B-ZIP Proteins Encoded by the Drosophila Genome: Evaluation of Potential Dimerization Partners (original) (raw)
Abstract
The basic region-leucine zipper (B-ZIP) (bZIP) protein motif dimerizes to bind specific DNA sequences. We have identified 27 B-ZIP proteins in the recently sequenced Drosophila melanogaster genome. The dimerization specificity of these 27 B-ZIP proteins was evaluated using two structural criteria: (1) the presence of attractive or repulsive interhelical g↔e‘ electrostatic interactions and (2) the presence of polar or charged amino acids in the ‘a’ and ‘d’ positions of the hydrophobic interface. None of the B-ZIP proteins contain only aliphatic amino acids in the‘a’ and ‘d’ position. Only six of the Drosophila B-ZIP proteins contain a “canonical” hydrophobic interface like the yeast GCN4, and the mammalian JUN, ATF2, CREB, C/EBP, and PAR leucine zippers, characterized by asparagine in the second ‘a’ position. Twelve leucine zippers contain polar amino acids in the first, third, and fourth ‘a’ positions. Circular dichroism spectroscopy, used to monitor thermal denaturations of a heterodimerizing leucine zipper system containing either valine (V) or asparagine (N) in the ‘a’ position, indicates that the V–N interaction is 2.3 kcal/mole less stable than an N–N interaction and 5.3 kcal/mole less stable than a V–V interaction. Thus, we propose that the presence of polar amino acids in novel positions of the ‘a’ position of Drosophila B-ZIP proteins has led to leucine zippers that homodimerize rather than heterodimerize.
Basic region-leucine zipper (B-ZIP) transcription factors bind as dimers to sequence-specific DNA and regulate gene expression. The transcriptional potential of B-ZIP proteins is often regulated by posttranslational phosphorylation in response to cellular signals (Hurst 1995). The recent completion of the Drosophila melanogaster genome sequence (Adams et al. 2000) provides the opportunity to identify the complete list of B-ZIP proteins in a complex eukaryote. Previously, a genomewide analysis using the Automated InterPro Motif Identification Resource identified a B-ZIP domain in 29 genes in Drosophila (Rubin et al. 2000). This number compares with 31 B-ZIP proteins identified in the Caenorhabditis elegans genome, 17 in the Saccharomyces cerevisiae genome, 71 in the Arabidopsis thaliana genome (Riechmann et al. 2000), and 65 in the human genome (Tupler et al. 2001). Knowing all the B-ZIP proteins in a genome allows us to predict all the dimerization partners of a particular B-ZIP protein, something that has eluded investigators in the past. A prediction of potential dimerization partners of B-ZIP proteins should focus the efforts of Drosophila geneticists as they examine possible dimerization between B-ZIP containing genes.
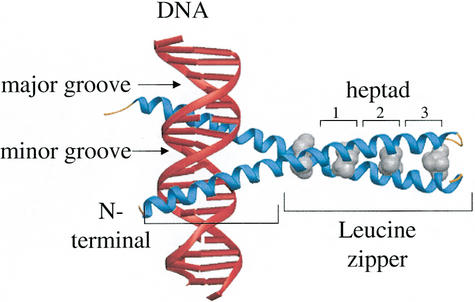
When bound to DNA, B-ZIP monomers are long α-helices, the N-terminal half binds in the major groove to sequence-specific double-stranded DNA, and the C-terminal half mediates dimerization to form a parallel leucine zipper coiled coil (Landschultz et al. 1988; Vinson et al. 1989; Ellenberger et al. 1992) (Fig. 1). The leucine zipper dimerization domain is typically composed of four to five heptad repeats of amino acids, with the seven unique positions in the heptad labeled ‘a’, ‘b’, ‘c’, ‘d’, ‘e’, ‘f’, and ‘g’ (McLachlan and Stewart 1975). The ‘g’, ‘a’, ‘d’, and ‘e’ positions are critical for dimerization stability and specificity. The shorter leucine zippers have less protein sequence flexibility because amino acids must be optimized for dimerization stability. Longer leucine zippers allow better regulation of dimerization specificity because they can contain amino acids that are suboptimal for stability but favor interaction with a particular partner.
Figure 1.
