Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation (original) (raw)
Abstract
tob is a member of an emerging family of genes with antiproliferative function. Tob is rapidly phosphorylated at Ser 152, Ser 154, and Ser 164 by Erk1 and Erk2 upon growth-factor stimulation. Oncogenic Ras-induced transformation and growth-factor-induced cell proliferation are efficiently suppressed by mutant Tob that carries alanines but not glutamates, mimicking phosphoserines, at these sites. Wild-type Tob inhibits cell growth when the three serine residues are not phosphorylated but is less inhibitory when the serines are phosphorylated. Because growth of Rb-deficient cells was not affected by Tob, Tob appears to function upstream of Rb. Intriguingly, cyclin D1 expression is elevated in serum-starved _tob_−/− cells. Reintroduction of wild-type Tob and mutant Tob with serine-to-alanine but not to glutamate mutations on the Erk phosphorylation sites in these cells restores the suppression of cyclin D1 expression. Finally, the S-phase population was significantly increased in serum-starved _tob_−/− cells as compared with that in wild-type cells. Thus, Tob inhibits cell growth by suppressing cyclin D1 expression, which is canceled by Erk1- and Erk2-mediated Tob phosphorylation. We propose that Tob is critically involved in the control of early G1 progression.
Keywords: Cell cycle entry, Tob phosphorylation, Ras/MAPK pathway
Cell cycle progression is controlled by systematic activation and/or inactivation of proliferative and antiproliferative genes. Abnormalities of these genes contribute to the development of cancer. One of the best examples is that genes encoding receptor protein tyrosine kinases (RTKs) are often activated by amplification, rearrangement, or point mutation. RTKs become activated and phosphorylate themselves as well as their cytoplasmic substrates in response to growth factor (Ullrich and Schlessinger 1990). RTK activation initiates a signal transduction cascade involving Ras and mitogen-activated protein kinases (MAPK or Erk, for extracellular signal-regulated protein kinases), which transmit growth-regulating signals to the nucleus (Marshall 1994). Ras is a central component of mitogenic signaling and is essential for cells to progress from the quiescent state (G0) through the G1/S transition (Stacey and Kung 1984). MAPK is activated downstream of Ras, and sustained activation of Erk1 and Erk2 (Erk1/2) MAPKs allows fibroblasts to pass through the G1 restriction point and enter the S phase (Brondello et al. 1995). Erk1/2 phosphorylate and activate a variety of transcription factors such as Elk1 (Davis 1995), c-Ets1, and c-Ets2 (Coffer et al. 1994), and kinases such as p90rsk1 (Dalby et al. 1998), MNK1, and MNK2 (Fukunaga and Hunter 1997; Waskiewicz et al. 1997). However, information on the Ras-dependent molecular events that directly link mitogenic signals to transition from the G0 to the G1 phase is limited. Also, none of the known substrates of Erk1/2 are clearly linked to regulation of G0/G1 transition.
Tob is a member of an emerging family of antiproliferative proteins comprising, in humans, Tob, Tob2, ANA (BTG3 in mouse), BTG1, and BTG2 (PC3 in rat and TIS21 in mouse; Bradbury et al. 1991; Fletcher et al. 1991; Rouault et al. 1992, 1996; Matsuda et al. 1996; Guehenneux et al. 1997; Yoshida et al. 1998; Ikematsu et al. 1999). The tob cDNA was identified by screening an expression library by means of protein–protein interaction with an ErbB2 probe (Matsuda et al. 1996). The biological significance of the interaction between Tob and ErbB2 remains to be elucidated. Importantly, mice lacking the tob gene frequently develop tumors (Y. Yoshida, unpubl.) and show an osteopetrotic phenotype that is caused by the facilitated growth and differentiation of osteoblasts (Yoshida et al. 2000). The data strongly suggest the importance of tob in cell growth regulation. The btg1 gene was first identified near the breakpoint of a chromosomal translocation found in a B-cell chronic lymphocytic leukemia. btg1 expression is correlated with suppression of cell proliferation (Rouault et al. 1992). tis21 is an immediate early gene induced by tumor promoter treatment of Swiss3T3 cells, and pc3 is induced along with differentiation of nerve growth factor-treated rat PC12 cells (Bradbury et al. 1991; Fletcher et al. 1991; Iacopetti et al. 1999). Expression of btg2 is induced via a p53-dependent mechanism in response to DNA damage and is thus likely to be relevant to cell cycle control and cellular responses to DNA damage (Rouault et al. 1996). In addition, two _tob_-related cDNAs, ANA and tob2, were cloned by a PCR-mediated cloning procedure (Yoshida et al. 1998; Ikematsu et al. 1999).
The Tob family proteins share a couple of structural and functional characteristics. First, the N-terminal 120 residues of the proteins are homologous, and two short stretches of ∼20 amino acids in the homologous domain define the family signature (Guehenneux et al. 1997). Second, overexpression of these gene products suppresses cell cycle progression (Rouault et al. 1992; Matsuda et al. 1996; Montagnoli et al. 1996; Rouault et al. 1996; Guehenneux et al. 1997; Lim et al. 1998; Yoshida et al. 1998; Ikematsu et al. 1999; Guardavaccaro et al. 2000). Intriguingly, ectopic expression of PC3 down-regulates cyclin D1 transcription, resulting in suppression of G1 progression (Guardavaccaro et al. 2000), but whose underlying mechanism and physiological relevance remain to be addressed. Third, the proteins of this family are associated with transcription factors such as Caf1, Hoxb9, and Smad (Bogdan et al. 1998; Rouault et al. 1998; Ikematsu et al. 1999; Guardavaccaro et al. 2000; Prevot et al. 2000; Yoshida et al. 2000). So far, all of the reported data implicate Tob family proteins in the regulation of cell growth. However, the underlying mechanisms of the growth regulation as well as the biological significance of this gene family have yet to be elucidated.
We report here that growth-factor stimulation of the RTK/Ras/MAPK pathway results in phosphorylation of Tob. Further, we provide evidence suggesting that Tob, unless phosphorylated by Erk1/2, suppresses cyclin D1 expression, thus inhibiting cell proliferation. We propose that Tob functions as a molecular switch that regulates cell cycle progression through early G1.
Results
Tob inhibits transition from G0/G1 to S phase
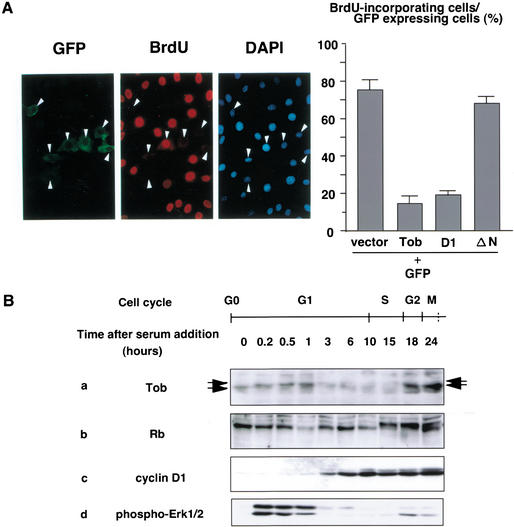
We showed previously that exogenously expressed Tob showed antiproliferative activity (Matsuda et al. 1996). To examine the effect of Tob expression on cell cycle progression, we microinjected a Tob expression plasmid or a control plasmid together with a green fluorescent protein (GFP) expression plasmid into serum-starved NIH3T3 cells. After cells were incubated with serum and BrdU, cells that entered S phase were identified by their reactivity to anti-BrdU antibody. Representative data for cells injected with the Tob expression plasmid are shown in Figure 1A (left). Whereas 161 of 220 (73%) GFP-expressing cells microinjected with control plasmid incorporated BrdU, only 56 of 420 (13.3%) GFP-expressing cells microinjected with Tob expression plasmid displayed BrdU incorporation (P < 0.05; Fig. 1A, right). We also found that only 17.7% of the cells microinjected with cDNA encoding the N-terminal half of Tob (D1 in Fig. 3A, see below) incorporated BrdU (Fig. 1A, right). The C-terminal half of Tob (TobΔN; amino acids 166–345) had no effect (Fig. 1A, right). The data indicated that Tob overexpression inhibited cell cycle progression from G0/G1 to S phase and that the N-terminal half of Tob, which included a domain conserved in all Tob family members (Matsuda et al. 1996; Guehenneux et al. 1997), was sufficient for its antiproliferative activity.
Figure 1.
Effect of Tob overexpression on cell cycle progression. (A) Serum-starved NIH3T3 cells microinjected with pME-Tob and GFP expression plasmids were refed with serum and examined for GFP expression (left), immunostained for BrdU (middle), or stained with DAPI to visualize nuclei (right). Arrowheads indicate GFP-expressing cells. The graph shows the percentage of GFP-expressing cells showing BrdU incorporation in response to expression of either vector containing no insert (vector), pME-Tob (Tob), pME-TobD1 (D1) (see Fig. 3A), or pME-TobΔN (ΔN). At least 70 cells were scored for each experiment. Values are shown as the mean ± SD of four experiments. (B) Time course of Tob, Rb, and cyclin D1 expression and Erk1/2 activation. Serum-starved NIH3T3 cells were stimulated with serum for the indicated period. Equal amounts of cell lysates were immunoblotted for Tob (a), Rb (b), cyclin D1 (c), and phospho-Erk1/2 (d). Arrows indicate slow-migrating and fast-migrating forms of Tob. Stages of the cell cycle are estimated from parallel FACS analyses (data not shown) and are shown at the top.
