishop laboratory for helpful discussions. We thank William Muller (McGill University) for making the MMTV-PyMT transgenic mice available and Yosef Refaeli (National Jewish Medical Research Center, Denver, CO) for providing the pMIG vector. We are grateful to Kirk Jones for consultation with histology, Thomas Link for assistance in reading CT scans, Patrick Brown for assistance in interpreting microarray data, Philip Bernard and Charles Perou for access to the UNC/Utah data set, and Ami Bhatt and Charles Craik for access to data before publication and discussions about MT-SP1. This work was supported by the G. W. Hooper Research Foundation, National Institutes of Health Grant CA44338 (to J.M.B.), University of California Discovery Grant bio02-10300 with Gamma Medica IDEAS, Inc., as a private sponsor (to B.H.H.), and National Institutes of Health Bioengineering Research Partnership Grant 5 R01 EB000348 (to B.H.H.). A.L.W. was supported by Susan G. Komen Breast Cancer Foundation Grant PDF0201190. J.B.S. is supported by a predoctoral fellowship from the National Science Foundation. C.T. is supported by a predoctoral bioengineering fellowship from the Whitaker Foundation. D.S.A.N and M.J.d.V are supported by Dutch Cancer Society Grant NKB 2002-2575.
Abbreviations
MSP
macrophage-stimulating protein
MT-SP1
membrane-type serine protease 1
MST1R
macrophage-stimulating 1 receptor
PyMT
Polyomavirus middle T antigen
MMTV
mouse mammary tumor virus
CT
computed tomography
TRAP
tartrate-resistant acid phosphatase.
Note Added in Proof.
Overexpression of murine Ron was recently shown to induce mammary tumor formation and metastasis to lung and liver (39), providing further evidence of the importance of the MSP pathway in breast cancer.
Footnotes
The authors declare no conflict of interest.
References
- 1.Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, Feuer EJ, Thun MJ. CA Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 2.Leonard EJ. Ciba Found Symp. 1997;212:183–191. discussion 192–187. [PubMed] [Google Scholar]
- 3.Lutz MA, Correll PH. J Leukocyte Biol. 2003;73:802–814. doi: 10.1189/jlb.0602319. [DOI] [PubMed] [Google Scholar]
- 4.Wang MH, Skeel A, Leonard EJ. J Clin Invest. 1996;97:720–727. doi: 10.1172/JCI118470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatt AS, Welm AL, Farady CJ, Vasquez M, Wilson K, Craik C. Proc Natl Acad Sci USA. 2007;104:5771–5776. doi: 10.1073/pnas.0606514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santoro MM, Gaudino G, Marchisio PC. Dev Cell. 2003;5:257–271. doi: 10.1016/s1534-5807(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 7.Wang MH, Montero-Julian FA, Dauny I, Leonard EJ. Oncogene. 1996;13:2167–2175. [PubMed] [Google Scholar]
- 8.Wang D, Shen Q, Chen YQ, Wang MH. Oncogene. 2004;23:1668–1680. doi: 10.1038/sj.onc.1207282. [DOI] [PubMed] [Google Scholar]
- 9.Peace BE, Hughes MJ, Degen SJ, Waltz SE. Oncogene. 2001;20:6142–6151. doi: 10.1038/sj.onc.1204836. [DOI] [PubMed] [Google Scholar]
- 10.Wang MH, Wang D, Chen YQ. Carcinogenesis. 2003;24:1291–1300. doi: 10.1093/carcin/bgg089. [DOI] [PubMed] [Google Scholar]
- 11.Chen YQ, Zhou YQ, Fisher JH, Wang MH. Oncogene. 2002;21:6382–6386. doi: 10.1038/sj.onc.1205783. [DOI] [PubMed] [Google Scholar]
- 12.Kang JY, Dolled-Filhart M, Ocal IT, Singh B, Lin CY, Dickson RB, Rimm DL, Camp RL. Cancer Res. 2003;63:1101–1105. [PubMed] [Google Scholar]
