Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless (original) (raw)
Abstract
The DNA-binding transcription factor Suppressor of Hairless [Su(H)] functions as an activator during Notch (N) pathway signaling, but can act as a repressor in the absence of signaling. Hairless (H), a novel Drosophila protein, binds to Su(H) and has been proposed to antagonize N signaling by inhibiting DNA binding by Su(H). Here we show that, in vitro, H directly binds two corepressor proteins, Groucho (Gro) and dCtBP. Reduction of gro or dCtBP function enhances H mutant phenotypes and suppresses N phenotypes in the adult mechanosensory bristle. This activity of gro is surprising, because it is directed oppositely to its traditionally defined role as a neurogenic gene. We find that Su(H)-H complexes can bind to DNA with high efficiency in vitro. Furthermore, a H-VP16 fusion protein causes dominant-negative phenotypes in vivo, a result consistent with the proposal that H functions in transcriptional repression. Taken together, our findings indicate that “default repression” of N pathway target genes by an unusual adaptor/corepressor complex is essential for proper cell fate specification during Drosophila peripheral nervous system development.
Keywords: Hairless, Suppressor of Hairless, Notch, Groucho, CtBP, corepressors, transcriptional repression, adaptor
The Notch (N) signaling pathway, named after the receptor protein Notch, is conserved across bilaterian phyla, and is employed in a diverse array of cell fate decisions in all three germ layers during animal development (for review, see Artavanis-Tsakonas et al. 1999). Ligand-receptor interactions trigger the proteolytic cleavage of N, releasing the intracellular domain of the receptor (NIC), which binds to the transcription factor Suppressor of Hairless [Su(H)]/Lag-1/RBP-Jκ/CBF1 and acts as a transcriptional coactivator. Su(H), the only known transducing transcription factor for the N pathway, directly activates N target genes in response to signaling (e.g., Bailey and Posakony 1995; Furukawa et al. 1995; Jarriault et al. 1995; Lecourtois and Schweisguth 1995).
In the absence of N signaling, Su(H) represses N target genes (Dou et al. 1994; Hsieh and Hayward 1995; Waltzer et al. 1995; Barolo et al. 2000; Morel and Schweisguth 2000), and Su(H)-mediated repression is essential for proper cell fate specification (Barolo et al. 2000). In mammalian cells, Su(H) binds to the corepressors SMRT and CIR, which are components of histone deacetylase complexes, thereby repressing Su(H) target genes (Kao et al. 1998; Hsieh et al. 1999). In invertebrates, however, the mechanism of Su(H)-mediated repression is not well understood.
Hairless (H), a novel Drosophila protein, is a potent antagonist of N pathway activity, and is required for the specification of many cell fates that are antagonized by N signaling (Bang et al. 1991, 1995; Bang and Posakony 1992; Schweisguth and Posakony 1994; Lyman and Yedvobnick 1995). Moreover, loss of H has been shown to increase the expression of N target genes (Barolo et al. 2000; Klein et al. 2000; Morel et al. 2001). H interacts directly with Su(H) in vitro, and has been proposed to counteract N signaling in vivo by interfering with DNA binding by Su(H) (Brou et al. 1994). However, in light of the subsequent discovery that Su(H)-mediated repression is important for specification of a H-dependent cell fate (Barolo et al. 2000), the notion that H acts primarily by inhibiting Su(H)-DNA interaction seems unlikely, because this model makes incorrect predictions concerning the phenotypic effect of loss or excess of H activity (Bang et al. 1991; Bang and Posakony 1992; Barolo et al. 2000).
An alternative model, in which H acts as a corepressor for Su(H), is consistent with genetic experiments indicating that H antagonizes Su(H)-mediated activation, but not Su(H)-mediated repression (Bang and Posakony 1992; Schweisguth and Posakony 1994; Bailey and Posakony 1995; Barolo et al. 2000). A corepressor model is further supported by findings that misexpression of H causes the transcriptional repression of N target genes (Bailey and Posakony 1995; Barolo et al. 2000; Furriols and Bray 2000). Furthermore, a recent study by Morel et al. (2001) found that H binds to the corepressor C-terminal binding protein (dCtBP) in vitro, although no role was determined for dCtBP in H function or in N-mediated cell fate decisions.
In this report, we present genetic loss-of-function, gain-of-function, and biochemical experiments suggesting that H opposes N pathway activity by acting as an adaptor between Su(H) and the corepressors Groucho (Gro) and dCtBP. Gro and dCtBP, both of which contact histone deacetylase repression complexes, are recruited by a wide variety of DNA-binding transcriptional repressors in flies and vertebrates (for review, see Courey and Jia 2001). We show that H directly interacts with Gro in vitro via an “engrailed homology 1” (eh1)/octapeptide-like motif, and with dCtBP via a consensus binding site at the extreme C-terminus of the H protein. Genetically, reduction of the activity of these corepressors exacerbates the H loss-of-function phenotype and the Su(H) derepression phenotype, and suppresses the effects of reduced N signaling, during Drosophila mechanosensory bristle development. In contrast to an influential study by Brou et al. (1994), we find that H-Su(H) complexes bind to DNA efficiently in vitro. Finally, we demonstrate that a H-VP16 fusion protein, in which the corepressor-binding domains of H have been replaced by a transcriptional activation domain, acts as a dominant-negative form of H in vivo. These in vivo experiments, together with our genetic and biochemical results, support a model in which H recruits the corepressors Gro and dCtBP to Su(H), and thereby mediates transcriptional repression of N target genes. Such “default repression” of N target genes in the absence of signaling appears to be as important as signal-mediated activation for the proper specification of cell fates in the Drosophila peripheral nervous system.
Results
H contains consensus corepressor interaction domains
H is a novel protein, with no known vertebrate homologs. However, the H gene has been identified in three members of the order Diptera: Drosophila melanogaster (Bang and Posakony 1992), Drosophila hydei (Marquart et al. 1999), and most recently the mosquito Anopheles gambiae (Celera Genomics/NIAID). H is surprisingly poorly conserved among these three species (Fig. 1B): It shares 63% identity between D. melanogaster and D. hydei (diverged ∼65 Mya; Beverley and Wilson 1984), and 33% identity between Drosophila and Anopheles (diverged ∼260 Mya; Gaunt and Miles 2002). The rapid divergence of the H protein sequence readily allows the identification of short conserved motifs, which are presumably important for H function. Two such regions occur in a part of H that is required for its interaction with Su(H) in vitro (Fig. 1A,B; Brou et al. 1994; Maier et al. 1997).
Figure 1.
H interacts directly with Gro and with dCtBP in vitro. (A) Domain diagram of the H protein. The Su(H)-interaction domain is shown in yellow; motifs that resemble Gro-binding and CtBP-binding domains are shown in red. The C-terminal region of H deleted in the H22 mutation is indicated by the purple bracket; the H fragments used in panels C and E are represented by the black bars; the region of H included in the HC peptide is represented by the blue bar. (B) The proposed Gro-interaction and dCtBP-interaction domains of H are conserved in dipteran evolution. Protein sequence identity between D. melanogaster H and its apparent orthologs in D. hydei and in the mosquito Anopheles gambiae is shown in 20-residue blocks. Below, the sequences of the proposed Gro-interaction and dCtBP-interaction motifs are aligned with the corresponding sequences in D. hydei and A. gambiae, along with similar motifs from other Gro- and CtBP-binding repressor proteins. (C) In vitro-translated _35_S-Gro binds to purified GST-H (lane 7), but not to GST alone (lane 5), or appreciably to GST-Su(H) (lane 13). GST-H618–723, which includes the YSIHSLLG motif, binds directly to Gro (lane 9); GST-H618–723m, a mutated fusion protein in which this motif has been changed to AAAHSAAG, does not bind Gro (lane 11). Gro-GST-Hairy interaction (lane 3) serves as a positive control for Gro binding, while in vitro-translated _35_S-Emc (even-numbered lanes) serves as a negative control. Approximate amounts of purified protein used: GST-Hairy, 0.2 μg; GST, 31 μg; GST-H, 0.8 μg; GST-H618–723 and GST-H618–723m, 15 μg; GST-Su(H), 3 μg. (D) Interaction between a biotinylated peptide (HC) representing the C-terminal 24 aa of H, including the PLNLS motif, and GST-dCtBP beads. Interaction was assayed by retention of 35S-streptavidin (SA; see Materials and Methods). Amounts of HC peptide used: lane 16, 100 μg; lane 17, 1 μg; lane 18, 100 ng; lane 19, 10 ng; lane 20, 1 ng; lane 21, no peptide. (E) Purified GST-dCtBP binds to an in vitro-translated, 6xHis-tagged C-terminal H fragment (H1036–1077; lane 26), but not to a fragment in which the PLNLSKH motif has been deleted (H1036–1070; lane 27).
