A Fine Physical Map of the Rice Chromosome 4 (original) (raw)
Abstract
As part of an international effort to completely sequence the rice genome, we have produced a fine bacterial artificial chromosome (BAC)-based physical map of the Oryza sativa japonica Nipponbare chromosome 4 through an integration of 114 sequenced BAC clones from a taxonomically related subspecies O. sativa indica Guangluai 4 and 182 RFLP and 407 expressed sequence tag (EST) markers with the fingerprinted data of the Nipponbare genome. The map consists of 11 contigs with a total length of 34.5 Mb covering 94% of the estimated chromosome size (36.8 Mb). BAC clones corresponding to telomeres, as well as to the centromere position, were determined by BAC-pachytene chromosome fluorescence in situ hybridization (FISH). This gave rise to an estimated length ratio of 5.13 for the long arm and 2.9 for the short arm (on the basis of the physical map), which indicates that the short arm is a highly condensed one. The FISH analysis and physical mapping also showed that the short arm and the pericentromeric region of the long arm are rich in heterochromatin, which occupied 45% of the chromosome, indicating that this chromosome is likely very difficult to sequence. To our knowledge, this map provides the first example of a rapid and reliable physical mapping on the basis of the integration of the data from two taxonomically related subspecies.
[The following individuals and institutions kindly provided reagents, samples, or unpublished information as indicated in the paper: S. McCouch, T. Sasaki, and Monsanto.]
Rice is the staple food crop for more than one-half of the world's population and has been used as the major model system in cereal genome research because of its small genome size and high synteny to other monocots (Ahn and Tanksley 1993; Moore et al. 1995; Gale and Devos 1998). For a model plant, rice also has an efficient transformation system (Hiei et al. 1994), a high-density genetic linkage map (Harushima et al. 1998), and a comprehensive rice transcript map with 6591 expressed sequence tag (EST) sites (Wu et al. 2002). The rice genome, estimated to be ∼430 Mb (Arumuganathan and Earle 1991), is the smallest one among major cereals, but it is by far the largest plant genome to be sequenced. Cultivated rice (Oryza sativa) is classified into two subspecies, indica and japonica (Oka 1991), which include most of the rice cultivars grown in the world. An international consortium, the International Rice Genome Sequencing Project (IRGSP) (http://rgp.dna.affrc.go.jp/cgi-bin/statusdb/seqcollab.pl) has adopted a “clone by clone” strategy to completely and accurately sequence the entire genome of the rice O. sativa ssp. japonica cultivar Nipponbare (Sasaki and Burr 2000). This strategy requires a comprehensive physical map of the entire japonica Nipponbare genome.
Approaches based on BAC fingerprinting, iterative hybridization, and sequence tag connector (STC, which is to use BAC-end sequences for connecting BAC clones by sequence identity) have been successfully applied in the physical mapping of large complex genomes, including human and Arabidopsis thaliana (Marra et al. 1997, 1999; Mozo et al. 1999; The International Human Genome Mapping Consortium 2001). In addition to sequencing the genome of the O. sativa ssp. indica cultivar Guangluai 4, we are sequencing the Nipponbare chromosome 4 as part of the international effort. Here we present the construction of a sequence-ready BAC physical map of the chromosome 4 of O. sativa ssp. japonica Nipponbare through an integrated approach using a large set of sequence scaffolds from a taxonomically relative subspecies O. sativa indica variety Guangluai 4 as a starting point. This approach, based on the assumption that the indica and japonica genomes are collinear, included the experimental hybridization and STC approaches in parallel with the sequencing project and can serve as a model system with publicly available genome sequence data from one species to construct the genome physical map of a taxonomically related species.
