The SXT Conjugative Element and Linear Prophage N15 Encode Toxin-Antitoxin-Stabilizing Systems Homologous to the tad-ata Module of the Paracoccus aminophilus Plasmid pAMI2 (original) (raw)
Abstract
A group of proteic toxin-antitoxin (TA) cassettes whose representatives are widely distributed among bacterial genomes has been identified. These cassettes occur in chromosomes, plasmids, bacteriophages, and noncomposite transposons, as well as in the SXT conjugative element of Vibrio cholerae. The following four homologous loci were subjected to detailed comparative studies: (i) tad-ata from plasmid pAMI2 of Paracoccus aminophilus (the prototype of this group), (ii) gp49-gp48 from the linear bacteriophage N15 of Escherichia coli, (iii) s045-s044 from SXT, and (iv) Z3230-Z3231 from the genomic island of enterohemorrhagic Escherichia coli O157:H7 strain EDL933. Functional analysis revealed that all but one of these loci (Z3230-Z3231) are able to stabilize heterologous replicons, although the host ranges varied. The TA cassettes analyzed have the following common features: (i) the toxins are encoded by the first gene of each operon; (ii) the antitoxins contain a predicted helix-turn-helix motif of the XRE family; and (iii) the cassettes have two promoters that are different strengths, one which is located upstream of the toxin gene and one which is located upstream of the antitoxin gene. All four toxins tested are functional in E. coli; overexpression of the toxins (in the absence of antitoxin) results in a bacteriostatic effect manifested by elongation of bacterial cells and growth arrest. The toxins have various effects on cell viability, which suggests that they may recognize different intracellular targets. Preliminary data suggest that different cellular proteases are involved in degradation of antitoxins encoded by the loci analyzed.
Bacterial plasmids have modular structures, since it is possible to dissect them into several functional cassettes. The core region of the plasmid backbone is composed of a set of conserved modules, coding for replication and stability functions, which are crucial for plasmid maintenance. Comparative analyses of plasmid genomes have revealed that the same modules or closely related modules can be found in various combinations in different plasmids hosted by phylogenetically distinct bacteria. The plasticity of plasmid genomes is the result of horizontal gene transfer, as well as numerous recombinational events, which take place in diverse bacterial hosts. Recent advances in bacterial genomics have revealed that shuffling of modules is not limited to plasmids but is a much more common phenomenon, occurring in all bacterial mobile genetic elements. These elements may thus be considered combinations of exchangeable functional cassettes (78).
Shuffling of modules, initially defined in plasmids, bacteriophages, and transposable elements, has resulted in (i) the generation of novel mobile elements, such as conjugative transposons (containing phage-related recombination modules plus plasmid-borne modules for conjugational transfer) (64), and (ii) the appearance of chimeric elements exhibiting new, unusual properties, including transposable bacteriophage Mu (containing a transposition module) (23) and temperate bacteriophages P1 and N15 of Escherichia coli (carrying a set of plasmid-derived modules responsible for replication and stable maintenance). The modular composition of P1 and N15 allows them to lysogenize their hosts as autonomous, circular (P1) or linear (N15) plasmid-like forms (54, 67).
P1 and typical low-copy-number plasmids include three well-defined modules responsible for their stable maintenance: (i) active partitioning systems (par) involved in efficient segregation of plasmid copies to daughter cells during cell division, (ii) multimer resolution systems (mrs) that increase the copy number of plasmid monomers to optimize their segregation, and (iii) toxin-antitoxin (TA) systems (also called addiction systems) responsible for postsegregational elimination of plasmidless cells from a bacterial population (for a review, see reference 42).
The addiction systems encode two components: (i) a toxin, which recognizes a specific cellular target; and (ii) an antitoxin, which counteracts the toxin. In all cases, the toxins are proteins, whereas the antidote can be a protein (which forms tight nontoxic complexes with the toxin) or antisense RNA (which inhibits formation of the toxin by binding to its mRNA) (31). This plasmid stabilization system functions through the different stabilities of toxins and antitoxins; the antitoxin component is less stable and is degraded by cellular protease Lon or Clp in the case of proteins (49, 80).
All plasmid-encoded TA systems act using a common scheme. A cell carrying a plasmid with a TA module produces both stable toxin and labile antitoxin, and the fact that an excess of the latter is always synthesized precludes interaction of the toxin with its cellular target. Once a bacterium loses the plasmid, the antidote is degraded, and the released toxin binds to its target. This results in growth arrest or cell death of plasmidless segregant cells, which consequently increases the number of plasmid-containing cells in the bacterial population (for a review, see reference 42).
The great majority of TA modules encode proteic components; some examples are ccdAB of plasmid F (63), parDE of RK2 (68), kis-kid of R1 (17), higBA of plasmid Rts1 (76), axe-txe of pRUM (35), and phd-doc of bacteriophage P1 (49). These genes constitute small, tightly regulated operons, in which the genes are generally in the same orientation and the gene encoding the antitoxin precedes the gene encoding the toxin. So far, an opposite gene orientation has been found only in the higBA module (76). In a few cases, including ω-ɛ-ζ of pSM19035 (83) and pasABC of pTF-FCII (72), the TA cassette encodes an additional element that performs regulatory functions.
Interestingly, TA modules are also found in bacterial chromosomes (1, 34). The recent study of Pandey and Gerdes (65) demonstrated that such cassettes are abundant and widespread in genomes of free-living bacteria (both gram-negative and gram-positive bacteria), as well as in archaea. TA loci are thought to be involved in the stringent response, helping cells to survive under nutritional stress conditions by limiting various metabolic activities, as shown by Christensen et al. (24) for the relBE locus of E. coli, and/or programmed cell death, as suggested for mazEF of E. coli (1). In the light of these findings, it is believed that plasmid TA cassettes originated from chromosomal loci which, when transferred to mobile genetic elements, were adopted as stabilizing systems (33).
In this report we describe a novel stabilizing system (tad-ata) encoded by plasmid pAMI2 of Paracoccus aminophilus JCM 7686. Our analysis revealed that the tad-ata locus may be considered the prototype of a widely distributed group of related modules linked with various genetic elements, including plasmids, bacteriophages, noncomposite transposons, and the SXT conjugative element, as well as bacterial chromosomes. We demonstrated that the modules encoded by bacteriophage N15 and the SXT element are functional stabilization systems, which act on the toxin-antitoxin principle.
The identification of this novel group of TA cassettes provides an additional example of global shuffling of structural modules, which has a great impact on the evolution of bacterial mobile genetic elements.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were grown in Luria-Bertani (LB) medium (69) at 30°C (Paracoccus spp. strains, Agrobacterium tumefaciens, and Rhodobacter sphaeroides) or 37°C (E. coli) or in TY medium (13) at 30°C (Rhizobium leguminosarum). When necessary, the medium was supplemented with antibiotics at the following concentrations: kanamycin, 50 μg ml−1; rifampin, 50 μg ml−1; ampicillin, 30 μg ml−1 (strains carrying pNDM220 derivatives and pOU82 derivatives) or 100 μg ml−1 (strains carrying pGEM-T Easy); tetracycline, 20 μg ml−1; streptomycin, 400 μg ml−1; and chloramphenicol, 20 μg ml−1. The Paracoccus spp., A. tumefaciens, R. sphaeroides, and R. leguminosarum strains formed colonies on solid media after 48 h of incubation.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or features | Reference or source |
|---|---|---|
| Strains | ||
| Paracoccus alcaliphilus JCM 7364R | Rifr derivative of wild-type strain JCM 7364 | 10 |
| Paracoccus alkenifer DSM 11593R | Rifr derivative of wild-type strain DSM 11593 | 9 |
| Paracoccus aminophilus JCM 7686 | Wild type | 79 |
| Paracoccus aminovorans JCM 7685R | Rifr derivative of wild-type strain JCM 7685 | 10 |
| Paracoccus denitrificans LDM 22.21R | Rifr derivative of wild-type strain LDM 22.21 | 11 |
| Paracoccus methylutens DM12R | Rifr derivative of wild-type strain DM12 | 10 |
| Paracoccus pantotrophus KL100 | Rifr derivative of wild-type strain DSM 11073 | 10 |
| Paracoccus solventivorans DSM 11592R | Rifr derivative of wild type strain DSM 11592 | 11 |
| Paracoccus thiocyanathus IAM 12816R | Rifr derivative of wild-type strain IAM 12816 | 10 |
| Paracoccus versutus UW225 | Rifr derivative of wild-type strain UW1 | 5 |
| Rhodobacter sphaeroides 2.4.1R | Rifr derivative of wild-type strain 2.4.1 | 11 |
| Agrobacterium tumefaciens GMI 9023 | Rifr | 47 |
| Rhizobium leguminosarum 1062 | Strr | 43 |
| Escherichia coli BR825 | polA::Tn_10 trp_ | 55 |
| Escherichia coli DH5Δlac | deoR thi1 relA1 supE44 endA1 gyrA96 recA1 hsdR17 Δ(argF lac)U169 Nalr | M. Yarmolinski |
| Escherichia coli EC16816 | hsdR2 galK2 galT22 supE44; contains N15; source of the gp49-gp48 TA cassette | V. Rybchin |