Another conserved motif in the H protein is YSIxxLLG, which is perfectly conserved from Drosophila to Anopheles (Fig. 1B). This sequence resembles certain examples of the “eh1” type of Gro-binding domain found in many transcriptional repressor proteins (Fig. 1B; Smith and Jaynes 1996). Among eh1 domains, the “octapeptide” motifs in the Pax 2/5/8 proteins, which have been shown to directly mediate repression by recruiting Gro-family corepressors (Eberhard et al. 2000), show the greatest similarity to this region of H (Fig. 1B).
In addition, the extreme C-terminal sequence of H, PLNLSKH, includes a match to the consensus binding site for the CtBP corepressor, Px(D/N)LS (Postigo and Dean 1999). The PLNLS motif, fully conserved from Drosophila to Anopheles, exactly matches motifs found in four vertebrate CtBP-binding transcription factors (Fig. 1B; Turner and Crossley 2001).
H also contains three lengthy alanine-repeat domains: AAAVAAAAAAAAA, AAAAAAAAAA, and AAVAAA AAAAAA. Alanine repeats and alanine-rich regions are common in transcriptional repression domains, and are found in many repressor proteins. However, these repeats are reduced or absent in the D. hydei and A. gambiae H proteins, which suggests that they may not make an essential contribution to H function.
H interacts directly with Gro and dCtBP in vitro
Given the presence of a putative Gro-binding domain in the H protein, we tested whether H and Gro interact directly in a GST pull-down experiment. We found that 35S-labeled Gro protein is bound efficiently by a GST-H fusion protein, but not by a ∼40-fold greater molar excess of GST alone (Fig. 1C). H618–723, a fragment of H containing the YSIHSLLG motif, also interacts with Gro in vitro; mutating this motif to AAAHSAAG (H618–723m) abolishes the interaction (Fig. 1C). Interestingly, GST-Su(H) failed to interact with Gro to any substantial extent in this assay (Fig. 1C).
The presence in the H protein sequence of a conserved motif matching a CtBP consensus binding site raised the possibility of a direct physical interaction between these two proteins. Indeed, Morel et al. (2001) reported that H binds to dCtBP in vitro. We tested both the sufficiency and the necessity of the extreme C-terminus of H for this interaction. Sufficiency was investigated using a novel derivative of the standard GST pull-down assay. Beads bearing either GST-dCtBP or GST alone were incubated with a biotinylated synthetic peptide, designated HC, representing the C-terminal 24 aa of H (including the PLNLS motif; Fig. 1A), followed by the addition of 35S-labeled streptavidin (see Materials and Methods). Retention of 35S-streptavidin was assayed as a measure of the amount of peptide retained on the beads. We found that the HC peptide binds efficiently to GST-dCtBP, but not to GST alone, indicating that the extreme C-terminus of H is sufficient to interact with dCtBP in vitro (Fig. 1D).
We used a more traditional GST pull-down assay to specifically assess the necessity of the PLNLSKH motif for H-dCtBP interaction. We generated two 35S-labeled, in vitro-translated fragments of the H protein (Fig. 1A): H1036–1077, corresponding to the C-terminal 41 aa of H, including PLNLSKH; and H1036–1070, identical except for the deletion of the PLNLSKH motif. H1036–1077 binds to GST-dCtBP and not to GST alone, whereas H1036–1070 binds to neither (Fig. 1E), demonstrating that dCtBP interacts with H via the PLNLSKH motif.
Reduction of gro or dCtBP function enhances H loss-of-function phenotypes during bristle development
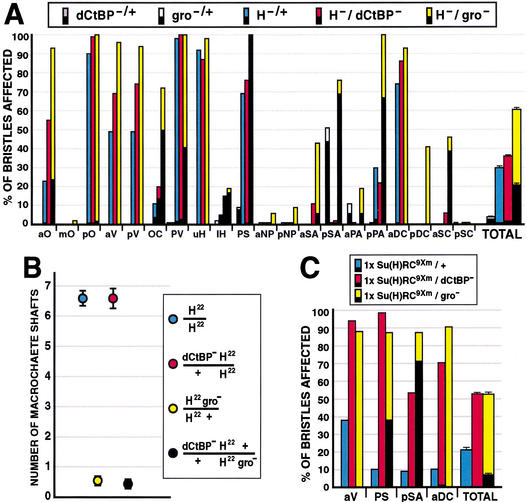
The development of the Drosophila adult mechanosensory bristle is the most thoroughly studied setting for N signaling, and the effects of gain or loss of N, Su(H), and H activities on cell fates in the bristle lineage are well documented (for review, see Posakony 1994). H null heterozygotes (_H_−/+) exhibit defects in mechanosensory bristle development resembling an increase in N signaling: conversion of the shaft cell to the socket fate, or, less frequently, conversion of the sensory organ precursor (SOP) cell to the epidermal fate, resulting in bristle loss (Fig. 2A; Bang et al. 1991). We quantified the H heterozygote phenotype (Bang et al. 1991) by scoring 20 pairs of macrochaete bristles for developmental defects (shaft-to-socket conversions or bristle loss); an average of 29.8% ± 0.9% of bristles are affected in _H_−/+ flies. Flies heterozygous for null mutations in both H and dCtBP show a highly significant enhancement of the H phenotype (36.3% ± 0.7% of bristles affected; P < 2 × 10−5; Fig. 2A). At certain individual bristle positions, the H phenotype is especially strongly enhanced in the _dCtBP_−/+ background [e.g., anterior orbitals (aO), 23% vs. 55% affected, _P_ < 5 × 10−6; Fig. 2A]. Such enhancement is observed with null alleles of H and dCtBP (Fig. 2A), as well as with weaker hypomorphic alleles of both genes (not shown). Enhancement of the H phenotype by reduction of dCtBP activity is almost entirely due to shaft-to-socket transformations; dCtBP's effect on bristle loss is comparatively mild (0.9% vs. 1.8%; P < 0.02; Fig. 2A).
Figure 2.
Genetic interactions among gro, dCtBP, H, and Su(H) during adult mechanosensory bristle development. (A,B) Percentage of bristles affected at 20 macrochaete positions on the adult head and thorax. The lower (black) part of each bar represents bristle loss, or balding, events; the upper part represents shaft-to-socket transformation phenotypes. “TOTAL” bars combine data from all 20 positions; error bars indicate standard error. At least 52 bristles were scored for each genotype at each position. (A) Genetic interactions between null alleles of dCtBP, gro, and H. (B) Effects of heterozygous gro and/or dCtBP mutations in flies homozygous for H22, an allele that deletes the dCtBP interaction domain of the H protein (see Fig. 1). The _y_-axis represents the number of normal shaft structures observed at 42 macrochaete bristle positions (the 20 pairs shown in A, plus the posterior sternopleurals) per fly. Twenty-five flies of each genotype were scored. Error bars indicate standard error. (C) Interactions between dCtBP or gro and a derepressed Su(H) transgene that produces a gain-of-function Su(H) phenotype (Barolo et al. 2000). Data are presented in the same manner as described for A.
Bristles of dCtBP null heterozygotes (dCtBP87De-10/+) occasionally show a dominant double-socket phenotype (Fig. 2A). More severely hypomorphic animals (dCtBP87De-10/dCtBP03463) die as pharate adults and exhibit more frequent shaft defects and shaft loss (not shown). Even in a H+ background, then, dCtBP contributes to the specification of the shaft cell fate.
