Expression Status of p16 Protein Is Associated with Human Papillomavirus Oncogenic Potential in Cervical and Genital Lesions (original) (raw)
Abstract
The p16 protein (p16) is a cyclin-dependent kinase (CDK) inhibitor that decelerates the cell cycle by inactivating the CDKs that phosphorylate retinoblastoma (Rb) protein. Recent biological studies have revealed that p16 expression is markedly influenced by the status of Rb expression, and p16 overexpression has been demonstrated in cervical cancers because of functional inactivation of Rb by human papillomavirus (HPV) E7 protein. To clarify the relationship between p16 overexpression and HPV infection in cervical carcinogenesis, immunohistochemical analysis of p16 and detection of HPV by in situ hybridization and polymerase chain reaction were performed on 139 formalin-fixed and paraffin-embedded samples of cervical and genital condylomatous and neoplastic lesions. Marked overexpression of p16 protein, ie, diffuse and strong immunostaining, was observed in all cervical cancers and preneoplastic lesions with infection by high- and intermediate-risk HPVs, ie, subtypes 16, 18, 31, 33, 52, and 58. Condylomata acuminata and low-grade squamous intraepithelial lesions with infection by low-risk HPV such as HPV-6/11 showed focal and weak immunohistochemical staining for p16. Our results clearly showed that the mode of p16 expression in lesions with high- and intermediate-risk HPVs differed from its expression in lesions with low-risk HPVs and thus might be attributable to differences in functional inactivation of Rb protein by different HPVs.
Many studies have shown that human papillomavirus (HPV) infection plays an important role in cervical carcinogenesis. 1 In fact, HPV infection has been detected in almost all preneoplastic and neoplastic lesions of the cervix. Recent extensive studies have revealed the existence of more than 70 subtypes of HPV, of which approximately 20 can infect the cervical epithelium and give rise to various lesions of the cervix. 1-3 Moreover, each HPV subtype has been shown to be associated with a different risk of neoplastic transformation by cervical epithelial cells, and the HPV subtypes have been classified into three categories according to the risk: high, intermediate, and low. 2,3 HPV-16 and -18 are representative of high-risk HPVs and are the most clinically important HPV subtypes, because infection by these viruses has a marked influence on outcome. HPV-31, -33, -35, -51, -52, and -58 are associated with intermediate risk for development of cervical cancer and high-grade squamous intraepithelial lesions (HSILs), whereas HPV-6 and -11 are classified as low-risk HPVs and are usually associated with benign hyperplastic lesions such as condylomata acuminata and low-grade squamous intraepithelial lesions (LSILs). 1-3
The reasons for the differences in oncogenic potential of each HPV subtype in cervical carcinogenesis remain unresolved. However, oncogenes E6 and E7 of HPV have been suggested to play an important role in the differences in oncogenic potential of each HPV subtype in cervical carcinogenesis. 4,5 The oncoproteins encoded by the E6 and E7 genes have the ability to bind host cell regulatory proteins, especially tumor suppressor gene products p53 and hypophosphorylated retinoblastoma (Rb) protein (pRb). 6,7 These changes may lead to degradation of p53 by the E6 oncoprotein and to functional inactivation of pRb through binding to the E7 gene product. 6,7 As a result of loss of tumor suppressor function, a decrease in the p21 protein level and liberation of the transcriptional factor E2F-1 from the E2F-Rb complex may occur, allowing activation of cyclin-dependent kinase and transcriptional activation of target promoters, respectively. 8-11
The CDKN2A gene product, p16 protein (p16), is a tumor suppressor protein that inhibits cyclin-dependent kinases (CDK)-4 and -6, which regulate the G1 checkpoint. 12,13 The CDKs phosphorylate pRb, which results in a conformational change and release of E2F from the pRb. Thus, inactivation of either p16 or pRb function allows the cell to enter the S phase after only a pause at the G1 checkpoint. Recent studies have revealed that pRb inactivation is usually reciprocal with p16 expression in various cancers. 14-23 However, our previous immunohistochemical study clearly demonstrated that both pRb and p16 are co-expressed in cervical preneoplastic and neoplastic lesions, in contrast to the neoplastic lesions of other organs. 24 In the present study, moreover, we demonstrated the interrelationship between immunohistochemical expression of p16 and infection with different HPV subtypes in genital and cervical preneoplastic and neoplastic lesions.
Materials and Methods
Cell Lines and Tissue Specimens
Five established cell lines (four neuroblastomas and the cervical cancer cell line SiHa) were fixed with 10% buffered formalin for 24 hours at 4°C and embedded in paraffin. The cell blocks embedded in paraffin were used to examine the specificity of the anti-p16 antibodies used in this study. Two of the four neuroblastoma cell lines, TGW and LAN1, were known to be positive for p16 expression, but the other two cell lines, LAN2 and NB69, were devoid of p16 expression due to transcriptional inactivation by methylation of the promoter region or for unknown reasons. 25 Overexpression of p16 in the HPV-16-positive SiHa cells was previously confirmed by Western blotting. 26
A total of 139 lesions, including 28 genital condylomata acuminata, 57 low- and high-grade squamous intraepithelial lesions (20 LSILs and 37 HSILs) of the cervix, and 54 invasive cervical carcinomas were randomly selected from the pathology files of Gunma University Hospital. The sites and numbers of the condylomata acuminata were as follows: anus, 3; penis, 13; vulva, 10; and vagina, 2. Of the 54 invasive carcinomas, 27 were invasive squamous cell carcinomas of either the keratinizing or nonkeratinizing large-cell type, 12 were microinvasive squamous cell carcinomas, 9 were adenocarcinomas, and the remaining 6 were adenosquamous carcinomas.