RESULTS AND DISCUSSION
Identification of the Chromosome 4 Seed BACs from the Rice Subspecies Indica Guangluai 4
We initiated a genome-sequencing project of the O. sativa indica Guangluai 4 in 1993. Two BAC libraries of the Guangluai 4 were constructed with restriction enzymes _Hin_dIII or _Bam_HI designated as OSIGBAC-H (Tao et al. 1994) and OSIGBAC-B, respectively. OSIGBAC-H clones were fingerprinted and assembled into BAC contigs to cover the entire genome (Hong et al. 1997). Because of a low genome coverage of the OSIGBAC-H library (<4X) and limited unique markers available for BAC anchoring, the actual genome coverage of the contigs was less than estimated. To construct a more complete physical map of the Guangluai 4 chromosome 4, we took a total of 182 genetic restriction fragment length polymorphism (RFLP) markers of the Nipponbare chromosome 4, provided by RGP (http://rgp.dna.affrc.go.jp/publicdata/geneticmap2000), as probes to hybridize the Guangluai 4 BAC libraries. Collectively, 566 BACs were identified by hybridization with 108 RFLP markers and 50 contigs assembled. The anchored BAC contigs were formed as a regional physical map of the Guangluai 4 chromosome 4. We also generated 13,000 BAC-end sequences from the OSIGBAC-H library for the STC approach to extend the anchored BAC contigs. The relationship within the contigs was further confirmed by restriction enzyme digestion and PCR analysis. We selected 84 BACs identified by at least two adjacent genetic markers, according to the genetic linkage map (Harushima et al. 1998), as seed BACs for complete sequencing. Overall, 114 Guangluai 4 BACs from the chromosome 4 with an average overlap of 21.9% have been sequenced. The total length of the sequenced Guangluai 4 BAC clones is ∼8.4 Mb. All of the sequenced Guangluai 4 BACs and the corresponding markers are listed in a table (at http://www.ncgr.ac.cn/rice/chr4/indica.htm).
Construction of a Fine Chromosome 4 Physical Map of the Rice Subspecies japonica Nipponbare
During the course of this study, the Clemson University Genomics Institute (CUGI) made the fingerprint data of two Nipponbare BAC libraries (20X coverage) publicly available (by FTP at ftp://ftp.genome.clemson.edu/pub/rice/fpc/) (Soderlund et al. 2000), as well as assembled contigs by a fingerprinting analysis (http://www.genome.clemson.edu/projects/rice/fpc) and 127,459 BAC-end sequences (http://www.genome.clemson.edu/projects/rice/rice_bac_end ) (Mao et al. 2000). Integration of the fingerprinted contig map with the genetic map and the Monsanto working draft data (Barry 2001) resulted in a comprehensive physical map of the entire rice genome with an estimated genome coverage of 89% (Chen et al. 2002). The chromosome 4 map had a coverage of 87.7% with 23 gaps (Chen et al., 2002). As part of the IRGSP effort to sequence the rice genome, we have made an effort to construct a sequence-ready Nipponbare chromosome 4 physical map.
Because O. sativa indica and japonica are closely related subspecies, a synteny is expected to be very high (Gale and Devos 1998). Therefore, we could use genetically anchored and fully sequenced indica BACs to search the Nipponbare BAC-end sequences for locating BAC contigs on its chromosome 4. To implement this strategy, we first assessed the sequence synteny of the two subspecies. A sequenced OSIGBAC-H0102C09, anchored by marker R2502, was used to search the Nipponbare BAC-end databases. A BAC clone OSJNBa0068L06 was subsequently identified on the basis of its end sequence identity to OSIGBAC-H0102C09 and then completely sequenced. We performed sequence alignment analysis between them. The result showed that they are almost collinear and identical except for four insertions or deletions (indels) of three miniature inverted transposable elements (MITEs) and one retrotransposon (Han, Q. Feng, Y.J. Zhang, Q. Zhao, et al., in prep.). The sequence identity between their collinear regions is about 99%. We also compared other chromosomal regions and found that the major differences between the two subspecies were caused by retrotransposons and transposons (Han, Q. Feng, Y.J. Zhang, Q. Zhao, et al., in prep.). These repetitive sequences could be identified and marked from unique sequences, which indicates that the large segments of sequences that contain continued unique sequences from one of the subspecies could be used confidently to locate the sequences of collinear regions from another subspecies. This enabled us to use the known indica sequence scaffolds to rapidly and accurately construct a fine physical map of the Nipponbare chromosome 4 through the integration of the fingerprinted data from the CUGI. Conversely, the japonica sequence scaffolds could also used for anchoring the BACs from the Guangluai 4 libraries by searching their BAC-end sequences. This approach has at least three advantages. First, the large number of the Nipponbare BAC contigs will be anchored rapidly on the chromosome 4 by use of the fully sequenced indica chromosome 4 BACs. Second, the integration of large segments of indica sequences with the Nipponbare BAC ends and fingerprinted BAC contigs will produce a minimum tiling path of the BACs for complete sequencing of the chro mosome 4. Third, the localization of identified BAC contigs will be unambiguous. To further cover the chromosome, a total of 182 genetic and 407 EST markers of the chromosome 4 from Nipponbare (Wu et al. 2002) was also used for the physical mapping by an in silico anchoring (Yuan et al. 2000). Anchored contigs were extended by chromosome walking through a re peated colony hybridization, as well as the in silico approach.