X-ray structure of GCN4 B-ZIP motif bound to a TRE DNA sequence (Ellenberger et al. 1992). The DNA is in red. The B-ZIP α-helices are in blue with the leucines in the ‘d’ position shown in gray. The N-terminus of the protein is labeled. The basic region and leucine zipper are labeled. The first three heptads of the leucine zipper are highlighted.
Amino acids in the ‘a’ and ‘d’ positions are typically hydrophobic and are on the same side of the α-helix, creating a hydrophobic interface that contributes to dimerization stability (Landschulz et al. 1989; Moitra et al. 1997). Typically, the ‘d’ position is occupied by leucine and the ‘a’ position by valine. An exception in B-ZIP leucine zippers is the second heptad ‘a’ position that often contains an asparagine. Asparagine in the ‘a’ position from one monomer can hydrogen bond interhelically with asparagine in the ‘a’ position of the second monomer to promote dimerization and prevent higher order oligomerization (Harbury et al. 1993). In contrast, asparagine does not form stable interactions with isoleucine in the ‘a’ position of leucine zipper proteins, preventing heterodimerization (Zeng et al. 1997). Conversely, charged amino acids in the ‘a’ position inhibit homodimerization and promote heterodimerization (unpublished data from Vinson group). An example is the Myc‖Max leucine zipper, in which a Myc homodimer is unstable because of an E in the ‘a’ position.
The g and e positions of the leucine zipper flank the hydrophobic interface and frequently contain charged amino acids (Cohen and Parry 1990; Vinson et al. 1993). X-ray structures of leucine zipper coiled-coil proteins reveal interhelical interactions between oppositely charged amino acids in the g position and the following e‘ position in the dimer (O’Shea et al. 1991; Glover and Harrison 1995; Chen et al. 1998; Lavigne et al. 1998; Day and Alber 2000). We refer to this interaction as g↔e‘; the prime (‘) indicates a residue on the second α-helix of the leucine zipper. Interacting amino acids in the g and e‘ positions lie across the hydrophobic interface such that their side-chain methylene groups pack with amino acids in the ‘a’ and ‘d’ positions of the hydrophobic core (Alber 1992). The g↔e‘ interactions between oppositely charged amino acids are attractive and promote dimerization (Vinson et al. 1993; Krylov et al. 1994; Zhou et al. 1994; Krylov et al. 1998), whereas g↔e‘ interactions between similarly charged amino acids, for example, E↔E or R↔R, are repulsive and inhibit homodimerization. For example, in the mammalian FOS protein, repulsive glutamate g↔e‘ interactions (E↔E) prevent homodimerization and thus help drive heterodimerization with JUN (Nicklin and Casari 1991; O’Shea et al. 1992).
Twelve Drosophila melanogaster B-ZIP genes have been isolated, including Vri (George and Terracol 1997), sis-A (Erickson and Cline 1993), crc (Hewes et al. 2000), cap‘n’collar (cnc) (Mohler et al. 1991), giant (gt) (Capovilla et al. 1992), slbo (Rorth and Montell 1992), pdp1 (Zhang et al. 1990), crebB-17A (Usui et al. 1993), crebA (Smolik et al. 1992), A3–3 (Heitzeberg 1999), kay, and Jra (Perkins et al. 1988, 1990; Zhang et al. 1990).
In this study, we have refined the estimate of the number of B-ZIP proteins in Drosophila melanogaster to 27 members using sophisticated search strategies and have subsequently inspected each potential B-ZIP protein for characteristics that affect dimerization specificity. Mammalian counterparts were identified for 21 Drosophila B-ZIP proteins, and conservation both throughout the entire protein and within the B-ZIP domain was evaluated. 13 Drosophila melanogaster leucine zippers contain a conserved asparagine in the second heptad ‘a’ position, as observed in mammalian B-ZIP proteins. Eight proteins contain asparagines in the first, third, or fourth heptad ‘a’ positions. We quantitate experimentally that the heterotypic interaction between asparagine and valine in the ‘a’ position is less stabilizing than either homotypic interaction. Coupling this additional insight into dimerization specificity with our knowledge of g↔e‘ interactions, we have predicted dimerization partners among the Drosophila melanogaster B-ZIP proteins.