HPV Typing by in Situ Hybridization and Polymerase Chain Reaction
All samples were investigated for the presence of HPV types 6 and 16 DNAs by the highly sensitive in situ hybridization (ISH) technique as described in our previous paper. 27 Briefly, formalin-fixed and paraffin-embedded blocks that had been used for routine pathological diagnosis were used for the ISH study. DNA probes for HPV-6 and -16 were prepared by nick translation of purified pBR322 plasmids containing HPV-6 or -16 DNA (kindly provided by Dr. H zur Hausen, German Cancer Research Center, Heidelberg, Germany) with biotin-11-deoxyuridine triphosphate (Bioprobe, ENZO, NY). After hybridization, the streptavidin-peroxidase reaction was performed and followed by signal amplification with biotinylated tyramide. 27,28 Finally, the hybridization signals were visualized by the benzidine reaction.
The ISH signal patterns for HPV were classified as three different morphological types, according to Cooper et al: 29,30 diffuse, dot, and mixed. Diffuse, dot, and mixed ISH signal patterns have been reported to correspond to episomal, integrated, and both episomal and integrated forms of HPV DNA, respectively. 29
After the ISH analysis of all samples was completed, those that failed to show either HPV-6 or -16 DNA were further studied for the presence of HPV DNA by the polymerase chain reaction (PCR) with consensus primers in formalin-fixed and paraffin-embedded tissues. Details regarding DNA extraction from the paraffin blocks and the PCR assay for detection and typing of HPVs are described elsewhere. 31 The consensus primers, L1C1/L1C2 for the L1 region of HPV (L1C1; 5′-CGTAAACGTTTTCCCTATTTTTTT-3′ and L1C2; 5′-TACCCTAAATACCCTATATTG-3′), which allowed amplification and identification of at least nine types of genital HPV DNA, were used in this study. 31,32 The amplified HPV fragments were typed on the basis of restriction fragment length polymorphism (RFLP) by using three restriction enzymes, _Rsa_I, _Dde_I, and _Hae_III. 32
Immunohistochemistry for p16
Sections 4 μm thick on silane-coated glass slides were dewaxed by passage through xylene, and the endogenous peroxidase activity was blocked with 0.3% H2O2/methanol. The slides were then rehydrated with 0.01 mol/L sodium phosphate/citrate buffer, pH 8.0, microwaved at 90°C for 15 minutes for antigen retrieval, and then left to cool for 15 minutes. After rinsing in 0.01 mol/L phosphate-buffered saline (PBS), pH 7.4, nonspecific antibody binding was reduced by incubating the sections with 10% normal horse serum in PBS for 30 minutes. After decanting excess serum, sections were incubated overnight at 4°C with a mouse monoclonal antibody that recognizes an epitope in the first ankyrin repeat (amino acids 1 to 32) of the p16 protein (JC8, 1:500 dilution with PBS). 24,33,34 After washing thoroughly with PBS, the slides were incubated with biotinylated horse anti-mouse IgG for 30 minutes followed by a 1:100 dilution of the avidin-biotin-peroxidase complex (Vectastain, Vector Laboratories, Burlingame, CA) for an additional 30 minutes. Peroxidase was then visualized with 0.02% 3–3′-diaminobenzidine tetrahydrochloride containing 0.005% H2O2 in 50 mmol/L ammonium acetate/citric acid buffer, pH 6.0. Finally, the sections were counterstained lightly with hematoxylin.
The staining pattern for p16 was graded in each lesion as follows: negative, 0 to 5% cells stained positive; focal/scattered positive, fewer than 80% cells stained positive; diffuse positive, more than 80% cells stained positive. This scoring system was devised based on the results in normal or reactive cervical specimens where anti-p16 antibody did not stain at all or only weakly stained a small subset of proliferating cells, such as fibroblasts, capillary endothelial cells, and histiocytes. 24
Three anti-p16 antibodies, JC8 plus two commercially available antibodies (polyclonal anti-p16 from Pharmingen, San Diego, CA; DCS-50.1, a mouse monoclonal anti-p16 from NeoMarkers, Fremont, CA), were used to immunohistochemically check 4-μm-thick sections prepared from the cell lines for the specificity of p16 staining.
Results
Immunohistochemical Staining for p16 in Cell Lines
To check the specificity of the three antibodies for p16, we performed immunostaining for p16 in five cell lines in which the status of p16 expression has been well characterized. Strong and diffuse p16 immunoreactivity was demonstrated with JC8 monoclonal antibody in three cell lines, SiHa (Figure 1A) ▶ , TGW, and LAN1 (Figure 1B) ▶ , but LAN2 cells were completely devoid of p16 immunoreactivity (Figure 1C) ▶ . Most NB69 cells were negative for p16, but a small number of p16-positive NB69 cells were seen in clusters (Figure 1D) ▶ . Immunohistochemical staining for p16 with a mouse monoclonal anti-p16 antibody (NeoMarkers) showed results similar to those obtained with JC8 antibody. All cell lines stained diffusely for p16 with polyclonal antibodies purchased from PharMingen. Therefore, JC8 was found to be reliable in its immunohistochemical specificity and was therefore used for analysis of all clinical specimens.