Thus far, we have integrated 114 sequenced seed BACs of Guangluai 4 with the Nipponbare fingerprinted BAC-contig data, the BAC-end sequences, and the genome physical map (ftp://ftp.genome.clemson.edu/pub/rice/fpc/) (Chen et al., 2002) to construct the chromosome 4 physical map ready for BAC clone selection for its complete sequencing. Taken together, 27 Nipponbare BAC contigs from the CUGI database (http://www.genome.clemson.edu/projects/rice/fpc) were anchored in silico. Twenty contigs were initially anchored by the Guangluai 4 sequence scaffolds. Comparison between the Guangluai 4 sequence scaffolds and the Nipponbare BAC ends enabled us to quickly make a large minimum-tiling path of BACs from the large sets of candidate clones. Locations of the selected Nipponbare BACs were unambiguously confirmed by combining the information of the fingerprinted contigs and the sequence identity between the Nipponbare BAC ends and the Guangluai 4 BAC sequence scaffolds (http://www.ncgr.ac.cn/rice/chr4/physicalmap/index.htm). The sequenced RFLP and EST markers of Nipponbare were also directly searched against the Nipponbare BAC-end database and 23 contigs identified by 25 RFLP and 93 EST markers with five contigs not previously known on the chromosome 4. The five contigs were anchored by the RFLP markers with three of them also anchored by the EST markers. Two more contigs (contigs 273 and 403) were identified through contig extension by chromosome walking. The 27 contigs were eventually merged into 11 (designated as CTG4–1 to CTG4–11) after the identification of several bridging clones. The location and length of the contigs and gaps are shown in Figure 1 and Table 1 together with the original CUGI's contigs. The detailed map of CTG4–1 as an example contig from the whole-chromosome map is shown in Figure 2. It shows the integrated data used for anchoring and joining the CUGI's fingerprinted contigs in this merged larger contig. The contigs were merged with the following three approaches (Table 1). First, the contigs were connected by contig-end sequence walk. Therefore, five bridging clones were identified, allowing a merge of seven contigs. Second, two Nipponbare contigs were merged by use of Guangluai 4 BACs. CTG4–8 originally consisted of three contigs (418, 88, and 89); the gap between contigs 418 and 88 was covered by a fully sequenced OSIGBAC-B0812A04 based on the overlaps with the sequenced Nipponbare BAC clones OSJNBa0072F16 and OSJNBa0060P14. Initially, we were unable to identify a Nipponbare BAC clone to bridge the two clones because no BAC-end sequence hits were detected in the CUGI and Monsanto BAC-end databases. Third, 11 gaps between the CUGI's contigs were joined after the sequencing analysis. Contigs 88 and 89 were finally merged after the completion of the two adjacent BAC sequences from both contigs (see the map of CTG4–8, http://www.ncgr.ac.cn/rice/chr4/physicalmap/ctg4-8.htm). The bridging BACs are described in Table 1. They were not previously assembled in the contigs because of either poor quality of the fingerprinting data or little overlap with the connected clones. The relationship between the fully sequenced BACs of the Guangluai 4 and the anchored Nipponbare contigs can be accessed at (http://www.ncgr.ac.cn/rice/chr4/physicalmap/index.htm). In addition, the sequence scaffolds from the two subspecies were used for extending contigs by searching against the BAC-end sequences from either the Guangluai 4 or Nipponbare variety by an iterative approach. In summary, the total length of the 11 BAC contigs was estimated to be ∼34.5 Mb (Fig. 1; Table 1). Therefore, on the basis of an estimated 36.8 Mb of the rice chromosome 4 (Saji et al. 2001), the physical map constructed here has a 94% chromosomal coverage and is the most covered chromosome 4 map obtained so far.
