A molecular link between gene-specific and chromosome-wide transcriptional repression (original) (raw)
Abstract
Gene-specific and chromosome-wide mechanisms of transcriptional regulation control development in multicellular organisms. SDC-2, the determinant of hermaphrodite fate in Caenorhabditis elegans, is a paradigm for both modes of regulation. SDC-2 represses transcription of X chromosomes to achieve dosage compensation, and it also represses the male sex-determination gene her-1 to elicit hermaphrodite differentiation. We show here that SDC-2 recruits the entire dosage compensation complex to her-1, directing this _X_-chromosome repression machinery to silence an individual, autosomal gene. Functional dissection of her-1 in vivo revealed DNA recognition elements required for SDC-2 binding, recruitment of the dosage compensation complex, and transcriptional repression. Elements within her-1 differed in location, sequence, and strength of repression, implying that the dosage compensation complex may regulate transcription along the X chromosome using diverse recognition elements that play distinct roles in repression.
Keywords: Dosage compensation, sex determination, transcriptional repression, chromatin, C. elegans
The complex patterns of gene expression that control development are established by multiple regulatory mechanisms that operate locally on individual genes or globally across entire chromosomes or subchromosomal domains. These gene-specific and chromosome-wide modes of regulation are generally controlled by different protein complexes; however, a small number of global repressors have also been shown to function as local silencers of individual genes (Dawes et al. 1999; Moazed 2001; Nielsen et al. 2001). For example, the Sir proteins of Saccharomyces cerevisiae repress transcription at individual silent mating-type loci as well as telomeric regions of chromosomes (Aparicio et al. 1991; Moretti et al. 1994). The SDC-2 (sex-determination and dosage compensation) chromatin-binding protein of Caenorhabditis elegans has the versatility to repress transcription of an autosomal sex-determination gene by 20-fold and the entire X chromosome by twofold (Nusbaum and Meyer 1989; Trent et al. 1991; Dawes et al. 1999; Meyer 2000). Such dual-function regulatory components provide unique opportunities to explore mechanisms of local and global regulation by analyzing the more tractable mechanisms of gene-specific regulation.
SDC-2 coordinates all hermaphrodite-specific aspects of C. elegans development (Nusbaum and Meyer 1989). Acting as a gene-specific repressor, SDC-2 induces hermaphrodite sexual differentiation in XX animals by repressing transcription of the male (XO) sex-determining gene her-1 (Trent et al. 1991; Dawes et al. 1999). Acting simultaneously as a chromosome-wide repressor, SDC-2 activates dosage compensation by triggering the assembly of a specialized protein complex onto hermaphrodite X chromosomes to reduce _X_-linked gene expression by half (Dawes et al. 1999). The dosage compensation complex (Chuang et al. 1996; Lieb et al. 1996, 1998) resembles the condensin complex, which drives mitotic chromosome compaction in vitro (Koshland and Strunnikov 1996; Hirano 2000). _X_-chromosome repression is essential and equalizes gene expression between the sexes. Failure to dosage compensate causes hermaphrodite lethality.
Here we show that SDC-2 recruits all known components of the _X_-chromosome dosage compensation complex to the autosomal her-1 gene, allowing us to perform a detailed dissection in vivo of DNA recognition elements essential for SDC binding, recruitment of the dosage compensation complex, and transcriptional regulation. Within her-1, diverse DNA elements specify SDC binding and distinct levels of repression. Thus, a chromosome-wide repression complex can achieve different degrees of repression by associating with diverse DNA targets.
Results
SDC proteins function as a complex in vivo to repress her-1 and X chromosomes
How does SDC-2 discriminate between different targets and repress them to different degrees? To answer this question, we first identified proteins that function with SDC-2 at her-1. sdc-2 was known to interact genetically with sdc-1 and sdc-3 to implement sex determination and dosage compensation in XX animals (Villeneuve and Meyer 1990; Davis and Meyer 1997; Dawes et al. 1999), but the precise molecular roles of SDC-1 and SDC-3 were not understood. Here we present three lines of evidence that the three SDC proteins form a complex in vivo to repress her-1 and X chromosomes directly.
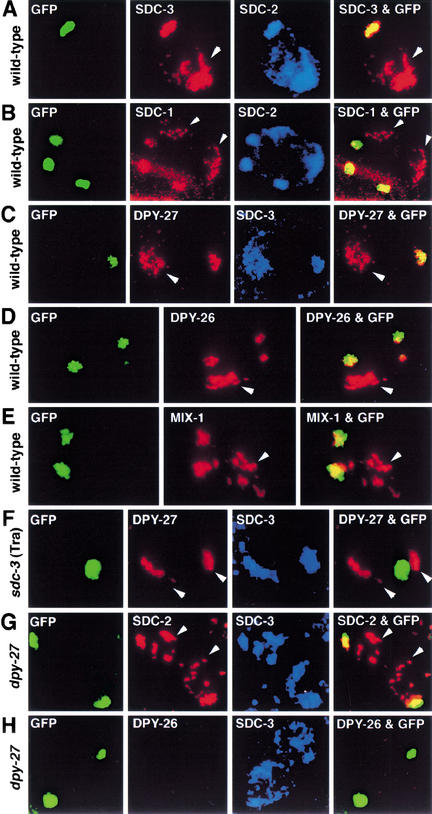
First, SDC-1, SDC-2, and SDC-3 all colocalize to X chromosomes and to her-1 regulatory regions in vivo. Hermaphrodites carrying multiple tandem copies of her-1 regulatory regions on GFP-tagged extrachromosomal arrays were stained with affinity-purified SDC antibodies (see Materials and Methods). SDC protein localization was assessed in adult intestinal nuclei, whose large size and polyploid DNA content facilitate the assay. The highly charged SDC-2 protein, which bears a coiled-coil motif, localized to X chromosomes and her-1 arrays in adult gut cells (Fig. 1A), as shown previously in embryos (Dawes et al. 1999), thus validating the assay. The zinc-finger proteins SDC-1 and SDC-3 (Nonet and Meyer 1991; Klein and Meyer 1993) colocalized with SDC-2 at her-1 and on X chromosomes (Fig. 1A,B), consistent with a direct role for these proteins in her-1 repression and dosage compensation. In XX animals carrying an sdc-3(Tra) mutation, localization of SDC-1, SDC-2, and SDC-3 to her-1 was greatly reduced, but localization to the X chromosome appeared unaffected (Fig. 1F; data not shown), consistent with the mutation impairing sex determination but not dosage compensation (DeLong et al. 1993). sdc-3(Tra) derepresses her-1 transcription, causing 100% of XX animals to be severely masculinized by disrupting a putative ATP-binding motif in SDC-3 (DeLong et al. 1993; Klein and Meyer 1993). Therefore, SDC-1, SDC-2, and SDC-3 are localized appropriately to achieve both gene-specific and chromosome-wide repression.