Figure 1.
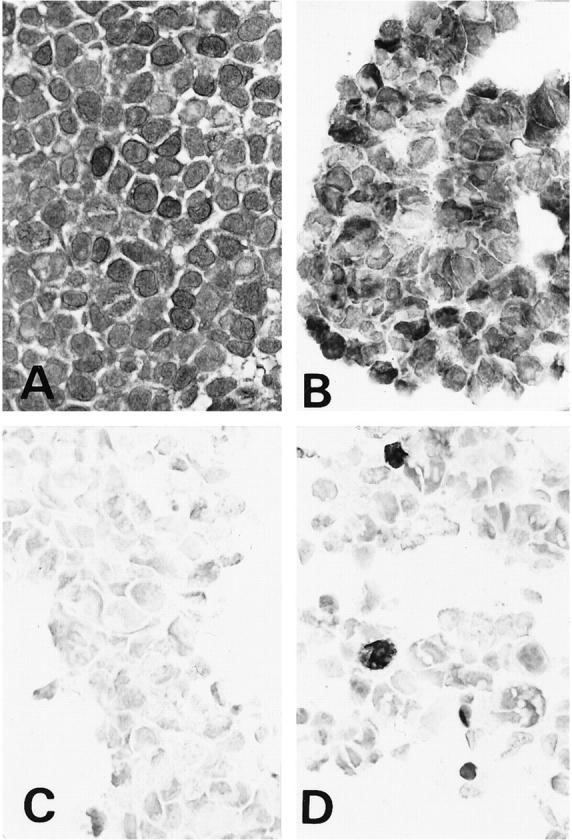
Immunohistochemical staining using anti-p16 antibody JC8 in cell lines. A and B: Diffuse and strong staining was observed in SiHa (A) and LAN1 cells (B) with intact p16 expression. C: LAN2 cells with inactivated p16 gene by hypermethylation showed no positive staining. D: NB69 cells that had lacked p16 expression by Western and Northern blotting showed almost no expression, but a few positive cells were observed.
Detection and Typing of HPV in Cervical and Genital Lesions
ISH for HPV-6 and -16 DNAs was carried out on all specimens, and as shown in Table 1 ▶ , 56 demonstrated signals for HPV-16 DNA in the nuclei of the tumor cells and 34 demonstrated signals for HPV-6 DNA. ISH for HPV-6 always detected HPV-11 DNA. Only one HSIL showed positive signals for both HPV-6 and -16 in different areas of the section.
Table 1.
Results of HPV Typing in Condylomas and Cervical Neoplasms
| Cervical lesions (n) | HPV-6/11 | HPV-16 | Other types | HPV negative |
|---|---|---|---|---|
| Condylomata acuminata (28) | 28 (28) | 0 | ND | ND |
| LSIL (20) | 6 (6) | 7 (7) | 7* | ND |
| HSIL (37) | 0 | 28 (28)† | 9‡ | ND |
| Cervical cancer (54) | 0 | 25 (21) | 12§ | 17 |
ISH signals in the nuclei showed various patterns but could be morphologically classified into diffuse, dots, and mixed patterns. ISH signals for HPV-16 DNA showed dots (Figure 2A) ▶ or mixed patterns in all invasive cervical cancers. Among the squamous intraepithelial lesions showing positive signals for HPV-16 DNA by ISH, all LSILs and 12 of the 28 HSILs showed diffuse signals, which were usually seen in nuclei located from the middle to the superficial layer of the cervical dysplastic epithelium, especially in the cells showing koilocytotic changes or with marked nuclear pleomorphism. However, the remaining 16 HSILs positive for HPV-16 DNA had dotted or mixed signal patterns (Figure 2C) ▶ . The ISH signals in the LSIL and condylomatous lesions positive for HPV-6/11 DNA, on the other hand, showed a diffuse or mixed pattern (Figure 2E) ▶ .
Figure 2.