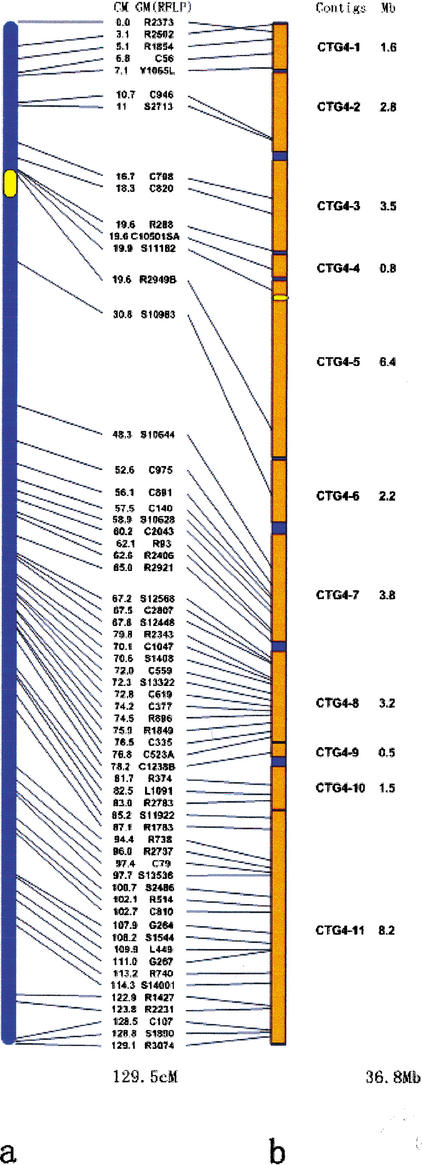
Figure 1.
Comparison between the genetic and physical maps of the rice chromosome 4. (A) A genetic linkage map. Genetic markers (GM), genetic distance (cM), and centromere (in yellow) are indicated. (B) A physical map. The portions of the 11 BAC contigs covering 34.5 Mb of the chromosome are in orange and the gaps in plain. The physical distance is indicated (Mb). The maps also show a discrepancy of two markers R2502 (3.1 cM) and R2373 (0 cM) between the genetic and physical maps revealed by DNA sequencing and fluorescence in situ hybridization (FISH) analysis (cross lines).
Table 1.
Status of the Rice Chromosome 4 Map
| Genetic Position (cM) | Physical length (Mb) | CUGl's contigs | Gap size (Mb) |
|---|---|---|---|
| CTG4-1 | 0.0–7.1 | 1.6 | contig 297, 78, 79 |
| 0.2 | |||
| CTG4-2 | 7.9–12.2 | 2.8 | contig 281, 80 |
| 0.3 | |||
| CTG4-3 | 13.1–19.6 | 3.5 | contig 273, 81, 84 |
| 0.2 | |||
| CTG4-4 | 19.6 | 0.8 | contig 404 |
| 0.2 | |||
| CTG4-5 | 19.6–24.9 | 6.4 | contig 82, 254, 83, 421 |
| 0.1 | |||
| CTG4-6 | 25.4–30.8 | 2.2 | contig 257 |
| 0.5 | |||
| CTG4-7 | 41.5–65.3 | 3.8 | contig 295, 85, 86, 322 |
| 0.4 | |||
| CTG4-8 | 67.2–77.9 | 3.2 | contig 418, 88, 89 (partial) |
| 0.1 | |||
| CTG4-9 | 78.0–78.2 | 0.5 | contig 269 |
| 0.4 | |||
| CTG4-10 | 81.7–84.1 | 1.5 | contig 403, 90, 91 |
| 0.1 | |||
| CTG4-11 | 84.5–129.6 | 8.2 | contig 92, 59 |
| Total | 34.5 | 27 | ∼2.5 |
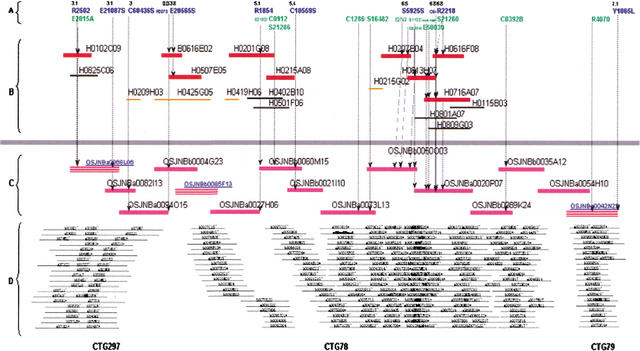
Figure 2.