Figure 1.
The _X_-chromosome dosage compensation machinery localizes to her-1 regulatory regions in vivo. Confocal images of an individual gut nucleus (A_–_F) or an embryonic nucleus (G,H) from wild-type or mutant [sdc-3(Tra) or _dpy-27_] XX animals immunostained with SDC, DPY, or MIX antibodies, as indicated in each panel. The nuclei contain extrachromosomal DNA arrays carrying multiple copies of her-1 regulatory regions (plasmid pHD25 of Fig. 3A), lac operator repeats (lacO), and a transgene encoding a LacI–GFP fusion protein. LacI–GFP repressor binding to lacO permits array detection by GFP autofluorescence. Colocalization (yellow) between arrays (green) and antibodies (red) in the merged images (right panels) showed association of the protein with her-1 regulatory sequences. Arrowheads mark the X chromosomes. Consistent with sdc-3(Tra) causing derepression of her-1, it blocks SDC and DPY proteins from associating with her-1.
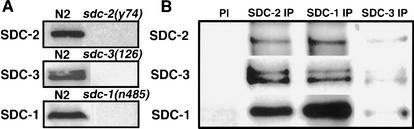
Second, the SDC proteins interact physically to form a complex. Antibodies to any one of the SDC proteins coimmunoprecipitated all three SDC proteins from wild-type embryonic extracts (Fig. 2B). These precipitation reactions were specific, because none of the preimmune sera precipitated any of the SDC proteins (Fig. 2B), and none of the SDC antibodies precipitated (data not shown) or identified (Fig. 2A) their cognate proteins from extracts of the respective sdc null mutants.
Figure 2.
The SDC proteins form a complex in vivo. (A) Detection of SDC proteins in embryonic extracts. Western blots of extracts from wild-type (N2) or sdc (null) mutant embryos carrying a deletion or nonsense mutation in the sdc gene were probed with the SDC antibody indicated on the left. Proteins of 250 kD, 240 kD (a doublet), and 140 kD were detected by SDC-2, SDC-3, or SDC-1 antibodies, respectively, in wild-type but not sdc (null) extracts, showing antibody specificity. (B) Antibody to any one SDC protein coimmunoprecipitated all three SDC proteins. Coimmunoprecipitations were performed with each SDC antibody on wild-type embryonic extracts. The coimmunoprecipitated material was separated by SDS-PAGE and immunoblotted with the antibody indicated on the left. (PI) Preimmune sera for SDC-1 antibody, (IP) immunoprecipitation.
Third, the SDC complex represses transcription of her-1, since ectopic production of SDC proteins in XO animals (normally males) induced hermaphrodite sexual development. SDC-2 is normally expressed only in XX animals, and ectopic expression of SDC-2 transformed 36% of XO animals into hermaphrodites (Dawes et al. 1999), a sexual transformation that required wild-type sdc-3 activity. Given the incomplete feminization with SDC-2 alone, we simultaneously overexpressed SDC-2 with either SDC-1 or SDC-3 to assess their combined contributions toward hermaphrodite development. Overexpression of only SDC-1 (data not shown) or SDC-3 (Davis and Meyer 1997) failed to feminize XO animals. However, overexpression of both SDC-2 and SDC-1 greatly enhanced the XO feminization, causing ∼88% of these XO animals to be sexually transformed (Table 1). Nearly all were self-fertile, in contrast to transformed XO animals that expressed only SDC-2. Increasing the level of SDC-3 in XO animals, as verified by antibody staining, did not enhance the feminization caused by SDC-2 (31% feminization with both proteins), indicating that SDC-3 was not limiting (Table 1). The synergy between SDC-1 and SDC-2 in feminizing XO animals in an SDC-3-dependent manner provides functional evidence that all three SDC proteins act together to repress her-1 directly.
Table 1.
sdc-1 enhances sdc-2 in feminizing XO animals
| Maternal genotypea | Paternal genotypea | Extent of XO feminizationb | No. of XO males | No. of XO intersexesc | No. of XO hermaphrodites |
|---|---|---|---|---|---|
| dpy-27d | _dpy-27; yIs30_[dpy-30::sdc-2(+)]d | 36%d | 408d | 31d | 200d |
| _dpy-27; yIs3_[sdc-3(+)] | _dpy-27; yIs30_[dpy-30::sdc-2(+)] | 31% | 84 | 1 | 36 |
| _dpy-27; yEx31_[sdc-1(+)] | _dpy-27; yIs30_[dpy-30::sdc-2(+)] | 88% | 23 | 5 | 162 |
SDC proteins recruit the X-chromosome dosage compensation complex to her-1
Because SDC-2 and SDC-3 play pivotal roles in assembling the dosage compensation complex onto X chromosomes (Chuang et al. 1996; Davis and Meyer 1997; Dawes et al. 1999), we asked whether SDC proteins recruit this complex to her-1. The dosage compensation complex includes the dosage compensation-specific protein DPY-27 and the dual-function proteins DPY-26 and MIX-1, which also act in meiosis and mitosis, respectively (Chuang et al. 1996; Lieb et al. 1996, 1998). The dosage compensation proteins resemble components of the widely conserved condensin complex, which drives mitotic chromosome condensation in vitro, implying that regulation of _X_-chromosome expression involves modulation of chromatin structure (Koshland and Strunnikov 1996; Hirano 2000). All dosage compensation proteins except SDC-2 require SDC-3 for their localization to the X chromosome (Chuang et al. 1996; Davis and Meyer 1997), and SDC-3, in turn, requires SDC-2 for its localization to the X chromosome (Davis and Meyer 1997). SDC-2 can localize to the X chromosome without other dosage compensation proteins (Dawes et al. 1999), suggesting that it recognizes the X chromosome and confers chromosome specificity to dosage compensation. Not all of the dosage compensation components appear essential for her-1 repression, because the rare dpy-26, dpy-27, or dpy-28 XX mutants that escape lethality develop as hermaphrodites (Plenefisch et al. 1989). Therefore, discovery of the complete dosage compensation machinery on her-1 would show that SDC-2 targets this machinery to the chromatin it binds.