RESULTS
Identifying Drosophila B-ZIP Domains
We searched for the B-ZIP protein motif (Vinson et al. 1989) in the recently completed Drosophila melanogaster DNA genome sequence (Adams et al. 2000). Previously, B-ZIP proteins have been identified in the yeast genome using a query based on the most conserved part of the B-ZIP motif, the basic region (Fernandes et al. 1997). We have used a modification of this query (Methods) to identify B-ZIP proteins in Drosophila melanogaster. Eighteen potential B-ZIP proteins were identified after searching the 14,100 predicted Drosophila open reading frames. Because the query represents the basic region without a leucine zipper, each of the 18 sequences (gi# 7290135, 7290320, 7290774, 7291080, 7291250, 7293451, 7294270, 7296965, 7298587, 7298780, 7300970, 7301182, 7301826, 7302191, 7302252, 7302350, 7302542, and 7303798) was inspected for an amphipathic α-helix located at an invariant distance in the C terminal direction from the basic region. Three sequences (gi7291080, gi7302191, and gi7302542) were discarded based on the absence of a satisfactory leucine zipper or basic regions, or the presence of α-helix breaking prolines within the motif.
To retrieve additional B-ZIP domains that may not conform precisely to the basic region query, we chose four sequences at random and subjected them to PSI-BLAST analysis (Altschul et al. 1997) performed to convergence. These were gi7290320 and gi729077, both PAR family members, gi7290135, an ATF3 homolog, and gi7298028. All hits with E values above the threshold of 0.001 were compared with the original set of 15 B-ZIP sequences. Eleven new sequences were identified (gi7291773, 7294768, 7295189, 7296431, 7297639, 7298028, 7300452, 7302966, 7298025, 7298026, and 7296993). After discarding one sequence (gi7296431) based on the absence of a satisfactory basic and leucine zipper region, the expanded set contained 25 sequences.
Three of the 15 original sequences were not reidentified by PSI-BLAST analysis and were thus considered the most distinctive. To identify other less related members of the B-ZIP family, we then used these three outlying sequences (gi7298587, gi7301182, and gi7302350) as queries in PSI-BLAST searches. PSI-BLAST analysis of gi7301182 retrieved only itself, and gi7298587 retrieved known sequences; however, gi7302350 retrieved five novel sequences. Multiple alignment of these sequences allowed four of the five new sequences to be eliminated based on the absence of satisfactory zipper or basic regions, leaving a total of 26 sequences in the set.
A separate regular expression query was performed against the same database, this time using the B-ZIP “regular expression” that contains both basic region and leucine zipper constraints (see Methods). Nine sequences were identified (gi7290320, 7290774, 7292623, 7293451, 7294270, 7295189, 7300970, 7302966, and 7303798), but only one of these (gi7292623, SisA) was a new addition to the existing set of B-ZIP motif proteins. To retrieve distant relatives of SisA, we used it as a query in a PSI-BLAST search. However, the search retrieved only SisA itself. Thus, the final tally for the number of B-ZIP proteins in Drosophila melanogaster is 27.
Mammalian Homologies
Mammalian counterparts were identified for 21 of the 27 Drosophila B-ZIP sequences using BLAST analysis with the B-ZIP region as the query. Table 1 presents the Drosophila name, synonyms, and the most related mammalian B-ZIP protein. In most cases, the most closely related human and mouse sequences are listed. This listing is intended to be representative rather than exhaustive.
Table 1.