| Escherichia coli EDL933 | Wild-type enterohemorrhagic E. coli O157:H7 strain, source of the Z3230-Z3231 TA cassette | 62 |
| Escherichia coli HW220 | K-12 derivative (CAG18439) containing SXT element of V. cholerae, source of the s045-s044 TA cassette | 44 |
| Escherichia coli MC1000 | araD 139 Δ(araABC-leu)7679 galU galK Δ(lac)X74 rpsL thi | 21 |
| Escherichia coli SG22025 | Kmr, Δ_lac rcsA_ 166::aphA | 50 |
| Escherichia coli SG22093 | Kmr Cmr, Δ_lac rcsA_ 166::aphA clpP 1::cat | 50 |
| Escherichia coli SG22095 | Kmr Tcr, Δ_lac rcsA_ 166::aphA lon 146::tet | 50 |
| Escherichia coli TG1 | supE hsd_Δ_5 thi Δ(lac proAB) F′ (traD36 proAB+_lacI_q_lacZ_ΔM15) | Laboratory collection |
| Plasmids | ||
| pAMI2 | Natural plasmid (approx. 18.6 kb) of P. aminophilus JCM 7686 | 4 |
| pAMI204 | Kmr, mini-pAMI2 | This study |
| pABW1 | Kmr, ori pMB1, mobilizable cloning vector, oriT RK2 | 6 |
| pMDB209 | pABW1 carrying a DNA region of pAMI2 present in pAMI204 | This study |
| pMDB300 | pMDB209 with diminished region of pAMI204 | This study |
| pABW1-REP | pABW1 carrying REP module of pAMI2 (amplified by PCR with primers LLDKREP and RREPAMI) inserted into HindIII site | This study |
| pABW1-REP+PAR | pABW1 carrying REP and PAR modules of pAMI2 (amplified by PCR with primers LLDKREP and RLDKPAR) inserted into HindIII site | This study |
| pABW1-REP+PAR+TA | pABW1 carrying REP, PAR, and TA modules of pAMI2 (amplified by PCR with primers LLDKREP and RKILHIII) inserted into HindIII site | This study |
| pUC4K | Source of kanamycin resistance cassette | 81 |
| pBGS18 | Kmr, ori pMB1, cloning vector | 73 |
| pBGS-AMI | pBGS18 carrying tad-ata (amplified by PCR with primers LLDZ2547 and RLDZ3578) inserted into SmaI site | This study |
| pBGS-SXT | pBGS18 carrying s044-s045 (amplified by PCR with primers LSXT and RSXT) inserted into SmaI site | This study |
| pBGS-N15 | pBGS18 carrying gp49-gp48 (amplified by PCR with primers LN15 and RN15) inserted into SmaI site | This study |
| pBGS-EDL | pBGS18 carrying Z3230-Z3231 (amplified by PCR with primers LEDL and REDL) inserted into SmaI site | This study |
| pMUT1 | pBGS-AMI with mutated tad | This study |
| pMUT2 | pMUT1 with mutated ata | This study |
| pFH450 | Cmr, ori R1, ori bacteriophage P1, segregational stability test vector for E. coli | 41 |
| pFH-AMI | pFH450 carrying tad-ata | This study |
| pFH-SXT | pFH450 carrying s045-s044 | This study |
| pFH-N15 | pFH450 carrying gp49-gp48 | This study |
| pFH-EDL | pFH450 carrying Z3230-Z3231 | This study |
| pABW3 | Kmr, ori pMB1, ori pTAV1, oriT RK2, segregational stability test vector for Paracoccus spp. | 8 |
| pABW3-AMI | pABW3 carrying tad-ata | This study |
| pABW3-SXT | pABW3 carrying s045-s044 | This study |
| pABW3-N15 | pABW3 carrying gp49-gp48 | This study |
| pABW3-EDL | pABW3 carrying Z3230-Z3231 | This study |
| pCF430 | Tcr, ori RK2, oriT RK2, P_BAD_ promoter | 60 |
| pTOX-tad | pCF430 carrying tad (amplified by PCR with primers TADL and TADR) inserted between PstI and XbaI sites | This study |
| pTOX-s045 | pCF430 carrying s045 (amplified by PCR with primers LTSXT and RTSXT) inserted between PstI and XbaI sites | This study |
| pTOX-gp49 | pCF430 carrying gp49 (amplified by PCR with primers LTN15 and RTN15) inserted between PstI and XbaI sites | This study |
| pTOX-Z3230 | pCF430 carrying Z3230 (amplified by PCR with primers LTEDL and RTEDL) inserted between PstI and XbaI sites | This study |
| pNDM220 | Apr, ori R1, pA1/O4/O3, _lacI_q | 34 |
| pANTI-ata | pNDM220 carrying ata (amplified by PCR with primers 2LAAMIBH and 2RAAMIER) inserted between EcoRI and BamHI sites | This study |
| pANTI-s044 | pNDM220 carrying s044 (amplified by PCR with primers 2LASXTBH and 2RASXTER) inserted between EcoRI and BamHI sites | This study |
| pANTI-gp48 | pNDM220 carrying gp48 (amplified by PCR with primers LAN15BHI and RAN15ERI) inserted between EcoRI and BamHI sites | This study |
| pANTI-Z3231 | pNDM220 carrying Z3231 (amplified by PCR with primers LAEDLBHI and RAEDLERI) inserted between EcoRI and BamHI sites blunt ended with Klenow | This study |
| pOU82 | Apr, ori R1, lacZYA | 30 |
| pOU-SXT | pOU82 carrying s045-s044 | This study |
| pOU-N15 | pOU82 carrying gp49-gp48 | This study |
| pCM132 | Kmr, ori RK2, lacZ reporter gene fusion vector | 56 |
| pCM-P1AMI | pCM132 carrying P1 promoter of tad-ata | This study |
| pCM-P1SXT | pCM132 carrying P1 promoter of s045-s044 (amplified by PCR with primers LP1SXT and RP1SXT2) inserted between EcoRI and BglII sites | This study |
| pCM-P1N15 | pCM132 carrying P1 promoter of gp49-gp48 (amplified by PCR with primers LP1N15 and RP1N15) inserted between EcoRI and BglII sites | This study |
| pCM-P1EDL | pCM132 carrying P1 promoter of Z3230-Z3231 (amplified by PCR with primers LP1EDL2 and RP1EDL2) inserted between EcoRI and BglII sites | This study |
| pCM-P2AMI | pCM132 carrying P2 promoter of tad-ata (amplified by PCR with primers LP2AMI and RP2AMI) inserted between EcoRI and BglII sites | This study |
| pCM-P2SXT | pCM132 carrying P2 promoter of s045-s044 (amplified by PCR with primers LP2EDL and RP2EDL) inserted between EcoRI and BglII sites | This study |
| pCM-P2N15 | pCM132 carrying P2 promoter of gp49-gp48 (amplified by PCR with primers LP2N15 and RP2N15) inserted between EcoRI and BglII sites | This study |
| pCM-P2EDL | pCM132 carrying P2 promoter of Z3230-Z3231 (amplified by PCR with primers LP2EDL and RP2EDL) inserted between EcoRI and BglII sites | This study |
| pRS551 | Kmr, ori pMB1, lacZ reporter gene fusion vector | 71 |
| pRS-P1AMI | pRS551 carrying P1 promoter of tad-ata | This study |
| pRS-P1SXT | pRS551 carrying P1 promoter of s045-s044 (amplified by PCR with primers LP1SXT and RP1SXT2) inserted between EcoRI and BamHI sites | This study |
| pRS-P1N15 | pRS551 carrying P1 promoter of gp49-gp48 (amplified by PCR with primers LP1N15 and RP1N15) inserted between EcoRI and BamHI sites | This study |
| pRS-P1EDL | pRS551 carrying P1 promoter of Z3230-Z3231 (amplified by PCR with primers LP1EDL2 and RP1EDL2) inserted between EcoRI and BamHI sites | This study |
| pRS-P2AMI | pRS551 carrying P2 promoter of tad-ata (amplified by PCR with primers LP2AMI and RP2AMI) inserted between EcoRI and BamHI sites | This study |
| pRS-P2SXT | pRS551 carrying P2 promoter of s045-s044 (amplified by PCR with primers LP2SXT and RP2SXT) inserted between EcoRI and BamHI sites | This study |
| pRS-P2N15 | pRS551 carrying P2 promoter of gp49-gp48 (amplified by PCR with primers LP2N15 and RP2N15) inserted between EcoRI and BamHI sites | This study |
| pRS-P2EDL | pRS551 carrying P2 promoter of Z3230-Z3231 (amplified by PCR with primers LP2EDL and RP2EDL) inserted between EcoRI and BamHI sites | This study |
| pGEM-T Easy | Apr, cloning vector | Promega |
| pBluescript II KS | Apr, cloning vector | Stratagene |
| pRK2013 | Kmr, helper plasmid carrying genes for conjugal transfer of RK2 | 27 |
DNA manipulations and PCR conditions.
Plasmid DNA was isolated as described by Birnboim and Doly (15) and, when required, was purified by CsCl-ethidium bromide gradient centrifugation. Common DNA manipulation methods were performed as described by Sambrook and Russell (69). Amplification by PCR was performed with a Mastercycler (Eppendorf) using synthetic oligonucleotides (Table 2), Pfu or OptiTaq polymerase (with the buffer supplied; Eurx), and appropriate template DNAs. PCR products were analyzed by electrophoresis on 0.8% or 2% agarose gels and, when necessary, purified with a Gel Out kit (A&A Biotechnology) and cloned into the pGEM-T Easy vector (Promega).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| LLDKREP | _AAGCTT_AGCTCATCGCGGAGGACAAG |
| RREPAMI | _AAGCTT_CCTTTTGCGTGCAGAACGCT |
| RLDKPAR | _AAGCTT_CCCATGCTCAACCTATACCTG |
| RKILHIII | _AAGCTT_CATCAAGCGGCTTCGGCAAT |
| LLDZ2547 | CCAACAAGACGGCCAAGACC |
| RLDZ3578 | CATCAAGCGGCTTCGGCAAT |
| LEDL | CTGGCGAAGAAGTTCAGGTG |
| REDL | GAAGAAGTTGCTGGAGAAGATGT |
| LN15 | CTCGCCTTTCCCTTGTTACT |
| RN15 | GCCATAACTCATACAGCTCG |
| LSXT | GCGTTAAGCCACTTGCAGAA |
| RSXT | GTGAAGGATGCCTTGGGATT |
| TADL | TC _CTGCAG_AGGTTGAGCATGGGTCAAGA |
| TADR | CG _TCTAGA_CGATCCCATCAACGATGCG |
| LTEDL | TC _CTGCAG_CTAAGGATGGAGTAATGAGA |
| RTEDL | CG _TCTAGA_CACACACTATCAAAGGTTTC |
| LTN15 | TC _CTGCAG_CATCAGGAGGTTATACTATG |
| RTN15 | CG _TCTAGA_TTCATTAGCTCATCTCCTTC |
| LTSXT | TC _CTGCAG_AAGTTAAGGATACCCATGAG |
| RTSXT | CG _TCTAGA_TGGCTAAAGCACGTTGTTT |
| 2LAAMIBH | T _GGATCC_GAGGCTCTGGCAAAGGAGAT |
| 2RAAMIER | C _GAATTC_AGCGGCTTCGGCAATATCCA |
| LAEDLBHI | T _GGATCC_AGAACTTGTTCAGGAGCGGA |
| RAEDLERI | C _GAATTC_GCACTAAGCAGCATGTGGAT |
| LAN15BHI | T _GGATCC_GTCTGAAGGAGATGAGCTAA |
| RAN15ERI | C _GAATTC_TCGCCTTTCCCTTGTTACTT |
| 2LASXTBH | T _GGATCC_CTTGCCAGAGATAGACAAAAAGAGGT |
| 2RASXTER | C _GAATTC_GCGTTAAGCCACTTGCAGAAGA |
| LP2AMI | C _GAATTC_AGTGGTTGAGAACGACGTGG |
| RP2AMI | T _GGATCC_GGGCAACACCAGATCAGCAA |
| LP1EDL2 | C _GAATTC_TGCGTAAGGATGGCGAGATG |
| RP1EDL2 | T _GGATCC_TGCGCGTAAATCGTCAAGAC |
| LP2EDL | C _GAATTC_GATCTATCGCGTCATGTATG |
| RP2EDL | T _GGATCC_CATCCCACACACACTATCAAAG |
| LP1N15 | C _GAATTC_GCCATAACTCATACAGCTCG |
| RP1N15 | T _GGATCC_CGCGTAGCTCATCAGGTA |
| LP2N15 | C _GAATTC_GCGTTATCGGTGGTGGC |
| RP2N15 | T _GGATCC_TAGCTCATCTCCTTCAGACG |
| LP1SXT | C _GAATTC_ATGCCTTGGGATTGACA |
| RP1SXT2 | T _GGATCC_TAGACTCAGTATGAGGAGGC |
| LP2SXT | C _GAATTC_GCTCAAGAAGGTATTGGTAG |
| RP2SXT | T _GGATCC_AAGCTGAACTCACTCTCAAG |
| GP1SXT2 | TTCAAAGCCACATGCGTGAG |
| GP2SXT2 | TAGAAGAGTTGACCAACTTGGATTG |
| GP3SXT2 | AGTCCTGCCTTTGTTCTCATGG |
| GP3SXT | CATAGACTCAGTATGAGGAGGC |
| AAP | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG |
| AUAP | GGCCACGCGTCGACTAGTAC |
Introduction of DNA into bacterial cells.