DPY-26, DPY-27, and MIX-1 all colocalized with SDC proteins on both her-1 arrays and X chromosomes (Fig. 1C–E). Furthermore, localization of the three dosage compensation proteins to her-1 was dependent on SDC proteins because the sdc-3(Tra) mutation disrupted the localization to her-1 but not to the X chromosome (Fig. 1F; data not shown). The localization of SDC-2 and SDC-3 was not dependent on DPY-27 (Fig. 1G), but DPY-26 required DPY-27 for its localization to her-1 (Fig. 1H), as it does for its localization to the X chromosome. Thus, assembly of known dosage compensation components onto her-1 resembles their assembly onto the X chromosome. Moreover, SDC-2 recruits the dosage compensation machinery to its chromatin targets, even though some components may be dispensable for repression.
The SDC/dosage compensation complex associates with three different chromatin targets within her-1
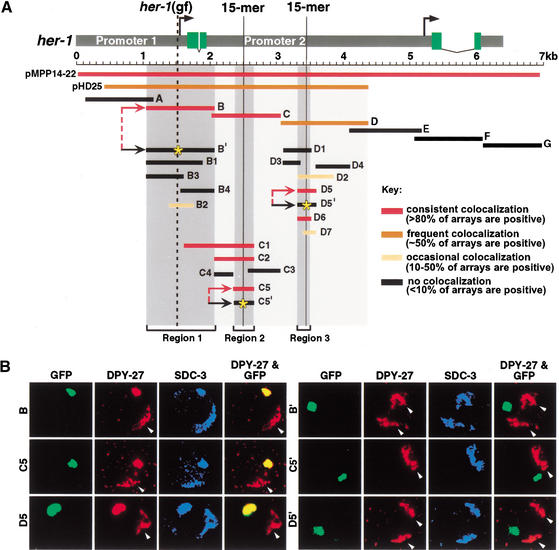
We determined the exact sites within her-1 that recruit the dosage compensation machinery. Using the extrachromosomal array assay to examine individual 1-kb fragments across her-1, we found that SDC-1, SDC-2, SDC-3, DPY-26, DPY-27, and MIX-1 all colocalized with three different regions defined by fragments B, C, and D (Fig. 3A,B). Localization of all proteins was disrupted by an sdc-3(Tra) mutation (data not shown), showing that binding was specific.
Figure 3.
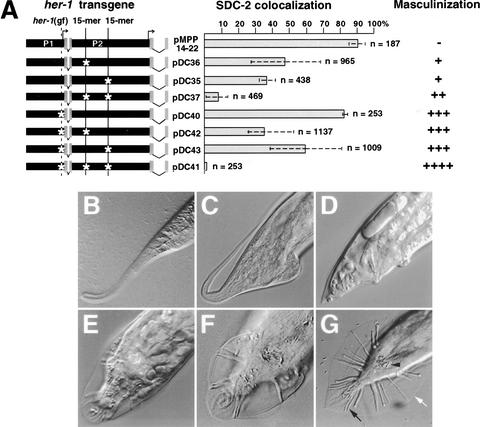
SDC-2 localization to her-1 is specified by three distinct DNA recognition elements whose binding capacity is disrupted by specific mutations. (A) Schematic of the her-1 gene and summary of subregions tested for SDC-2 colocalization by the array assay. Transcription from the P1 promoter produces the functional male-specific her-1 transcript, including four exons (green). A promoter (P2) resides within the second intron of her-1. P2 is coregulated with P1 and makes a 0.8-kb transcript of unknown function that includes the last two exons of her-1 (Trent et al. 1991; Perry et al. 1993). The degree of SDC-2 colocalization with her-1 regions is shown by color, with the key on the right. The three smallest regions with strong SDC-2 colocalization (region 1 [B], region 2 [C5], and region 3 [D6]) are shown by dark gray shading. C5 and D6 share an identical 15-bp element (solid vertical lines) and 50% overall sequence identity. B has no obvious similarity with C5 or D5 but contains the site of her-1(gf) (dashed vertical line). SDC-2 colocalization was completely disrupted by a randomized 15-mer in either C5 or D5 and by the G → A transition of her-1(gf) in B (yellow stars in B‘, C5′, and D5′). (B) Confocal images of an individual gut nucleus from a wild-type XX animal bearing GFP-tagged extrachromosomal arrays (green) with either wild-type (B, C5, and D5) or mutant (B‘, C5′, and D5′) versions of regions 1–3. Animals were immunostained with antibodies to DPY-27 (red) or SDC-3 (blue). Colocalization between the array and protein appears as yellow in the merged image. Arrowheads indicate X chromosomes.
Fragment B includes the P1 promoter that produces the 1.2-kb functional her-1 transcript (Trent et al. 1991; Perry et al. 1993). This regulatory region was first implicated in her-1 repression by a gain-of-function mutation, her-1(gf), that partially derepresses her-1 transcription, causing substantial, but incomplete masculinization of XX mutants (Trent et al. 1988). The location of her-1(gf) 2 bp before the transcriptional start site prompted us to test whether her-1(gf) interferes with binding of the repression complex (Perry et al. 1994). Indeed, SDC-2, SDC-3, and DPY-27 failed to associate with a fragment (B‘) harboring the A → T transition of her-1(gf), showing that derepression of her-1 transcription is caused at least in part by disrupting repressor binding (Fig. 3A,B; data not shown). The her-1(gf) mutation appears to eliminate rather than reduce SDC binding, because overexpression of SDC-2 failed to suppress the XX masculinization caused by her-1(gf) (see Materials and Methods).