Closest Human Counterpart to Drosophila B-ZIP Proteins
| Drosophila Sequence db designation | Synonyms | Mammalian Sequence (human, mouse) | % identity | |||
|---|---|---|---|---|---|---|
| basic region | zipper (L1-L5) | g-a-d-e positions | ||||
| CG7786 | gi7302966 | DBP (gi1706312#, gi8393240) | 64 | 48 | 70 | |
| CG17888a | gi7295189 | Pdp1, Par-domain protein 1 | HLF (gi4504421#, gi8394435) | 88 | 43 | 60 |
| CG3136* | gi7302252 | ATF6 (gi3953531, gi8393190) | 76 | 26 | 40 | |
| CG4575 | gi7290774 | HLF (gi4504421#, gi8394435) | 76 | 23 | 40 | |
| CrebB17-A | gi7293451 | CREB, dCREB2, CG6103 | CREM (gi8393194, gi479997) | 92 | 75 | 81 |
| CG8669 | gi7298780 | crc | ATF-4 (gi14779030, gi6753128) | 68 | 34 | 40 |
| crebA | gi7294270 | dCREB-A, CG7450 | Oasis (gi14211949, gi6754918) | 76 | 37 | 55 |
| CG12850 | gi7291864 | ATF-4 (gi14779030, gi6753128) | 44 | 26 | 30 | |
| CG9954* | gi7302350 | MAFF (gi7513139, gi2696885) | 76 | 26 | 35 | |
| gt | gi7290320 | giant, CG7952 | HLF (gi4504421#, gi8394435) | 84 | 26 | 45 |
| slbo/CEBP | gi7291773 | DmC/EBP, slow border cells, CG4354 | C/EBP+D55 β & α (gi4885131, gi109606) | 76 | 23 | 30 |
| CG10034* | gi7298587 | MAF (gi4885447#, gi1708910) | 72 | 37 | 35 | |
| CG13624 | gi7301182 | HYPOTHETICAL (gi18559858) | 92 | 34 | 35 | |
| vri | gi7296965 | vrilie, CG14029 | NFIL-3 (gi4885517#, gi8393832) | 80 | 31 | 23 |
| CG9415 | gi7291250 | XBP (gi105867, gi7305633) | 76 | 17 | 30 | |
| CG17836 | gi7300452 | none | na | na | na | |
| CG18619* | gi7297639 | CRE-BP-like 2 (gi4503035) | 100 | 40 | 50 | |
| CG15479 | gi7298028 | none | na | na | na | |
| sisA | gi7292623 | CG1641 | none | na | na | na |
| CG14014 | gi7296993 | none | na | na | na | |
| CG16813 | gi7298025 | none | na | na | na | |
| CG16815 | gi7298026 | none | na | na | na | |
| Jra* | gi7303798 | Djun, Jun-related antigen, CG2275 | JUND (gi18590942#, gi6680512) | 84 | 49 | 60 |
| CG6272* | gi7294768 | C/EBP (gi4885129#, gi6680916) | 36 | 28 | 30 | |
| A3-3 | gi7290135 | CG11405 | ATF3 (gi226728, gi13562096) | 72 | 45 | 65 |
| cnc* | gi7300970 | CNC_DROME, CG4578 | NFE2 (gi5453774, gi6754834) | 80 | 14 | 20 |
| kay | gi7301826 | D-Fos, Fos-related antigen, Fra, CG15509 | FOS (gi4885241#, gi6753894) | 60 | 37 | 55 |
Six B-ZIP proteins score the highest matches in reciprocal queries against the databases and also align over >50% of the length of the sequence and have been tentatively designated orthologs (Table 1). These include Pdp1, an HLF ortholog; CG3136, an ATF4 ortholog; CG12850, an ATF2 ortholog; CG9954 and CG10034, both possible MAF orthologs; Jra, a Jun homolog; and CG6272, a C/EBP homolog.
Table 1 also presents several measures of the relatedness between a Drosophila B-ZIP protein sequence and the closest related human protein sequence. The existence of an identical mouse counterpart to the human sequence is indicated by a # sign in column 4, showing evolutionary conservation within vertebrates. To represent conservation between the homologous Drosophila and human sequences, we calculated % identities for (1) the basic region, (2) the first five heptads of the leucine zipper region, and (3) the ‘g’, ‘a’, ‘d’, and ‘e’ positions of the leucine zipper that are critical for dimerization stability and specificity. The basic regions are more highly conserved than the leucine zipper. Within the leucine zipper, the ‘g’, ‘a’, ‘d’, and ‘e’ positions are more conserved than the entire leucine zipper, indicating that the determinants of dimerization specificity were actively conserved during the divergence of the insects and mammals. CREB is the most conserved B-ZIP domain, with 75% conservation throughout the leucine zipper region.