Heterozygous reduction of gro function (_gro_−/+) enhances the _H_−/+ phenotype (Bang 1993; Price et al. 1997) even more strongly than does _dCtBP_−/+, more than doubling the number of affected bristles (60.3% ± 1.1% of bristles affected; P < 1 × 10−60; Fig. 2A). As with dCtBP, certain bristle positions are extremely sensitive to the dose of gro in an _H_−/+ background [e.g., ocellars (OC), 11% vs. 72% affected, _P_ = 1 × 10−14; Fig. 2A]. However, unlike dCtBP, gro mutations significantly enhance not only the H shaft-to-socket phenotype, but also the H bristle loss phenotype (0.9% vs. 20.9% bristle loss; P < 1 × 10−85; Fig. 2A), which has been shown previously to be due to loss of the SOP fate, presumably by transformation to the epidermal fate (Bang et al. 1991). These observations indicate that gro+ functionally antagonizes N signaling in at least two different cell fate decisions during bristle development.
The finding that gro plays a previously unsuspected role in establishing the SOP fate is supported by our observation that _gro_−/+ flies exhibit a mild haploinsufficient bristle loss phenotype, even in a H+ background (Fig. 2A). This dominant gro phenotype, also observed with another strong gro allele, groE48 (not shown), has not been described previously.
Genetic interaction between H and dCtBP (but not between H and gro) requires a C-terminal domain of H
H22 is a partial loss-of-function allele of H which, when homozygous, causes shaft-to-socket transformations and bristle loss (Bang et al. 1991). The H22 mutation is a small deletion that removes the C-terminal 69 aa of H, including the PLNLS motif, replacing this sequence with six extraneous residues due to frameshift (Maier et al. 1997); the Gro-interaction motif is still present in the mutant protein (Fig. 1A). We have shown here that a small C-terminal fragment of H, including the PLNLS motif, is both necessary and sufficient to bind dCtBP in vitro (see Fig. 1D,E). If the effects of dCtBP on bristle cell fates in vivo are mediated by interaction with H via its PLNLS domain, then reduction of dCtBP function would not be expected to enhance the H22 homozygous phenotype as it does the H null heterozygote phenotype. We quantified the H22/H22 phenotype by scoring the number of normal bristle shafts present at 42 macrochaete positions (Fig. 2B). The loss of one dose of gro significantly enhances the H22/H22 phenotype (6.6±0.2 shafts in H22/H22 vs. 0.6±0.2 in _H22 gro_−/H22; P < 1 × 10−21), indicating that the effect of reduced _gro_ function on the _H_ phenotype does not require the extreme C-terminus of H. In contrast, the loss of one dose of _dCtBP_ fails to affect the _H22_/_H22_ phenotype (6.6±0.2 shafts in _H22_/_H22_ vs. 6.6±0.3 in _H22 dCtBP_−/_H22_; _P_>0.9). Reduction of dCtBP function also had no significant effect in a _H22 gro_−/H22 background (Fig. 2B). Thus, the genetic interaction between H and dCtBP depends on the C-terminus of the H protein, which includes a dCtBP interaction domain. The genetic interaction of gro and H, however, does not require this domain.
Reduction of gro or dCtBP function enhances the effects of Su(H) derepression
We recently identified an autoregulatory enhancer in the Su(H) gene that drives high levels of transcription specifically in the socket cells of external sensory organs (Barolo et al. 2000). Su(H) autoactivates via this enhancer in socket cells, which respond to N signaling, but autorepresses in shaft cells, which do not respond to N signaling. Mutating the nine autoregulatory Su(H) binding sites in a Su(H) rescue construct [_Su(H)RC9Xm_] derepresses Su(H) expression in shaft cells, and results in a moderate frequency of shaft-to-socket cell fate transformations (Barolo et al. 2000). If the corepressors Gro and dCtBP normally contribute, via H, to repression by Su(H), then the Su(H) derepression phenotype might be expected to be sensitive to reductions in the dosage of gro and dCtBP. Figure 2C shows the results of experiments to test this expectation.
One copy of a derepressed mutant Su(H) transgene [_Su(H)RC9Xm_] was placed in a _dCtBP_−/+ or _gro_−/+ background; all flies in these experiments have two wild-type endogenous copies of Su(H). Genetic interactions were assayed by scoring the frequencies of shaft-to-socket transformations and bristle loss (Fig. 2C). The loss of one dose of dCtBP highly significantly enhances the Su(H) derepression phenotype [21% ± 1% of bristles affected in _Su(H)RC9Xm/_+ vs. 53% ± 1% in _Su(H)RC9Xm/dCtBP_−; P < 1 × 10−92]. Likewise, reduction of gro activity strongly enhances the effects of Su(H) derepression [52% ± 1% of bristles affected in _Su(H)RC9Xm /gro_−; P < 1 × 10−67]. As with _H_−/+ (see Fig. 2A), the weak bristle loss phenotype associated with Su(H)RC9Xm is significantly enhanced by the loss of one dose of gro (0.2% vs. 7.0%; P < 1 × 10−43), but not by the loss of one dose of dCtBP (0.2% vs. 0.1%; p = 0.32), again suggesting a role for gro (but perhaps not dCtBP) in maintaining the SOP fate.
Reduction of H, gro, or dCtBP function suppresses N loss-of-function phenotypes in the bristle lineage
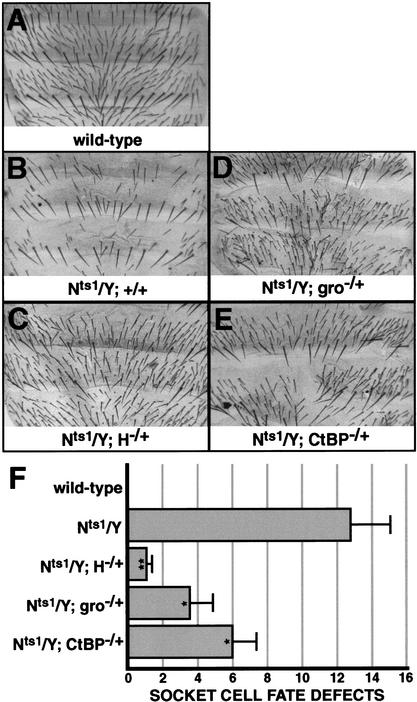
The genetic data presented thus far demonstrate that phenotypes conferred by both loss of H function and lack of Su(H) autorepression are sensitive to the dosages of both gro and dCtBP. We asked a similar question for the oppositely directed phenotypes resulting from loss of N function. Male flies bearing the temperature-sensitive allele Nts1 (Nts1/Y) display a variety of bristle defects when shifted to a nonpermissive temperature after sensory organ precursor (SOP) cell fate specification, due to the loss of various _N_-dependent cell fates in the bristle lineage. These defects include “double-shafting” due to socket-to-shaft cell fate conversions, as well as “balding” due to pIIA-to-pIIB fate conversions (Fig. 3A,B,F; Hartenstein and Posakony 1990). We find that loss of one dose of H causes a highly significant suppression of the Nts1 phenotype (Fig. 3C,F). Similarly, the loss of one dose of either gro or dCtBP partially but significantly suppresses the Nts1 phenotype (Fig. 3D–F), again suggesting that both corepressors contribute to the specification of _N_-independent cell fates (e.g., pIIB, shaft) in the bristle lineage.
Figure 3.
Reduction of H, gro, or dCtBP function suppresses N loss-of-function phenotypes affecting the socket/shaft and pIIA/pIIB cell fate decisions in the bristle lineage. (A_–_E) Abdomens of adult male flies of the following genotypes: wild type (A), Nts1/Y (B), and Nts1/Y flies lacking one dose of H (C), gro (D), or dCtBP (E), shifted to a restrictive temperature (31°C) at 22 h after puparium formation. (F) Quantification of Nts1 effects on the socket-shaft cell fate decision. The _x_-axis represents the average number of socket cell fate defects on the dorsal abdomen of at least eight flies of each genotype. Asterisks denote the statistical significance of differences between Nts1/Y flies and each of the other genotypes, by Student's _t_-test: *, P < 0.05; **, P < 0.005. Error bars indicate standard error.