Fragments C and D are within the large second intron of her-1. Unlike P1, this specific region had not been implicated in her-1 repression by gain-of-function mutations. However, the partial masculinization of XX animals by her-1(gf) compared with the nearly complete masculinization by sdc-3(Tra) suggested that SDC-mediated her-1 repression requires sequences outside the gf region. Moreover, indirect experiments suggested a possible involvement of the second intron in her-1 repression (Li et al. 1999).
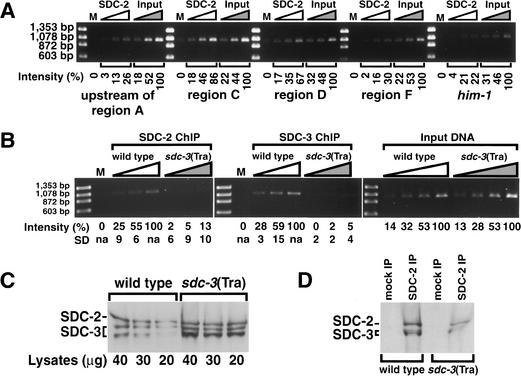
We explored whether the protein–DNA interactions observed with fragments C and D on extrachromosomal arrays also occurred at the endogenous her-1 gene by performing chromatin immunoprecipitations (ChIP) from lysates of formaldehyde-treated wild-type embryos. SDC-2 antibodies were used to immunoprecipitate the SDC complex with its associated DNA, and the DNA was analyzed for enrichment of her-1 fragments using primers to regions A–F in separate PCR reactions. Primers flanking him-1, a gene on a different chromosome, and genomic fragments just upstream of region A were used as controls. Only DNA from regions C and D was specifically enriched by threefold to fourfold relative to the negative control (Fig. 4A). In parallel ChIPs performed with SDC-2 or SDC-3 antibodies, only DNA from wild-type, but not sdc-3(Tra) lysates was enriched for fragment C (Fig. 4B). This result confirmed the specificity of the ChIP by showing that it correctly reflects the disruption of SDC binding to her-1 caused by sdc-3(Tra). In controls, equivalent levels of region C DNA were detected in PCRs using DNA extracted from wild-type and mutant lysates (Fig. 4B). Likewise, comparable levels of SDC proteins were detected in both lysates with Western blots (Fig. 4C) and IP experiments (Fig. 4D). Together these experiments show that the SDC complex associates with regions C and D in the endogenous her-1 gene. The inability to detect region B by this assay suggested that region B has a lower capacity for SDC binding than regions C and D, as shown below and hypothesized previously (Li et al. 1999).
Figure 4.
Chromatin immunoprecipitation experiments (ChIP) show the interaction of SDC proteins with the endogenous her-1 gene. (A) PCR analysis of DNA from a ChIP performed with SDC-2 antibodies and lysates of formaldehyde cross-linked XX embryos. Primer sets specific to her-1 regions or a control gene (him-1) were used for PCRs with mock-precipitated DNA (M) and twofold serial dilutions of SDC-2-precipitated DNA (SDC-2) or input DNA (Input). The intensity of the PCR band produced by each primer pair from IP-enriched DNA was normalized to the corresponding PCR band produced from the highest concentration of input DNA. Regions D and C of her-1 were specifically enriched above him-1 control DNA by threefold or fourfold, respectively. (Primers flanking him-1 produced a PCR product from IP-enriched DNA of 22% normalized intensity, whereas primers flanking regions D and C produced bands of 67% and 86%, respectively, normalized intensity.) (B) PCR analysis with region C primers was performed on twofold serial dilutions of DNA from a ChIP using SDC-2 or SDC-3 antibodies and lysates of formaldehyde-treated wild-type or sdc-3(Tra) XX embryos. The intensity of each PCR band was normalized to the intensity of the PCR band made from the highest concentration of IP-enriched DNA from the wild-type lysate. The average intensities and standard deviations were calculated from four sets of PCR analyses on material from two independent ChIP experiments. The specificity of the ChIP protocol was shown by the precipitation of region C DNA from wild-type but not mutant lysates. Similar levels of region C DNA were detected by PCR using twofold serial dilutions of wild-type and sdc-3(Tra) input lysates. (C,D) Similar levels of SDC-2 and SDC-3 were detected by Western blot analysis of either (C) whole lysates or (D) SDC-2 IP material from lysates of formaldehyde-treated wild-type and sdc-3(Tra) embryos.
Diverse DNA recognition elements reside within the three distinct her-1 chromatin targets
Having shown that fragments B, C, and D are true SDC targets, we defined the DNA sequence requirements for SDC binding more precisely. Of four region B subfragments, only B2 supported significant SDC-2 colocalization (Fig. 3A). However, localization to B2 was less consistent than localization to B, suggesting that strong SDC-2 binding requires more than one discrete element within region B. In contrast, the robust localization to regions C and D was narrowed to a 303-bp fragment (C5) of C and to a 192-bp fragment (D6) of D (Fig. 3A,B). SDC-1, SDC-3, DPY-26, DPY-27, and MIX-1 all colocalized with SDC-2 on these fragments, showing that all the information required for SDC proteins to interact with chromatin and recruit the dosage compensation complex can be specified by 192 bp of DNA (Fig. 3A,B; data not shown) that has been removed from its native chromosomal context.
Very limited similarity in DNA sequences was found between B (region 1) and either C5 (region 2) or D6 (region 3). In contrast, C5 and D5 (a 287-bp fragment that includes D6) share 50% overall identity and a 15-bp stretch (CAAAAACTGAGCCTG) of complete identity on the antisense strand of C5 and the sense strand of D5. An exact copy of this element is not found on X or elsewhere in the genome. Randomizing the 15-bp element to ACAGACTGCAGATAC (for C5′ and D5′) or GA CAGACGTCAATAC (for D5′) prevented SDC-2, SDC-3, and DPY-27 proteins from localizing to arrays with the mutant fragments (Fig. 3A,B; data not shown). The 15-bp repeated sequence is therefore necessary for targeting SDC and DPY proteins to regions 2 and 3. The 15-mer is not sufficient, however, because SDC-2 failed to associate with arrays carrying multiple copies of random DNA and either the 15-mer or a 28-bp element that includes the 15-mer and neighboring common sequences (data not shown). Other _cis_-acting sequences must be essential. Thus, the three DNA elements used to target the dosage compensation machinery to her-1 are diverse. Either SDC proteins themselves have flexibility in sequence recognition or other cellular components help confer sequence specificity and binding.