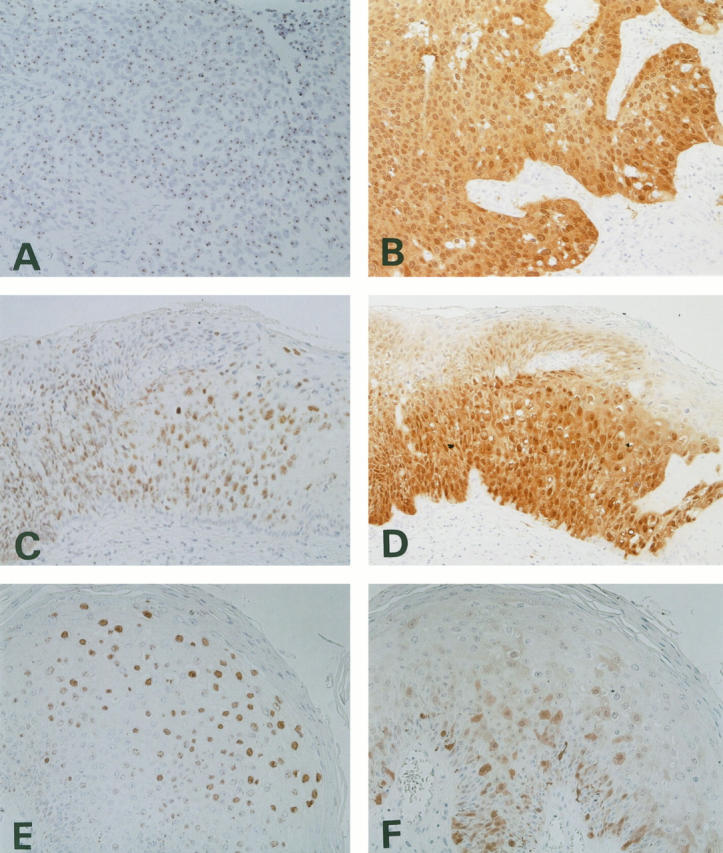
ISH for HPV (A, C, and E) and immunohistostaining for p16 (B, D, and F). A and B, C and D, and E and F are in the same areas of each lesion. A and B: HPV-16-positive cervical cancer case. A: ISH pattern showing the dot-type signals. B: Clear and distinct positive staining for p16 was seen throughout the nuclei and cytoplasm of the tumor cells. No positive staining was seen in the stroma. C and D: HPV-16-positive HSIL case. C: ISH pattern showing a mixture of dotted and diffuse signals. D: Intense and diffuse p16 reactivity in both nuclei and cytoplasm was confined to the dysplastic lesion. E and F: HPV-6-positive condyloma acuminatum case. E: ISH pattern showing a mixture of many diffuse signals and a few dots. F: Positive stained tumor cells were heterogeneously scattered among the many negative cells within the lesion.
Among the remaining 49 lesions, in which neither HPV-6/11 nor -16 DNA could be detected by ISH, a combination of PCR and RFLP analysis revealed various HPV subtypes, including HPV-16. There were four cervical cancer lesions with HPV-16, six lesions with HPV-52, five with HPV-33, six with HPV-18, two with HPV-31, two with HPV-58, and seven with unknown HPV types. No HPV DNA was detected in the remaining 17 cancers by PCR in this study, and they were therefore regarded as HPV negative. The 17 HPV-negative cancers included 9 adenocarcinomas (7 endocervical-gland-type adenocarcinomas and 1 clear-cell adenocarcinoma and mucinous carcinoma each) and 1 adenosquamous cell carcinoma. The relationship between the histopathological diagnosis of the lesions and their HPV subtype is summarized in Table 1 ▶ .
Immunohistochemical Staining for p16 in Cervical and Genital Lesions
Immunohistochemical analysis for p16 was performed in all 139 specimens investigated with JC8 in this study, and the results in relation to HPV subtype are summarized in Table 2 ▶ . In the lesions judged to be diffusely positive, almost all neoplastic cells were strong and homogeneously positive for p16, whereas most lesions considered to be focal/scattered positive contained fewer than 30% neoplastic cells showing weak p16 immunostaining. In general, immunoreactivity for p16 was not detected in the non-neoplastic epithelia or in mesenchymal cells, but variable weak immunoreactivity for p16 was occasionally seen only in the nuclei and cytoplasm of proliferating fibroblasts, endothelial cells, some inflammatory cells surrounding the cancer tissue, and hyperplastic endocervical glands. These non-neoplastic cells were always weakly stained, and immunoreactivity was seen in only a small subset of the cells, but this served as an internal positive control when lesions were judged negative for p16 immunostaining. However, strong and diffuse immunoreactivity for p16 was uniformly observed in both the nuclei and cytoplasm of all HSILs and invasive cancer lesions as well as those of many LSILs, in marked contrast with non-neoplastic tissues (Figure 2, B and D) ▶ . In most condylomas and approximately one-third of LSILs, p16 immunoreactivity was focal and scattered (Figure 2F) ▶ .
Table 2.
Results of Immunohistochemical Analysis of p16 in Association with HPV Type
| Number of cases | Diffuse | Focal/scattered | Negative | |
|---|---|---|---|---|
| HPV-6/11 | 34 | 0 | 32 | 2 |
| HPV-16 | 60 | 60 | 0 | 0 |
| Other types | 28 | 27 | 1* | 0 |
| HPV negative | 17 | 12 | 3 | 2 |
Strong and diffuse immunostaining for p16 was seen in various cervical lesions infected with HPV-16 and other HPV subtypes, namely, HPV-18, -31, -33, -52, and -58 and unclassified subtypes. As an exception, only one unclassified HPV subtype was detected in one LSIL in which p16 immunoreactivity was focal and weak. On the other hand, 32 of 34 lesions positive for HPV-6 showed heterogeneous focal immunostaining of the tumor cells for p16. Weakly p16-positive tumor cells were sparsely scattered or sometimes focally clustered within these lesions. The remaining two specimens were completely negative for p16. Diffuse, strong p16 staining, similar to that seen in the lesions positive for HPV-16 and other HPV subtypes, was not observed in HPV-6-positive lesions. In a unique HSIL case in which ISH demonstrated simultaneous HPV-6 and -16 infection in different areas, p16 immunoreactivity was clearly shown to be strong and diffuse in the HPV-16-positive area but completely absent in the HPV-6-positive area. The border between the areas of infection by the different HPV subtypes demonstrated by ISH and p16 protein immunoreactivity was clear and distinct (Figure 3) ▶ .