Su(H) can bind to H and to DNA simultaneously in vitro
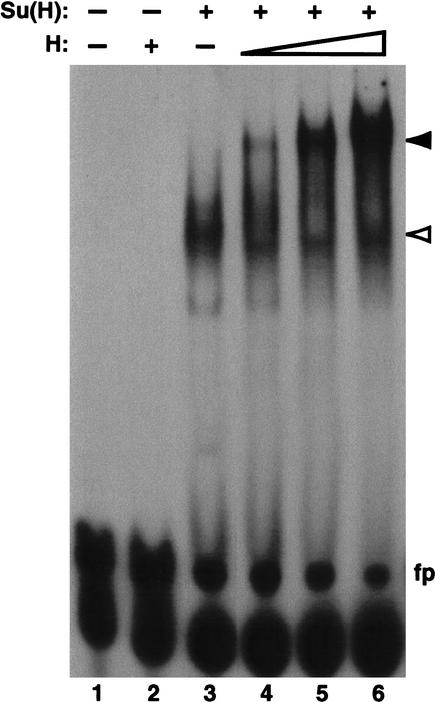
A previous report on the biochemical function of H (Brou et al. 1994) concluded that H can diminish Su(H)-DNA interactions in a gel retardation assay, and proposed that H antagonizes N signaling by preventing Su(H) from binding to its target promoters. This mode of action for H—inhibiting the DNA-binding activity of Su(H)—is clearly inconsistent with the observation that H function is specifically required for the fates of bristle cells that also depend on Su(H)-mediated repression of N target genes (see Discussion). We wondered whether a lower ratio of H to Su(H) protein than the fivefold molar excess used by Brou et al. (1994) might reveal the binding of a H/Su(H) complex to DNA. We therefore revisited the gel retardation experiment, using H:Su(H) molar ratios ranging from 1:30 to 1:6. We found that in the presence of increasing amounts of H, the retarded band corresponding to Su(H)-DNA complexes is progressively replaced by a complex of much lower mobility (Fig. 4), indicating that H/Su(H) complexes can bind DNA with high efficiency.
Figure 4.
Su(H)-H protein complexes bind efficiently to DNA in vitro. Gel retardation analysis of a 32P-labeled oligonucleotide probe containing a high-affinity Su(H) binding site (5′-CGT GTGAA-3′) from the Su(H) autoregulatory enhancer. Mutation of a single base pair of this sequence (to CGTGTCAA) abolishes binding of Su(H) to the probe, indicating that Su(H) binding is sequence-specific (Barolo et al. 2000). His-tagged Su(H) and H proteins were expressed in bacteria and affinity-purified. H alone does not bind to the probe (lane 2). Su(H) alone binds to a large percentage of the probe (lane 3, white arrowhead). The addition of both proteins produces a band of substantially lower mobility, or supershift (lanes 4_–_6, black arrowhead). Approximate amounts of protein used: His-Su(H), 180 ng (lanes 3_–_6); His-H, 10 ng (lane 4), 25 ng (lane 5), 50 ng (lanes 2,6). fp: free probe.
The PLNLSKH motif of H provides some, but not all, of H's ability to convert cell fate in vivo
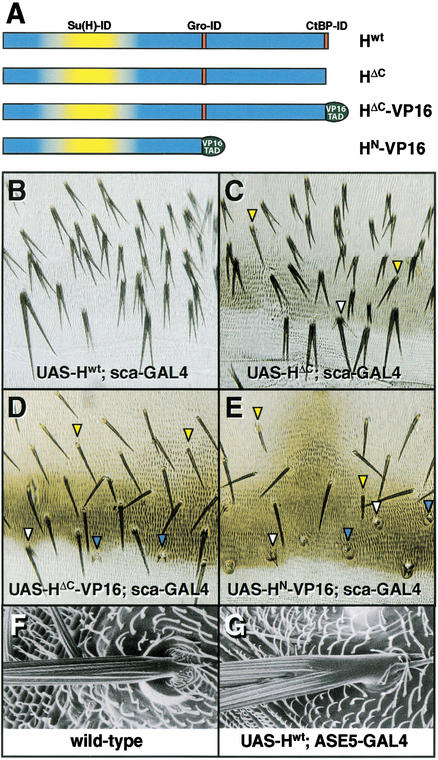
To test the possibility that H participates in transcriptional repression, we designed a series of in vivo GAL4/UAS misexpression constructs in which putative repression domains of H were deleted or replaced with a heterologous transcriptional activation domain (Fig. 5A). Flies carrying these UAS-H transgenes were crossed to flies bearing sca-GAL4, which drives expression of the GAL4 activator in proneural cluster cells, most strongly in SOPs, and in the bristle lineage.
Figure 5.
Replacement of putative repression domains of H with a transcriptional activation domain creates dominant-negative effects in vivo. (A) Diagram of transgenic GAL4-inducible H expression constructs. Protein interaction domains of H are indicated. _UAS-H_wt contains wild-type H driven by five UAS sites. UAS-HΔC encodes a H protein lacking the C-terminal-most seven aa (PLNLSKH), deleting the identified dCtBP interaction domain (see Fig. 1). UAS-HΔC-VP16 encodes a H protein containing the transcriptional activation domain (TAD) of the VP16 protein in place of the PLNLSKH residues of H. UAS-HN-VP16 encodes a H protein lacking both putative repression domains, containing instead the VP16 TAD fused to an N-terminal fragment of H. (B_–_E) Dorsal abdomens of adult (C_–_E) or pharate adult (B; note reduced pigmentation in posterior tergites and in bristle shafts) flies carrying both sca-GAL4 and one of the UAS-H transgenes described above. (F,G) The effects of H misexpression on the socket cell fate are cell-autonomous. Shown are SEMs of head macrochaetes of a wild-type fly (F) and a _UAS-H_wt ASE5-GAL4 fly, in which H is misexpressed specifically in socket cells (G).
Misexpression of wild-type H (Hwt) with sca-GAL4 causes an extremely expressive socket-to-shaft cell fate transformation phenotype (Fig. 5B), in which the converted socket cells and normal shaft cells produce shafts of nearly equal length. Our observation that only socket cells display cell fate transformation effects under this misexpression regimen, while earlier N-mediated cell fate decisions in bristle development (epidermal/SOP, pIIA/pIIB) are unaffected, is in keeping with many previous observations that the socket/shaft decision is by far the most sensitive cell fate decision in bristle development with respect to the dose of H (Bang et al. 1991; Bang and Posakony 1992; Schweisguth and Posakony 1994), perhaps because of Su(H) autoregulation in the socket and shaft cells (Barolo et al. 2000).
Misexpression of HΔC, a mutated form of H lacking its C-terminal seven residues (PLNLSKH), causes socket-to-shaft conversions with high frequency (Fig. 5C), indicating that H can function to affect cell fate without the identified dCtBP-interaction domain. However, deletion of this domain does appear to debilitate the activity of HΔC, in two respects. First, many socket cells are only partially converted to the shaft fate, as revealed by obvious asymmetry between the two shafts (Fig. 5C, yellow arrowheads). We also observe sockets that produce multiple small shafts (Fig. 5C, white arrowhead), a common feature of mild socket-to-shaft conversions. Second, UAS-Hwt; sca-GAL4 flies die as pharate adults, before eclosion from the pupal case (Fig. 5B), while UAS-HΔC; sca-GAL4 flies survive to adulthood. We therefore conclude that the dCtBP-binding region of H is required for some, but not all, of the function of H in vivo.
Replacing putative repression domains of H with an activation domain causes dominant-negative effects in vivo
Transcriptional repressor proteins with discrete repression domains can often be converted to activators by replacing repression domains with a transcriptional activation domain from the herpesvirus protein VP16 (VP16-TAD). Expression of these chimeric proteins can generate dominant-negative phenotypes by activating target genes normally repressed by the transcription factor (e.g., Waltzer et al. 1995; Jimenez et al. 1996; Gao et al. 2001; Kudoh and Dawid 2001). We tested the proposition that H acts to promote transcriptional repression by replacing putative repression domains of H with the VP16 transcriptional activation domain (Fig. 5A).
Replacement of the PLNLSKH motif with VP16-TAD (HΔC-VP16) results in a dramatic suppression of the socket-to-shaft conversions observed with Hwt and HΔC (Fig. 5D; cf. panels B,C). Most sockets appear normal (Fig. 5D, yellow arrowheads), although some partial socket-to-shaft conversion events are observed (white arrowhead), including some sockets with multiple small shafts from which the normal shaft has broken off (blue arrowheads), a common feature of very mild socket-to-shaft conversions (Wang et al. 1997; S. Barolo and J. Posakony, unpubl.).