- 13.Maggiora P, Marchio S, Stella MC, Giai M, Belfiore A, De Bortoli M, Di Renzo MF, Costantino A, Sismondi P, Comoglio PM. Oncogene. 1998;16:2927–2933. doi: 10.1038/sj.onc.1201812. [DOI] [PubMed] [Google Scholar]
- 14.Lee WY, Chen HH, Chow NH, Su WC, Lin PW, Guo HR. Clin Cancer Res. 2005;11:2222–2228. doi: 10.1158/1078-0432.CCR-04-1761. [DOI] [PubMed] [Google Scholar]
- 15.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 16.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 17.Guy CT, Cardiff RD, Muller WJ. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW. Am J Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guy CT, Muthuswamy SK, Cardiff RD, Soriano P, Muller WJ. Genes Dev. 1994;8:23–32. doi: 10.1101/gad.8.1.23. [DOI] [PubMed] [Google Scholar]
- 20.Rauh MJ, Blackmore V, Andrechek ER, Tortorice CG, Daly R, Lai VK, Pawson T, Cardiff RD, Siegel PM, Muller WJ. Mol Cell Biol. 1999;19:8169–8179. doi: 10.1128/mcb.19.12.8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webster MA, Hutchinson JN, Rauh MJ, Muthuswamy SK, Anton M, Tortorice CG, Cardiff RD, Graham FL, Hassell JA, Muller WJ. Mol Cell Biol. 1998;18:2344–2359. doi: 10.1128/mcb.18.4.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bieche I, Onody P, Tozlu S, Driouch K, Vidaud M, Lidereau R. Int J Cancer. 2003;106:758–765. doi: 10.1002/ijc.11273. [DOI] [PubMed] [Google Scholar]
- 23.Welm AL, Kim S, Welm BE, Bishop JM. Proc Natl Acad Sci USA. 2005;102:4324–4329. doi: 10.1073/pnas.0500470102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Degen SJ, McDowell SA, Waltz SE, Gould F, Stuart LA, Carritt B. DNA Seq. 1998;8:409–413. doi: 10.3109/10425179809020903. [DOI] [PubMed] [Google Scholar]
- 25.Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van't Veer LJ, Bartelink H, et al. Proc Natl Acad Sci USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox D. J R Stat Soc B. 1972;34:187–220. [Google Scholar]
- 28.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perreard L, Fan C, Quackenbush JF, Mullins M, Gauthier NP, Nelson E, Mone M, Hansen H, Buys SS, Rasmussen K, et al. Breast Cancer Res. 2006;8:R23. doi: 10.1186/bcr1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peace BE, Toney-Earley K, Collins MH, Waltz SE. Cancer Res. 2005;65:1285–1293. doi: 10.1158/0008-5472.CAN-03-3580. [DOI] [PubMed] [Google Scholar]
- 31.Lin EY, Pollard JW. Novartis Found Symp. 2004;256:158–168. discussion 168–172:259–269. [PubMed] [Google Scholar]
- 32.Lin EY, Pollard JW. Br J Cancer. 2004;90:2053–2058. doi: 10.1038/sj.bjc.6601705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin EY, Nguyen AV, Russell RG, Pollard JW. J Exp Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD. Nature. 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 35.Ortega N, Behonick DJ, Colnot C, Cooper DN, Werb Z. Mol Biol Cell. 2005;16:3028–3039. doi: 10.1091/mbc.E04-12-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurihara N, Iwama A, Tatsumi J, Ikeda K, Suda T. Blood. 1996;87:3704–3710. [PubMed] [Google Scholar]
- 37.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, Brown PO. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zinser GM, Leonis MA, Toney K, Pathrose P, Thobe M, Kader SA, Peace BE, Beauman SR, Colllins MH, Waltz SE. Cancer Res. 2006;66:11967–11974. doi: 10.1158/0008-5472.CAN-06-2473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information