Figure 3.
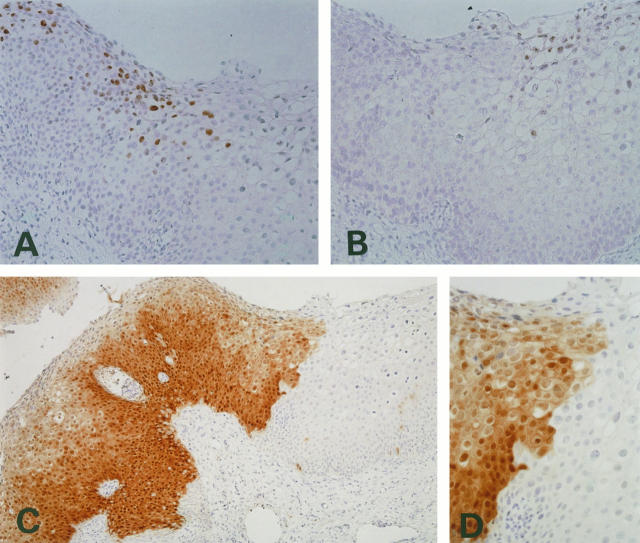
HSIL case double positive for HPV-16 and -6 in different areas within the same section. A: ISH for HPV-16. B: ISH for HPV-6. C and D: Immunostaining for p16 in the same lesion. Diffuse and intense staining corresponding to the HPV-16-positive lesion was seen (left side of the lesion), whereas the HPV-6-positive lesion was negative for p16 expression (right side; C). D: Higher magnification of the border between the positive and negative staining.
On the other hand, 12 of the 17 HPV-negative cancer cases also demonstrated strong and diffuse p16 protein immunoreactivity, and the remaining 5 cases showed focal to negative staining for p16. Interestingly, these five cases that lacked p16 protein immunoreactivity consisted of four adenocarcinomas (including one case each of clear-cell adenocarcinoma and mucinous carcinoma) and one adenosquamous carcinoma of the cervix.
Discussion
The p16/cyclin D1/cdk4/pRb cell cycle regulatory cascade is central to regulation of the G1-to-S phase transition and to understanding human cancers. HPV infection is critically involved in cervical carcinogenesis and plays an important role in the p16/cyclin D1/cdk4/pRb cell cycle regulatory cascade by binding of its oncoproteins E6 and E7 to tumor suppressor genes p53 and pRb, respectively. 4-7 In cervical cancers, loss of p53 function is the most frequently observed and best characterized epigenetic alteration caused by p53 protein degradation as a result of HPV E6 protein binding. 6 These p53 alterations have been shown to influence the p16/cyclin D1/cdk4/pRb cell cycle regulatory cascade indirectly by altering p21 function. In the p16/cyclin D1/cdk4/pRb cell cycle regulatory cascade, the correlation between pRb and p16 is obvious in various cancers. Various cancers with mutation or deletion of the Rb gene show overexpression of p16, and a reciprocal correlation between pRb and p16 has been established. 14-17,21,22 The Rb gene in cervical cancers has been shown to be functionally inactivated as a consequence of HPV E7 protein expression. 7 Rb and CDKN2A/p16 gene alterations are infrequent events in cervical cancer. 11,35-37 Experimental studies have demonstrated not only p16 overexpression in established cervical cancer cell lines but also marked enhancement of p16 expression in immortalized human ectocervical cells by HPV-16 and -18. 26,38 These studies have also suggested that inactivation of the p16/cyclin D1/cdk4/pRb cell cycle regulatory cascade by HPV occurs during the early immortalization step in cervical carcinogenesis, but not during late malignant transformation. 38
The present and our previous immunohistochemical studies have clearly demonstrated that p16 is strongly expressed in almost all cervical cancers and that immunoreactivity for pRb is also preserved. 24 This finding in cervical cancer is in contrast to the results of previous studies in other cancers: immunoreactivity for p16 has been reported to be inversely correlated to immunoreactivity for pRb in cancer of the lung, breast, and bladder. 14-17,21,22 Cervical cancers are different from other cancers in regard to the status of Rb tumor suppressor function; ie, Rb tumor suppressor function is thought to be functionally inactivated by HPV oncoproteins, resulting in p16 overexpression in cervical cancer. 26,38 Recently, HPV-associated tonsillar carcinomas have also been demonstrated to overexpress p16 by both Western blotting and immunohistochemical methods. 39
The monoclonal antibody JC8 used in this study has already been used for immunohistochemical analysis of p16 expression in glioblastomas and lung cancers, in which p16-positive immunostaining can exclude inactivation of the p16 gene by deletion and methylation. 33,34 Our immunohistochemical study using this antibody clearly demonstrated strong p16 expression in cervical and genital lesions, including cervical cancers. Also, the immunohistochemical specificity of JC8 against p16 was clearly certified using five known cancer cell lines, in which the status of p16 expression had already been determined by molecular biological techniques. 25,26 Interestingly, JC8 showed focal strong reactivity in some NB69 cells that did not express p16 on Northern and Western blots. 25 The results seemed to be due to the high sensitivity of JC8 immunohistochemistry on individual cells beyond the limit of detection by molecular techniques. The results suggest that heterogeneity of p16 expression among tumor cells is present even in established cell lines; however, additional studies are required to explain this phenomenon. In this study, monoclonal antibody JC8 failed to detect p16 immunoreactivity in most normal cells. Immunoreactivity for p16 in the nuclei and cytoplasm of tumor cells was more distinctive and intense than in the non-neoplastic mucosa and glands within the same sections. However, the lack of p16 immunoreactivity does not necessarily imply loss of p16 expression; ie, lack of p16 immunoreactivity may not reflect gene inactivation but may represent a physiological normal state in non-neoplastic or nonproliferating cells. 33,40 In most normal cells, p16 expression is known to be low at both the mRNA and protein levels. 14,22,33,40 In this regard, expression of p16 may be detectable immunohistochemically only when p16 is physiologically up-regulated, especially in cervical and genital lesions. The p16 immunonegativity only in proliferating neoplastic cells may indicate inactivation of the gene by deletion or methylation, although the presence of a low level of p16 cannot be ruled out completely.