Figure 3.
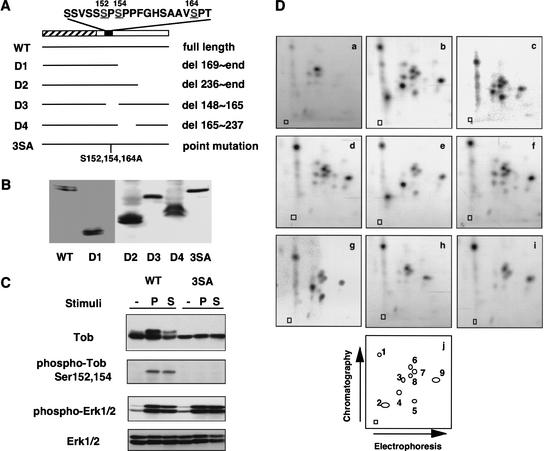
Determination of phosphorylation sites. (A) Schematic representation of Tob mutants. The hatched box indicates the homologous region among Tob family members. The black box indicates the serine-rich region, and the amino acid sequence of this region is shown above. The bars represent the portion of Tob that is expressed by wild-type (WT) or mutant constructs (D1–D4 and 3SA). (B) Western blot analysis of Tob mutants shown in A, which were expressed in NIH3T3 cells. (C) Detection of phosphorylated Tob with phosphorylation site-specific antibodies. NIH3T3 cells transfected with plasmids encoding either wild-type Tob (WT) or the Tob3SA mutant (3SA) were serum-starved and treated with PDGF (P) or serum (S). Cell lysates were immunoblotted for Tob, phospho-Tob, phospho-Erk1/2, and Erk1/2. (D) Two-dimensional chymotryptic phosphopeptide maps of in vivo phosphorylated Tob. NIH3T3 cells were transfected with Tob expression plasmid encoding wild-type Tob (a,b), or Ser152Ala (c), Ser154Ala (d), Ser164Ala (e), Ser152,154Ala (f), Ser152,164Ala (g), Ser154,164Ala (h), or 3SA (i) mutants, were labeled with 32P-orthophosphate and stimulated with PDGF. Panel a is a phosphopeptide map of wild-type Tob from unstimulated cells. Panels b_–_i are phosphopeptide maps of wild-type Tob and Tob mutants from PDGF-stimulated cells. Panel j is a schematic illustration showing the position of the various phosphopeptides detected in panels a_–_i. Phosphopeptide positions not numbered in panel j indicate partially digested fragments because they disappeared by additional digestion with chymotrypsin (data not shown). Squares indicate origins.
Tob expression during the cell cycle
To elucidate the mechanism by which Tob inhibited cell proliferation, expression of Tob throughout the cell cycle was analyzed. Growth of NIH3T3 cells was arrested by serum starvation, and the quiescent cells were stimulated with serum to initiate synchronous growth. After incubating the cells for varying periods in the presence of serum, whole-cell extracts were subjected to immunobiochemical analysis (Fig. 1B). Synchronous cell cycle progression was monitored by cyclin D1 expression, Rb phosphorylation, and Erk1 and Erk2 activation. As reported previously (Baldin et al. 1993; Lavoie et al. 1996), cyclin D1 was detected 3 h after serum stimulation (mid-G1) and continued to increase until S phase (Fig. 1B, panel c). Rb inactivation, as evidenced by its phosphorylation, became apparent 15–18 h after stimulation (S phase). At this time point, cyclin D1 expression was maximal. Hypophosphorylated Rb appeared again 24 h after stimulation (G2/M; Fig. 1B, panel b). Activation of Erk1/2, which was detected by anti-phospho-Erk1/2 antibodies, was evident in early G1 phase and G2/M phase (Fig. 1B, panel d). The Tob protein was detected in G0-arrested cells, and the level of its expression remained constant until 1 h after stimulation (Fig. 1B, panel a). In G1 through S phase, expression of Tob declined significantly and remained barely detectable. Subsequently, at G2 phase, Tob expression resumed. In addition to the alteration of Tob expression level during the cell cycle, we detected a slow-migrating form of Tob that appeared 10 min after serum stimulation and lasted for at least 1 h. Interestingly, appearance of the slow-migrating form of Tob correlated with activation of Erk1/2. These results showed that Tob is rapidly modified upon mitogenic stimulation, possibly by phosphorylation (see below), and subsequently becomes deteriorated at mid-G1 phase.
Induction of Tob phosphorylation upon mitogenic stimulation
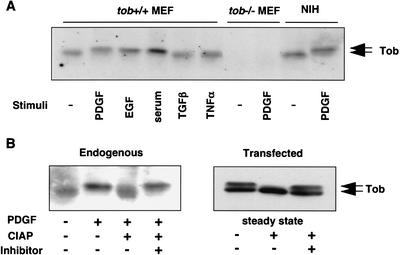
To confirm that the slow-migrating form of Tob was generated upon stimulation of cell proliferation, mouse embryonic fibroblasts (MEFs) were serum-starved for 48 h and then exposed to epidermal growth factor (EGF), platelet-derived growth factor (PDGF), serum, transforming growth factor β (TGFβ), or tumor necrosis factor α (TNFα; Fig. 2A). Mitogenic stimulation of cells with PDGF, EGF, or serum resulted in the appearance of the slow-migrating form of Tob. In contrast, treatment of cells with TNFα and TGFβ did not induce the slow-migrating form. Identical results were obtained when serum-starved NIH3T3 cells were treated with PDGF (Fig. 2A).
Figure 2.
Tob phosphorylation in response to mitogenic stimulation. (A) Effect of growth-factor treatment on Tob mobility in SDS-polyacrylamide gels. Serum-starved MEFs (tob+/+ MEF and _tob_−/− MEF) and NIH3T3 (NIH) cells were incubated with the indicated stimulants for 15 min. Cell lysates were immunoblotted for Tob. (B) Effect of phosphatase treatment on Tob mobility in SDS-polyacrylamide gels. Lysates prepared from unstimulated (−) or PDGF-stimulated (+) NIH3T3 cells or pME-Tob-transfected asynchronously growing (steady-state) NIH3T3 cells were incubated with CIAP in the absence (−) or presence (+) of phosphatase inhibitors. Samples were immunoblotted for Tob. Arrows indicate hyperphosphorylated and hypophosphorylated Tob.
To characterize the nature of the slow-migrating form of Tob, its sensitivity to calf intestine alkaline phosphatase (CIAP) was analyzed. The results showed that the slow-migrating form of Tob was sensitive to CIAP treatment (Fig. 2B, left). In asynchronously growing Tob-transfected cells, both slow-migrating and fast-migrating forms of Tob were detected (Fig. 2B, right). Again, only the slow-migrating form of Tob was sensitive to CIAP. From these data, we concluded that the slow-migrating form of Tob was generated by phosphorylation, and the phosphorylation event was induced by mitogenic stimulation. In addition, because Tob immunoprecipitated from NIH3T3 cells showed no reactivity with anti-phospho-tyrosine antibody (data not shown), Tob seemed to be phosphorylated at serine/threonine residues.
Determination of Tob phosphorylation sites
To identify the phosphorylation sites, we performed two types of experiments: deletion analysis and phosphopeptide mapping. First, because the slow migration of Tob was caused by its phosphorylation (Fig. 2), mutant Tob lacking phosphorylation sites was expected to be a single band on SDS-polyacrylamide gels. Therefore, to help identify the phosphorylation sites, we expressed various Tob deletion mutants (Fig. 3A) in NIH3T3 cells and examined their mobility in SDS-polyacrylamide gels. As shown in Figure 3B, Tob deletion mutant D3 lacking amino acids 148–165 migrated as a single band. In contrast, Tob deletion mutants D1, D2, and D4 as well as wild-type Tob appeared as doublets. The data suggested that slow-migration-related phosphorylation of Tob occurred in the region deleted in the D3 mutant. Among eight serine residues present in this region (Fig. 3A), only three (Ser 152, Ser 154, and Ser 164) match the consensus phosphorylation site for proline-directed kinases such as Cdks and MAPKs (Hall and Vulliet 1991; Adam and Hathaway 1993). Because MAPK seemed likely to be involved in Tob phosphorylation (see below), we converted the three serine residues to alanines by site-directed mutagenesis. The mutated Tob protein (Tob3SA) appeared as a single band on SDS-polyacrylamide gels and had relative mobility similar to that of fast-migrating, wild-type Tob (Fig. 3B). Furthermore, unlike wild-type Tob, mitogenic stimulation of serum-starved, Tob3SA-expressing NIH3T3 cells did not produce the slow-migrating form of Tob (Fig. 3C). Note that the expression level of endogenous Tob, in comparison to that of exogenous Tob, was too low to be detected in this assay. Mitogenic stimulation was monitored by immunoblotting with anti-phospho-Erk1/2 and anti-Erk1/2 antibodies (Fig. 3C). These data suggested that growth factor stimulation induced phosphorylation of Tob at Ser 152, Ser 154, and/or Ser 164.