Transformation of E. coli TG1 was performed by the method of Kushner (48), and electroporation of Paracoccus spp. cells was carried out as previously described (7). For triparental mating, the donor strain E. coli TG1 carrying a mobilizable vector, E. coli DH5α carrying the helper plasmid pRK2013, and a suitable recipient strain were mixed at a ratio of 1:2:1. One hundred microliters of this mixture was spread onto a plate containing solidified TY or LB medium, depending on the recipient strain. After overnight incubation at 30°C, the bacteria were washed off the plate, and suitable dilutions were plated on selective media containing rifampin or streptomycin (selectable markers of the recipient strains) and kanamycin to select for transconjugants. The plasmid content of transconjugants was verified by screening several colonies using a rapid alkaline extraction procedure and agarose gel electrophoresis. Spontaneous resistance of the recipient strains to the antibiotics used for selection was not detectable under these experimental conditions.
Plasmid stability assay.
The stability of plasmids during growth under nonselective conditions was tested as described previously (10). Briefly, stationary-phase cultures were diluted in fresh medium without antibiotic selection and cultivated for approximately 10, 20, and 30 generations. Samples were diluted and plated onto solid medium in the absence of selective antibiotics. One hundred colonies were tested for the presence of the Kmr markers (pABW3 derivatives) or Cmr markers (pFH450 derivatives) by replica plating. Plasmid retention was determined by using the percentage of kanamycin- or chloramphenicol-resistant colonies. In the case of pOU82 and its derivatives samples were plated onto medium containing 5-bromo-4-chloro-3-indolyl-d-galactopyranoside (X-Gal) and isopropyl β-d-thiogalactoside (IPTG). Blue and white colonies were scored as plasmid-containing and plasmidless colonies, respectively. All the stability assays were performed at least in triplicate.
Determination of transcription start sites by 5′-RACE analysis and reverse transcription-PCR.
Mapping of transcription start sites was performed using the rapid amplification of 5′ cDNA ends (5′-RACE) system (Invitrogen) as recommended by the manufacturer. Briefly, total RNA was extracted from an E. coli TG1 culture carrying pBGS-SXT using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Contaminating DNA was digested for 15 min at 37°C with 20 U of RNase-free DNase I (Roche). First-strand cDNA was synthesized from 1 μg RNA in a reaction primed with 1 pmol of primer GP1SXT2 using 200 U of SuperScript II reverse transcriptase (Invitrogen); the reaction mixture was incubated at 42°C for 50 min. The GP1SXT2 primer binds approximately 570 bp downstream of the transcription start site of the s045 gene and 270 bp downstream of the transcription start site of the s044 gene (identified in this study). After purification and dC tailing, the first strand was amplified using the 5′ RACE anchor primer (AAP) and SXT-specific internal primer GP2SXT2. The primary PCR products were reamplified using the AUAP primer (Invitrogen) together with SXT-specific internal nested primer GP3SXT2 or GP3SXT. PCR products were then purified, cloned in the pGEM-T Easy vector (Promega), and sequenced.
For reverse transcription-PCR, DNA-free total RNA from E. coli TG1 carrying pBGS-SXT was prepared and reverse transcribed as described above and subsequently PCR amplified using primers LTSXT (which anneals 15 bp upstream of the start codon of s044) and RP2SXT (which anneals 82 bp downstream of the s045 coding region) or primers LTSXT and 2RASXTER (which anneals 55 bp downstream of the s044 coding region).
Analysis of the influence of toxins on E. coli cell growth.
E. coli MC1000 strains carrying the pCF430 vector or its derivatives belonging to the pTOX series (containing toxin genes under P_BAD_ promoter control) were grown at 37°C in LB medium in the presence of tetracycline and glucose. The overnight cultures were diluted in fresh medium to obtain an initial optical density at 600 nm (OD600) of 0.06. After 1 h of growth (OD600, ∼0.1) each of the cultures was divided into two equal portions; one portion was supplemented with glucose and the other portion was supplemented with arabinose, both at a final concentration of 0.2%. The growth of the cultures was then monitored for a further 3 h. Viable cell counts were determined by plating sample dilutions (taken at hourly intervals) on LB agar plates containing glucose (0.2%) and tetracycline.
Toxicity rescue experiments.
E. coli MC1000 strains carrying the pCF430 vector or its derivatives belonging to the pTOX series (containing toxin genes under P_BAD_ promoter control) were transformed with the pNDM200 vector or derivatives of this vector belonging to the pANTI series (carrying antidotes of the TA cassettes analyzed cloned under IPTG-inducible promoter control). The resulting two-plasmid strains were grown overnight and diluted in fresh LB medium as described above. After 1 h of growth (OD600, ∼0.1) the cultures were supplemented with arabinose at a final concentration of 0.2%. After an additional 1 h of incubation the cultures were divided into two equal portions, one of which was supplemented with IPTG at a final concentration of 2 mM. The growth of the cultures was then monitored for an additional 3 h. To determine the viable cell counts, culture samples were taken at hourly intervals, diluted, and plated on LB agar plates containing glucose (0.2%), tetracycline, and ampicillin.
Assays for β-galactosidase activity.
The β-galactosidase activities in E. coli DH5Δlac and Paracoccus versutus UW225 were measured by examining the conversion of _o_-nitrophenyl-β-d-galactopyranoside into nitrophenol as described by Miller (58). The assays for β-galactosidase activity were repeated five times.
In vitro transcription and translation.
In vitro coupled transcription-translation was carried out using the E. coli S30 extract system for circular DNA as described by the supplier (Promega). Proteins synthesized in the reactions were labeled by incorporation of [35S]methionine, separated by 16.5% Tricine—sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described by Schagger and von Jagow (70), and visualized by autoradiography.
Electron microscopy.
Samples of cultures of E. coli MC1000 carrying the pCF430 vector (control) or derivatives of this vector containing genes for toxins (pTOX-tad, pTOX-gp49, pTOX-s045, or pTOX-Z3230) were taken after 3 h of incubation in the presence of glucose (for repression of toxin expression) or arabinose (for induction of toxin expression). Cells from 500 μl of each culture were collected by centrifugation and fixed in 1 ml of 4% paraformaldehyde for 30 min at room temperature. The samples were then centrifuged again, washed three times in phosphate-buffered saline (pH 8.0), and dehydrated in a graduated ethanol series (25%, 50%, 75%, and 96% ethanol; 15 min in each). The processed cell suspensions were mounted on microslides and dried. Microslides carrying the samples were placed onto stubs and coated with colloidal gold particles (thickness, 7 to 8 nm; Polaron SC7620). Samples were then viewed using a LEO 1430VP scanning electron microscope equipped to capture digital images and measure cells.
DNA sequencing and bioinformatics.
The nucleotide sequence of the basic replicon of pAMI2 was determined in the DNA Sequencing and Oligonucleotide Synthesis Laboratory at the Institute of Biochemistry and Biophysics, Polish Academy of Sciences, using a dye terminator sequencing kit and an automated sequencer (ABI 377; Perkin-Elmer). A combination of vector-specific primers and primer walking was used to obtain the entire nucleotide sequence of the pAMI2 basic replicon.
The nucleotide sequence of pAMI2 was analyzed using the ORF FINDER program available at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Similarity searches were performed using the BLAST programs (3) provided by the NCBI (http://www.ncbi.nlm.nih.gov/BLAST). Sequence alignment was performed using the programs CLUSTALW (75) at the BCM Search Launcher (http://searchlauncher.bcm.tmc.edu/multi-align/Options/clustalw.html) and MULTALIN (26) at the INRA website (http://prodes.toulouse.inra.fr/multalin/multalin.html). Protein domain architecture was analyzed using the SMART database (51) (http://smart.embl-heidelberg.de/). Helix-turn-helix prediction was performed using the HELIX-TURN-HELIX MOTIF PREDICTION program (28) at the PBIL-IBCP Lyon-Gerland website (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_hth.html).
Nucleotide sequence accession number.
The nucleotide sequence of the basic replicon of pAMI2 has been annotated and deposited in the GenBank database under accession number DQ855273.
RESULTS
Basic replicon of pAMI2 is functional in different Alphaproteobacteria hosts but not in E. coli.
The initial aim of this study was to characterize the maintenance region of plasmid pAMI2 (18.6 kb) of P. aminophilus JCM 7686. For this purpose a pAMI204 minireplicon was constructed, containing a 7-kb PstI fragment of pAMI2 and a kanamycin resistance cassette. All attempts to introduce pAMI204 (isolated from the paracoccal host) into E. coli failed, suggesting that the region analyzed does not function in this host as a replicon. Further trimming of the cloned region revealed that the basic replicon of pAMI2 was encoded in a 4.2-kb EcoRI-SalI restriction fragment (Fig. 1A). To facilitate further analysis, we constructed an E. coli-Paracoccus spp. shuttle plasmid, pMDB300, composed of the EcoRI-SalI fragment and a mobilizable _E. coli_-specific (unable to replicate in Paracoccus spp.) vector, pABW1. pMDB300 was readily transferred by conjugation into nine species of Paracoccus (Table 1), as well as into R. leguminosarum 1062, R. sphaeroides 2.4.1R, and A. tumefaciens GMI 9023. Since the pABW1 vector cannot replicate in the strains tested, it appeared that the replication system of pAMI2 is functional in different representatives of the Alphaproteobacteria.
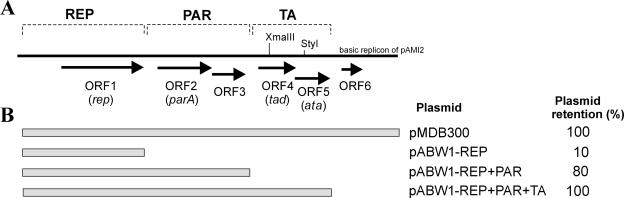
FIG. 1.
(A) Genetic organization of the maintenance region of plasmid pAMI2 (present in shuttle plasmid pMDB300). The predicted plasmid modules responsible for replication (REP), active partitioning (PAR), and postsegregational killing (TA) are indicated. The arrows indicating predicted coding regions also indicate the direction of transcription. Restriction sites used for mutational analysis are indicated. (B) Segregational stability of shuttle plasmids containing the maintenance modules of pAMI2. The shaded boxes represent DNA segments of pAMI2 cloned in vector pABW1 (unable to replicate in Paracoccus spp.). The values on the right indicate levels of retention of the plasmids in P. versutus UW225 after approximately 30 generations of growth under nonselective conditions (see Materials and Methods for details).