No HPV DNA was detected in 17 cervical cancers by either PCR or ISH in this study, and 5 of these cancers did not show strong, diffuse immunostaining for p16. The high rate of HPV negativity (17/54) may be due to the bias in our series, which included many cases of adenocarcinoma or related subtypes. All of the 17 HPV-negative cases were confirmed immunohistochemically to express Rb, 24 and thus an inverse correlation between p16 and Rb was seen only in the 5 cases in which diffuse p16 staining was not seen, which may indicate inactivation of the p16 gene. There were no p16-negative cases in the high-risk HPV group. Overexpression of p16 in the remaining 12 cancers negative for HPV may have been due to false negative results for HPV in this study caused by loss of a subgenomic region on the HPV DNA, by a very low HPV copy number that was below the limit of detection by our PCR method, or by the presence of a novel unknown HPV subtype. 31,32,41
In this study, we demonstrated that p16 immunostaining patterns are markedly different in cervical cancers infected with low-risk and high- or intermediate-risk HPVs. The strong immunoreactivity for p16 in various cervical lesions suggested that deletion or mutational inactivation of the p16 gene is infrequent and that up-regulation or overexpression of the p16 gene occurs in cervical carcinogenesis. Our observation that p16 immunostaining differed in cervical lesions with low-risk and high-risk HPVs may be attributable to the difference in functional inactivation of pRb. The results of previous studies have indicated that viral oncoproteins of low-risk HPV such as HPV-6 have no effect on p16/cyclin D1/cdk4/pRb complexes, because the affinity of HPV-6 E7 protein for cellular pRb is 10-fold lower than that of HPV-16 E7 for pRb. 42-45
Our immunohistochemical study suggested the possible usefulness of p16 protein as a marker to differentiate high- and intermediate-risk-HPV-related neoplastic lesions from low-risk-HPV-related lesions in the cervix. It was particularly noteworthy that p16 staining appears to be most useful in LSIL cases, which may contain low- or high-risk HPV, because the lesions would progress to HSIL or cancer if they were caused by high-risk HPV. 2,3,32 In this study, diffuse and strong p16 immunoreactivity was so highly specific and sensitive among the high- and intermediate-risk-HPV-related lesions that its overexpression in cervical precursor lesions, especially in LSILs, may provide valuable information regarding the HPV subtype infecting the lesions without the need for molecular techniques such as PCR, Southern blotting, or ISH.
In conclusion, p16 is overexpressed in most cervical lesions, and the status of its immunoreactivity allows differentiation between infection with low-risk HPVs and high- or intermediate-risk HPVs.
Acknowledgments
We thank Dr. Jim Koh, Massachusetts General Hospital Cancer Center, and Dr. David N. Louis, the Molecular Neuro-Oncology Laboratory, Massachusetts General Hospital, Boston, MA, for providing the monoclonal antibody JC8 against human p16 protein.
Footnotes
Address reprint requests to Dr. Takaaki Sano, Second Department of Pathology, Gunma University School of Medicine, 3-39-22, Showa-machi, Maebashi, Gunma, 371-8511, Japan. E-mail: sanot@akagi.sb.gunma-u.ac.jp.