To directly confirm the phosphorylation sites, wild-type Tob and a series of mutants that carried Ser152Ala, Ser154Ala, Ser164Ala, Ser152,154Ala, Ser152,164Ala, Ser154,164Ala, or Ser152,154,164Ala (3SA) substitution were expressed in NIH3T3 cells. After serum starvation, cells were metabolically labeled with [32P]orthophosphate and stimulated with PDGF. Wild-type Tob and all the mutants incorporated 32P in response to PDGF (data not shown). Chymotryptic phosphopeptide mapping of wild-type Tob revealed four major (p1, p2, p4, and p9) and three minor (p3, p5, and p7) phosphopeptides that were dependent on PDGF stimulation (Fig. 3D, panels a and b). Appearance of phosphopeptides p6 and p8 was independent of PDGF stimulation. Relative to wild-type Tob (Fig. 3D, panel b), phosphopeptide p2 did not appear in the map of the Ser152Ala mutant, and phosphopeptide p3 did not appear in the map of the Ser164Ala mutant (Fig. 3D, panels c and e). These results indicate that phosphopeptide p2 contains phospho-Ser 152 and phosphopeptide p3 contains phospho-Ser 164. In the map of the Ser154Ala mutant, three phosphopeptides (p2, p4, and probably p5) disappeared or became less detectable (Fig. 3D, panel d). Based on charge differences of the peptides and relative positions in the chromatographic dimension, we concluded that p2 and p4 were phosphoisomers of the same peptide; p2 was phosphorylated on both Ser 152 and Ser 154, and p4 was phosphorylated on only Ser 154. Interestingly, a peptide phosphorylated only on Ser 152 was undetectable in the maps of the Ser154Ala and Ser154,164Ala mutants (Fig. 3D, panels d and h). When compared with the map of the Ser154Ala mutant, no phosphopeptides were eliminated from the map of the Ser152,154Ala mutant (Fig. 3D, panels d and f). The data suggested that Ser 154 phosphorylation was a prerequisite for Ser 152 phosphorylation. The signal for the peptide containing phosphorylated Ser 164 was weaker than signals for Ser 152 and Ser 154, suggesting that Ser 164 phosphorylation was minor. Alternatively, it appeared possible that Ser 164 phosphorylation was more susceptible to phosphatases.
To further confirm that Ser 152 and Ser 154 are phosphorylated in vivo, rabbit polyclonal antibodies for the amino acid sequence containing phosphorylated Ser 152 and Ser 154 were raised. Cell lysates of PDGF-stimulated Tob-transfectants were analyzed by immunoblotting with these antibodies. The results revealed a single immunoreactive protein having the same mobility as phosphorylated Tob, indicating specific reactivity of the antibodies with phosphorylated Tob (Fig. 3C). The antibodies did not recognize Tob3SA (Fig. 3C) from PDGF-treated cells by immunoblot analysis. Finally, appearance of three peptides (p1, p7, and p9) in the map of Tob3SA (Fig. 3D, panel i) suggested that Tob was phosphorylated at least at three serine/threonine residues other than Ser 152, 154, and 164 upon mitogenic stimulation. These three additional phosphorylation sites are to be determined.
Inhibition of Tob phosphorylation by MEK inhibitor PD98059
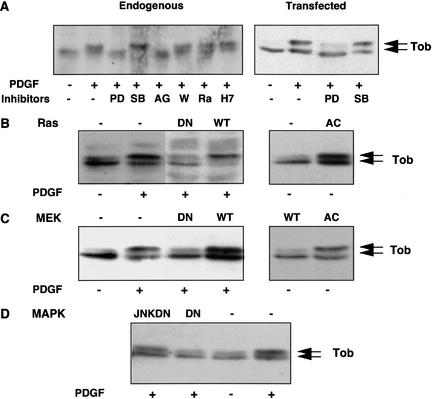
PDGF stimulation activates various signaling pathways such as the Ras/MAPK and PI3-kinase/Akt pathways (Burgering et al. 1994). To search for a kinase relevant to Tob phosphorylation, we examined the level of Tob phosphorylation in the cells treated with various inhibitors for signal transduction (Fig. 4A). PD98059, which blocks activation of MEK1/2 by inhibiting Raf or MEKK1, inhibited phosphorylation of both endogenous and exogenous Tob. Pretreatment of the cells with tyrphostin AG1296, which is a specific inhibitor of PDGF receptor (PDGFR), also inhibited phosphorylation of Tob. In contrast, SB203580, which is a p38MAPK-specific inhibitor that does not inhibit Erk1/2, did not affect Tob phosphorylation. Moreover, the PI3-kinase inhibitor wortmannin, the p70 S6 kinase inhibitor rapamycin, and the protein kinase C inhibitor H7 did not affect Tob phosphorylation. These results suggested that RTK-induced MEK activation was involved in Tob phosphorylation.
Figure 4.
The Ras/MEK/MAPK pathway is involved in Tob phosphorylation. (A) Untransfected (left) or Tob-transfected (right) NIH3T3 cells were serum-starved, pretreated with the indicated inhibitors for 3 h, and stimulated with PDGF for 10 min. (B_–_D) NIH3T3 cells were cotransfected with the indicated expression plasmid encoding Ras (B), MEK (C), and JNK or MAPK (D) together with pME-Tob. Cells were serum-starved and lysed before or after PDGF stimulation. Cell lysates were analyzed by immunoblotting. (PD) PD98059, (SB) SB203580, (AG) tyrphostin AG1296, (W) wortmannin, (Rap) rapamycin, (DN) dominant-negative form, (AC) constitutively active form, (WT) wild-type, (JNKDN) dominant-negative form of JNK. Arrows indicate hyperphosphorylated and hypophosphorylated Tob.
Tob phosphorylation by MAPK
The above findings led us to hypothesize that Ras, MEK, and MAPK, which are downstream of PDGFR, were responsible for Tob phosphorylation. To examine this possibility, we cotransfected dominant-negative mutants of Ras, MEK, or MAPK with the Tob expression plasmid and stimulated resting cells with PDGF. We found that PDGF-induced phosphorylation of Tob was inhibited by expression of each dominant-negative mutant (Fig. 4B–D). In contrast, transfection of cells with plasmids encoding wild-type MEK, Ras, and a dominant-negative mutant of Jun N-terminal kinase had little effect on PDGF-induced Tob phosphorylation. Furthermore, constitutively activated MEK and Ras induced Tob phosphorylation in a manner independent of PDGF stimulation (Fig. 4B,C). Thus, we concluded that the Ras/MEK/MAPK pathway participated in mitogen-induced Tob phosphorylation.
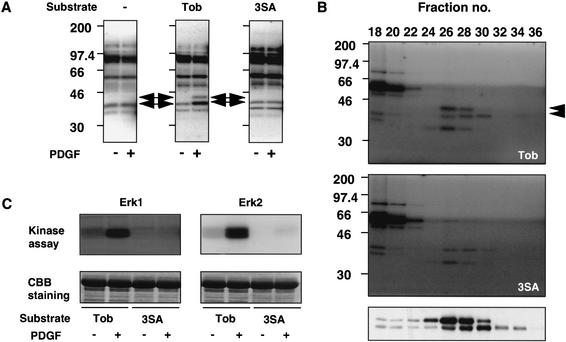
To identify kinases that phosphorylated Tob upon PDGF stimulation, we performed an in-gel kinase assay of cell extracts from unstimulated or PDGF-stimulated NIH3T3 cells using GST-fusion proteins possessing the N-terminal 168 amino acids of Tob (GST-TobN) or Tob3SA (GST-TobN3SA) as substrates. Two kinases with molecular masses of ∼42 kD (p42) and 44 kD (p44) phosphorylated GST-TobN in a PDGF-stimulation-dependent manner (Fig. 5A). Neither kinase phosphorylated GST-TobN3SA before and after PDGF stimulation (Fig. 5A, right). Thus, p42 and p44 were candidate kinases that phosphorylated Tob specifically at Ser 152, Ser 154, and/or Ser 164 in response to PDGF stimulation. Based on the molecular masses of these kinases, we speculated that p44 was Erk1 and p42 was Erk2.
Figure 5.