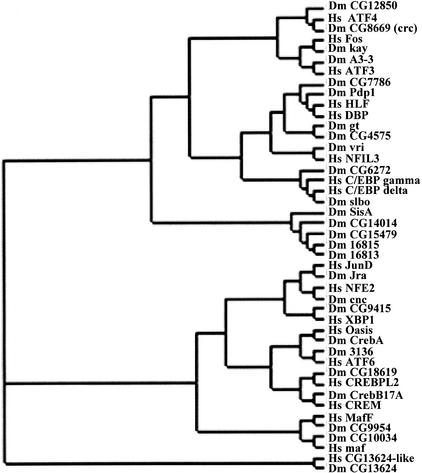
Figure 2 presents a phylogenetic analysis of an alignment of the Drosophila B-ZIP proteins and their mammalian counterparts based only on their B-ZIP motif protein sequence. Each Drosophila sequence clusters very closely with its mammalian counterpart. This indicates that the Drosophila B-ZIP proteins are more closely related to their human counterpart than they are to other Drosophila B-ZIP proteins. This is not true for the four PAR proteins that are more closely related to each other than they are to any human protein. The five Drosophila sequences lacking any mammalian relative cluster together. These are unusual B-ZIP sequences and the question of whether they are true B-ZIP proteins is considered later in the discussion.
Figure 2.
Rectangular cladogram representing the phylogenetic relationship among the Drosophila B-ZIP proteins and their closest human counterparts. The tree was made from a multiple alignment of 25 amino acids from the basic region and the first four heptads of the leucine zipper region.
Alignment of the Protein Sequences of the 27 Drosophila B-ZIP Domains
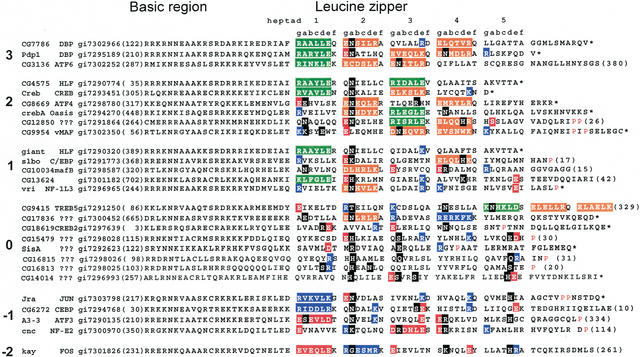
The protein sequence alignment of the 27 identified Drosophila melanogaster B-ZIP motifs is shown in Figure 3. The sequences begin four amino acids at the N terminus of the conserved asparagine (N) in the basic region (Vinson et al. 1989) and continue to the natural C terminus of the protein or until the leucine zipper contains a proline or glycine that is predicted to terminate the α-helix. We have highlighted ‘a’ and ‘d’ positions that contain polar or charged amino acids (black boxes) and the g↔e‘ interactions are color coded (green, orange, blue, or red), as described in the figure legend. Figure 4 presents a schematic of a coiled-coil dimer that graphically describes the color code used in Figure 3. A similar analysis has been done for the 53 identified human B-ZIP proteins (Vinson et al. 2002).
Figure 3.
Alignment of 27 identified Drosophila melanogaster B-ZIP motifs using the single letter amino acid code. The proteins are arranged into groups based on the number of attractive g↔e‘ interactions minus repulsive g↔e‘ interactions ranging from 3 to –2 pairs. The column starts with the name of the protein. Next is the name of the closest mammalian homolog, followed by the gi# for the Drosophila melanogaster sequence. The protein sequence of the B-ZIP motif follows. The number of amino acids from the predicted N terminus of the protein to the B-ZIP motif is given in parentheses. The C terminus of each sequence is either the natural C terminus denoted by an asterisk (*), or a truncation with the number of amino acids to the C terminus in parentheses. To help visualize the potential g↔e‘ interactions, we grouped heptads (gabcdef). If both the ‘g’ and ‘e’ positions contain charged amino acids, we color both of these amino acids and the intervening ones (gabcde). We use green for the attractive basic–acidic pairs (R↔E and K↔E, K↔D), orange for the attractive acidic–basic pairs (E↔R, E↔K, D↔K, and D↔R), red for the repulsive acidic pairs (E↔E, D↔E, and E↔D), and blue for the repulsive basic pairs (K↔K, R↔K, K↔R, and R↔R). If only one of the two amino acids in the g↔e‘ pair is charged, we color only that amino acid: red if it is acidic and blue if it is basic. If the ‘a’ or ‘d’ positions contain polar or charged amino acids, they are colored black. The α-helix breaking prolines, indicative of the C terminus of the leucine zipper, are colored red.