References
- 1.zur Hausen H: Papillomavirus infections: a major cause of human cancers. Biochim Biophys Acta 1996, 1288:F55-F78 [DOI] [PubMed] [Google Scholar]
- 2.Lorincz AT, Reid R, Bennett Jenson A, Greenberg MD, Lancaster W, Kurman RJ: Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet Gynecol 1992, 79:328-337 [DOI] [PubMed] [Google Scholar]
- 3.Matsukura T, Sugase M: Identification of genital human papillomaviruses in cervical biopsy specimens: segregation of specific virus types in specific clinicopathologic lesions. Int J Cancer 1995, 61:13-22 [DOI] [PubMed] [Google Scholar]
- 4.Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT: HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J 1989, 8:3905-3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munger K, Phelps WC, Bubb V, Howley PM, Schlegel R: The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol 1989, 63:4417-4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Werness BA, Levine AJ, Howley PM: Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 1990, 248:76-79 [DOI] [PubMed] [Google Scholar]
- 7.Dyson N, Howley PM, Munger K, Harlow E: The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 1989, 243:934-937 [DOI] [PubMed] [Google Scholar]
- 8.Bagchi S, Raychaudhuri P, Nevins JR: Adenovirus E1A proteins can dissociate heteromeric complexes involving the E2F transcription factor: a novel mechanism for E1A trans-activation. Cell 1990, 62:659-669 [DOI] [PubMed] [Google Scholar]
- 9.Lechner MS, Mack DH, Finicle AB, Crook T, Vousden KH, Laimins LA: Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J 1992, 11:3045-3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phelps WC, Yee CL, Munger K, Howley PM: The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 1988, 53:539-547 [DOI] [PubMed] [Google Scholar]
- 11.Amortegui AJ, Meyer MP, Elborne VL, Amin RM: P53, retinoblastoma gene product, and cyclin protein expression in human papillomavirus virus DNA-positive cervical intraepithelial neoplasia and invasive cancer. Mod Pathol 1995, 8:907-912 [PubMed] [Google Scholar]
- 12.Serrano M, Hannon GJ, Beach D: A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993, 366:704-707 [DOI] [PubMed] [Google Scholar]
- 13.Koh J, Enders GH, Dynlacht BD, Harlow E: Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature 1995, 375:506-510 [DOI] [PubMed] [Google Scholar]
- 14.Shapiro GI, Edwards CD, Kobzik L, Godleski J, Richards W, Sugarbaker DJ, Rollins BJ: Reciprocal Rb inactivation and p16 expression in primary lung cancers and cell lines. Cancer Res 1995, 55:505-509 [PubMed] [Google Scholar]
- 15.Sakaguchi M, Fujii Y, Hirabayashi H, Yoon HE, Komoto Y, Kusafuka T, Okada A, Matsuda H: Inversely correlated expression of p16 and Rb protein in non-small cell lung cancers. Int J Cancer 1996, 65:442-445 [DOI] [PubMed] [Google Scholar]
- 16.Kratzke RA, Greatens TM, Rubins JB, Maddaus MA, Niewoehner DE, Niehans GA, Geradts J: Rb and p16INK4a expression in resected non-small cell lung tumors. Cancer Res 1996, 56:3415-3420 [PubMed] [Google Scholar]
- 17.Kinoshita I, Dosaka-Akita H, Mishima T, Akie K, Nishi M, Hiroumi H, Hommura F, Kawakami Y: Altered p16 and retinoblastoma protein status in non-small cell lung cancer. Cancer Res 1996, 56:5557-5562 [PubMed] [Google Scholar]
- 18.Piccinin S, Doglioni C, Maestro R, Vukosavljevic T, Gasparotto D, D’Orazi C, Boiocchi M: P16/CDKN2 and CDK4 gene mutations in sporadic melanoma development and progression. Int J Cancer 1997, 74:26-30 [DOI] [PubMed] [Google Scholar]
- 19.Papadimitrakopoulou V, Izzo J, Lippman SM, Lee JS, Fan YH, Clayman G, Ro JY, Hittelman WN, Lotan R, Hong WK, Mao L: Frequent inactivation of p16INK4a in oral premalignant lesions. Oncogene 1997, 14:1799-1803 [DOI] [PubMed] [Google Scholar]
- 20.Reed AL, Califano J, Cairns P, Westra WH, Jones RM, Koch W, Ahrendt S, Eby Y, Sewell D, Nawroz H, Bartek J, Sidransky D: High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res 1996, 56:3630-3633 [PubMed] [Google Scholar]
- 21.Geradts J, Wilson PA: High frequency of aberrant p16 INK4a expression in human breast cancer. Am J Pathol 1996, 149:15-20 [PMC free article] [PubMed] [Google Scholar]
- 22.Geradts J, Kratzke RA, Niehans GA, Lincoln CE: Immunohistochemical detection of the cyclin-dependent kinase inhibitor 2/multiple tumor suppressor gene 1 (CDKN2/MTS1) product p16 in archival human solid tumors: correlation with retinoblastoma protein expression. Cancer Res 1995, 55:6006-6011 [PubMed] [Google Scholar]
- 23.Wadayama B, Toguchida J, Shimizu T, Ishizaki K, Sasaki MS, Kotoura Y, Yamamuro T: Mutation spectrum of the retinoblastoma gene in osteosarcomas. Cancer Res 1994, 54:3042-3048 [PubMed] [Google Scholar]
- 24.Sano T, Oyama T, Kashiwabara K, Fukuda T, Nakajima T: Immunohistochemical overexpression of p16 protein associated with intact retinoblastoma protein expression in cervical cancer and cervical intraepithelial neoplasia. Pathol Int 1998, 48:580-585 [DOI] [PubMed] [Google Scholar]