Basic replicon of pAMI2 encodes two stabilizing systems.
The complete nucleotide sequence of the basic replicon of pAMI2 consisted of 4.2 kb and had a G+C content of 60.5%. It was predicted to include six putative open reading frames (ORFs) transcribed in the same direction (Fig. 1A). Based on the in silico analysis, the following two putative modules were distinguished: (i) REP, responsible for plasmid replication, which contains ORF1 coding for a protein with the highest level of similarity to a probable replication protein of A. tumefaciens plasmid pAgK84 (accession no. YP_086770); and (ii) PAR, whose structure is similar to the structure of the partitioning system of plasmid pTAR of A. tumefaciens (46), which contains ORF2 and ORF3, whose predicted products exhibit the highest levels of similarity to the ParA-like protein of Erythrobacter sp. (accession no. ZP_01039613) and a hypothetical protein of Sinorhizobim meliloti (accession no. CAC48461), respectively (Fig. 1A). The putative products of ORF4, ORF5, and ORF6 exhibit similarity to a phage-related protein of Polaromonas naphthalenivorans (accession no. ZP_01019465), a transcriptional regulator of the XRE family of Nitrosospira multiformis (accession no. YP_413493), and a conserved hypothetical protein of S. meliloti (accession no. CAC47048), respectively.
To confirm that the predicted REP and PAR regions are functional, the DNA fragments carrying the REP module or the REP and PAR modules were amplified by PCR and cloned into the vector pABW1, which yielded pABW1-REP and pABW1-REP+PAR, respectively. Both plasmids were transferred by triparental mating into plasmidless P. versutus UW225 (routinely used by us as a host strain for paracoccal plasmids) and were able to replicate, which proved that the REP module contained genetic information sufficient for plasmid replication (therefore, it may be considered a minimal replicon). Figure 1B shows that the presence of the PAR module, adjacent to REP, significantly increased plasmid stability (plasmid pABW1-REP+PAR). However, a high level of stability of the entire basic replicon was restored only when the PAR module was accompanied by ORF4 and ORF5 (plasmid pABW1-REP+PAR+TA) (Fig. 1B).
To confirm the role of ORF4 and ORF5 in plasmid stabilization, the DNA region containing both ORFs was amplified by PCR and cloned into the low-copy-number shuttle vector pABW3, which is unstable in paracocci (Kmr; carries the heterologous replication system of plasmid pTAV1 of P. versutus UW1 [8]). The resulting plasmid (pABW3-AMI) was stably maintained, since no plasmidless cells were detected after approximately 30 generations of growth under nonselective conditions (data not shown). In contrast, the pABW3 vector, tested under analogous conditions, was present in approximately 4% of the cells after the same growth period (8).
In a further experiment, the growth of P. versutus UW225 carrying pABW3 (control) or pABW3-AMI was monitored in two parallel liquid cultures (without a selective antibiotic). The OD600 of the cultures were determined after 9 h of incubation. At that time the cultures were diluted appropriately and plated on LB agar and LB agar supplemented with kanamycin. We observed that the growth of the strain carrying pABW3-AMI (OD600, 0.42) was slower than the growth of UW225 carrying the empty vector pABW3 (OD600, 0.67), which was in agreement with the numbers of CFU obtained on antibiotic-free LB agar (4.4 × 108 CFU/ml for the former strain and 1.6 × 109 CFU/ml for the latter strain). Interestingly, we observed similar numbers of Kmr colonies for the two strains tested, and there were no Kms cells of UW225(pABW3-AMI) that were able to form colonies on solid medium (data not shown). All the observations described above might reflect the elimination (or growth arrest) of segregants lacking pABW3-AMI (a characteristic feature of toxin-antitoxin systems). The small size of the ORF4 and ORF5 genes and their genetic organization were also typical of TA modules.
In summary, in addition to PAR we identified a second locus determining the stable maintenance of the basic replicon of pAMI2, which exhibits no homology at the nucleotide or amino acid sequence (products of ORF4 and ORF5) level to any previously described stabilizing system.
Mutational analysis suggests that ORF4 and ORF5 constitute a toxin-antitoxin cassette.
The overlapping ORFs ORF4 and ORF5 (19-bp overlap) encoded predicted proteins with molecular masses of 13.8 kDa (pI 7.0) and 12.2 kDa (pI 9.2), respectively. To investigate the role of these proteins in plasmid stabilization, a mutational analysis was performed. Two out-of-frame insertions (4 bp) were attempted at the XmaIII (ORF4) or StyI (ORF5) sites (Fig. 1A) of the locus (present in plasmid pBGS-AMI). The mutated modules were then cloned into the unstable vector pABW3, and the resulting plasmids were transferred into P. versutus UW225, where their stability was tested. The results of this analysis were as follows: (i) a mutant with a mutation in ORF4 was obtained (pMUT1), and the mutation eliminated the ability of the system to stabilize pABW3 in the UW225 strain (data not shown); (ii) in spite of repeated attempts, no mutant with a single mutation in ORF5 was isolated (a pool of clones carrying deletion plasmids was consistently obtained); and (iii) mutation of ORF5 was possible, but only after prior mutation of ORF4, and the double mutant module (pMUT2) was also unable to stabilize pABW3 (data not shown). These results are compatible with the hypothesis that this locus functions as an addiction system. Thus, it seems that ORF4 of the basic replicon of pAMI2 codes for a potential toxin and ORF5 codes for an antitoxin (mutation of ORF5 results in manifestation of the toxic activity of the ORF4 product). The presence of two protein components was confirmed by using the coupled in vitro _E. coli-_derived transcription-translation system (data not shown). Taking into account the findings described above, ORF4 and ORF5 were designated tad (_t_oxin of _ad_diction system) and ata (_a_nti_t_oxin of _a_ddiction system), respectively.
tad-ata homologs are widely distributed among bacterial genomes.
Sequence database comparisons were carried out to identify loci homologous to pAMI2 tad-ata. Pairs of genes of the tad-ata type are conserved and widely distributed in bacterial genomes. In all cases a gene homologous to tad (encoding the toxin) precedes a gene encoding the antitoxin, and the genes overlap (in most cases the overlap is only 4 bp).
We identified more than 140 homologous loci (listed in Table S1 in the supplemental material) in various members of the Proteobacteria (alpha, beta, and gamma subgroups), Bacteroidetes, Cyanobacteria, and Chlorobi, as well as the Acidobacteria and Firmicutes. Most of the modules are present in bacterial chromosomes, but modules were also identified in (i) plasmids (pXF51 of Xylella fastidiosa [accession no. AE003851], plasmids 1 and 3 of Nitrosospira multiformis [accession no. NC_007615 and NC_007617, respectively], pDC3000B of Pseudomonas syringae [18], pRV500 of Lactobacillus sakei [2], and pSRQ800 Lactococcus lactis [16]); (ii) linear bacteriophages (N15 of E. coli [accession no. NC_001901] and φKO2 of Klebsiella oxytoca [22]); (iii) conjugative element SXT of Vibrio cholerae (12); and (iv) noncomposite transposon Tn_5503_ (Tn_3_ family) occurring in plasmid Rms149 of Pseudomonas aeruginosa (39) and a related unnamed transposon of plasmid pND6-1 from Pseudomonas sp. strain ND6 (52) (data not shown).
Sequence similarity searches showed that Tad (toxin) and Ata (antitoxin) and relatives of the molecules identified here belong to large protein families classified in the COG4679 (Tad relatives) and COG5606 and COG1396 (Ata relatives) clusters of orthologous groups (74). Analyses performed using the SMART database (50) revealed that the COG5606 and COG1396 clusters are members of a large group of transcription factors belonging to the helix-turn-helix XRE family, typified by the repressor protein of the defective prophage PBSX of Bacillus subtilis (57) (data not shown). The presence of DNA binding domains is typical for antitoxin proteins, which often play a major role in regulating expression of the TA operons (for a review, see reference 33).
Besides tad-ata, three of the homologous modules, encoded by various genetic elements, were chosen for further analysis. These modules originated from (i) bacteriophage N15 (gp49-gp48; accession no. NP_046945 and NP_046944), (ii) the SXT element (s045-s044; accession no. AAL59719 and AAL59720), and (iii) the genomic island present in the chromosome of enterohemorrhagic E. coli strain O157:H7 strain EDL933 (Z3230-Z3231; accession no. NP_288570 and NP_288571).
pAMI2, N15, and SXT encode stabilizing systems with different host ranges.
The four _tad-ata-_like loci were initially tested to determine their stabilizing activities in two evolutionarily distinct hosts, E. coli (Gammaproteobacteria) and P. versutus (Alphaproteobacteria). For this purpose the conserved gene pairs were amplified by PCR and cloned into the plasmid stability test vectors pFH450 (used for analysis in E. coli) and pABW3 (functional in P. versutus). The resulting pFH450 derivatives (designated pFH-AMI, pFH-SXT, pFH-N15, and pFH-EDL) were transferred into E. coli BR825 (which has a mutation in the DNA polymerase I [_polA_] gene), where their stability was tested. pFH450 is a hybrid plasmid containing two replication systems, the ColE1 replicon and prophage P1, that does not have auxiliary stabilization mechanisms (41). When plasmid pFH450 is harbored by E. coli BR825 (polA), it is a low-copy-number replicon due to the activity of the P1 replication system (the ColE1 replicon is nonfunctional in this host) and is therefore very unstable, resulting in a high frequency of plasmidless cells when cells divide (Fig. 2A). Only two of the modules tested, derived from the SXT element and prophage N15, were able to promote segregational stability of pFH450 under nonselective conditions (Fig. 2A); therefore, they encode functional stability cassettes, which can stabilize heterologous replicons in E. coli.
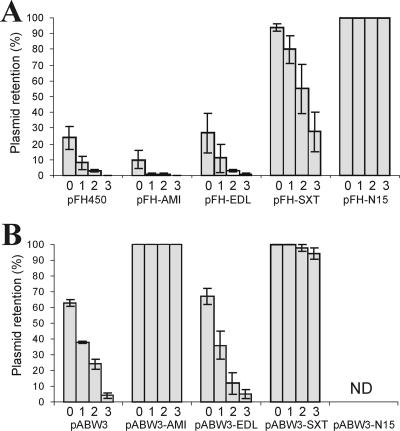
FIG. 2.