Differential recruitment of SDC-2 to her-1 by individual recognition elements
Identification of mutations that eliminated SDC binding to individual sites in her-1 allowed us to assess and correlate in vivo the functional contribution of each site toward overall SDC binding and repression of her-1. We introduced the her-1(gf) mutation of region 1 and the randomized 15-mers of regions 2 and 3 together or separately into full-length _her-1_-rescuing constructs to test the role of each site. XX animals expressing a single construct from a GFP-tagged extrachromosomal array were examined for the frequency of SDC-2 localization to arrays and for sexual transformation to the male fate, an indicator of transcriptional derepression (see Materials and Methods). The functional significance of a site could then be inferred by comparing the change in SDC localization with the degree of sexual transformation caused by disrupting that site (Fig. 5A).
Figure 5.
The relative contributions of the three her-1 recognition elements differ for SDC binding and her-1 repression. (A) Mutation of specific DNA sequences within full-length her-1 transgenes disrupts SDC-2 localization to her-1 and repression of her-1. her-1 constructs included in transgenic arrays are depicted by diagrams on the left, with mutations indicated by white stars. The percent SDC-2 localization to transgenic arrays is represented by gray bars. The standard deviation between lines assayed is represented by a dotted line. (n) Total number of nuclei scored in all lines. Masculinization caused by the full-length her-1 transgenes was quantified (see Materials and Methods) and then rated as (−) none, (+) weak, (++) moderate, (+++) strong, or (++++) severe. (B_–_G) Examples of masculinization caused by derepression of her-1. DIC photomicrographs of tails from (B) a wild-type XX hermaphrodite, (C_–_F) XX animals masculinized by full-length her-1 transgenes, and (G) a wild-type XO male. Lateral views (B_–_D) and ventral views (E_–_G). (White arrow) male fan; (black arrow) male sensory rays; (black arrowhead) male spicules.
Regions 2 and 3 had approximately equivalent SDC-2 binding activity in the context of the full-length her-1 gene, and these regions supported more robust binding than region 1 (Fig. 5A). SDC-2 localized to 90% of arrays carrying a wild-type her-1 gene. The localization was reduced only slightly, to 85%, by the her-1(gf) lesion in region 1, even though this lesion abolished SDC-2 localization to a fragment containing only region 1. The remaining SDC binding must have occurred through regions 2 and 3. Indeed, in transgenes with a wild-type region 1, randomization of either 15-mer decreased SDC-2 localization to 20%–40%, and randomization of both 15-mers decreased SDC-2 localization to 10%. SDC localization was not significantly reduced by disrupting region 1 on a transgene already mutant for either region 2 or region 3, but was mildly reduced by disrupting region 1 on a transgene mutant for both regions 2 and 3. The comparatively weak SDC binding affinity for region 1 in the context of the full-length her-1 transgene is consistent with the difficulty in detecting region 1 by ChIP analysis on the endogenous gene.
Complete repression of her-1 requires the participation of all three SDC-binding regions
Is the strength of SDC binding to a region correlated with effectiveness in repressing her-1? Analysis of sexual phenotype in XX animals with wild-type or modified full-length her-1 transgenes revealed that complete repression of her-1 required the participation of all three SDC-binding regions. However, region 1, the weakest in SDC-binding activity, made the greatest single contribution to repression (Fig. 5A–G). Regions 2 and 3 contributed repression activity in the absence of region 1, but repression was less effective than from region 1 (Fig. 5A–G). The degree of her-1 repression from regions 2 and 3 may be more comparable to the repression of _X_-linked genes that occurs during dosage compensation.
These conclusions were drawn from the following observations (Fig. 5A–G): XX animals carrying wild-type her-1 transgenes showed very low levels (−) of masculinization, indicating strong repression. Mutation of either region 2 or 3 caused weak masculinization (+) that was correlated with intermediate disruption of SDC-2 localization. Mutation of both regions 2 and 3 caused moderate masculinization (++), despite causing strong disruption of SDC-2 localization. Finally, mutation of region 1 caused strong masculinization (+++), despite causing only a slight reduction in SDC localization. This masculinization was not enhanced by disrupting only region 2 or 3 but was enhanced by disrupting both regions 2 and 3, causing severe masculinization (++++).
The weak binding of SDC proteins to region 1 on a full-length her-1 transgene raised the concern that SDC proteins may not mediate repression from region 1. Therefore, we assessed the impact of disrupting SDC binding specifically to region 1 by introducing an sdc-3(Tra) mutation into genotypically her-1(−) animals carrying full-length her-1(+) transgenes with mutations in regions 2 and 3. A twofold to threefold increase in the number of masculinized animals was found, showing that SDC proteins contribute to repression from region 1 in vivo.
The involvement of regions 2 and 3 in SDC-mediated her-1 repression in vivo is further reinforced by interpreting previous genetic data in the context of the SDC-binding data. Our results show that the her-1(gf) mutation eliminates SDC binding to region 1 but not to regions 2 and 3, whereas the sdc-3(Tra) mutation severely reduces SDC binding to all three regions. Thus, the greater degree of masculinization in XX animals caused by the sdc-3(Tra) mutation (100% of mutants) compared with the her-1(gf) mutation (30% of mutants) must result from reduction of SDC binding to regions 2 and 3. Therefore, regions 2 and 3 contribute substantially to SDC-mediated repression of the endogenous her-1 gene, along with region 1.
Discussion
Gene-specific repression
The work presented here reports a detailed dissection of DNA recognition elements required for SDC binding in vivo and for recruitment of the C. elegans dosage compensation complex to the autosomal gene target her-1. Within her-1, three SDC-binding sites contribute to her-1 repression: one overlaps the start point of transcription, and two reside within the second intron. Given the locations of the binding sites, more than one mechanism of SDC-mediated repression is likely to control her-1 transcription. A repression complex bound at any one of the sites could prevent transcription initiation by damping the action of transcriptional activators or by interfering with binding of the basal transcriptional machinery. If such interference were achieved through regions 2 and 3, it would occur over a distance of 1–2 kb and likely be accomplished by a distinct mechanism from that at region 1. Alternatively, repressors bound to regions 2 and 3 might use another strategy to achieve repression, such as impairing transcriptional elongation, altering chromatin structure, or assisting repressor binding to region 1, for example, by looping DNA or acting as repressor storage sites. Through different distances and potentially different mechanisms, the three SDC-binding sites act in concert to repress her-1 by 20-fold.