Identification of Tob kinase. (A) In-gel kinase assay. Lysates prepared from unstimulated (−) or PDGF-stimulated (+) NIH3T3 cells were tested for phosphorylating activity in gels containing no substrate (−), GST–TobN (Tob), or GST–TobN3SA (3SA). Arrows indicate the positions in which Tob-phosphorylating activities were detected. (B) Lysates of PDGF-stimulated NIH3T3 cells were fractionated by gel-filtration chromatography. Each fraction was analyzed for the phosphorylating activity of GST–TobN (top) or GST–TobN3SA (middle) and was also assayed for Erk1/2 activity by immunoblotting with anti-phospho-Erk1/2 antibodies (bottom). Arrowheads indicate Tob-phosphorylating activities. Positions of molecular size markers are indicated on the left. (C) In vitro kinase assays of Erk1 (left) or Erk2 (right) immunoprecipitated from lysates of unstimulated (−) or PDGF-stimulated (+) NIH3T3 cells. Assays were performed with GST–TobN (Tob) and GST–TobN3SA (3SA) fusion proteins as substrates. (Lower panels) Exogenously added substrates monitored by Coomassie brilliant blue (CBB) staining.
To confirm this hypothesis, whole-cell lysates from PDGF-stimulated NIH3T3 cells were fractionated by gel filtration chromatography. Proteins in each fraction were probed with anti-phospho-Erk1/2 antibodies and subjected to in-gel kinase assay for TobN or TobN3SA protein phosphorylation activity. The gel filtration profile of Erk1 activity corresponded well with p44-mediated Tob phosphorylation, and the profile of Erk2 activity corresponded to p42-mediated Tob phosphorylation (Fig. 5B). All phosphorylation signals detected in the gel containing GST-Tob3SA were due to autophosphorylated, unidentified kinases, because these signals were observed in control experiments with no substrate in the gel (data not shown). An autophosphorylated 42-kD protein at a peak fraction (fraction 28) from the gel filtration chromatography had a retention time that was shorter than that of Erk2 (fraction 30; Fig. 5B, bottom panel). These findings strongly suggested that Erk1/2 catalyzed phosphorylation of Tob upon growth factor stimulation.
Erk1/2 immunoprecipitated from PDGF-stimulated NIH3T3 cells consistently phosphorylated GST-TobN but not GST-Tob3SA (Fig. 5C). In addition, chymotryptic phosphopeptide mapping of the GST-TobN fusion protein with or without Ser152Ala, Ser154Ala, Ser164Ala, Ser152,154Ala, Ser152,164Ala, Ser154,164Ala, or Ser152,154,164Ala substitution that were incubated with [γ-32P]ATP and Erk2 revealed that serine residues 152, 154, and 164 of GST-TobN were phosphorylated (data not shown). The result also suggested that Tob was phosphorylated at these residues by Erk1/2 in vivo. Note that the three peptides that did not contain Ser 152, Ser 154, and Ser 164 (Fig. 3D, p1, p7, and p9) were undetectable in these assays using GST-TobN. This suggested that the other phosphorylation sites were not present in the TobN sequence and were thus located in the C-terminal region of Tob. Alternatively, the activity of kinases that phosphorylate these sites was undetectable.
Reduction of Tob antiproliferative activity by phosphorylation
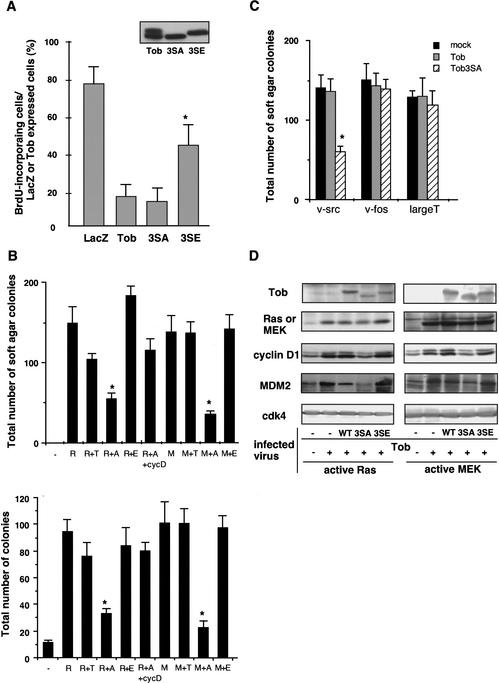
To examine the biological relevance of mitogen-induced Tob phosphorylation, we constructed Tob mutants in which serines at the Erk1/2 phosphorylation sites (Ser 152, Ser 154, and Ser 164) were substituted with glutamic acids. Glutamate can mimic phosphoserine (Morrison et al. 1993; Maciejewski et al. 1995); this is supported by the observation that migration of the mutant (Ser152,154,164Glu mutant: Tob3SE) and phosphorylated wild-type Tob in SDS-polyacrylamide gels was similar (Fig. 6A, inset). The Tob3SE protein thus appeared to mimic Tob phosphorylated at these same sites. Tob3SE as well as wild-type Tob and Tob3SA were expressed in serum-starved NIH3T3 cells, and BrdU incorporation was examined after serum refeeding. Cells microinjected with Tob3SE showed higher BrdU incorporation than those microinjected with Tob or Tob3SA (Fig. 6A). The expression levels of each mutant were similar among the transfected NIH3T3 cells (Fig. 6A, inset), suggesting that the stability as well as the efficiency of translation of each mutant were similar. Therefore, phosphorylation of Tob at Ser 152, Ser 154, and Ser 164 was likely to diminish its antiproliferative activity.
Figure 6.
Effects of wild-type and mutant Tob on Ras/MAPK-dependent proliferation and transformation. (A) Serum-starved NIH3T3 cells microinjected with pME-Tob, pME-Tob3SE, pME-Tob3SA, or pME-β-galactosidase were refed with serum and then immunostained for Tob, β-galactosidase, or BrdU. The graph shows the percentage of BrdU incorporation in NIH3T3 cells that expressed β-galactosidase (LacZ), Tob (Tob), Tob3SA (3SA), and Tob3SE (3SE). At least 70 cells were scored for each experiment. Values are shown as the mean ± SD of five experiments. (Inset) Immunoblot of the lysates from NIH3T3 cells transfected with plasmid encoding wild-type or mutant Tob proteins. (B) NIH3T3 cells infected with the retrovirus carrrying active Ras (R) or active MEK (M) together with retrovirus for Tob wild-type (T), 3SA (A), 3SE (E), or cyclin D1 (cycD). The cells were seeded in soft agar to examine anchorage-independent growth (upper). NIH3T3 cells infected with the indicated retrovirus and cultured in DMEM containing 1% serum in the presence of drug were examined for anchorage-dependent growth, and the number of drug-resistant colonies was counted (lower). Values are shown as the mean ± SD of four experiments. *P < 0.02 as compared with wild-type Tob. (C) Soft agar colony formation was performed as in B for the cells coinfected with the retrovirus carrying v-src, v-fos, or large T cDNA and the virus for wild-type Tob (gray) or Tob3SA (striped) or the control virus (black). *P < 0.02 as compared with wild-type Tob. (D) Aliquots of NIH3T3 cells infected with the indicated retrovirus were lysed, and the lysates were analyzed by immunoblotting with antibodies for the indicated proteins.
It should be noted, however, that wild-type Tob, like Tob3SA, inhibited G1 progression (Figs. 1,6). It appeared that microinjection of the cDNAs resulted in expression of large amounts of the protein products. Therefore, most of the wild-type Tob proteins expressed from microinjected plasmids were not phosphorylated upon serum stimulation because of the limitation of endogenous Erk1/2. In support of this, a large fraction of transfected Tob remained unphosphorylated upon mitogenic stimulation, whereas most endogenous Tob under the same condition was well phosphorylated (Figs. 2,3). A significant amount of exogenously introduced unphosphorylated Tob should have been able to suppress G1 progression. Moreover, the mutant Tob3SE was still able to suppress progression from the G0 phase through the G1/S transition to a certain extent. We assume that the inhibitory activity of Tob is negatively regulated by phosphorylation as well as degradation (see Discussion). The Tob protein overproduced by microinjection may remain present at the unusually high level in the mid-G1 phase, where the level of endogenous Tob becomes low, and, therefore, could be inhibitory to cell growth. Indeed, microinjection of a lesser amount of DNA (1/10 of that used in Fig. 6A) revealed Tob3SE had little inhibitory activity. Wild-type Tob has less inhibitory activity (66% BrdU incorporation as compared with control), whereas Tob3SA was still strongly inhibitory (21.5% BrdU incorporation as compared with control) in the same experimental condition (data not shown).