Segregational stability of stability probe vectors and their derivatives containing TA cassettes of pAMI2 (tad-ata), E. coli EDL933 (Z3230-Z3231), SXT (s045-s044), and N15 (gp49-gp48) determined in E. coli BR825 (A) and P. versutus UW225 (B). Plasmid retention is expressed as the percentage of Kmr colonies. The bars for each plasmid tested indicate the results for an overnight culture grown in the presence of kanamycin (bar 0) and for cultures grown under nonselective conditions after approximately 10, 20, and 30 generations (bars 1, 2, and 3, respectively). (A) Stability of the probe vector pFH450 and its derivatives with inserted tad-ata (pFH-AMI), Z3230-Z3231 (pFH-EDL), s045-s044 (pFH-SXT), and gp49-gp48 (pFH-N15) genes. (B) Stability of the probe vector pABW3 and its derivatives with inserted tad-ata (pABW3-AMI), Z3230-Z3231 (pABW3-EDL), gp49-gp48 (pABW3-N15), and s045-s044 (pABW3-SXT) genes. ND, not determined (selection of mutated clones; see text for details).
In the analysis performed with P. versutus UW225 with the three pABW3 derivatives (pABW3-SXT, pABW3-N15, and pABW3-EDL), stability was observed only with the module derived from the SXT element (Fig. 2B). The module with a chromosomal origin, encoded by E. coli O157:H7 strain EDL933, was unable to stabilize plasmids in either of the hosts (Fig. 2).
Introduction of pABW3-N15 into P. versutus UW225 resulted in selection of only individual transconjugants, compared with the 106 transconjugants obtained with the pABW3 vector. The stability of pABW3-N15 in three UW225 transconjugants (tested after approximately 30 generations of growth under nonselective conditions) did not differ from the stability determined for the empty pABW3 vector (data not shown). The highly reduced number of transconjugants might have resulted from toxicity of the TA cassette for plasmid-containing P. versutus cells. Thus, it is probable that these transconjugants contained mutations affecting the expression or activity of the toxin or the cellular target recognized by the toxin. This phenomenon was not investigated further in this study.
All the toxins analyzed recognize a cellular target in E. coli.
The toxic activities of Tad and its three relatives (from SXT, N15, and E. coli EDL933) were tested with E. coli. For this purpose the genes encoding the predicted toxins (tad, s045, Z3230, and gp49) were amplified by PCR and cloned in the pCF430 vector under control of the P_BAD_ promoter, derived from the arabinose operon of E. coli. The vector also carried the araC gene. The araC_-P_BAD regulatory system allows highly regulated expression, with tight repression in the absence of arabinose and strong induction in its presence (38). The repression is increased by growth in the presence of glucose.
The pCF430 vector and its four derivatives (pTOX-tad, pTOX-s045, pTOX-Z3230, and pTOX-gp49) were introduced into E. coli MC1000 (Δ_ara_ Δ_lac_). Each of the strains was grown in a medium supplemented with either glucose (for repression of P_BAD_) or arabinose (for induction of toxin synthesis) as described in the legend to Fig. 3.
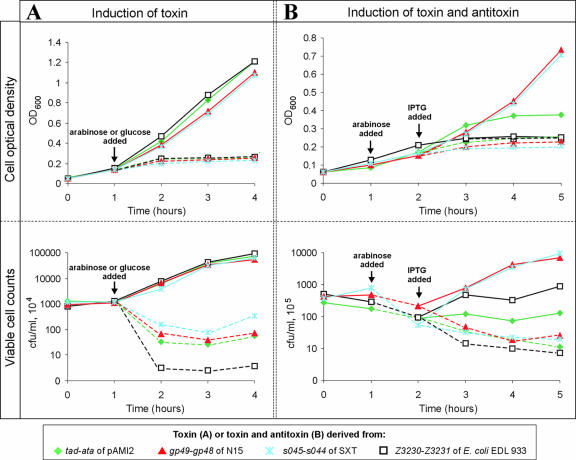
FIG. 3.
Growth rates and viable cell counts for E. coli MC1000 overexpressing toxins (A) or toxins together with their cognate antitoxins (B) derived from the TA cassettes of pAMI2 (tad-ata), E. coli EDL933 (Z3230-Z3231), SXT (s045-s044), and N15 (gp49-gp48). (A) The E. coli MC1000 strains harboring pCF430 derivatives containing genes for toxins cloned under control of an arabinose-inducible promoter (pTOX-tad, pTOX-Z3230, pTOX-s045, and pTOX-gp49) were cultured as described in the Results. After 1 h of growth each of the cultures was divided into two equal portions, one of which was supplemented with glucose (which repressed expression of toxins) (solid lines) and one of which was supplemented with arabinose (which induced expression of toxins) (dashed lines). The culture OD600 was determined at hourly intervals (top panel). The ability to form colonies was also determined at hourly intervals by plating appropriate dilutions of the cultures on LB agar supplemented with glucose (bottom panel). Colonies were counted after 24 h of incubation at 37°C. (B) E. coli MC1000 strains harboring pTOX plasmids (see above) together with pNDM200 derivatives containing genes for antitoxins under control of an IPTG-inducible promoter [MC1000(pTOX-tad/pANTI-ata), MC1000(pTOX-Z3230/pANTI-Z3231), MC1000(pTOX-s045/pANTI-s044), and MC1000(pTOX-gp49/pANTI-gp48)] were grown as described in the Results (solid lines). After 1 h of growth the cultures were supplemented with arabinose (which induced expression of toxins) (dashed lines). After 2 h the cultures were divided into two equal portions, one of which was supplemented with IPTG (which induced expression of antitoxins) (solid lines). The culture OD600 was determined at hourly intervals (top panel). The ability to form colonies was also determined at hourly intervals by plating appropriate dilutions of the cultures on LB agar supplemented with glucose (bottom panel). Colonies were counted after 24 h of incubation at 37°C.
With the control strain (carrying pCF430), we observed no significant differences in the growth rates of cultures grown in the presence of glucose or arabinose (data not shown). As shown in Fig. 3A, overproduction of a toxin (induced by arabinose) resulted in each case in efficient inhibition of cell growth, which was accompanied by a reduction in the number of CFU (Fig. 3A). Interestingly, the lowest final number of CFU was observed with the chromosomally encoded Z3230 toxin of E. coli EDL933 (approximately 10,000-fold reduction in cell viability, compared with the approximately 100-fold reduction caused by the s045-encoded toxin of SXT), which was derived from the module that was unable to stabilize plasmids in the hosts tested.
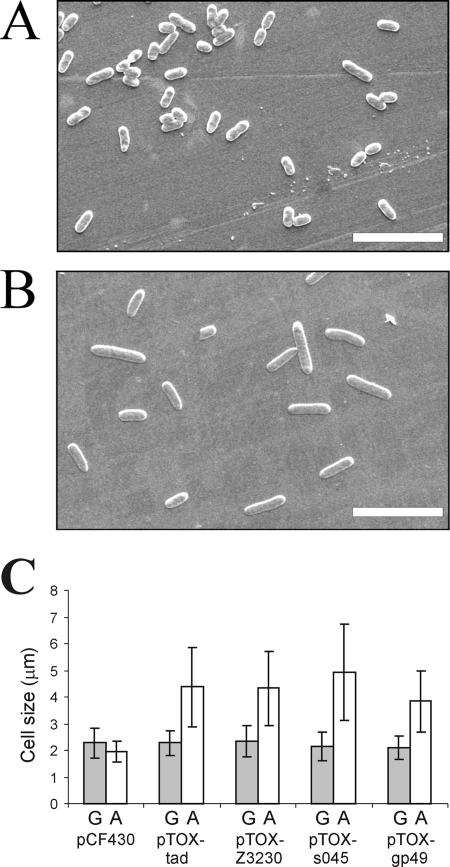
Microscopic examination of the cultures 3 h after arabinose induction revealed elongation of cells overproducing the toxins (Fig. 4B) (also visible after 8 h of growth [data not shown]). Such an effect was not observed in the repressed cultures or in cells carrying the empty vector pCF430 (Fig. 4A and C). The elongated cells were approximately twice as long as the cells in the control culture (Fig. 4C), which indicates that the toxins may specifically inhibit cell division.
FIG. 4.
Morphology of E. coli MC1000 cells overproducing TA cassette toxins. (A and B) Scanning electron microscopy examination of E. coli MC1000 cells containing vector pCF430 (control) (A) or pTOX-tad (carrying the tad gene under control of the P_BAD_ promoter) (B). Samples were taken 3 h after addition of arabinose (for induction of toxin expression). Bars = 10 μm. (C) Comparison of the sizes of E. coli MC1000 cells containing the pCF430 vector or its derivatives carrying genes for toxins, including tad (pTOX-tad), Z3230 (pTOX-Z3230), s045 (pTOX-s045), and gp49 (pTOX-gp49). The cells were grown in LB medium containing glucose (for repression of toxin expression) (bars G) or arabinose (for induction of toxin expression) (bars A). One hundred cells were measured in each case.
Evidence for interaction of antitoxins with their cognate toxins.
We next tested whether the predicted antitoxins encoded by the four loci analyzed were able to block the actions of their cognate toxins. The antitoxin genes (ata, s044, Z3231, and gp48) were amplified by PCR and cloned in vector pNDM220 downstream of the IPTG-inducible promoter pA1/O4/03 (34). The plasmids obtained, pANTI-ata, pANTI-s044, pANTI-Z3231, and pANTI-gp48, were introduced into E. coli MC1000 carrying compatible toxin-expressing plasmids pTOX-tad, pTOX-s045, pTOX-Z3230, and pTOX-gp49, respectively. The resulting two-plasmid strains were examined in growth experiments performed with medium containing arabinose (for induction of toxins) and IPTG (for induction of antitoxins) (Fig. 3B).
Induction of the putative antitoxin genes gp48 (from N15) and s044 (from SXT) resulted in a rapid reversal of the effects of the cognate toxins, which was shown by continuous increases in the OD600 of the cultures (Fig. 3B). In both cases, the initial number of CFU was restored by approximately 1 h after IPTG induction (Fig. 3B).
Induction of the Z3231-encoded antitoxin (derived from the chromosome of E. coli EDL933) apparently did not reverse the toxic effect resulting from overexpression of the Z3230 protein, judging by the nearly identical OD600 of cultures grown in the presence and in the absence of IPTG (Fig. 3B). This effect was observed even after 7 h of incubation of the cultures following IPTG induction (data not shown). Despite this negligible effect on culture growth, induction of the Z3231 antitoxin resulted in a significant increase in cell viability (Fig. 3B).
Similar results were obtained for the tad-ata system. In this case, induction of the Ata protein slightly stimulated growth of the culture, but only over a short period of time (approximately 1 h), as judged from the higher OD600 (Fig. 3B), which corresponded to the increased number of CFU (Fig. 3B). After this short-term stimulation, growth was apparently arrested, in a manner similar to the manner observed for the Z3230-Z3231 system (Fig. 3B). Again, the effect was visible even after 7 h of incubation in the presence of IPTG (data not shown).
The results described above suggested that although the antitoxins encoded by the tad-ata and Z3230-Z3231 modules gradually restored the viability of the cells (which suggests that their proteic components interact in vivo), they were not able to fully counteract the growth arrest produced by the toxins. This observation may explain the inability of both of these systems to stabilize plasmids in E. coli.
Similar experiments were performed to determine the abilities of the antitoxin proteins encoded by all the TA systems analyzed to interact with noncognate toxin proteins. Inhibition of the toxic effect was not observed following induction of any of the antitoxins, so it seems that these proteins are not able to cross-react in vivo with heterologous toxins (data not shown).