The BAC contig map of CTG4-1 near the short arm telomere together with its collinear Guangluai 4 region. (A) Markers associated with the map. Fifteen EST (in green) and 11 RFLP (in blue) markers used to anchor contigs are shown together with their genetic positions (numbers in cM). (B) Anchored Guangluai 4 BAC contigs by the markers. Fully sequenced BACs (red lines), BACs identified using the Nipponbare sequence scaffold against the Guangluai 4 BAC ends (orange lines), and other clones (black lines) in the contigs are indicated. (C) Fifteen tiled and completed sequenced Nipponbare BACs. Three bridging clones OSJNBa0094015 and OSJNBb0004G23 for joining CUGI contigs 297 and 78 and OSJNBa0054H10 for CUGI contigs 78 and 79 are shown. (D) Display of three original fingerprinted CUGI contigs 297, 78, and 79. CTG4-1 is composed of 295 fingerprinted CUGI BACs and three extended bridge clones. Contig 79 was anchored by marker Y1065L by in silico anchoring. Contigs 297 and 78 were anchored by the Guangluai 4 sequence scaffolds as indicated in syntenic positions.
Confirmation of the Chromosome 4 Contigs by Fluorescence In Situ Hybridization (FISH) and Cytogenetic Analysis
To validate the contig relationship of the physical map, we conducted cytogenetic analysis with BAC clones within the assigned contigs. Eight CUGI's contigs (297, 78, and 79 covered by CTG4–1; 404 by CTG4–4; 83 and 421 by CTG4–5; 257 by CTG4–6; and 59 by CTG4–11) were initially confirmed on their chromosome locations by FISH of meiotic pachytene chromosomes (http://www.ncgr.ac.cn/rice/chr4/FISH/index.htm). The rest of six merged contigs (CTG4–2, 3, 7, 8, 9, and 10) were eventually identified (their locations on the chromosome 4) by FISH analysis (data not shown). OSJNBb0085F13 from CTG4–1 and OSJNBb0020J19 from CTG4–11 were mapped to distal regions close to the telomeres of the short and long arms, respectively (Fig. 3). To further determine its telemeric location, two additional BACs within CTG4–1 were subject to pachytene FISH analysis (Fig. 4). The orientations of the BAC clones showed that the order of the genetic markers R2373 (0 cM) and R2502 (3.1 cM) was misidentified on the genetic map. Their correct order on the physical map was further confirmed by sequence analysis of the marker-containing BACs (Fig. 2). The CUGI's contig 83 was about 2 Mb long, representing the largest contig identified by Chen et al. (2002) but anchored to 19.9–20.9 cM by DNA sequences of only three RFLP (E679S, E31045S, and E2466S) and two EST (R10666 and C53760) markers through in silico anchoring. No sequenced indica BACs were available at this region either. To verify this location, three BACs (OSJNBa0019J05, OSJNBb0068N06, and OSJNBb0056F09) from CTG83 were used for FISH analysis. The results showed that this contig was located near the pericentromeric region of the chromosome 4, and its location was therefore confirmed independently. All of the FISH results can be accessed at http://www.ncgr.ac.cn/rice/chr4/FISH/index.htm. The centromere of the chromosome 4 was physically mapped with a rice centromere-specific satellite DNA pRCS2 (Dong et al. 1998) as a probe (Fig. 3). DAPI staining of the chromosome 4 at the pachytene stage showed that the entire short arm and the pericentromeric region of the long arm are rich in heterochromatin (Cheng and Gu 1994). This region represented about 30% of the chromosome when examined visually (Fig. 3). By calculating 50 chromosome 4 pachytene spreads, we measured the length ratio of the long arm versus the short arm to be 5.13 (data not shown) (Cheng et al. 2001), but this ratio was measured to be 2.9 according to the physical map (Fig. 1B). Taken together, the results showed that the heterochromatic region of the chromosome 4 appears highly condensed at the pachytene stage and could be larger than that estimated by DAPI staining. It likely occupied 45% of the chromosome 4 as calculated by the physical map.
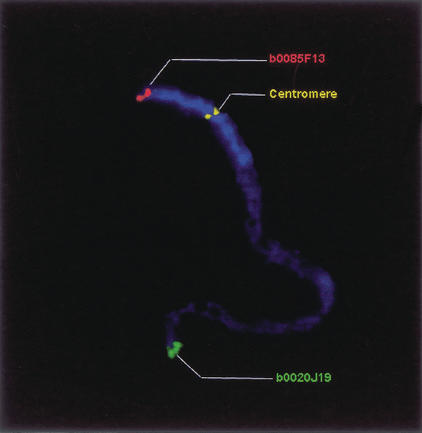
Figure 3.