Our experiments have raised interesting questions. First, why does region 1 appear to be the most important region for her-1 regulation in vivo, yet have the lowest affinity for SDC complex binding? A similar observation was made with the regulation of the Drosophila master sex-determination gene Sxl. The site most effective in activating Sxl transcription in vivo had the lowest affinity for the activators (Yang et al. 2001). In the case of her-1, overlap between region 1 and the start site of transcription raises the possibility that repression from region 1 is more complex than repression from other regions, possibly involving unidentified repressor molecules, in addition to the SDC proteins. Such additional repressor proteins could act independently of SDC proteins or act in conjunction with them, for example, by assisting SDC binding. Alternatively, the her-1(gf) mutation might bolster the inherent rate of transcription initiation, making the task of repression more difficult and the role of region 1 appear greater than the roles of regions 2 and 3.
Second, what is the functional consequence of recruiting the entire dosage compensation complex to her-1? Previous work established that the 20-fold repression of her-1 has different genetic requirements from the twofold repression of the X chromosome. For example, the sdc-3(Tra) mutation prevents assembly of SDC and dosage compensation DPY proteins on her-1 but not on the X chromosome, thereby selectively disrupting her-1 repression. Moreover, the DPY proteins were thought to play a direct role in repressing only _X_-linked genes but not her-1, because rare dpy XX mutants that escape lethality develop as hermaphrodites. However, numerous genetic experiments have shown that dpy-26, dpy-27, or dpy-28 mutations can affect sexual fate in selective genetic backgrounds (Hodgkin 1987; Miller et al. 1988; Trent et al. 1988; Plenefisch et al. 1989; DeLong et al. 1993). These previous genetic observations, together with our discovery of DPY proteins localized to her-1 in vivo, suggest that DPY proteins may modulate her-1 repression directly.
Chromosome-wide repression
The behavior of SDC proteins at her-1 has important implications for their function in _X_-chromosome-wide gene repression during dosage compensation. SDC proteins have the unexpected ability to associate with diverse DNA elements whose binding capacity can be disrupted by specific mutations. At her-1, the sequence and location of each different DNA element specified a distinct level of repression, indicating that repression by the complex is flexible and dependent on the DNA context of its target. The diversity of sequences recognized by the SDC proteins at her-1 suggests that a specific DNA sequence may not be solely responsible for recruiting the dosage compensation machinery to sites along the X chromosome. Although no recognition elements have yet been identified on X (Lieb et al. 2000), the regulation of her-1 suggests that reduction of _X_-linked gene expression has the potential to occur at the level of individual genes, through either short- or long-range mechanisms. Dosage compensation might therefore proceed by combining gene-specific repression strategies with global repression mechanisms acting at the level of chromatin structure, mediated by the condensin-like dosage compensation complex.
Materials and methods
Constructs
Transgenic lines with extrachromosomal arrays carrying her-1 sequences were created as previously described (Dawes et al. 1999; Lieb et al. 2000) with lacO repeat plasmid pSV2-dhFr8.32 (50 μg/mL; Straight et al. 1996), hsp16-2::lacI-gfp plasmid pPD49–78 (100 μg/mL), rol-6(su1006) marker plasmid pRF4 (100 μg/mL), and her-1 DNA (50 μg/mL).
The 1-kb regions of her-1 (designated A–G ) and subregions of B, C, and D were PCR-amplified from the full-length her-1 construct pMPP14-22 (Perry et al. 1993) and blunt-end-ligated into pBluescript. For C4, D1, D2, D3, and D4, PCR products were purified and injected directly into worms. No differences were detected in SDC-2 colocalization assays with extrachromosomal arrays made from her-1 PCR products or plasmids. Altered full-length her-1 constructs were made by sequential site-directed mutagenesis of pMPP14-22 using PCR. DNA sequence analysis confirmed the sequence changes.
Antibodies
SDC-3 antibodies were produced against a GST–SDC-3 fusion protein containing amino acids 1067–1340 of SDC-3 and purified against a thioredoxin–SDC-3 fusion protein containing the same SDC-3 region. SDC-1 antibodies were produced against a his-tagged–SDC-1 fusion protein containing amino acids 615–1034 of SDC-1 and purified against a GST–SDC-1 fusion protein containing amino acids 624–981. SDC-2 antibodies used for immunostaining were raised and affinity-purified against a C-terminal 35-amino-acid peptide plus a CGG linker, CGGDAEESI EDPLDIVEMTLKRALPRSMSPSSKRRRMR. Affinity-purified rabbit SDC-2 antibodies used for IP and ChIP reactions were made to a fusion protein containing the first 455 amino acids of SDC-2 and six tandem histidine residues (Dawes et al. 1999). Rat SDC-2 antibodies made to the same fusion protein were used for detecting SDC-2 in the ChIP Westerns. Immunostaining of gravid adults followed (Lieb et al. 2000), except SDC-1 antibodies were incubated with a 1-mg/mL protein lysate from sdc-1(n485) embryos, and SDC-2 antibodies were prepared as in Dawes et al. (1999). The sdc-1(n485) mutation causes a C → T transition at nucleotide 2634, resulting in a stop codon at amino acid 136. her-1 extrachromosomal arrays carrying lacO and hsp16-2::lacI-gfp were identified as in Carmi et al. (1998) and Lieb et al. (2000), except worms were heat-shocked at 33°C for 30 min and allowed to recover at room temperature for 45 min.