Replacement of the C-terminal third of H, which contains the Gro- and dCtBP-interaction domains, with VP16-TAD (HN-VP16) abolishes the ability of H to cause socket-to-shaft conversions (Fig. 5E). Instead, this chimeric activator causes failures of the shaft cell fate, generating a spectrum of defects ranging from moderately shortened shafts (Fig. 5E, yellow arrowheads) to tiny, unpigmented shafts (white arrowheads) to complete shaft-to-socket transformations (blue arrowheads). These defects in the establishment of the shaft cell fate mimic H loss-of-function (or N gain-of-function) phenotypes. Thus, the normal direction of H's cell fate effects in vivo (promoting the N-independent shaft fate) can be reversed (promoting the N-dependent socket fate) by replacement of its putative transcriptional repression domains with an activation domain. H therefore shares with many transcriptional repressors the property that such substitution creates a dominant-negative form of the protein.
H cell-autonomously converts socket cells to the shaft cell fate
Overexpression of H under control of a heat-shock promoter has long been known to cause socket-to-shaft cell fate conversions (Bang and Posakony 1992). However, the ubiquitous expression pattern of heat-shock constructs leaves open the question of whether H can act cell-autonomously, or whether its effects on the socket cell are mediated by a nearby cell(s) that is also affected by H overexpression. The sca-GAL4 driver, though more restricted in its spatial activity pattern, likewise drives expression in cells neighboring the socket cell. The proposal that H antagonizes N-dependent cell fates by repressing N-activated target genes via Su(H) requires that H can function cell-autonomously, and we sought to demonstrate this explicitly. We constructed a socket cell-specific GAL4 driver, ASE5-GAL4. This construct uses a 372-bp autoregulatory socket enhancer of Su(H), called ASE5 (Barolo et al. 2000), to drive high levels of GAL4 expression exclusively in socket cells of external sense organs. In UAS-Hwt; ASE5-GAL4 flies, all bristles lack normal sockets and include ectopic shaft structures (Fig. 5F,G), indicating that the cell fate effects of H overexpression are in fact cell-autonomous. This finding is consistent with our proposal that H acts as a transcription factor, effecting, through Su(H), the direct repression of N pathway target genes. The effects of Hwt, HΔC, HΔC-VP16, and HN-VP16 on the socket cell fate when driven by ASE5-GAL4 were similar to those seen with sca-GAL4, except that, as expected, no effects on the shaft cell fate were observed (data not shown).
Discussion
Links between H and transcriptional repression
A gel retardation experiment reported by Brou et al. (1994), indicating that H can inhibit the binding of Su(H) to DNA in vitro, has strongly influenced interpretations of genetic studies of H, Su(H), and N. A DNA-binding-inhibition model of H function is indeed consistent with both loss- and gain-of-function genetic data demonstrating that H affects cell fate in a manner antagonistic to N signaling, including the N-stimulated transcriptional activation function of Su(H) (Bang et al. 1991, 1995; Bang and Posakony 1992; Schweisguth and Posakony 1994; Bailey and Posakony 1995; Lyman and Yedvobnick 1995). However, the recent discovery of Su(H)-mediated transcriptional repression (Barolo et al. 2000; Morel and Schweisguth 2000) has forced a reconsideration of this simple model, since it makes incorrect predictions about the effect of H on a cell fate that is dependent on the repression function of Su(H) (see below; Barolo et al. 2000). We propose that the genetic data on cell fate are instead consistent with a different role for H: facilitating transcriptional repression by Su(H).
We have reported here that the H protein contains two conserved transcriptional repression domains: an eh1/octapeptide-like motif that binds the Gro corepressor, and a Px(D/N)LS-type motif that binds the dCtBP corepressor. We found that both the dCtBP-binding domain and the region of H containing the Gro-binding domain contribute to H function in vivo. Taken together, these data are consistent with the proposal that H promotes the repression of N pathway target genes by recruiting the corepressors Gro and dCtBP to Su(H).
Gro and dCtBP promote a N-inhibited cell fate
During the socket/shaft cell fate decision in adult mechanosensory bristle development, the cell that responds to N signaling takes the socket fate, while its sister cell, in which N signal transduction is blocked by the Numb protein, takes the shaft fate (for review, see Posakony 1994). Overexpression of Su(H), or loss of H function, during the socket/shaft decision causes both cells to adopt the socket fate; conversely, overexpression of H, or loss of Su(H) function, results in two shaft cells (Bang et al. 1991; Bang and Posakony 1992; Schweisguth and Posakony 1994). Our recent work on transcriptional regulation of Su(H) established that autorepression by Su(H) in shaft cells is important for maintaining the shaft cell fate (Barolo et al. 2000). In the present study, we demonstrated that the corepressors Gro and dCtBP are important for specification of the shaft cell, a fate which is inhibited by N signaling and depends on both H activity and Su(H)-mediated repression. We find that reduction of gro or dCtBP function strongly enhances the effects of both reduction of H activity and loss of Su(H) repression, and suppresses the effects of reduced N signaling in the bristle lineage. We therefore conclude that Gro and dCtBP, along with H and transcriptional repression mediated by Su(H), act in the opposite direction from the N signaling pathway during the socket/shaft cell fate decision, in that they promote the fate (shaft) that is inhibited by N signaling. Our observation that both gro and dCtBP heterozygotes show a weak dominant (haploinsufficient) shaft-to-socket cell fate conversion phenotype is further confirmation of an important role for both corepressors in promoting the shaft cell fate. These results represent the first in vivo functional evidence for the involvement of Gro and dCtBP in transcriptional repression mediated by Su(H).
Gro: A “double agent” in lateral inhibition
Our genetic analyses show that gro loss-of-function mutations enhance the effects of reduced H activity on two N-mediated cell fate decisions, the socket/shaft decision and the epidermal/SOP decision, while reduction of gro activity suppresses the effects of N loss of function on the socket/shaft and pIIA/pIIB cell fate decisions. In addition, we find that gro has a weak haploinsufficient bristle loss phenotype, resembling an excess of N signaling. A role for gro in promoting the SOP cell fate is surprising, because gro was originally identified as a “neurogenic” gene that acts to inhibit the SOP fate downstream of N signaling, in its capacity as a corepressor for bHLH transcriptional repressor proteins encoded by N target genes in the Enhancer of split gene complex [E(spl)-C]. In fact, gro was named after the phenotype of flies homozygous for gro1, a weak hypomorphic allele: bushy tufts of bristles over the eyes caused by a failure of N-mediated lateral inhibition of the SOP fate (for review, see Parkhurst 1998). At least one E(spl)-C bHLH repressor gene appears to be directly repressed by Su(H) in SOPs (B. Castro, S.B., and J.W.P., unpubl.); we are currently testing our proposal that Gro promotes the SOP fate by cooperating with H to repress N target genes in this cell. If proved, this would represent a novel and complex form of regulation, in which Gro inhibits the SOP fate in all but one cell of the proneural cluster by partnering with the E(spl)-C bHLH repressors, and simultaneously promotes the SOP fate in one neighboring cell by preventing the expression of its own partners.
DNA binding by a H/Su(H) complex in vitro
Potent inhibition of Su(H)'s DNA binding activity by H, as proposed by Brou et al. (1994), is clearly incompatible with the proposition that a H/Su(H) complex directly represses N/Su(H) target genes. The mechanosensory organ lineage offers a particularly clear experimental example of this conflict. We showed recently that proper specification of the shaft cell fate requires autorepression by Su(H); loss of this repression causes the shaft cell to transform its fate at substantial frequency to that of its sister, the socket cell (Barolo et al. 2000). If H functions primarily to antagonize DNA binding by Su(H), then reduction of H activity should if anything lead to an increase in Su(H) autorepression, which should in turn stabilize the shaft fate. Instead, the shaft cell is highly sensitive to decreased H function, which readily causes its transformation to a socket cell (Lees and Waddington 1942; Bang et al. 1991). Thus, the genetic data on H's role in preserving the bristle shaft cell fate are irreconcilable with a simple Su(H)-DNA-binding-inhibition model for H function.