Inhibition of Ras/MAPK signaling by hypophosphorylated Tob
To further address the role of Tob phosphorylation in the regulation of cell growth, we tested the effects of Tob expression on cell transformation by oncogenic Ras and oncogenic MEK. In this experiment, we tried to express low levels of Tob so that all exogenous Tob was phosphorylated in the presence of active Ras or MEK. To do this, we used a retrovirus-mediated expression system. As shown in Figure 6D, only the hyperphosphorylated form of Tob was detected in NIH3T3 cells infected with retrovirus expressing wild-type Tob together with retrovirus expressing active Ras or active MEK. Similar levels of expression of Tob3SA and Tob3SE mutants were also achieved with the retrovirus system. Interestingly, wild-type Tob and Tob3SE had little or only a slight effect on active Ras- or active MEK-mediated cell transformation, which was assayed by the anchorage-independent growth of the cells (Fig. 6B, top). In contrast, Tob3SA largely inhibited the transformation. Similarly, by examining the effects of Tob on anchorage-dependent growth of the active Ras- or MEK-expressing cells, we showed that Tob3SA, but not wild-type or Tob3SE, significantly inhibited proliferation of these cells (Fig. 6B, bottom). The data indicated that phosphorylation of Tob diminished its otherwise antiproliferative activity, and that reduction of the antiproliferative activity was critical for active Ras- or MEK-mediated cell proliferation and transformation.
In similar experiments, we showed that Tob3SA suppressed v-_src_-dependent, but not v-_fos_- and _large T_-dependent cell transformation and cell proliferation (Fig. 6C; data not shown). Wild-type Tob was not inhibitory to the v-_src_-mediated cell transformation. There is a report that transformation of NIH3T3 cells by v-src is dependent on Ras activation (Stacey et al. 1991). This well explains why nonphosphorylatable Tob3SA but not wild-type Tob, which becomes phosphorylated through the Ras/MAPK pathway, could suppress the v-_src_-induced cell transformation. Inability of Tob to inhibit _large T_-dependent cell transformation suggested that the inhibitory activity of Tob was mediated through Rb, because _large T_-mediated cell transformation was directly coupled to Rb inactivation (DeCaprio et al. 1988).
Tob suppresses cyclin D1 expression
To address the molecular mechanism by which the Tob3SA mutant mimicking a hypophosphorylated form of Tob suppressed active Ras/MEK-mediated cell growth and transformation, we examined the expression levels of effector molecules downstream of Ras/MEK in the presence or absence of the wild-type and mutant Tob proteins. As reported previously (Liu et al. 1995; Ries et al. 2000), levels of cyclin D1 and MDM2 expression were elevated in active Ras- or MEK-transformed NIH3T3 cells (Fig. 6D). Importantly, retrovirus-mediated expression of the Tob3SA mutant significantly suppressed active Ras/MEK-induced cyclin D1 and MDM2 expression, whereas wild-type Tob and Tob3SE mutant showed little effect on their expression (Fig. 6D).
We then examined whether increment of cyclin D1 expression antagonized the inhibitory effect of Tob. As shown in Figure 7A, Tob was unable to impair G1 progression in the presence of exogenously coexpressed cyclin D1. Expression of cyclin E, which can functionally replace cyclin D1 (Geng et al. 1999), also rescued the cells from Tob-mediated G1 arrest. In contrast, cyclin B1, the M-phase cyclin, was unable to antagonize the antiproliferative activity of Tob. We also showed that suppression of Ras-induced transformation by the Tob3SA mutant was largely canceled by coexpression of exogenous cyclin D1 (Fig. 6B). Provided that Tob suppressed cell growth by blocking cyclin D1 expression, its antiproliferative activity would be lost in cells lacking Rb protein. Indeed, S-phase entry of _rb_−/− cells was little affected by Tob expression (Fig. 7A, right). As previously reported (Peeper et al. 1997; Guardavaccaro et al. 2000), S-phase entry of _rb_−/− cells was affected by p27 but not by PC3. Furthermore, S-phase entry of the _rb_−/− cells microinjected with Rb expression plasmid together with Tob plasmid was equivalent to 24% of those microinjected with Rb and LacZ plasmids (data not shown). Thus, reintroduction of Rb into the _rb_−/− cells restored Tob-mediated G1 arrest. Our data suggested that Tob was a molecular switch for the expression of targets of the Ras/MAPK pathway such as cyclin D1: phosphorylation of Tob at Ser 152, 154, and 164 was required for efficient induction of the cyclin D1 gene.
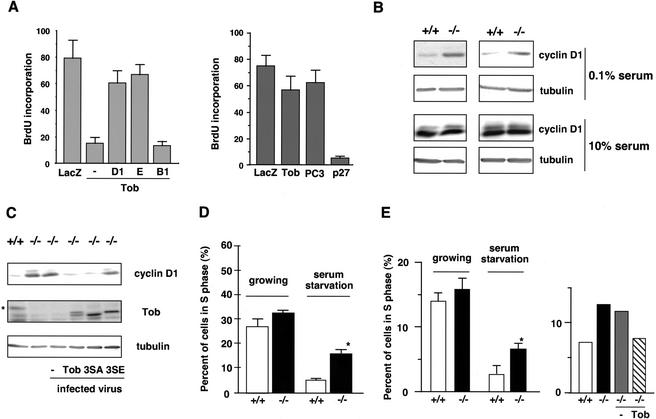
Figure 7.
Relation between cyclin D1 expression and Tob-induced growth arrest, and defects in _tob_−/− cells. (A) Serum-starved NIH3T3 cells (left) and _rb_−/− MEFs (right) were injected with the indicated expression vectors. After serum addition, BrdU incorporation was examined as shown in Figure 6A. (D1) cyclin D1, (E) cyclin E, (B1) cyclin B1. (B) Cell lysates prepared from serum-starved or asynchronously growing wild-type (+/+) or _tob_−/− (−/−) primary MEFs were immunoblotted for cyclin D1. Anti-α-tubulin immunoblotting was performed for loading control. (C) Cell lysates prepared from serum-starved wild-type (+/+) or _tob_−/− (−/−) MEFs were immunoblotted for cyclin D1, Tob, and α-tubulin. _tob_−/− MEFs were infected with Tob wild-type, 3SA, or 3SE retrovirus. An asterisk shows endogenous Tob proteins. Note that the apparent molecular weight of mouse Tob is higher than that of human Tob. (D) Established MEF cell lines (at passage 20) from wild-type (+/+) or _tob_−/− (−/−) primary MEFs were serum-starved for 72 h (0.1%). The graph shows the percentage of cells in S phase as determined by FACS. *P < 0.02 as compared with wild-type MEFs (n = 4). (E) S-phase population in primary osteoblast culture prepared from wild-type (+/+) and _tob_−/− (−/−) mice (left) was analyzed in the serum-starved condition. The S-phase population in _tob_−/− osteoblasts infected with control retrovirus (−) or the retrovirus carrying wild-type Tob was analyzed after 72 h of serum starvation (right). The average of two experiments is shown.
Increased cyclin D1 expression in tob−/− MEFs
Because Tob expression inhibited cyclin D1 expression unless it was phosphorylated, we hypothesized that cyclin D1 might be expressed in quiescent cells that lack the expression of Tob. To examine this possibility, we analyzed expression levels of cyclin D1 in embryonic fibroblasts from wild-type mice (wt MEFs) and _tob_−/− mice (_tob_−/− MEFs) by immunoblotting. In the serum-starved condition, cyclin D1 expression was hardly detectable in wild-type MEFs, whereas the level of cyclin D1 was significantly elevated in _tob_−/− MEFs (Fig. 7B). Expression of wild-type Tob and hypophosphorylated Tob3SA, but not Tob3SE mimicking phosphorylated Tob, by retrovirus-mediated gene transfer restored the suppression of cyclin D1 expression in the serum-starved condition (Fig. 7C). These data suggested that Tob was involved in suppression of cyclin D1 expression in the quiescent cells and mitogen-induced Tob phosphorylation canceled the suppression. Under growing conditions, expression of cyclin D1 in _tob_−/− MEFs was equivalent to that in wild-type MEFs (Fig. 7B).
Furthermore, we found that established cell lines from _tob_−/− MEFs (_tob_−/− 3T3 cells at the 20th passage) entered into S phase even in the serum-starved condition much more frequently than did the established cell lines from wild-type MEFs (Fig. 7D). Reintroduction of Tob in the _tob_−/− 3T3 cells resulted in only a partial recovery of growth arrest. This might be because of the complementary function of other Tob family members that might have been inactivated in the _tob_−/− 3T3 cells. It was also possible that other growth regulatory genes (p53, Rb, etc.) were inactivated in the _tob_−/− 3T3 cells. The complementation was also suggested by our finding that the frequency of S-phase entry of _tob_−/− MEFs was similar to that of wild-type MEFs at their early passages. Because expression of tob2 is low in osteoblasts, and because osteoblast proliferation and differentiation were abrogated in _tob_−/− mice (Yoshida et al. 2000), we analyzed cell cycle distribution in _tob_−/− osteoblasts. In the serum-starved condition, the population of _tob_−/− primary osteoblasts in S phase was significantly higher than that of wild-type osteoblasts (Fig. 7E). Reexpression of Tob in _tob_−/− primary osteoblasts resulted in the decrease of S-phase cells to a level similar to that of wild-type osteoblasts. These data suggested that Tob drove cells in quiescence in the absence of growth stimulation by suppressing cyclin D1 expression.