BAC-FISH mapping of a pachytene chromosome 4. FISH of OSJNBb0085F13 (red signal) and OSJNBb0020J19 (green signal) revealed that they are near the telomeres of the short and long arms, respectively. The centromere of the chromosome 4 was detected by a rice centromere-specific satellite DNA pRCS2 as described previously (Dong et al. 1998) and indicated in yellow. The short arm and the pericentromeric region of the long arm are deeply stained by DAPI and therefore represent the heterochromatin-rich regions, which occupied almost one-third of the chromosome when visually measured.
Figure 4.
FISH analysis of CTG4-1. Three BAC clones, OSJNBa0068L06 (red signal), OSJNBb0085F13 (green signal), and OSJNBa0042N22 (pink signal), within CTG4-1 were chosen for FISH analysis to confirm the contig localization and orientation. RFLP markers R2502 (3.1 cM) and R2373 (0 cM) matched OSJNBa0068L06 and OSJNBb0004G23, respectively. Because OSJNBb0004G23 and OSJNBb0085F13 are overlapped, the order of the two markers was confirmed on the physical map by FISH analysis. Therefore, their genetic order is reversed on the physical map.
Relationship between Genetic Recombination Frequency and Physical Distance on Chromosome 4
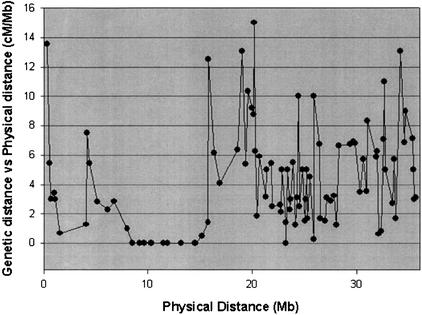
To reveal the relationship between the genetic and physical distances, we calculated the genetic distance (cM) per megabase and found a significant variation along the chromosome (Fig. 5). For example, CUGI's CTG83 covered by CTG4–5 has been estimated to be 2 Mb long in physical distance, but its genetic distance is only 1 cM with a ratio of 0.5 cM/Mb, whereas a ratio of 5.4 cM per Mb is found for CUGI's CTG257 covered by CTG4–6. The average ratio of the euchromatic region was estimated to be 4.8 cM/Mb and that of the heterochromatic region to be 2.3 cM/Mb (Fig. 5). A substantial reduction of recombination was found to occur in the regions of the short arm and the pericentromeric region in agreement with the DAPI staining result (Fig. 3).
Figure 5.
A relationship between the genetic recombination frequency and the physical distance of the rice chromosome 4. X axis shows the physical distance in cM along the chromosome 4. Y axis indicates the ratio of the genetic distance versus the physical distance (cM/Mb).
The fine BAC physical map of the Nipponbare chromosome 4 can be accessed at http://www.ncgr.ac.cn/rice/chr4/physicalmap/index.htm along with the integrated genome map of the CUGI at http://www.genome.clemson.edu/projects/rice/fpc/integration. The BACs and BAC contigs of indica that have been sequenced or identified with the sequenced ends are shown with the map.
In summary, we rapidly constructed a fine physical map of rice O. sativa japonica Nipponbare chromosome 4 through the data integration. Eleven BAC-based contigs covered 94% of the entire chromosome, including euchromatin and heterochromatin. The centromere was covered by the contig CTG4–5. Therefore, the centromere region was cloned and subsequently sequenced. The total length of the remaining gaps was estimated to be 2.5 Mb. We also identified that the chromosome 4 is a highly heterochromatic one by a detailed cytogenetic analysis. Therefore, a combined approach including iterative STC and hybridization and cytogenetic methods are required for its physical mapping. Because of highly repetitive sequences in chromosome 4, sequence scaffolds from related subspecies are very useful to efficiently identify the collinear clones by searching the clone-end sequences. Until now, 222 BAC clones representing 80% of the Nipponbare chromosome 4 had been completely sequenced and deposited into the EMBL database (accession nos. can be accessed at http://www.ncgr.ac.cn/rice/chr4/japonica.htm ). The comparison of the available RFLP and EST marker sequences with the sequenced clones fully supported that the physical map constructed in this study is an accurate one.