More generally, it has become apparent that repression of N targets by Su(H) is just as general and important a mechanism in Drosophila as it is in vertebrates (Barolo et al. 2000; Klein et al. 2000; Morel and Schweisguth 2000; Furriols and Bray 2001). If Su(H)-mediated repression of N target genes is essential for proper specification of N-independent cell fates, as we have shown for the shaft cell (Barolo et al. 2000), then preventing Su(H)-DNA interaction by overexpressing H would have the same effect as deleting Su(H) binding sites in N target genes: namely, their derepression, leading to N gain-of-function phenotypes (such as shaft-to-socket conversions). Instead, however, overexpression of H has been shown to repress the activity of N-regulated genes (Bailey and Posakony 1995), and the phenotypic effects of gain and loss of H function suggest a strictly antagonistic relationship between H and N signaling. Therefore, we believe that the accumulated genetic data point to a role for H in facilitating Su(H)-mediated repression of N target genes.
We have shown here that purified Su(H) can bind efficiently to both purified H and DNA simultaneously, allowing the possibility that a H/Su(H) complex may function as a transcription factor. Morel et al. (2001) reported a weak supershift of Su(H)/DNA complexes by full-length H, and a stronger supershift by an N-terminal H fragment (H1–293). We observe a robust supershift of Su(H)/DNA complexes with full-length H, strongly supporting the notion that H/Su(H)/DNA complexes may form efficiently in vivo. The discrepancy between our results and those of previous studies may reflect different experimental conditions, such as buffer composition or protein purification protocols, or may be due to the relatively low H:Su(H) molar ratios used in our experiments. Further work will be needed to reject or confirm the possibility that H antagonizes the Su(H)-DNA interaction under physiological conditions. However, given the genetic arguments outlined above, and the consistency of our results with a view of H as a transcriptional repressor, it seems likely that DNA-binding inhibition is not the primary mechanism by which H contributes to the specification of N-independent cell fates.
In vivo experiments consistent with transcriptional repression by H/Su(H) complexes
Misexpression of wild-type and modified forms of H in the adult bristle lineage has led us to three conclusions about the function of the H protein in vivo. First, the replacement of the putative repression domains of H with a transcriptional activation domain results in a dominant-negative form of H that elicits N gain-of-function phenotypes, a result consistent with normal repression of N target genes by H. Conversion to an antimorphic form by the addition of an activation domain is a common property of transcriptional repressor proteins (e.g., Waltzer et al. 1995; Jimenez et al. 1996; Gao et al. 2001; Kudoh and Dawid 2001).
Second, the significant, but somewhat weakened, effects of HΔC relative to wild-type H suggests that the C-terminal region of H including a dCtBP-interaction domain is important for some, but not all, of H's activity in vivo. This conclusion is supported by analysis of the H22 allele, which produces mutant H protein lacking its C-terminal 69 aa, including the PLNLS motif (Maier et al. 1997). H22, unlike H null alleles, is a homozygous viable mutation, and its effects on N-mediated cell fate decisions, though strong, are milder than those of H null mutations (Bang et al. 1991). The fact that the H22/H22 phenotype, unlike the H null heterozygote phenotype, is not enhanced by loss of dCtBP, as well as the absence of any other consensus CtBP binding sites in the H protein, are consistent with the idea that H interacts with dCtBP solely via this C-terminal motif. If this is indeed the case, then both the mildness of the H22 mutant phenotype and the potency of misexpressed HΔC protein indicate that the dCtBP corepressor contributes some, but not all, of the repressive activity of the H protein.
Third, the dominant-negative activity of H-VP16 depends on the removal of the region of H containing the Gro-binding domain, suggesting that this region contributes some of the wild-type repression function of the H protein. These last two conclusions conflict with the assertion of Morel et al. (2001) that the C-terminal dCtBP binding site is wholly responsible for H-mediated repression. Overall, our misexpression experiments provide evidence that both the region of H that binds to dCtBP and the region that binds to Gro contribute to the function of H in vivo. This is consistent with the strong phenotypic interaction between H and the genes encoding these two corepressors.
An adaptor model for H function: Bridging the gap between Su(H) and its corepressors
Our results support the hypothesis that H antagonizes N signaling by acting as an adaptor molecule between the transcription factor Su(H) and the corepressor proteins Gro and dCtBP (Fig. 6). This model entails an unusual mechanism of repression: DNA-binding transcriptional repressors that recruit CtBP or the Gro family of corepressors generally do so via direct protein–protein interactions (for review, see Courey and Jia 2001), although evidence for CtBP recruitment by non-DNA-binding proteins has been reported (Turner and Crossley 1998; Melhuish and Wotton 2000; Vo et al. 2001). In mammalian cells, the corepressors SMRT and CIR bind directly to the Su(H) homolog CBF1 (Kao et al. 1998; Hsieh et al. 1999).
Figure 6.
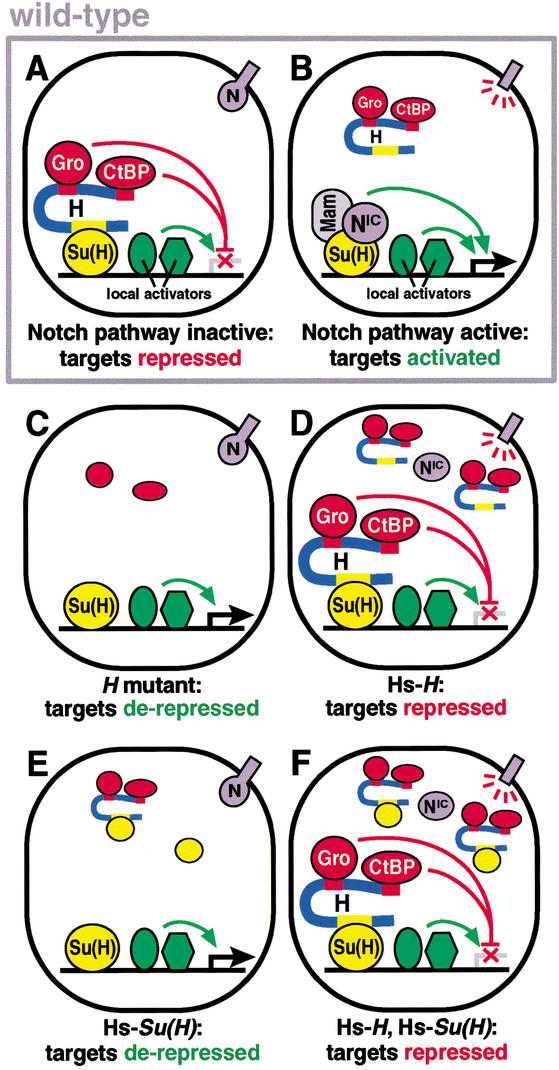
Proposed transcriptional roles for Su(H), H, N, Gro, and dCtBP during socket/shaft cell fate determination. (A) In the absence of N signaling, Su(H) binds H, which in turn binds the corepressors Gro and dCtBP, repressing N target genes and promoting the shaft cell fate (“default repression”). A similar H repression complex, including H and Gro but perhaps not dCtBP, promotes the SOP cell fate during lateral inhibition. (B) In the presence of N signaling, a NIC activation complex, probably including Mastermind (Mam), displaces the H repression complex and activates N target genes, promoting the socket cell fate. “Local activators” are transcription factors, bound to N target promoters, that cooperate with Su(H)/NIC to activate transcription during N signaling, and which must be counteracted by Su(H)/H repression complexes in the absence of signaling (Barolo et al. 2000). (C_–_F) Proposed mechanisms underlying phenotypes caused by experimental changes in levels of H and/or Su(H). (C) In H mutant flies, N/Su(H) target genes are derepressed, leading to N gain-of-function phenotypes. (D) Upon H overexpression, H repression complexes displace NIC activation complexes, repressing N target genes and causing N loss-of-function phenotypes. (E) Su(H) overexpression causes titration of H repression complexes, thereby derepressing N target genes. (F) Su(H) overexpression strongly enhances the H overexpression phenotype, due to an increased number of Su(H)/H repression complexes.
If protein complexes containing H are important for Su(H)-mediated repression, why is H not found in vertebrates? Several possible explanations are apparent. First, vertebrate homologs of Su(H) may not make use of an adaptor/corepressor complex, but rather may recruit all corepressors (possibly including Gro and CtBP) directly, as in the case of SMRT and CIR. A second, related possibility is that vertebrate versions of Su(H) do not utilize Gro and CtBP as corepressors; in this view, H may have appeared exclusively in the protostome lineage to add to Su(H)'s corepressor repertoire. Third, it is possible that vertebrates employ not a homolog, but an analog, of H, one evolved independently after the divergence of protostomes and deuterostomes. Finally, the fact that the predicted D. melanogaster H protein is only 33% identical to its apparent ortholog in A. gambiae suggests that the vast majority of the H sequence is not under selective constraint. Thus, an ortholog of H may indeed exist in vertebrates, but be so highly diverged as to be unrecognizable by typical sequence analyses. This possibility seems less likely, since at least the Su(H)-interaction domain might be expected to be well conserved, given the strong evolutionary conservation of Su(H) itself (Schweisguth and Posakony 1992).