Antitoxins encoded by N15 and SXT are degraded by different cellular proteases.
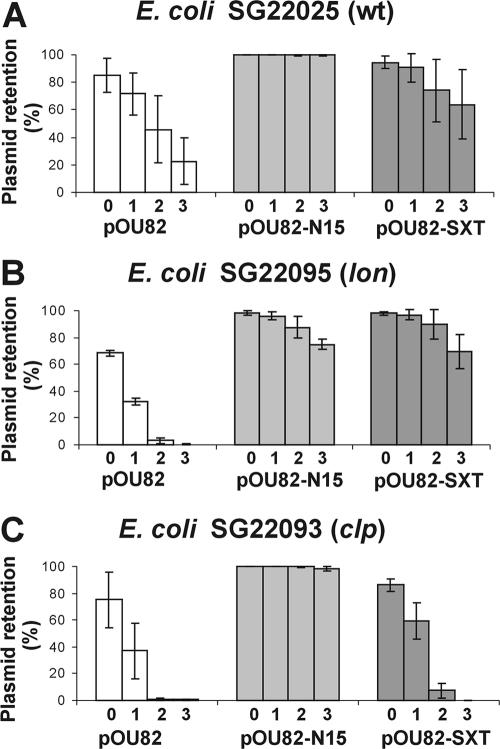
As mentioned above, it has been shown that antitoxins are less stable than toxins and are degraded by the cellular protease Lon or Clp (50, 80). To identify the proteases responsible for degradation of the Ata-like antitoxins, we studied the abilities of the TA loci derived from N15 and SXT to stabilize plasmids in three E. coli strains, SG22025 and two mutated derivatives of this strain, SG22093 (clp mutant defective in ClpAP and ClpXP proteases) and SG22095 (lon mutant defective in the Lon protease) (Fig. 5). To achieve this, EcoRI-BamHI fragments containing the TA modules of N15 and SXT were recovered from plasmids pBGS-N15 and pBGS-SXT, respectively, cloned into the segregational stability test vector pOU82 (to obtain pOU-N15 and pOU-SXT, respectively), and introduced into the strains indicated above. As shown in Fig. 5B and C, plasmid stabilization by gp49-gp48 was decreased in a lon mutant, while stabilization by the s045-s044 module was abolished in a clp strain. Since the TA plasmid stabilization systems function through the differential stabilities of toxins and antitoxins, these preliminary results strongly suggest that the antitoxins analyzed might be degraded by different cellular proteases.
FIG. 5.
Segregational stability of stability probe vector pOU82 and its derivatives containing TA cassettes of N15 (gp49-gp48) and SXT (s045-s044) determined in E. coli SG22025 (wild type) (A), E. coli SG22095 (lon) (B), and SG22093 (clp) (C). The four bars for each plasmid tested indicate the results for overnight cultures grown in the presence of ampicillin (bar 0) and for cultures grown under nonselective conditions for approximately 10, 20, and 30 generations (bars 1, 2, and 3, respectively).
Cassettes of the tad-ata type have two promoters.
To identify promoters in the four systems analyzed and to compare their strengths, DNA sequences upstream of the genes for toxins and antitoxins were amplified by PCR or isolated on separate restriction fragments and inserted upstream of the promoterless reporter lacZ gene in a promoter probe vector to generate transcriptional fusions. To test the functionality of the putative promoters in E. coli DH5Δlac, as well as in P. versutus UW225, two test vectors were used: pRS551 (specific for E. coli) and pCM132 (functional in Paracoccus spp.). The 16 resulting plasmid constructs (Fig. 6) were introduced into the appropriate hosts, and β-galactosidase activity assays were used to examine promoter strength.
FIG. 6.
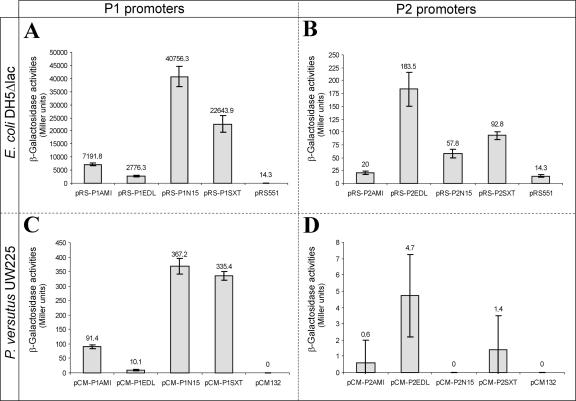
Comparison of the activities of the P1 and P2 promoters of TA cassettes from pAMI2, SXT, N15, and E. coli EDL933 determined in E. coli DH5Δlac (A and B) and P. versutus UW225 (C and D). (A and B) β-Galactosidase activities produced by promoter probe vector pRS551 (control) and its derivatives carrying lacZ transcriptional fusions with P1 (A) and P2 (B) promoters originating from the TA cassettes tad-ata (pRS-P1AMI and pRS-P2AMI), Z3230-Z3231 (pRS-P1EDL and pRS-P2EDL), gp49-gp48 (pRS-P1N15 and pRS-P2N15), and s045-s044 (pRS-P1SXT and pRS-P2SXT). (C and D) β-Galactosidase activities produced by promoter probe vector pCM132 (control) and its derivatives carrying lacZ transcriptional fusions with P1 (C) or P2 (D) promoters originating from tad-ata (pCM-P1AMI and pCM-P2AMI), Z3230-Z3231 (pCM-P1EDL and pCM-P2EDL), gp49-gp48 (pCM-P1N15 and pCM-P2N15), and s045-s044 (pCM-P1SXT and pCM-P2SXT).
The results suggested that all four loci encode two promoters, termed P1 and P2, whose genes are located upstream of the toxin and antitoxin genes, respectively. As shown in Fig. 6A, all of the P1 promoters were functional in E. coli, although they exhibited different levels of activity; the most active P1 promoters were those of N15 and SXT, while the least active promoter was that of EDL933. Analogous results were obtained with P. versutus, but all promoters displayed much lower β-galactosidase activities in this host (Fig. 6C).
The activities of the P2 promoters were very low compared with the activities of their cognate P1 promoters (e.g., the level of expression of β-galactosidase driven by P1 of N15 in E. coli was approximately 925 times higher than the level of expression driven by P2) (Fig. 6A and B). The weakest promoter was P2 derived from the paracoccal TA module, while the strongest promoter was the EDL933 module P2 promoter (Fig. 6B and D). It also appeared that the P2 promoter of N15 was nonfunctional in P. versutus (see plasmid pCM-P2N15 in Fig. 6D). As shown in Fig. 6D, the β-galactosidase activities specified by all P2 promoters in P. versutus were extremely low. However, the results which we obtained are in agreement with preliminary observations which indicated that transconjugants of P. versutus carrying pCM-P2N15 (or the empty vector pCM132) formed white colonies on LB agar plates containing X-Gal, indicating that there was a lack of β-galactosidase expression. In contrast, blue colonies were formed by strains carrying pCM132 derivatives with cloned P2 promoters of pAMI2, SXT, and EDL933 (data not shown).
In summary, the activities of the promoters analyzed were variable and depended on their sources and the nature of the bacterial host, which might reflect the host ranges of the TA modules analyzed.
TA cassette of V. cholerae SXT conjugative element generates two transcripts.
5′-RACE was performed to map the transcription start sites in one of the systems analyzed, the broad-host-range TA module of SXT. Two PCR products were obtained (data not shown), and DNA sequencing allowed precise identification of the terminal sequence, which confirmed the generation of two transcripts and hence the presence of two promoters.
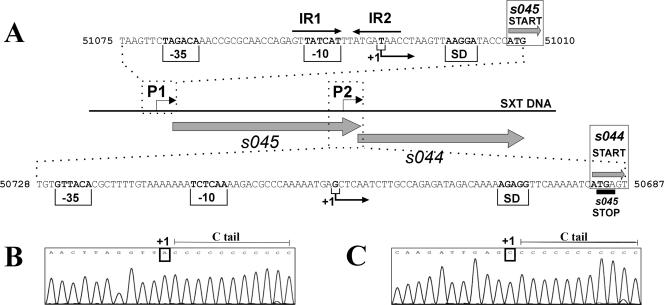
The first transcription start site was mapped to a thymidine residue located 21 nucleotides upstream of the start codon for the s045-encoded toxin (Fig. 7A and B). Based on its location, a putative promoter was identified (Fig. 7A), containing −35 and −10 regions (TAGACA and TATCAT, respectively) separated by an ideal spacing, 17 bp (residues which do not match the canonical promoter sequences [TTGACA and TATAAT] for the σ70 factor of E. coli [40] are underlined). As shown in Fig. 7A, perfect 8-bp inverted repeats overlap the −10 hexamer. Analogously located palindromes are also found in other TA loci. It has been suggested that these regions may be putative binding sites for transcriptional regulators (i.e., for antitoxins or antitoxin-toxin complexes) (37, 61).
FIG. 7.
(A) Genetic organization of the TA cassette (s045-s044) from the SXT element. The large shaded arrows represent the genes for the toxin (s045) and the antidote (s044) and indicate their transcriptional orientation. The nucleotide sequences of the P1 and P2 promoter regions are shown (reverse complement of the sequence deposited in the GenBank database; accession no. AY055428). The numbers at the ends of the sequences are the nucleotide positions according to the accession no. AY055428 sequence. The putative −35 and −10 motifs located upstream of the transcription start points (+1), determined by 5′-RACE, are indicated and are in bold type. The predicted ribosome-binding sites are also indicated (SD) and are in bold type. The start codons (ATG) for s045 and s044 (START) are also in bold type. The terminal codon for s044 (STOP) is underlined. Inverted repeats (IR1 and IR2) are indicated by arrows. (B and C) Electropherograms obtained from a 5′-RACE PCR experiment. The PCR fragments obtained were cloned into the pGEM-T Easy vector. The sequences in the electropherograms were obtained with a vector-specific primer and dC-tailed cDNAs. The last base upstream of the C tail corresponds to the first nucleotide transcribed. The corresponding T (upstream of the P1 promoter) (B) and G (upstream of the P2 promoter) (C) on the reverse-complement strands are indicated (+1).
The location of the second start point (a guanine residue 42 bp upstream of the start codon for the s044-encoded antitoxin) suggested putative promoter sequences, GTTACA (−35 region) and TCTCAA (−10 region) with 16-bp spacing (Fig. 7A and C). The predicted promoter is more divergent from the σ70 consensus, which may explain why it drives relatively low-level expression (as judged by the β-galactosidase activity assay [Fig. 6B]). Despite the presence of the internal promoter, the two genes of the s045-s044 module are cotranscribed, which was shown by reverse transcription-PCR analysis (data not shown).