- 25.Takita J, Hayashi Y, Kohno T, Yamaguchi N, Hanada R, Yamamoto K, Yokota J: Deletion map of chromosome 9 and p16(CDKN2A) gene alterations in neuroblastoma. Cancer Res 1997, 57:907-912 [PubMed] [Google Scholar]
- 26.Khleif SN, DeGregori J, Yee CL, Otterson GA, Kaye FJ, Nevins JR, Howley PM: Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc Natl Acad Sci USA 1996, 93:4350-4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sano T, Hikino T, Niwa Y, Kashiwabara K, Oyama T, Fukuda T, Nakajima T: In situ hybridization with biotinylated tyramide amplification: detection of human papillomavirus DNA in cervical neoplastic lesions. Mod Pathol 1998, 11:19-23 [PubMed] [Google Scholar]
- 28.Kerstens HMJ, Poddighe PJ, Hanselaar AGJM: A novel in situ hybridization signal amplification method based on the deposition of biotinylated tyramine. J Histochem Cytochem 1995, 43:347-352 [DOI] [PubMed] [Google Scholar]
- 29.Cooper K, Herrington CS, Stickland JE, Evans MF, McGee JOD: Episomal and integrated human papillomavirus in cervical neoplasia shown by non-isotopic in situ hybridization. J Clin Pathol 1991, 44:990-996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper K: Nonisotopic in situ hybridization in the detection of integrated HPV 16/18 in cervical cancers. Hum Pathol 1996, 27:745. [DOI] [PubMed] [Google Scholar]
- 31.Sano T, Sakurai S, Fukuda T, Nakajima T: Unsuccessful effort to detect human papillomavirus DNA in urinary bladder cancers by the polymerase chain reaction and in situ hybridization. Pathol Int 1995, 45:506-512 [DOI] [PubMed] [Google Scholar]
- 32.Yoshikawa H, Kawana T, Kitagawa K, Mizuno M, Yoshikura H, Iwamoto A: Detection and typing of multiple genital human papillomaviruses by DNA amplification with consensus primers. Jpn J Cancer Res 1991, 82:524-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burns KL, Ueki K, Jhung SL, Koh J, Louis DN: Molecular genetic correlates of p16, cdk4 and pRb immunohistochemistry in glioblastomas. J Neuropathol Exp Neurol 1998, 57:122-130 [DOI] [PubMed] [Google Scholar]
- 34.Kashiwabara K, Oyama T, Sano T, Fukuda T, Nakajima T: Correlation between methylation status of the p16/CDKN2 gene and the expression of p16 and Rb proteins in primary non-small cell lung cancers. Int J Cancer 1998, 79:215-220 [DOI] [PubMed] [Google Scholar]
- 35.Choo K-B, Chang KY: Absence of mutation in the p53 and the retinoblastoma genes in primary cervical carcinomas. Virology 1993, 193:1042-1046 [DOI] [PubMed] [Google Scholar]
- 36.Kelley MJ, Otterson GA, Kaye FJ, Popescue NC, Johnson BE, Dipaolo JA: CDKN2 in HPV-positive and HPV-negative cervical-carcinoma cell lines. Int J Cancer 1995, 63:226-230 [DOI] [PubMed] [Google Scholar]
- 37.Hirama T, Miller CW, Wilczynski SP, Koeffler HP: P16 (CDKN2/cyclin-dependent kinase-4 inhibitor/multiple tumor suppressor-1) gene is not altered in uterine cervical carcinomas or cell lines. Mod Pathol 1996, 9:26-31 [PubMed] [Google Scholar]
- 38.Nakao Y, Yang X, Yokoyama M, Ferenczy A, Tang S-C, Pater MM, Pater A: Induction of p16 during immortalization by HPV 16 and 18 and not during malignant transformation. Br J Cancer 1997, 75:1410-1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andl T, Kahn T, Pfuhl A, Nicola T, Erber R, Conradt C, Klein W, Helbig M, Dietz A, Weidauer H, Bosch FX: Etiological involvement of oncogenic human papillomavirus in tonsillar squamous cell carcinomas lacking retinoblastoma cell cycle control. Cancer Res 1998, 58:5-13 [PubMed] [Google Scholar]
- 40.Dublin EA, Patel NK, Gillett CE, Smith P, Peters G, Barnes DM: Retinoblastoma and p16 proteins in mammary carcinoma: their relationship to cyclin D1 and histopathological parameters. Int J Cancer 1998, 79:71-75 [DOI] [PubMed] [Google Scholar]
- 41.Choo KB, Lee HH, Pan CC, Wu SM, Liew LN, Cheung WF, Han SH: Sequence duplication and internal deletion in the integrated human papillomavirus type 16 genome cloned from a cervical carcinoma. J Virol 1988, 62:1659-1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gage JR, Meyers C, Wettstein FO: The E7 proteins of the nononcogenic human papillomavirus type 6b (HPV-6b), and of the oncogenic HPV-16 differ in retinoblastoma protein binding, and other properties. J Virol 1990, 46:723-730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeager T, Stadler W, Belair C, Puthenveettil J, Olopade O, Reznikoff C: Increased p16 levels correlate with pRb alterations in human urothelial cells. Cancer Res 1995, 55:493-497 [PubMed] [Google Scholar]
- 44.Reznikoff CA, Yeager TR, Belair CD, Savelieva E, Puthenveettil JA, Stadler WM: Elevated p16 at senescence and loss of p16 at immortalization in human papillomavirus 16 E6, but not E7, transformed human uroepithelial cells. Cancer Res 1996, 56:2886-2890 [PubMed] [Google Scholar]
- 45.Xiong Y, Kuppuswamy D, Li Y, Livanos EM, Hixon M, White A, Beach D, Tlsty TD: Alteration of cell cycle kinase complexes in human papillomavirus E6- and E7-expressing fibroblasts precedes neoplastic transformation. J Virol 1996, 70:999-1008 [DOI] [PMC free article] [PubMed] [Google Scholar]