In contrast to a DNA-binding inhibition model for H function, an adaptor/corepressor model explains why H counters NIC/Su(H)-mediated activation, but not Su(H)-mediated repression. Like previous views of H function, this model presumes competition between Su(H)-binding partners, in this case between NIC-containing activation complexes and H/Gro/dCtBP repression complexes. NIC activation complexes are likely to include the Mastermind (Mam) protein (Kitagawa et al. 2001 and references therein), and may also include the p300 coactivator (Oswald et al. 2001). In the presence of N signaling, Su(H)/NIC/Mam complexes presumably replace Su(H)/H/Gro/dCtBP complexes on target genes, and convert Su(H) from a repressor to an activator (Fig. 6B). Whether this occurs by simple affinity-based competition for binding to Su(H), or by a mechanism involving active impairment of the H/Su(H) interaction, is unknown. Under an adaptor/corepressor model, the H mutant phenotype results from derepression of Su(H)/N target genes in cells lacking N pathway activity, thus mimicking an increase in N signaling (Fig. 6C). The H overexpression phenotype may be explained by the displacement of NIC-containing activation complexes by an excess of H-containing repression complexes, thus repressing NIC/Su(H) target genes in cells that respond to the N signal (Fig. 6D).
Similarly, we propose that the Su(H) overexpression phenotype, which resembles a gain of N function, is caused by a titration of H repression complexes by excess Su(H), and the subsequent derepression of Su(H) target genes (Fig. 6E). The fact that overexpression of Su(H) strongly enhances the effect of H overexpression on lateral inhibition (Morel et al. 2001; F. Schweisguth, S. Barolo, and J.W. Posakony, unpubl.) supports this view (Fig. 6F), but is very much at odds with a DNA-binding-inhibition model for H function.
Default repression: A crucial counterpoint to N signaling
It has recently become apparent that the transcriptional target genes of at least six major developmental signaling pathways are in many cases subject to “default repression”; that is, binding sites for signal-regulated transcription factors, which mediate activation during signaling events, mediate repression in the absence of signaling (for review, see Barolo and Posakony 2002). Each of these pathways uses a different mechanism to switch from repression to activation upon stimulation of the pathway, but in each case, the effect seems to be the same: restricting the expression of pathway target genes to cells that receive active signaling. Our results strongly suggest that H contributes to default repression in the N pathway by directly recruiting the corepressors Gro and dCtBP to Su(H), and that formation of H/Su(H) repression complexes is crucial for the establishment of two N-inhibited cell fates, the SOP and shaft cell fates. Default repression, therefore, appears to be as important as signal-dependent activation for proper cell fate specification in this developmental context.
Materials and methods
Fly stocks, crosses, and temperature shifts
H, gro, and dCtBP loss of function
HE31 is a null allele (Schweisguth and Posakony 1994). groE73 is a genetically null allele (Preiss et al. 1988). dCtBP87De-10 is a strong hypomorphic or null allele (Hilliker et al. 1980; Poortinga et al. 1998).
Su(H) derepression
Su(H)RC9Xm is a Su(H) rescue transgene with single-base mutations in autoregulatory Su(H) binding sites (Barolo et al. 2000). In Figure 2A, dCtBP/+ flies are siblings of gro/+ flies, and H/+ flies are siblings of either H/dCtBP or H/gro flies. In Figure 2B, Su(H)RC9Xm/+ flies are siblings of either Su(H)RC9Xm/dCtBP or Su(H)RC9Xm/gro flies.
N loss of function
Nts1 females (Shellenbarger and Mohler 1978) were crossed to w1118/Y, HE31 e/TM6C, dCtBP87De-10/TM6C, or e4 groE73 tx/TM6C males. Non-Tubby progeny were kept at 25°C until 22 h after puparium formation, shifted to 31°C for 18 h in a thermal cycler, and then returned to 25°C. Male (Nts1/Y) adult or pharate adult progeny were examined.
In vitro protein interaction assays
H-Gro interaction
35S-Gro and 35S-Emc proteins were generated with the TNT Quick coupled transcription/translation kit (Promega), from cDNA clones in pBluescript and pNB40, respectively. Glutathione beads bound to various purified GST fusion proteins, or to GST alone, were incubated with translation lysate in 600 μL of HEMNK buffer (Dubnicoff et al. 1997) for 2 h at 4°C. Beads were washed with 4 × 600 μL HEMNK at 4°C. Samples were analyzed by SDS-PAGE followed by autoradiography.
H-dCtBP interaction
A biotinylated peptide (HC) corresponding to the extreme C-terminus of H (Biotin-spacer-SAMFHTSSLRNEQSSDLPLNLSKH-CONH2) was synthesized (SynPep). Various amounts of HC peptide resuspended in PBS were added to 50 μL of glutathione beads bearing purified GST-dCtBP or GST alone and incubated for 1 h at 4°C in 100 μL TLS buffer (150 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl pH 8.0, 0.5% NP-40). Beads were washed with 6 × 100 μL TLS. Ten microliters of 35S-streptavidin (Amersham Pharmacia) was incubated with the beads in 50 μL TLS for 30 min at 4°C, followed by 6 × 200 μL washes with TLS. Samples were analyzed by SDS-PAGE followed by autoradiography. The 35S-labeled, 6xHis-tagged H fragments H1036–1070 and H1036–1077 were generated from PCR products cloned into the pRSET vector (Invitrogen) and were produced in E. coli extract with the EcoPro coupled transcription/translation kit (Novagen).
DNA binding assays
Electrophoretic mobility shift assays (EMSAs) were performed as described by Bailey and Posakony (1995), using purified His-tagged Su(H) and H proteins. Oligonucleotide probe sequence is available upon request.
Transgenic expression constructs
UAS-H misexpression constructs were cloned into the pUAST vector (Brand and Perrimon 1993). UAS-HΔC contains a stop codon after codon 1070, eliminating the final seven codons (PLNLSKH). UAS-HΔC-VP16 contains the VP16 activation domain sequences (codons 414–491) fused downstream of H(1–1070), separated by a _Kpn_I site. UAS-HN-VP16 contains the same VP16 domain codons fused downstream of H(1–653).