Discussion
We show here that the Tob protein is a novel substrate of Erk1/2 MAPKs and that it plays an important role in controlling mitogenic-stimulation-dependent cell growth.Tob appears to function upstream of Rb, because Tob is unable to inhibit growth of _rb_−/− cells or large T-dependent cell transformation. Our data clearly show that Erk1/2 phosphorylates Ser 152, Ser 154, and Ser 164, which are critical for regulation of its antiproliferative activity (discussed below). In addition, phosphopeptide mapping of 32P-labeled Tob revealed a few more sites that are phosphorylated upon PDGF stimulation (Fig. 3D). Phosphorylation of the additional sites may also contribute to the regulation. Kinases responsible for phosphorylation of these additional sites have not been identified. p90rsk1, which is activated by Erk1/2 (Dalby et al. 1998), is one of the candidates, because it is able to phosphorylate Tob in vitro (Suzuki et al. 2001). Similar to the Rb protein, which is phosphorylated at multiple sites by several Cdks and thereby binds to distinct proteins (Knudsen and Wang 1996), Tob, being phosphorylated at multiple sites, may interact with various proteins to show several distinct roles.
Tob expression declines gradually after 3 h of serum stimulation (early G1; Fig. 1B). The decline in Tob expression may be caused by degradation or decreased production of Tob. Because Tob becomes stabilized by proteasome inhibitor (data not shown), ubiquitin/proteasome-dependent degradation appears to regulate the level of Tob. Our unpublished data show that okadaic acid treatment results in rapid degradation of Tob, suggesting that phosphorylation of Tob plays a part in controlling its stability. However, the significant time lag between Erk1/2-mediated Tob phosphorylation and the decrease in Tob expression (Fig. 1) suggests that growth-signal-induced rapid phosphorylation of Tob is not directly relevant to its degradation. Consistently, wild-type Tob and Tob3SA degraded with a similar half-life upon PDGF stimulation (data not shown). A likely scenario is that Tob might be first phosphorylated at multiple sites, including Ser 152, Ser 154, and Ser 164, upon growth factor stimulation and would be further phosphorylated at the other sites to induce its degradation. Our present data suggest that initial phosphorylation is an acute response by which the antiproliferative activity of Tob is regulated. Degradation of Tob may be necessary for cells to progress through late G1.
Growth-factor stimulation induces activation of RTK and subsequent activation of Ras. Multiple Ras effector pathways contribute to G1 progression by, for example, inactivating cyclin-dependent kinase inhibitor p27 (Rivard et al. 1999) or inducing G1 cyclin expression (Peeper et al. 1997). Apparently, the MEK/MAPK pathway, which is activated by Ras, is involved in the induction of a variety of immediate early genes such as fos and genes relevant to G1 progression. Activation of the Erk1/2 cascade is necessary and sufficient to fully induce cyclin D1 (Lavoie et al. 1996), which is required for Cdk4 activation. However, the precise molecular mechanism by which cyclin D1 expression is regulated has been to be elucidated. Recent evidence shows that ectopic expression of PC3 inhibits cyclin D1 transcription (Guardavaccaro et al. 2000), although the molecular mechanism underlying the inhibition is to be further examined. It remains obscure whether PC3 is involved in regulation of Ras/MAPK-mediated cyclin D1 expression in the context of growth-factor-induced cell growth. Rather, it is suggested that PC3 might play a role in DNA-damage-induced cyclin D1 suppression (Rouault et al. 1996; Guardavaccaro et al. 2000).
Our present findings suggest that Tob is an important molecule that links Ras/MAPK activities to regulation of cyclin D1 expression and eventually to cell cycle progression of growth-stimulated cells through G1. Three experimental observations support the idea that Tob downstream of Erk1/2 regulates cyclin D1 expression. First, the level of cyclin D1 is suppressed by a Tob mutant in which serines of the Erk1/2 target are mutated to alanines but is not suppressed by phosphorylated wild-type Tob or mutant Tob having phosphoserine-mimicking glutamates at the Erk1/2 target sites (Fig. 6B). Second, cyclin D1 expression is evident in _tob_−/− MEFs unlike its suppression in wild-type MEFs in a quiescent condition (Fig. 7B). However, the level of cyclin D1 in serum-starved _tob_−/− MEFs appeared to be not sufficient for their efficient progression into S phase, probably owing at least in part to functional redundancy of the Tob family proteins. Third, coexpression of cyclin D1 counteracted the Tob-mediated growth inhibition (Figs. 6B, 7A). Thus, inhibition of oncogenic Ras- or oncogenic MEK-induced cell transformation by hypophosphorylated mutant Tob (Tob3SA, Fig. 6B) could be largely caused by the suppression of cyclin D1. Consistently, growth of _rb_−/− cells was not affected by Tob (Fig. 7A). Furthermore, an S-phase population is significantly higher in _tob_−/− cells than in wild-type cells in a serum-starved condition (Fig. 7E). We conclude that Tob functions to maintain cells in a quiescent state in the absence of mitogenic stimulation. Consistent with this conclusion, tob mRNA was highly expressed in anergic T cells and quiescent T cells (Tzachanis et al. 2001). It remains to be considered whether some stresses such as DNA damage could induce Tob expression. Note that the level of Tob expression resumes at G2 and M phase. It is possible that the newly synthesized Tob protein is related to whether cells enter the next round of the cell cycle or cease to proliferate. Interestingly, Saccharomyces cerevisiae with a mutation in the gene encoding Caf1 (CCR4-associated factor 1), whose mammalian homolog interacts with Tob, is unable to proceed from M phase to G1 phase (Liu et al. 1997).
An important question to be addressed is how Tob suppresses cyclin D1 expression downstream of Ras/MAPK signaling. Tob would not function as a competitive inhibitor of the substrates for Erk-mediated phosphorylation, because Tob overexpression did not inhibit Erk phosphorylation of Elk1 or Ets1 (data not shown). Accumulating data show that Tob family proteins are associated with the transcription machinery. For example, they interact with Caf1 (Draper et al. 1995; Bogdan et al. 1998; Rouault et al. 1998; Ikematsu et al. 1999). Yeast Caf1 is an important component of transcriptional regulatory complex CCR4–NOT, which is conserved from yeast to humans (Albert et al. 2000). The CCR4–NOT complex is likely to participate either negatively or positively in the control of specific sets of genes including those involved in the late mitotic phase of the cell cycle (Liu et al. 1997, 1998; Bai et al. 1999). BTG1 and BTG2 interact with homeodomain-containing transcription factor Hoxb9 (Prevot et al. 2000), resulting in stimulation of Hoxb9-mediated transcription. BTG1 and BTG2 also regulate estrogen receptor α-mediated transcription (Prevot et al. 2001). In addition, Tob interacts with Smad family proteins, which are capable of binding to DNA (Yoshida et al. 2000). These observations strongly suggest that Tob family proteins are novel regulators of gene transcription. Tob could associate with a specific transcription factor or cofactor to regulate cyclin D1 expression. Intriguingly, BTG1 and BTG2 interact with PRMT (protein arginine N-methyltransferase), and the interaction positively modulates the activity of PRMT (Lin et al. 1996). Although the role of PRMT remains yet to be established, there are reports showing that methylation of histone H4 by PRMT1 facilitates activity of nuclear androgen receptor (Strahl et al. 2001; Wang et al. 2001). A possibility that Tob interacts with PRMT and regulates transcription of the cyclin D1 gene or other genes remains to be investigated.
Our data reveal that the N-terminal half of Tob is required for antiproliferative activity, and we provide evidence that phosphorylation of Tob by Erk1/2 results in loss of this activity. Ability of Tob to suppress cyclin D1 expression appears to be relevant to its antiproliferative activity. The importance of Tob function in cell growth regulation is strongly supported by our present observation that oncogenic Ras-induced cell transformation is suppressed by the hypophosphorylated form of Tob as well as our unpublished data showing that _tob_−/− mice develop various types of tumors with high frequency (Y. Yoshida, unpubl.). We propose here that Tob is a novel effector molecule functioning downstream of the Ras/MAPK pathway and regulating the G0/G1 transition.
Materials and methods
Cell culture assays and transgene expression
NIH3T3 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% calf serum. Wild-type, _tob_−/−, and _rb_−/− MEFs were grown in DMEM containing 10% fetal calf serum. Preparation and culture of primary osteoblasts were previously described (Yoshida et al. 2000). To obtain quiescent cells, cells were washed three times with serum-free DMEM followed by culture for 24–48 h in DMEM containing 0.05% serum. Cells were restimulated with 10% serum or growth factors (20 ng/mL PDGF-BB, 10 ng/mL EGF, 10 ng/mL TNFα, and 10 ng/mL TGFβ) for 15 min. For transfection, NIH3T3 cells in 60-mm dishes were transfected with 5 μg of DNA using SuperFect (QIAGEN) or GenePORTER (Gene Therapy Systems). Then, 16 h after transfection, the cells were serum-starved for an additional 24 h. The cells were next exposed to serum or PDGF for 10 min. NIH3T3 cells were infected with recombinant retrovirus and cultured in the presence of drug (G418 and puromycin). After drug selection, ∼103 cells were seeded in soft agar. Microinjection was carried out as described (Ikematsu et al. 1999). GST-fusion proteins were produced in Escherichia coli DH5α and purified according to the protocols suggested by the manufacturer.