DISCUSSION
In this study we identified a novel group of modules, widely distributed among genetic elements, that encode two proteic components, a toxin and an antitoxin. The prototype of this group is the tad-ata locus of plasmid pAMI2 of P. aminophilus JCM 7686. The basic replicon of pAMI2, besides the tad-ata locus, appears to encode two additional maintenance modules involved in plasmid replication and partitioning. The plasmid could be maintained in several species tested representing the genera Paracoccus, Agrobacterium, Rhizobium, and Rhodobacter (all classified as Alphaproteobacteria) but not in E. coli.
The tad-ata homologs are present in various genetic elements, although most are located in bacterial chromosomes. In the genomes of some bacteria, several copies of the modules are present; Photorhabdus luminescens subsp. laumondii TTO1 has eight homologous loci, while X. fastidiosa Ann-1 has six (data not shown). Careful inspection of the sequences adjacent to the modules revealed that one of the modules of P. luminescens (plu1223-plu1224; accession no. NC_005126) is flanked by two identical, divergently orientated insertion sequences (IS_Plu13_C and IS_Plu13_B) (data not shown), which suggests that it may be mobile as a part of a putative composite transposon.
In previous studies of TA cassettes the workers have focused mainly on analysis of a particular system identified in a given element (48, 76, 83). In this study we performed complex comparative analyses of four related loci derived from distinct genetic elements hosted by representatives of the Gammaproteobacteria (TA modules of E. coli bacteriophage N15, SXT element of V. cholerae, and genomic island of E. coli EDL933) and Alphaproteobacteria (TA module of P. aminophilus plasmid pAMI2). It appeared that all the loci derived from the mobile elements (N15, SXT, and pAMI2) had stabilization functions, although their host ranges varied. The TA modules of N15 and pAMI2 exhibited a narrow host range (they can stabilize heterologous replicons in their native hosts or closely related hosts [E. coli and P. versutus, respectively]), while the SXT TA module was functional in both species tested. It appears that the TA cassettes of N15 and pAMI2 are adapted to their specific hosts and that their host ranges reflect the host ranges of their carrier replicons. In contrast, SXT does not encode its own replication system, which could potentially limit its distribution. Therefore, the functionality of the TA module of SXT in distinct hosts seems to be justified from an evolutionary point of view. However, to draw more general conclusions, it is necessary to test the activity of the cassettes analyzed in a wider range of the host species.
It is important to point out that the _tad-ata_-like locus of the SXT element is the first stabilization system identified in the large group of integrative and conjugative elements (ICE). These elements can exist in bacterial cells in two forms: (i) linear and integrated within a chromosome or (ii) circular and excised from the donor site. Since the circular, plasmid-like forms are present as single copies and they are unable to replicate, cell division results in the production of ICE-less cells. Therefore, the presence of a stabilizing addiction system within SXT (eliminating cells lacking SXT from the population) seems to be evolutionarily advantageous for the element.
The presence of the toxin-antitoxin system might explain the high level of stability of SXT observed by Burrus and Waldor (19). These workers found that despite the relatively high frequency of excision of this element from the donor site (approximately 1.5 excised molecules per 100 cells), it was not possible to detect loss of the element, even after hundreds of generations of growth in the absence of selection pressure.
Interestingly, the s045-s044 module of SXT has been inserted into the conserved backbone of the element, in one of the four insertion hotspots that have been identified (20). Comparative studies of ICEs related to SXT (R392, R997, pMERPH) have revealed that none of them contains genes homologous to s044 or s045. However, in the hotspots they carry other unique genes or group of genes, which seem to confer element-specific properties (20). Their particular functions, as well as the mechanisms of their acquisition, have not been analyzed.
The homologues of Tad and Ata are currently described in the databases as putative conserved proteins with unknown functions. In the case of the Tad relatives, they are also designated phage-related proteins. However, our analyses revealed the presence of homologous proteins in just two bacteriophages (N15 and φKO2), in which they were probably first described.
N15 appears to be the second bacteriophage (after P1) for which the presence of the TA cassette has been confirmed. The TA cassettes of N15 and P1 are highly divergent, since no sequence similarities were observed at the nucleotide or amino acid sequence level (data not shown). Both N15 and P1 can lysogenize E. coli cells as autonomous forms imitating plasmids. In previous studies investigating loci responsible for the stable maintenance of N15 workers identified a module responsible for segregational stability, composed of the parA and parB genes, as well as numerous partitioning sites located throughout the phage genome (36). Our data show that the stability of the N15 prophage might also be controlled at the postsegregational level. A module related to tad-ata is also harbored by phage φKO2 of K. oxytoca (the closest relative of N15), but it is absent from the genome of another relative, the linear prophage PY54 of Yersinia enterocolitica (22), which exemplifies the plasticity of phage genomes.
We showed that loci of the tad-ata type encode two promoters, a strong promoter whose gene precedes the putative operon (P1) and a second, much weaker promoter whose gene is located upstream of the antitoxin gene (P2). The presence of two promoters is not unique among TA cassettes, since this is also the case for the higBA operon of plasmid Rts1 of Proteus vulgaris (76, 77). Interestingly, the two types of loci have the same gene arrangement; the toxin gene is upstream of the gene for the antidote, an order which is the reverse of the order in other known TA systems. It is thus probable that the occurrence of internal P2 promoters, which ensure that there is excess antitoxin in bacterial cells, is strongly linked with this unusual gene arrangement. Both types of loci also encode antitoxins containing a helix-turn-helix motif of the XRE family; however, their toxins do not show significant sequence similarities (data not shown). We suggest that the loci of the tad-ata type might be considered a distinct subclass of the higBA family.
It is obvious that a balance in the levels of expression of toxin and antitoxin proteins in a bacterial cell is vital for proper functioning of TA cassettes. Therefore, it seems probable that reduced activity of the P2 promoters in different hosts might result in a deficiency of antitoxin, which would lead to manifestation of the toxic effect. This is consistent with our observations with the host P. versutus, in which the P2 promoter of the N15-derived cassette seems not to be functional, as judged by promoter probe vector analysis. In this case, conjugal transfer of the shuttle plasmid pABW3-N15 (which contains the TA cassette of N15) from E. coli to P. versutus resulted in a dramatic reduction in the number of transconjugants (only individual colonies were obtained, compared with 106 transconjugants selected with control plasmids), which most probably was a result of the plasmid-specified toxicity expressed in the recipient cells. The observations described above suggest that the presence of a TA cassette in a plasmid might influence the host range of the carrier replicon.
As mentioned above, two of the TA cassettes tested, those derived from the chromosome of E. coli EDL933 (Z3230-Z3231) and from pAMI2 (tad-ata), were not able to stabilize plasmids in E. coli. Our results suggest that this was not due to the different activities of the P1 and P2 promoters in this host. In these cases we observed that the Z3230 and Ata antitoxins were unable (in contrast to the antitoxins of N15 or SXT) to cause a complete rapid reversal of a toxic effect exerted by the cognate toxins. Moreover, the Z3231 and Tad toxins had the most severe effect on E. coli cells (Fig. 3A), which suggests that these proteins may recognize different targets in E. coli or recognize the same target but with different affinities.
Little is currently known about the cellular targets of toxins encoded by various TA cassettes. Two plasmid-encoded toxins, CcdB and ParE, have been shown to inactivate topoisomerase II DNA gyrase (14, 45), which results in inhibition of crucial cellular processes, such as transcription and/or DNA replication. On the other hand, the chromosomally encoded RelE and MazF toxins of E. coli, the HigB-1 and Hig-B2 toxins encoded by a superintegron of V. cholerae (25), and the Kid toxin of plasmid R1 possess RNase activities (66, 59, 82). Since these toxins have bacteriostatic or bactericidal effects (32, 53), they have aroused interest as potential antibacterial agents. There is a need to develop specific compounds that activate or mimic bacterial toxins (29, 33), and it is also important to learn more about specific interactions of toxins with their cellular targets.
In this study we showed that although the toxins encoded by the four _tad-ata_-like TA cassettes have similar effects on E. coli cells (i.e., growth inhibition and formation of elongated cells), they have different effects on cell viability (Fig. 3). Identification of the intracellular targets of Tad and related toxins, which is essential for understanding this system, is the immediate goal of our future studies.
Supplementary Material
[Supplemental material]
Acknowledgments
We are grateful to C. Fuqua for providing plasmid pCF430; K. Gerdes for providing plasmid pNDM220 and E. coli strain MC1000; F. Hayes for providing plasmid pFH450 and E. coli strain BR825; M. E. Lidstrom for providing plasmid pCM132; J. Paciorek for providing total DNA of E. coli EDL933; N. V. Ravin and M. Lobocka for providing E. coli strain EC16816; D. Rawlings for providing plasmid pOU82 and E. coli strains SG22025, SG22093, and SG22095; M. Waldor for providing E. coli strain HW220; and M. Yarmolinski and M. Lobocka for providing E. coli strain DH5Δlac. We also acknowledge M. Putyrski for technical assistance.
This work was supported by the State Committee for Scientific Research, Poland (grant 2 P04A 073 29).
Footnotes
▿
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93**:**6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alpert, C. A., A. M. Crutz-Le Coq, C. Malleret, and M. Zagorec. 2003. Characterization of a theta-type plasmid from Lactobacillus sakei: a potential basis for low-copy-number vectors in lactobacilli. Appl. Environ. Microbiol. 69**:**5574-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25**:**3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baj, J., E. Piechucka, D. Bartosik, and M. Wlodarczyk. 2000. Plasmid occurrence and diversity in the genus Paracoccus. Acta Microbiol. Polon. 49**:**265-270. [PubMed] [Google Scholar]
- 5.Bartosik, D., J. Baj, E. Piechucka, and M. Wlodarczyk. 1992. Analysis of Thiobacillus versutus pTAV1 plasmid functions. Acta Microbiol. Polon. 42**:**97-100. [Google Scholar]
- 6.Bartosik, D., A. Bialkowska, J. Baj, and M. Wlodarczyk. 1997. Construction of mobilizable cloning vectors derived from pBGS18 and their application for analysis of replicator region of a pTAV202 mini-derivative of Paracoccus versutus pTAV1 plasmid. Acta Microbiol. Polon. 46**:**379-383. [PubMed] [Google Scholar]
- 7.Bartosik, D., J. Baj, and M. Wlodarczyk. 1998. Molecular and functional analysis of pTAV320, a _repABC_-type replicon of the Paracoccus versutus composite plasmid pTAV1. Microbiology 144**:**3149-3157. [DOI] [PubMed] [Google Scholar]
- 8.Bartosik, D., M. Szymanik, and E. Wysocka. 2001. Identification of the partitioning site within the _repABC_-type replicon of the composite Paracoccus versutus plasmid pTAV1. J. Bacteriol. 183**:**6234-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartosik, D., M. Witkowska, J. Baj, and M. Wlodarczyk. 2001. Characterization and sequence analysis of the replicator region of the novel plasmid pALC1 from Paracoccus alcaliphilus. Plasmid 45**:**222-226. [DOI] [PubMed] [Google Scholar]
- 10.Bartosik, D., J. Baj, A. A. Bartosik, and M. Wlodarczyk. 2002. Characterization of the replicator region of megaplasmid pTAV3 of Paracoccus versutus and search for plasmid-encoded traits. Microbiology 148**:**871-881. [DOI] [PubMed] [Google Scholar]
- 11.Bartosik, D., J. Baj, M. Sochacka, E. Piechucka, and M. Wlodarczyk. 2002. Molecular characterization of functional modules of plasmid pWKS1 of Paracoccus pantotrophus DSM 11072. Microbiology 148**:**2847-2856. [DOI] [PubMed] [Google Scholar]
- 12.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184**:**4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beringer, J. E. 1974. T factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84**:**188-198. [DOI] [PubMed] [Google Scholar]
- 14.Bernard, P., and M. Couturier. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226**:**735-745. [DOI] [PubMed] [Google Scholar]
- 15.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7**:**1513-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boucher, I., E. Emond, M. Parrot, and S. Moineau. 2001. DNA sequence analysis of three Lactococcus lactis plasmids encoding phage resistance mechanisms. J. Dairy Sci. 84**:**1610-1620. [DOI] [PubMed] [Google Scholar]
- 17.Bravo, A., T. G. de Morrontegui, and R. Diaz. 1987. Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol. Gen. Genet. 210**:**101-110. [DOI] [PubMed] [Google Scholar]
- 18.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100**:**10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burrus, V., and M. K. Waldor. 2003. Control of SXT integration and excision. J. Bacteriol. 185**:**5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burrus, V., J. Marrero, and M. K. Waldor. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55**:**173-183. [DOI] [PubMed] [Google Scholar]
- 21.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138**:**179-207. [DOI] [PubMed] [Google Scholar]
- 22.Casjens, S. R., E. B. Gilcrease, W. M. Huang, K. L. Bunny, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2004. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 186**:**1818-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaconas, G., and R. M. Harshey. 2002. Transposition of phage Mu DNA, p. 384-402. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology Press, Washington, DC.