To fill as many as possible remaining gaps in the Nipponbare map, we are currently trying to identify the clones from additional genomic libraries such as P1-derived artificial chromosome (PAC) libraries constructed by the RGP. In addition, other approaches including single chromosome isolation with optical tweezers are also being tested. To complete the Guangluai 4 chromosome 4 gap filling, two deep-coverage BAC and PAC genomic libraries have been constructed. The two maps are being constantly refined and can be accessed at http://www.ncgr.ac.cn/rice/chr4/physicalmap/index.htm.
Because of its high-resolution representation of the rice O. sativa japonica chromosome 4, the map is of high value for studies devoted to comparative genome analysis between the two subspecies, as well as their genome organization and function (Sasaki and Burr 2000). Our comparative physical mapping will also help understand hybrid heterosis between indica and japonica subspecies. In conclusion, the strategy described will be effective for constructing a physical map of the entire genomes between closely related subspecies or species in other plants, as well as in mammals. Although some of the genome sizes of the closely related species vary greatly, the gene order and content and other single copy DNA sequences are to be very conserved along the chromosomes. The genomic sequence scaffolds from one species are therefore to be efficiently used for constructing a physical map of another genome of closely related species. Similarly, this approach can also be applied in building up super contigs with draft sequences of one variety of rice according to a complete genome sequence or genome sequence scaffold from another variety of rice. It is obvious that once the reference genome sequence is completed, the genome research on closely related species will be much more effective. To our knowledge, this is the first report on the comparative physical mapping of the two major cultivated rice subspecies chromosomes.
METHODS
Genetic and EST Markers Used for Colony Hybridization and In Silico Anchoring
A total of 182 RFLP markers of Nipponbare cloned in pBSSK or pT7T3–18U or pCRII were obtained from the Rice Genome Program (RGP) in Japan (http://rgp.dna.affrc.go.jp/publicdata/geneticmap2000/chr04.html) (Harushima et al. 1998). Another 12 RFLP markers (RZ602, RZ656, RG788, RG476, CDO539X, RG161, RG169, RZ590, RZ250, RZ819, RG620, and CDO36) were provided by Susan McCouch at Cornell University. A total of 407 EST markers of Nipponbare were provided by RGP (http://rgp.dna.affrc.go.jp/publicdata/estmap2001/Chr04.html) (Wu et al., 2002). All of the genetic and EST markers known on chromosome 4 were searched against the database of BAC-end sequences (http://www.genome.clemson.edu/projects/rice/rice_bac_end/) to anchor the contigs. In Silico mapping was performed in BLASTN, searching with a stringent cutoff of >95% identity against the databases of BAC-end sequences, marker sequences, and BAC-draft sequences by use of proper sequences.
BAC Library Construction
An additional BAC library (OSIGBAC-B), constructed in the laboratory of the National Center for Gene Research, was constructed from _Bam_HI partially digested genomic DNA from O. sativa ssp. indica cultivar Guangluai 4 seedling tissue and cloned into the pBeloBAC11 vector as described previously (Tao et al. 1994).
Labeling and Hybridization
Radioactive and ECL (Random Prime Labeling and Detection System, version II, Amersham plc) labeling of the RFLP markers, PCR fragments, BAC-end sequences, and plasmids, as well as hybridization procedures, have been described previously (Hong et al. 1997) and followed according to the protocols of the ECL labeling kit.
Fluorescence In Situ Hybridization
Preparation of rice pachytene chromosome spreads on slides and the FISH procedures were performed according to Cheng et al. (2001).
WEBSITE REFERENCES
http://rgp.dna.affrc.go.jp; Rice Genome Research Program (RGP).
http://www.genome.clemson.edu; Clemson University Genomics Institute (CUGI)
http://www.ncgr.ac.cn; National Center for Gene Research.
http://www.tigr.org; The Institute for Genome Research.
Acknowledgments
We thank Takuji Sasaki for providing rice genetic and EST markers, the Monsanto for the rice working draft sequence data, Gerard Barry for his help, and Qifa Zhang for his critical discussion. This work was supported by grants from the Ministry of Science and Technology of the People's Republic of China, Chinese Academy of Sciences, and the Shanghai Municipal Commission of Science and Technology. The work at the CUGI was funded by Novartis, USDA-CREES, and NSF. The work at the University of Wisconsin was supported by a grant from the Consortium for Plant Biotechnology Research, Inc., Novartis Seeds, Inc., and Dow AgroSciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bhan@ncgr.ac.cn; FAX (86) 21-64825775.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.48902.