Acknowledgments
We thank Katie Medders for gathering phenotypic data. Adina Bailey constructed the GST-H expression plasmid. We thank Yutaka Nibu and Mike Levine for communicating unpublished results, and for generously providing the GST-dCtBP plasmid. We thank Lisa Johnson, Nick Reeves, and Par Towb for valuable comments and criticism. This work was supported by NIH grant GM62279 to J.W.P.; S.B. was supported in part by an NIH NRSA postdoctoral fellowship (GM20123).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
EMAIL: jposakony@ucsd.edu; FAX (858) 534-0566.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.987402.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of Hairless directly activates transcription of Enhancer of split Complex genes in response to Notch receptor activity. Genes & Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Bang AG. Genetic and molecular analysis of Hairless and its role in Drosphila adult peripheral nervous system development. Ph.D. Thesis. University of California San Diego; 1993. [Google Scholar]
- Bang AG, Bailey AM, Posakony JW. Hairless promotes stable commitment to the sensory organ precursor cell fate by negatively regulating the activity of the Notch signaling pathway. Dev Biol. 1995;172:479–494. doi: 10.1006/dbio.1995.8033. [DOI] [PubMed] [Google Scholar]
- Bang AG, Hartenstein V, Posakony JW. Hairless is required for the development of adult sensory organ precursor cells in Drosophila. Development. 1991;111:89–104. doi: 10.1242/dev.111.1.89. [DOI] [PubMed] [Google Scholar]
- Bang AG, Posakony JW. The Drosophila gene Hairless encodes a novel basic protein that controls alternative cell fates in adult sensory organ development. Genes & Dev. 1992;6:1752–1769. doi: 10.1101/gad.6.9.1752. [DOI] [PubMed] [Google Scholar]
- Barolo S, Posakony JW. Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes & Dev. 2002;16:1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- Barolo S, Walker RG, Polyanovsky AD, Freschi G, Keil T, Posakony JW. A Notch-independent activity of Suppressor of Hairless is required for normal mechanoreceptor physiology. Cell. 2000;103:957–969. doi: 10.1016/s0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- Beverley SM, Wilson AC. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J Mol Evol. 1984;21:1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Lecourtois M, Vandekerckhove J, Kourilsky P, Schweisguth F, Israel A. Inhibition of the DNA-binding activity of Drosophila Suppressor of Hairless and of its human homolog, KBF2/RBP-Jκ, by direct protein-protein interaction with Drosophila Hairless. Genes & Dev. 1994;8:2491–2503. doi: 10.1101/gad.8.20.2491. [DOI] [PubMed] [Google Scholar]
- Courey AJ, Jia S. Transcriptional repression: The long and the short of it. Genes & Dev. 2001;15:2786–2796. doi: 10.1101/gad.939601. [DOI] [PubMed] [Google Scholar]
- Dou S, Zeng X, Cortes P, Erdjument-Bromage H, Tempst P, Honjo T, Vales LD. The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol Cell Biol. 1994;14:3310–3319. doi: 10.1128/mcb.14.5.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnicoff T, Valentine SA, Chen G, Shi T, Lengyel JA, Paroush Z, Courey AJ. Conversion of dorsal from an activator to a repressor by the global corepressor Groucho. Genes & Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard D, Jimenez G, Heavey B, Busslinger M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000;19:2292–2303. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols M, Bray S. Dissecting the mechanisms of Suppressor of Hairless function. Dev Biol. 2000;227:520–532. doi: 10.1006/dbio.2000.9923. [DOI] [PubMed] [Google Scholar]
- ————— A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr Biol. 2001;11:60–64. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Kobayakawa Y, Tamura K, Kimura K, Kawaichi M, Tanimura T, Honjo T. Suppressor of Hairless, the Drosophila homologue of RBP-Jκ, transactivates the neurogenic gene E(spl)m8. Jpn J Genet. 1995;70:505–524. doi: 10.1266/jjg.70.505. [DOI] [PubMed] [Google Scholar]
- Gao X, Chandra T, Gratton MO, Quelo I, Prud'homme J, Stifani S, St-Arnaud R. HES6 acts as a transcriptional repressor in myoblasts and can induce the myogenic differentiation program. J Cell Biol. 2001;154:1161–1172. doi: 10.1083/jcb.200104058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Posakony JW. A dual function of the Notch gene in Drosophila sensillum development. Dev Biol. 1990;142:13–30. doi: 10.1016/0012-1606(90)90147-b. [DOI] [PubMed] [Google Scholar]
- Hilliker AJ, Clark SH, Chovnick A, Gelbart WM. Cytogenetic analysis of the chromosomal region immediately adjacent to the rosy locus in Drosophila melanogaster. Genetics. 1980;95:95–110. doi: 10.1093/genetics/95.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ-D, Hayward SD. Masking of the CBF1/RBPJκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Pinchin SM, Ish-Horowicz D. In vivo interactions of the Drosophila Hairy and Runt transcriptional repressors with target promoters. EMBO J. 1996;15:7088–7098. [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes & Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Oyama T, Kawashima T, Yedvobnick B, Kumar A, Matsuno K, Harigaya K. A human protein with sequence similarity to Drosophila Mastermind coordinates the nuclear form of Notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol Cell Biol. 2001;21:4337–4346. doi: 10.1128/MCB.21.13.4337-4346.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T, Seugnet L, Haenlin M, Martinez Arias A. Two different activities of Suppressor of Hairless during wing development in Drosophila. Development. 2000;127:3553–3566. doi: 10.1242/dev.127.16.3553. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Dawid IB. Role of the iroquois3 homeobox gene in organizer formation. Proc Natl Acad Sci. 2001;98:7852–7857. doi: 10.1073/pnas.141224098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the Enhancer of split Complex genes triggered by Notch signaling. Genes & Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- Lees AD, Waddington CH. The development of the bristles in normal and some mutant types of Drosophila melanogaster. Proc Roy Soc Ser B. 1942;131:87–110. [Google Scholar]
- Lyman DF, Yedvobnick B. Drosophila Notch receptor activity suppresses Hairless function during adult external sensory organ development. Genetics. 1995;141:1491–1505. doi: 10.1093/genetics/141.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D, Marquart J, Thompson-Fontaine A, Beck I, Wurmbach E, Preiss A. In vivo structure-function analysis of Drosophila Hairless. Mech Dev. 1997;67:97–106. doi: 10.1016/s0925-4773(97)00117-2. [DOI] [PubMed] [Google Scholar]
- Marquart J, Alexief-Damianof C, Preiss A, Maier D. Rapid divergence in the course of Drosophila evolution reveals structural important domains of the Notch antagonist Hairless. Dev Genes Evol. 1999;209:155–164. doi: 10.1007/s004270050239. [DOI] [PubMed] [Google Scholar]
- Melhuish TA, Wotton D. The interaction of the Carboxyl terminus-Binding Protein with the Smad corepressor TGIF is disrupted by a holoprosencephaly mutation in TGIF. J Biol Chem. 2000;275:39762–39766. doi: 10.1074/jbc.C000416200. [DOI] [PubMed] [Google Scholar]
- Morel V, Lecourtois M, Massiani O, Maier D, Preiss A, Schweisguth F. Transcriptional repression by Suppressor of Hairless involves the binding of a Hairless-dCtBP complex in Drosophila. Curr Biol. 2001;11:789–792. doi: 10.1016/s0960-9822(01)00224-x. [DOI] [PubMed] [Google Scholar]
- Morel V, Schweisguth F. Repression by Suppressor of Hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes & Dev. 2000;14:377–388. [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst SM. Groucho: Making its Marx as a transcriptional co-repressor. Trends Genet. 1998;14:130–132. doi: 10.1016/s0168-9525(98)01407-3. [DOI] [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: A Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posakony JW. Nature versus nurture: Asymmetric cell divisions in Drosophila bristle development. Cell. 1994;76:415–418. doi: 10.1016/0092-8674(94)90105-8. [DOI] [PubMed] [Google Scholar]
- Postigo AA, Dean DC. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss A, Hartley DA, Artavanis-Tsakonas S. The molecular genetics of Enhancer of split, a gene required for embryonic neural development in Drosophila. EMBO J. 1988;7:3917–3927. doi: 10.1002/j.1460-2075.1988.tb03278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JV, Savenye ED, Lum D, Breitkreutz A. Dominant enhancers of Egfr in Drosphila melangaster: genetic links between the Notch and Egfr signaling pathways. Genetics. 1997;147:1139–1153. doi: 10.1093/genetics/147.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F, Posakony JW. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-o. [DOI] [PubMed] [Google Scholar]
- ————— Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development. 1994;120:1433–1441. doi: 10.1242/dev.120.6.1433. [DOI] [PubMed] [Google Scholar]
- Shellenbarger DL, Mohler JD. Temperature-sensitive periods and autonomy of pleiotropic effects of l(1)Nts1, a conditional Notch lethal in Drosophila. Dev Biol. 1978;62:432–446. doi: 10.1016/0012-1606(78)90226-9. [DOI] [PubMed] [Google Scholar]
- Smith ST, Jaynes JB. A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2- and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development. 1996;122:3141–3150. doi: 10.1242/dev.122.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The CtBP family: Enigmatic and enzymatic transcriptional co-repressors. Bioessays. 2001;23:683–690. doi: 10.1002/bies.1097. [DOI] [PubMed] [Google Scholar]
- Vo N, Fjeld C, Goodman RH. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol Cell Biol. 2001;21:6181–6188. doi: 10.1128/MCB.21.18.6181-6188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltzer L, Bourillot PY, Sergeant A, Manet E. RBP-Jκ repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Younger-Shepherd S, Jan LY, Jan YN. Only a subset of the binary cell fate decisions mediated by Numb/Notch signaling in Drosophila sensory organ lineage requires Suppressor of Hairless. Development. 1997;124:4435–4446. doi: 10.1242/dev.124.22.4435. [DOI] [PubMed] [Google Scholar]