Coimmunoprecipitation reactions
Lysates were prepared by sonicating wild-type or mutant embryos in homogenization buffer (50 mM HEPES at pH 7.6, 200 mM KCl, 1 mM EDTA, 1 mM EGTA, 0.2% Triton X-100, 5% glycerol, and protease inhibitor cocktail [Calbiochem]). Cellular debris was cleared by centrifugation at 5000_g_ at 4°C for 5 min. Lysates were sonicated further and centrifuged at 20,000_g_ at 4°C for 10 min. The supernatant was incubated with primary antibody at 4°C for 4 h, cleared by centrifugation at 20,000_g_, and incubated with Protein A Sepharose at 4°C for 1 h. Immunocomplexes bound to Protein A Sepharose were washed with homogenization buffer and eluted with 0.15 M glycine (pH 2.5). Proteins were precipitated with trichloroacetic acid, washed with acetone, and resuspended in Laemmli sample buffer (Laemmli 1970). SDS-PAGE (Novex) and immunoblotting (BioRad) using chemiluminescent detection (ECL, Pharmacia) were performed on immunoprecipitated material.
Quantification of SDC-2 localization to her-1 arrays
To quantify the localization of SDC-2 to her-1 arrays (Fig. 3A), an average of 87 array-bearing nuclei (range 25–295) were scored for each construct in every experiment. Multiple independent lines showed consistent results for each construct. For the smallest fragments and their mutant forms, the following number of arrays were scored: B, n = 78; B‘, n = 40; C5, n = 569; C5′, n = 159; D5, n = 169; and D5′, n = 186. Colocalization between SDC-1, SDC-3, DPY-26, DPY-27, or MIX-1 on her-1 arrays was analyzed similarly, with at least 40 worms and >200 gut nuclei examined for each construct.
Effect of SDC-2 overexpression on her-1(n695gf) mutants
To assess whether overexpression of SDC-2 suppresses her-1(gf) mutants, yIs30_[dpy30::sdc-2(+); unc-76(+)]; him-8 IV; unc-76 V survivor males were crossed to unc-42(e270) her-1(n695_gf) V hermaphrodites, and non-Unc F1 hermaphrodites were allowed to self-fertilize. Overexpression of SDC-2 from yIs30 was confirmed by antibody staining. One-hundred Unc-42 F2 XX adults were scored for sexual phenotype, including masculinization of tail and soma, sterility, and egg-laying defects, and then picked for single-worm PCR. The presence of yIs30 was detected using the oligonucleotides HD-43 (CTC GCT GTC AGT GTT TTG TCC TG) and HD-44 (CAT CCA TCT CGA AAT CTC CGA G), which span the dpy-30–sdc-2 junction in the transgene, to create a PCR product of ∼500 bp. Of 100 F2 animals, 64 carried at least one copy of yIs30. Overexpression of SDC-2 from yIs30 did not rescue any of the sexual phenotypes associated with her-1(gf).
Chromatin immunoprecipitation analysis
We devised a protocol similar to that of Hecht and Grunstein (1999) using worm embryos that were fixed in M9 buffer with 2% formaldehyde at room temperature for 30 min. Excess formaldehyde was quenched and removed with a 0.1 M Tris-HCl (pH 7.5) wash and two M9 washes. Lysates were prepared as described above except ChIP buffer was used (50 mM HEPES-KOH at pH 7.6, 1 mM EDTA, 140 mM KCl, 0.5% NP-40, 10% glycerol, 5 mM DTT). Lysates were precleared against Protein A Sepharose or IgGsorb (The Enzyme Center). For each ChIP reaction, 3 mg of total protein was incubated with 5 μg of affinity-purified antibodies for 2 h. After clearing nonspecific aggregates by centrifugation at 16,000_g_, the immunocomplexes were captured with Protein A Sepharose and subjected to four 1-mL ChIP buffer washes at 100 mM KCl and two at 1 M KCl, and two TE (10 mM Tris-HCl at pH 8.0, 1 mM EDTA) washes. The precipitates were eluted with 10 mM Tris-HCl (pH 8) and 1% (w/v) SDS. For protein detection, the immunocomplexes were washed six times with 1 mL of ChIP buffer and eluted with 0.1 M glycine (pH 3). For ChIP analysis, formaldehyde cross links were reversed by incubation at 65°C overnight in 0.2 M NaCl. Proteins were removed by proteinase K digestion and phenol-chloroform extraction. Following ethanol precipitation, the DNA was resuspended in 100 μL of TE. For input DNA control, DNA was extracted from 3 mg of lysates as described above. PCR amplifications (25 cycles with 55°C annealing for 30 sec and 72°C extension for 90 sec in 50 μL of reaction volume) were carried out on 0.75, 1.5, and 3 μL of undiluted SDC precipitated DNA and input DNA diluted 3300-fold. The working concentration for each primer pair was optimized to produce similar amounts of PCR products from the input DNA. For all primers, PCR amplification was always linear in the dilution range of the DNA used. PCR products resolved on an agarose gel were visualized by ethidium bromide staining and quantitated on the Gel-Doc System (Bio-Rad) using the pixel value for each band to calculate relative intensity.
Quantification of her-1 repression from extrachromosomal arrays
Masculinization of XX animals carrying extrachromosomal arrays of full-length wild-type or mutant her-1 transgenes was used to evaluate derepression of her-1. Egg-laying defects, sterility, male gonad structure, and evidence of male tail formation (Fig. 5C–F) were indicators of masculinization (Trent et al. 1988). Percent masculinization (% Tra) is [(the number of masculinized transgenic animals per line)/(total number of transgenic animals per line)] × 100. Variability in masculinization (% Tra) between lines with the same construct was observed, and all lines generated animals with a range of masculinized tails. Variability may be caused by differential array silencing or stability and a different number of transgenes per array. Therefore, a qualitative rating for masculinization was devised using two criteria: (1) the maximum masculinization (% Tra) value per construct and (2) the degree of masculinization for most animals per construct. The qualitative ratings (shown in parentheses) correlated with maximum masculinization (% Tra): (−) none, 8% Tra; (+) weak, 41%–55% Tra; (++) moderate, 78% Tra, with most animals having tails C and D; (+++) strong, 83%–100% Tra, with most animals having tails C–E; and (++++) severe, 100% Tra, with most animals having tails E and F. The number of transgenic animals (n) scored from all lines of each construct was: pMPP14-22, n = 456, 5 lines; pDC35, n = 562, 5 lines; pDC36, n = 767, 3 lines; pDC37, n = 1008, 4 lines; pDC40; n = 121, 1 line; pDC41, n = 86, 2 lines; pDC42, n = 1281, 8 lines; pDC43, n = 282, 2 lines.