REFERENCES
- Ahn S, Tanksley SD. Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci. 1993;90:7980–7984. doi: 10.1073/pnas.90.17.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- Barry G. The use of the Monsanto draft rice genome sequence in research. Plant Physiol. 2001;125:1164–1165. doi: 10.1104/pp.125.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Presting G, Barbazuk WB, Goicoechea JL, Blackmon B, Fang G, Kim H, Frisch D, Yu Y, Sun S, et al. An integrated physical and genetic map of the rice genome. Plant Cell. 2002;14:537–545. doi: 10.1105/tpc.010485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZK, Gu MH. Karyotype analysis for pachytene chromosome of indica, japonica rice and their hybrid. Chinese J Genet. 1994;21:182–187. [Google Scholar]
- Cheng ZK, Buell CR, Wing RA, Gu M, Jiang J. Toward a cytological characterization of the rice genome. Genome Res. 2001;11:2133–2141. doi: 10.1101/gr.194601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Miller JT, Jackson SA, Wang GL, Ronald PC, Jiang J. Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci. 1998;95:8135–8140. doi: 10.1073/pnas.95.14.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Comparative genetics in the grasses. Proc Natl Acad Sci. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta T, Komari T, Kumashiru T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Hong GF, Qian YM, Yu SL, Hu X, Zhu J, Tao WH, Li W, Su C, Zhao HY, Qiu LF, et al. A 120 kilobase resolution contig map of the rice genome. DNA Seq. 1997;7:319–335. doi: 10.3109/10425179709034052. [DOI] [PubMed] [Google Scholar]
- Mao L, Wood TC, Yu Y, Budiman MA, Tomkins J, Woo S, Sasinowski M, Presting G, Frisch D, Goff S, et al. Rice transposable elements: A survey of 73,000 sequence-tagged-connectors. Genome Res. 2000;10:982–990. doi: 10.1101/gr.10.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Sekhon M, Hillier L, Martienssen R, Chinwalla A, Crockett J, Fedele J, Grover H, Gund C, et al. A map for sequence analysis of the Arabidopsis thaliana genome. Nat Genet. 1999;22:265–270. doi: 10.1038/10327. [DOI] [PubMed] [Google Scholar]
- Moore G, Devos KM, Wang Z, Gale MD. Cereal genome evolution. Grasses, line up and form a circle. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- Mozo T, Dewar K, Dunn P, Ecker JR, Fischer S, Kloska S, Lehrach H, Marra M, Martienssen R, Meier-Ewert S, et al. A complete BAC-based physical map of the Arabidopsis thaliana genome. Nat Genet. 1999;22:271–275. doi: 10.1038/10334. [DOI] [PubMed] [Google Scholar]
- Oka HI. Genetic diversity of wild and cultivated rice. In: Khush GS, Toenniessen GH, editors. Rice biotechnology. Wallingford, Oxon: CAB International; 1991. pp. 55–80. [Google Scholar]
- Saji S, Umehara Y, Antonio BA, Yamane H, Tanoue H, Baba T, Aoki H, Ishige N, Wu J, Koike K, et al. A physical map with yeast artificial chromosome (YAC) clones covering 63% of the 12 rice chromosomes. Genome. 2001;44:32–37. doi: 10.1139/gen-44-1-32. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Burr B. International rice genome sequencing project: The effort to completely sequence the rice genome. Curr Opin Plant Biol. 2000;3:138–141. doi: 10.1016/s1369-5266(99)00047-3. [DOI] [PubMed] [Google Scholar]
- Soderlund C, Humphray S, Dunham A, French L. Contigs built with fingerprints, markers and FPC V4.7. Genome Res. 2000;10:1772–1787. doi: 10.1101/gr.gr-1375r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao QZ, Zhao HY, Qiu LF, Hong GF. Construction of a full bacterial artificial chromosome (BAC) library of Oryza sativa genome. Cell Res. 1994;4:127–133. [Google Scholar]
- The International Human Genome Mapping Consortium. A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- Wu J, Maehara T, Shimokawa T, Yamamoto S, Harada C, Takazaki Y, Ono N, Mukai Y, Koike K, Yazaki J, et al. A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell. 2002;14:525–535. doi: 10.1105/tpc.010274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Liang F, Hsiao J, Zismann V, Benito M, Quackenbush J, Wing R, Buell R. Anchoring of rice BAC clones to the rice genetic map in silico. Nucleic Acids Res. 2000;28:3636–3641. doi: 10.1093/nar/28.18.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]