- 24.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98**:**14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen-Dalsgaard, M., and K. Gerdes. 2006. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 62**:**397-411. [DOI] [PubMed] [Google Scholar]
- 26.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16**:**10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host-range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77**:**7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18**:**5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engelberg-Kulka, H., B. Sat, M. Reches, S. Amitai, and R. Hazan. 2004. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 12**:**66-71. [DOI] [PubMed] [Google Scholar]
- 30.Gerdes, K., J. E. Larsen, and S. Molin. 1985. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 161**:**292-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerdes, K., A. Nielsen, P. Thorsted, and E. G. Wagner. 1992. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable hok, srnB and pndA effector messenger RNAs. J. Mol. Biol. 226**:**637-649. [DOI] [PubMed] [Google Scholar]
- 32.Gerdes, K., T. Thisted, and J. Martinussen. 1990. Mechanism of post-segregational killing by the hok/sok system of plasmid R1: sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol. Microbiol. 4**:**1807-1818. [DOI] [PubMed] [Google Scholar]
- 33.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3**:**371-382. [DOI] [PubMed] [Google Scholar]
- 34.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29**:**1065-1076. [DOI] [PubMed] [Google Scholar]
- 35.Grady, R., and F. Hayes. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47**:**1419-1432. [DOI] [PubMed] [Google Scholar]
- 36.Grigoriev, P. S., and M. B. Lobocka. 2001. Determinants of segregational stability of the linear plasmid-prophage N15 of Escherichia coli. Mol. Microbiol. 42**:**355-368. [DOI] [PubMed] [Google Scholar]
- 37.Gronlund, H., and K. Gerdes. 1999. Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J. Mol. Biol. 285**:**1401-1415. [DOI] [PubMed] [Google Scholar]
- 38.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177**:**4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haines, A. S., K. Jones, M. Cheung, and C. M. Thomas. 2005. The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J. Bacteriol. 187**:**4728-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15**:**2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayes, F. 1998. A family of stability determinants in pathogenic bacteria. J. Bacteriol. 180**:**6415-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301**:**1496-1499. [DOI] [PubMed] [Google Scholar]
- 43.Hirsch, P. R., M. van Montagu, A. W. B. Johnston, N. J. Brewin, and J. Schell. 1980. Physical identification of bacteriocinogenic, nodulation and other plasmids in strains of Rhizobium leguminosarum. J. Gen. Microbiol. 120**:**403-412. [Google Scholar]
- 44.Hochhut, B., and M. K. Waldor. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32**:**99-110. [DOI] [PubMed] [Google Scholar]
- 45.Jiang, Y., J. Pogliano, D. R. Helinski, and I. Konieczny. 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44**:**971-979. [DOI] [PubMed] [Google Scholar]
- 46.Kalnin, K., S. Stegalkina, and M. Yarmolinsky. 2000. pTAR-encoded proteins in plasmid partitioning. J. Bacteriol. 182**:**1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koekman, B. P., P. J. Hooykaas, and R. A. Schilperoort. 1982. A functional map of the replicator region of the octopine Ti plasmid. Plasmid 7**:**119-132. [DOI] [PubMed] [Google Scholar]
- 48.Kushner, S. R. 1978. An improved method for transformation of E. coli with ColE1-derived plasmids, p. 17-23. In H. B. Boyer and S. Nicosia (ed.), Genetic engineering. Elsevier/North-Holland, Amsterdam, The Netherlands.
- 49.Lehnherr, H., E. Maguin, S. Jafri, and M. B. Yarmolinsky. 1993. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233**:**414-428. [DOI] [PubMed] [Google Scholar]
- 50.Lehnherr, H., and M. R. Yarmolinsky. 1995. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc. Natl. Acad. Sci. USA 92**:**3274-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34**:**D257-D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, W., J. Shi, X. Wang, Y. Han, W. Tong, L. Ma, B. Liu, and B. Cai. 2004. Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 336**:**231-240. [DOI] [PubMed] [Google Scholar]
- 53.Lioy, V. S., M. T. Martin, A. G. Camacho, R. Lurz, H. Antelmann, M. Hecker, E. Hitchin, Y. Ridge, J. M. Wells, and J. C. Alonso. 2006. pSM19035-encoded z toxin induces stasis followed by death in a subpopulation of cells. Microbiology 152**:**2365-2379. [DOI] [PubMed] [Google Scholar]
- 54.Lobocka, M. B., D. J. Rose, G. Plunkett III, M. Rusin, A. Samojedny, H. Lehnherr, M. B. Yarmolinsky, and F. R. Blattner. 2004. Genome of bacteriophage P1. J. Bacteriol. 186**:**7032-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ludtke, D. N., B. G. Eichorn, and S. J. Austin. 1989. Plasmid-partition functions of the P7 prophage. J. Mol. Biol. 209**:**393-406. [DOI] [PubMed] [Google Scholar]
- 56.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147**:**2065-2075. [DOI] [PubMed] [Google Scholar]
- 57.McDonnell, G. E., and D. J. McConnell. 1994. Overproduction, isolation, and DNA-binding characteristics of Xre, the repressor protein from the Bacillus subtilis defective prophage PBSX. J. Bacteriol. 176**:**5831-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 59.Munoz-Gomez, A. J., M. Lemonnier, S. Santos-Sierra, A. Berzal-Herranz, and R. Diaz-Orejas. 2005. RNase/anti-RNase activities of the bacterial parD toxin-antitoxin system. J. Bacteriol. 187**:**3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Newman, J. R., and C. Fuqua. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227**:**197-203. [DOI] [PubMed] [Google Scholar]
- 61.Nieto, C., T. Pellicer, D. Balsa, S. K. Christensen, K. Gerdes, and M. Espinosa. 2006. The chromosomal relBE2 toxin-antitoxin locus of Streptococcus pneumoniae: characterization and use of a bioluminescence resonance energy transfer assay to detect toxin-antitoxin interaction. Mol. Microbiol. 59**:**1280-1296. [DOI] [PubMed] [Google Scholar]
- 62.O'Brien, A. D., T. A. Lively, M. E. Chen, S. W. Rothman, and S. B. Formal. 1983. Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (Shiga) like cytotoxin. Lancet i**:**702. [DOI] [PubMed] [Google Scholar]
- 63.Ogura, T., and S. Hiraga. 1983. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 80**:**4784-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Osborn, A. M., and D. Boltner. 2002. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid 48**:**202-212. [DOI] [PubMed] [Google Scholar]
- 65.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33**:**966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112**:**131-140. [DOI] [PubMed] [Google Scholar]
- 67.Ravin, N. V. 2006. N15: the linear plasmid prophage, p. 448-456. In R. Calendar and S. T. Abedon (ed.), The bacteriophages, 2nd ed. Oxford University Press, Oxford, United Kingdom.
- 68.Roberts, R. C., A. R. Strom, and D. R. Helinski. 1994. The parDE operon of the broad-host-range plasmid RK2 specifies growth inhibition associated with plasmid loss. J. Mol. Biol. 237**:**35-51. [DOI] [PubMed] [Google Scholar]
- 69.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 70.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166**:**368-379. [DOI] [PubMed] [Google Scholar]
- 71.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53**:**85-96. [DOI] [PubMed] [Google Scholar]
- 72.Smith, A. S., and D. E. Rawlings. 1997. The poison-antidote stability system of the broad-host-range Thiobacillus ferrooxidans plasmid pTF-FC2. Mol. Microbiol. 26**:**961-970. [DOI] [PubMed] [Google Scholar]
- 73.Spratt, B. G., P. J. Hedge, S. te Heesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41**:**337-342. [DOI] [PubMed] [Google Scholar]
- 74.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278**:**631-637. [DOI] [PubMed] [Google Scholar]
- 75.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22**:**4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tian, Q. B., M. Ohnishi, A. Tabuchi, and Y. Terawaki. 1996. A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 220**:**280-284. [DOI] [PubMed] [Google Scholar]
- 77.Tian, Q. B., T. Hayashi, T. Murata, and Y. Terawaki. 1996. Gene product identification and promoter analysis of hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 225**:**679-684. [DOI] [PubMed] [Google Scholar]
- 78.Toussaint, A., and C. Merlin. 2002. Mobile elements as a combination of functional modules. Plasmid 47**:**26-35. [DOI] [PubMed] [Google Scholar]
- 79.Urakami, T., H. Araki, H. Oyanagi, K. Suzuki, and K. Komagata. 1990. Paracoccus aminophilus sp. nov. and Paracoccus aminovorans sp. nov., which utilize N,_N_-dimethylformamide. Int. J. Syst. Bacteriol. 40**:**287-291. [DOI] [PubMed] [Google Scholar]
- 80.Van Melderen, L., P. Bernard, and M. Couturier. 1994. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol. Microbiol. 11**:**1151-1157. [DOI] [PubMed] [Google Scholar]
- 81.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19**:**259-268. [DOI] [PubMed] [Google Scholar]
- 82.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12**:**913-923. [DOI] [PubMed] [Google Scholar]
- 83.Zielenkiewicz, U., and P. Ceglowski. 2005. The toxin-antitoxin system of the streptococcal plasmid pSM19035. J. Bacteriol. 187**:**6094-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]