Acknowledgments
We thank D. Lapidus for anti-SDC-1 antibody; C. Tsai for anti-SDC-3 antibody; A. Lee for rat anti-SDC-2 fusion protein antibody; A. Chan for immunostaining protocols; D. King for peptide synthesis; M. Harrison for help with figures; and T. Cline, C. Hassig, and J. Rine for critical review. D.S.C. is a postdoctoral fellow of the American Cancer Society, and R.C.C. is a postdoctoral fellow of the Leukemia and Lymphoma Society. B.J.M. is an investigator of the Howard Hughes Medical Institute. This work was supported by NIH grant R37GM30702 to B.J.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bjmeyer@uclink4.berkeley.edu; FAX (510) 643-5584.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.972702.
References
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Carmi I, Kopczynski JB, Meyer BJ. The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature. 1998;396:168–173. doi: 10.1038/24164. [DOI] [PubMed] [Google Scholar]
- Chuang P-T, Lieb JD, Meyer BJ. Sex-specific assembly of a dosage compensation complex on the nematode X chromosome. Science. 1996;274:1736–1739. doi: 10.1126/science.274.5293.1736. [DOI] [PubMed] [Google Scholar]
- Davis TL, Meyer BJ. SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development. 1997;124:1019–1031. doi: 10.1242/dev.124.5.1019. [DOI] [PubMed] [Google Scholar]
- Dawes HE, Berlin DS, Lapidus DM, Nusbaum C, Davis TL, Meyer BJ. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science. 1999;284:1800–1804. doi: 10.1126/science.284.5421.1800. [DOI] [PubMed] [Google Scholar]
- DeLong LD, Plenefisch JD, Klein RD, Meyer BJ. Feedback control of sex determination by dosage compensation revealed through Caenorhabditis elegans sdc-3 mutations. Genetics. 1993;133:875–896. doi: 10.1093/genetics/133.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Grunstein M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- Hirano T. Chromosome cohesion, condensation, and separation. Annu Rev Biochem. 2000;69:115–144. doi: 10.1146/annurev.biochem.69.1.115. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Primary sex determination in the nematode C. elegans. Development. 1987;101 (Suppl.):5–15. doi: 10.1242/dev.101.Supplement.5. [DOI] [PubMed] [Google Scholar]
- Klein RD, Meyer BJ. Independent domains of the sdc-3 protein control sex determination and dosage compensation in C. elegans. Cell. 1993;72:349–364. doi: 10.1016/0092-8674(93)90113-5. [DOI] [PubMed] [Google Scholar]
- Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li W, Streit A, Robertson B, Wood WB. Evidence for multiple promoter elements orchestrating male-specific regulation of the her-1 gene in Caenorhabditis elegans. Genetics. 1999;152:237–248. doi: 10.1093/genetics/152.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Capowski EE, Meneely P, Meyer BJ. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science. 1996;274:1732–1736. doi: 10.1126/science.274.5293.1732. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Albrecht MR, Chuang P-T, Meyer BJ. MIX-1: An essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- Lieb JD, de Solorzano CO, Rodriguez EG, Jones A, Angelo M, Lockett S, Meyer BJ. The Caenorhabditis elegans dosage compensation machinery is recruited to X chromosome DNA attached to an autosome. Genetics. 2000;156:1603–1621. doi: 10.1093/genetics/156.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ. Sex in the worm: Counting and compensating X-chromosome dose. Trends Genet. 2000;16:247–253. doi: 10.1016/s0168-9525(00)02004-7. [DOI] [PubMed] [Google Scholar]
- Miller LM, Plenefisch JD, Casson LP, Meyer BJ. xol-1: A gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell. 1988;55:167–183. doi: 10.1016/0092-8674(88)90019-0. [DOI] [PubMed] [Google Scholar]
- Moazed D. Common themes in mechanisms of gene silencing. Mol Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes & Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoter. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Meyer BJ. Early aspects of Caenorhabditis elegans sex determination and dosage compensation are regulated by a zinc-finger protein. Nature. 1991;351:65–68. doi: 10.1038/351065a0. [DOI] [PubMed] [Google Scholar]
- Nusbaum C, Meyer BJ. The Caenorhabditis elegans gene sdc-2 controls sex determination and dosage compensation in XX animals. Genetics. 1989;122:579–593. doi: 10.1093/genetics/122.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MD, Li W, Trent C, Robertson B, Fire A, Hageman JM, Wood WB. Molecular characterization of the her-1 gene suggests a direct role in cell signaling during Caenorhabditis elegans sex determination. Genes & Dev. 1993;7:216–228. doi: 10.1101/gad.7.2.216. [DOI] [PubMed] [Google Scholar]
- Perry MD, Trent C, Robertson B, Chamblin C, Wood WB. Sequenced alleles of the Caenorhabditis elegans sex-determining gene her-1 include a novel class of conditional promoter mutations. Genetics. 1994;138:317–327. doi: 10.1093/genetics/138.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenefisch JD, DeLong L, Meyer BJ. Genes that implement the hermaphrodite mode of dosage compensation in Caenorhabditis elegans. Genetics. 1989;121:57–76. doi: 10.1093/genetics/121.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A, Belmont A, Robinett C, Murray A. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Trent C, Wood WB, Horvitz HR. A novel dominant transformer allele of the sex-determining gene her-1 of Caenorhabditis elegans. Genetics. 1988;120:145–157. doi: 10.1093/genetics/120.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent C, Purnell B, Gavinski S, Hageman J, Wood WB. Sex-specific transcriptional regulation of the C. elegans sex-determining gene her-1. Mech Dev. 1991;34:43–56. doi: 10.1016/0925-4773(91)90090-s. [DOI] [PubMed] [Google Scholar]
- Villeneuve AM, Meyer BJ. The role of sdc-1 in the sex determination and dosage compensation decisions in Caenorhabditis elegans. Genetics. 1990;124:91–114. doi: 10.1093/genetics/124.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Lu H, Hong Y, Jinks TM, Estes PA, Erickson JW. Interpretation of X chromosome dose at Sex-lethal requires non-E-box sites for the basic helix–loop–helix proteins SISB and daughterless. Mol Cell Biol. 2001;21:1581–1592. doi: 10.1128/MCB.21.5.1581-1592.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]