Antibodies, growth factors, and inhibitors
The anti-Tob polyclonal antibodies (Matsuda et al. 1996) were purified by GST–TobN-conjugated affinity column chromatography. Anti-Tob monoclonal antibody (4B1) raised by immunizing mice with the GST–TobN protein was obtained from IBL. The anti-Erk1 polyclonal antibodies (C-16), anti-Erk2 polyclonal antibodies (C-14), anti-Rb polyclonal antibodies (C-15), and anti-cyclin D1 monoclonal antibody (72–13G) were purchased from Santa Cruz Biotechnology. The anti-phospho-Erk1/2 polyclonal antibodies were from New England Biolabs. Anti-phospho-Tob antibodies were prepared by immunizing rabbits with synthetic peptides CASSVSSS*PS*PPFGHS (for Ser 152 and Ser 154), or CFGHSAAVS*PTFMPRS (for Ser 164; S* indicates phosphorylated serine residue) and were affinity-purified using a phosphopeptide-conjugated column. PD98059 and SB203580 were from Calbiochem; PDGF-BB was from Diaclone research; EGF, TNFα, TGFβ, AG1296, wortmannin, and rapamycin were from Sigma; and H7 was from Seikagaku Corporation. Cells were incubated with the kinase inhibitors for 3 h at the following concentrations: 50 μM PD98059; 25 μM SB203580; 25 μM AG1296; 10 μM H7; 1 μM wortmannin; and 1 μM rapamycin.
Construction of plasmids
The full-length tob cDNA was cloned into a pME18S vector (Ikematsu et al. 1999), and the resulting plasmid was termed pME-Tob. Tob point mutants were generated by site-directed mutagenesis following Kunkel's method (Kunkel and Soni 1988). Tob deletion mutants were generated by overlap extension PCR. Expression plasmids for the GST-fusion proteins were constructed in the pGEX-5X-1 vector (Pharmacia Biotech). Wild-type dominant-negative, and constitutive active MAPKK, and dominant-negative MAPK in pME18S, and dominant-negative JNK in pcDNA3 were provided by E. Nishida (Kyoto University, Kyoto, Japan) and Y. Gotoh (Tokyo University, Tokyo, Japan). The cDNAs of Ras (wild-type), RasS17N (dominant-negative), RasG12V (constitutive active), cyclin B1, cyclin D1, cyclin E, and Tob mutants were cloned into the pME18S vector. To obtain recombinant retroviruses carrying cDNAs for active MAPKK, active Ras, cyclin D1, and Tob and its mutants, each cDNA was inserted between the LTR sequences of expression plasmid. The resulting plasmids were transfected into platE cells.
Protein analysis
For immunoblotting, cells were lysed with RIPA buffer (50 mM Tris-HCl, 1% Triton X-100, 0.1% SDS, 0.15 M NaCl, 10 mM sodium pyrophosphate, 10 mM sodium fluoride, 0.1 mM sodium orthovanadate, 1 mM EDTA at pH 7.5) supplemented with protease inhibitor cocktails (Sigma). Cell lysates were electrophoresed in SDS-polyacrylamide gel and then transferred onto Immobilon-P membranes (Millipore). Immunoblots were probed with the appropriate antibodies as described (Ikematsu et al. 1999). To assay for Erk kinase activity, cells were lysed in RIPA buffer after serum starvation with or without PDGF stimulation. Erk1/2 immunoprecipitates from NIH3T3 cells were washed successively with lysis buffer, 50 mM HEPES-NaOH (pH 7.4), and kinase buffer (50 mM HEPES-NaOH, 10 mM MgCl2, 3 mM MnCl2, 25 mM β-glycerophosphate, 10 mM sodium fluoride, 1 mM sodium vanadate, 1 mM DTT at pH 7.4). Kinase reactions were carried out at 30°C for 30 min after adding [γ-32P]ATP (NEN, 0.2 mCi/mL) and GST-fusion proteins (final concentration at 0.2 mg/mL). For in-gel kinase assays, lysates from unstimulated or PDGF-stimulated NIH3T3 cells were separated by electrophoresis on SDS-polyacrylamide gel with resolving gels containing GST–TobN or GST–TobN3SA fusion proteins (final concentration at 1 mg/mL). The in-gel kinase assays were carried out as previously described (Fukunaga and Hunter 1997). For phosphatase treatment, cell lysates were diluted 1:3 with 50 mM Tris-HCl (pH 8.0) solution containing 7 U of CIAP (Boehringer Mannheim) and incubated at 37°C for 90 min. The reaction was inhibited with NaF (50 mM), sodium vanadate (1 mM), sodium diphosphate (10 mM), and okadaic acid (50 nM).
Metabolic labeling and peptide mapping analysis
NIH3T3 cells transfected with wild-type or mutant Tob expression plasmids were serum-starved and metabolically labeled with [32P]orthophosphate (NEN, 1 mCi/mL) in phosphate-free DMEM including 0.1% dialyzed calf serum for 6 h. Then cells were stimulated with PDGF for 10 min and lysed in RIPA buffer. Tob was immunoprecipitated from the lysates with anti-Tob monoclonal antibody, and the immunoprecipitates were separated by SDS-polyacrylamide gel electrophoresis. The 32P-labeled Tob proteins were excised from the SDS-polyacrylamide gel and digested with chymotrypsin (0.2 mg/mL; Worthington Biochemical) as described (Boyle et al. 1991). The chymotryptic peptide samples were electrophoresed for 40 min at 1.0 kV in pH 1.9 buffer with the HTLE7000 apparatus (CBS Scientific); the plates were air-dried and then placed in tanks for ascending chromatography using phosphochromo buffer. After ascending chromatography, the plates were air-dried and exposed to presensitized Kodak XAR film.
Acknowledgments
We thank J. Inoue, K. Semba, Y. Yamanashi, S. Matsuda, J. Fujimoto, and T. Miyasaka for their valuable discussions. We also thank T. Seito (Immuno-Biological Laboratories) for providing us with the anti-Tob monoclonal antibody; E. Nishida, Y. Gotoh, S. Yokoyama, and M. Shirouzu for expression constructs of Ras, MEK, and MAPKs; R. Weinberg for _rb_−/− MEFs and pBabe-LargeT; T. Akagi for pBabe-v-Src; T. Akiyama for cyclin D1 and cyclin E; T. Kishimoto for cyclin B1 cDNA; J.P. Rouault for pFBR; and H. Fujiki for okadaic acid. We thank M. Sudol for reviewing the manuscript. This work was supported in part by a grant for Advanced Cancer Research from the Ministry of Education, Science, Sports, and Culture of Japan and from the Organization for Pharmaceutical Safety and Research of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tyamamot@ims.u-tokyo.ac.jp; FAX 81-3-5449-5413.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.962802.
References
- Adam LP, Hathaway DR. Identification of mitogen-activated protein kinase phosphorylation sequences in mammalian h-Caldesmon. FEBS Lett. 1993;322:56–60. doi: 10.1016/0014-5793(93)81110-l. [DOI] [PubMed] [Google Scholar]
- Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, Timmers HT. Isolation and characterization of human orthologs of yeast CCR4–NOT complex subunits. Nucleic Acids Res. 2000;28:809–817. doi: 10.1093/nar/28.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol Cell Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes & Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- Bogdan JA, Adams-Burton C, Pedicord DL, Sukovich DA, Benfield PA, Corjay MH, Stoltenborg JK, Dicker IB. Human carbon catabolite repressor protein (CCR4)-associative factor 1: Cloning, expression and characterization of its interaction with the B-cell translocation protein BTG1. Biochem J. 1998;336:471–481. doi: 10.1042/bj3360471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Bradbury A, Possenti R, Shooter EM, Tirone F. Molecular cloning of PC3, a putatively secreted protein whose mRNA is induced by nerve growth factor and depolarization. Proc Natl Acad Sci. 1991;88:3353–3357. doi: 10.1073/pnas.88.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondello JM, McKenzie FR, Sun H, Tonks NK, Pouyssegur J. Constitutive MAP kinase phosphatase (MKP-1) expression blocks G1 specific gene transcription and S-phase entry in fibroblasts. Oncogene. 1995;10:1895–1904. [PubMed] [Google Scholar]
- Burgering BM, Freed E, van der Voorn L, McCormick F, Bos JL. Platelet-derived growth factor-induced p21ras-mediated signaling is independent of platelet-derived growth factor receptor interaction with GTPase-activating protein or phosphatidylinositol-3-kinase. Cell Growth Differ. 1994;5:341–347. [PubMed] [Google Scholar]
- Coffer P, de Jonge M, Mettouchi A, Binetruy B, Ghysdael J, Kruijer W. junB promoter regulation: Ras mediated transactivation by c-Ets-1 and c-Ets-2. Oncogene. 1994;9:911–921. [PubMed] [Google Scholar]
- Dalby KN, Morrice N, Caudwell FB, Avruch J, Cohen P. Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J Biol Chem. 1998;273:1496–1505. doi: 10.1074/jbc.273.3.1496. [DOI] [PubMed] [Google Scholar]
- Davis RJ. Transcriptional regulation by MAP kinases. Mol Reprod Dev. 1995;42:459–467. doi: 10.1002/mrd.1080420414. [DOI] [PubMed] [Google Scholar]
- DeCaprio JA, Ludlow JW, Figge J, Shew JY, Huang CM, Lee WH, Marsilio E, Paucha E, Livingston DM. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- Draper MP, Salvadore C, Denis CL. Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol Cell Biol. 1995;15:3487–3495. doi: 10.1128/mcb.15.7.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher BS, Lim RW, Varnum BC, Kujubu DA, Koski RA, Herschman HR. Structure and expression of TIS21, a primary response gene induced by growth factors and tumor promoters. J Biol Chem. 1991;266:14511–14518. [PubMed] [Google Scholar]
- Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Whoriskey W, Park MY, Bronson RT, Medema RH, Li T, Weinberg RA, Sicinski P. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell. 1999;97:767–777. doi: 10.1016/s0092-8674(00)80788-6. [DOI] [PubMed] [Google Scholar]
- Guardavaccaro D, Corrente G, Covone F, Micheli L, D'Agnano I, Starace G, Caruso M, Tirone F. Arrest of G(1)-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol Cell Biol. 2000;20:1797–1815. doi: 10.1128/mcb.20.5.1797-1815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guehenneux F, Duret L, Callanan MB, Bouhas R, Hayette S, Berthet C, Samarut C, Rimokh R, Birot AM, Wang Q, et al. Cloning of the mouse BTG3 gene and definition of a new gene family (the BTG family) involved in the negative control of the cell cycle. Leukemia. 1997;11:370–375. doi: 10.1038/sj.leu.2400599. [DOI] [PubMed] [Google Scholar]
- Hall FL, Vulliet PR. Proline-directed protein phosphorylation and cell cycle regulation. Curr Opin Cell Biol. 1991;3:176–184. doi: 10.1016/0955-0674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- Iacopetti P, Michelini M, Stuckmann I, Oback B, Aaku-Saraste E, Huttner WB. Expression of the antiproliferative gene TIS21 at the onset of neurogenesis identifies single neuroepithelial cells that switch from proliferative to neuron-generating division. Proc Natl Acad Sci. 1999;96:4639–4644. doi: 10.1073/pnas.96.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu N, Yoshida Y, Kawamura-Tsuzuku J, Ohsugi M, Onda M, Hirai M, Fujimoto J, Yamamoto T. Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene. 1999;18:7432–7441. doi: 10.1038/sj.onc.1203193. [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Wang JY. Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J Biol Chem. 1996;271:8313–8320. doi: 10.1074/jbc.271.14.8313. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- Lavoie JN, L'Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- Lim IK, Lee MS, Ryu MS, Park TJ, Fujiki H, Eguchi H, Paik WK. Induction of growth inhibition of 293 cells by downregulation of the cyclin E and cyclin-dependent kinase 4 proteins due to overexpression of TIS21. Mol Carcinog. 1998;23:25–35. doi: 10.1002/(sici)1098-2744(199809)23:1<25::aid-mc4>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- Liu HY, Toyn JH, Chiang YC, Draper MP, Johnston LH, Denis CL. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 1997;16:5289–5298. doi: 10.1093/emboj/16.17.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Chao JR, Jiang MC, Ng SY, Yen JJ, Yang-Yen HF. Ras transformation results in an elevated level of cyclin D1 and acceleration of G1 progression in NIH 3T3 cells. Mol Cell Biol. 1995;15:3654–3663. doi: 10.1128/mcb.15.7.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewski PM, Peterson FC, Anderson PJ, Brooks CL. Mutation of serine 90 to glutamic acid mimics phosphorylation of bovine prolactin. J Biol Chem. 1995;270:27661–27665. doi: 10.1074/jbc.270.46.27661. [DOI] [PubMed] [Google Scholar]
- Marshall CJ. MAP kinase kinase kinase, MAP kinase kinase and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Kawamura-Tsuzuku J, Ohsugi M, Yoshida M, Emi M, Nakamura Y, Onda M, Yoshida Y, Nishiyama A, Yamamoto T. Tob, a novel protein that interacts with p185erbB2, is associated with anti-proliferative activity. Oncogene. 1996;12:705–713. [PubMed] [Google Scholar]
- Montagnoli A, Guardavaccaro D, Starace G, Tirone F. Overexpression of the nerve growth factor-inducible PC3 immediate early gene is associated with growth inhibition. Cell Growth Differ. 1996;7:1327–1336. [PubMed] [Google Scholar]
- Morrison P, Takishima K, Rosner MR. Role of threonine residues in regulation of the epidermal growth factor receptor by protein kinase C and mitogen-activated protein kinase. J Biol Chem. 1993;268:15536–15543. [PubMed] [Google Scholar]
- Peeper DS, Upton TM, Ladha MH, Neuman E, Zalvide J, Bernards R, DeCaprio JA, Ewen ME. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature. 1997;386:177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- Prevot D, Voeltzel T, Birot AM, Morel AP, Rostan MC, Magaud JP, Corbo L. The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J Biol Chem. 2000;275:147–153. doi: 10.1074/jbc.275.1.147. [DOI] [PubMed] [Google Scholar]
- Prevot D, Morel AP, Voeltzel T, Rostan MC, Rimokh R, Magaud JP, Corbo L. Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: Involvement in estrogen receptor α signaling pathway. J Biol Chem. 2001;276:9640–9648. doi: 10.1074/jbc.M008201200. [DOI] [PubMed] [Google Scholar]
- Ries S, Biederer C, Woods D, Shifman O, Shirasawa S, Sasazuki T, McMahon M, Oren M, McCormick F. Opposing effects of Ras on p53: Transcriptional activation of mdm2 and induction of p19ARF. Cell. 2000;103:321–330. doi: 10.1016/s0092-8674(00)00123-9. [DOI] [PubMed] [Google Scholar]
- Rivard N, Boucher MJ, Asselin C, L'Allemain G. MAP kinase cascade is required for p27 downregulation and S phase entry in fibroblasts and epithelial cells. Am J Physiol. 1999;277:C652–C664. doi: 10.1152/ajpcell.1999.277.4.C652. [DOI] [PubMed] [Google Scholar]
- Rouault JP, Rimokh R, Tessa C, Paranhos G, Ffrench M, Duret L, Garoccio M, Germain D, Samarut J, Magaud JP. BTG1, a member of a new family of antiproliferative genes. EMBO J. 1992;11:1663–1670. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault JP, Falette N, Guehenneux F, Guillot C, Rimokh R, Wang Q, Berthet C, Moyret-Lalle C, Savatier P, Pain B, et al. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- Rouault JP, Prevot D, Berthet C, Birot AM, Billaud M, Magaud JP, Corbo L. Interaction of BTG1 and p53-regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J Biol Chem. 1998;273:22563–22569. doi: 10.1074/jbc.273.35.22563. [DOI] [PubMed] [Google Scholar]
- Stacey DW, Kung HF. Transformation of NIH 3T3 cells by microinjection of Ha-ras p21 protein. Nature. 1984;310:508–511. doi: 10.1038/310508a0. [DOI] [PubMed] [Google Scholar]
- Stacey DW, Roudebush M, Day R, Mosser SD, Gibbs JB, Feig LA. Dominant inhibitory Ras mutants demonstrate the requirement for Ras activity in the action of tyrosine kinase oncogenes. Oncogene. 1991;6:2297–2304. [PubMed] [Google Scholar]
- Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Matsuda S, Tsuzuku JK, Yoshida Y, Yamamoto T. A serine/threonine kinase p90rsk1 phosphorylates the anti-proliferative protein Tob. Genes Cells. 2001;6:131–138. doi: 10.1046/j.1365-2443.2001.00406.x. [DOI] [PubMed] [Google Scholar]
- Tzachanis D, Freeman GJ, Hirano N, van Puijenbroek AA, Delfs MW, Berezovskaya A, Nadler LM, Boussiotis VA. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat Immunol. 2001;2:1174–1182. doi: 10.1038/ni730. [DOI] [PubMed] [Google Scholar]
- Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Matsuda S, Ikematsu N, Kawamura-Tsuzuku J, Inazawa J, Umemori H, Yamamoto T. ANA, a novel member of Tob/BTG1 family, is expressed in the ventricular zone of the developing central nervous system. Oncogene. 1998;16:2687–2693. doi: 10.1038/sj.onc.1201805. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Tanaka S, Umemori H, Minowa O, Usui M, Ikematsu N, Hosoda E, Imamura T, Kuno J, Yamashita T, et al. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell. 2000;103:1085–1097. doi: 10.1016/s0092-8674(00)00211-7. [DOI] [PubMed] [Google Scholar]