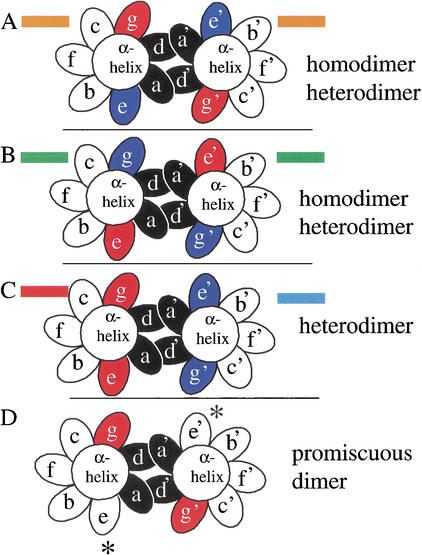
Figure 4.
End view, looking from N terminus to C terminus, of a coiled coil with the seven unique positions of the heptad presented as ellipses. The ‘a’ and ‘d’ positions are colored black. The four possible combinations of acidic and basic amino acids in the ‘g’ and ‘e’ positions are presented and color coded as used in Figure 2. (A) An α-helix with a g↔e‘ pair containing an acidic amino acid in the ‘g’ position and a basic amino acid in the following ‘e’ position (orange in Fig. 2) can form a homodimer or heterodimer with a similarly charged α-helix. (B) An α-helix with a g↔e‘ pair containing a basic amino acid in the ‘g’ position and an acidic amino acid in the following ‘e’ position (green in Fig. 2) can form a homodimer or heterodimer with a similarly charged α-helix. (C) A heterodimer between an acidic g↔e‘ pair (red in Fig. 2) and a basic g↔e‘ pair (blue in Fig. 2). (D) A dimer with an “incomplete” g↔e‘ pair resulting in promiscuous dimerization.
Surface Charge of Leucine Zippers: ‘g’ and ‘e’ Interactions
We have observed pronounced preferences in the frequency of charged and polar amino acids in the ‘a’, ‘d’, ‘e’, and ‘g’ positions for each heptad of the Drosophila B-ZIP proteins (Table 2). For the ‘g’ and ‘e’ positions, charged amino acids are concentrated in the first four heptads. The fifth heptad rarely contains either attractive or repulsive g↔e‘ interactions and may represent a natural limit for the length of the dimerization domain of B-ZIP proteins. An exception to this is CG9415, which contains attractive g↔e‘ interactions in the fifth, sixth, and seventh heptads but not in the first four heptads. An additional indication of the natural limit of the leucine zipper is the high frequency of α-helix breaking prolines and glycines in the fifth and sixth heptad of the leucine zippers (Fig. 3).
Table 2.
Number of Highlighted Amino Acids from Figure 2 in the g and e and the a and d Positions for Each Heptad of the Leucine Zipper
| Heptad # | 1 | 2 | 3 | 4 | 5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Coiled coil | g | e | g | e | g | e | g | e | g | e |
| Basic (K,R) | 13 | 4 | 3 | 11 | 5 | 10 | 3 | 12 | 5 | 2 |
| Acidic (D,E) | 5 | 11 | 14 | 0 | 9 | 6 | 9 | 1 | 0 | 6 |
| Coiled coil | a | d | a | d | a | d | a | d | a | d |
| Acidic (E) | 2 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 2 |
| Basic (K,R) | 1 | 1 | 3 | 0 | 1 | 1 | 4 | 1 | 3 | 2 |
| Polar (NTSHQ) | 3 | 1 | 19 | 1 | 5 | 2 | 4 | 4 | 2 | 8 |
Of the 216 ‘g’ and ‘e’ positions in the first four heptads of these 27 proteins (4 heptads × 2 positions per heptad × 27 sequences), 54% are occupied by a charged residue, equally distributed between basic and acidic amino acids. There are approximately twice as many arginines as there are lysines. The shorter aspartic acid is significantly underrepresented relative to glutamic acid, as has been observed for other coiled-coil proteins (Cohen and Parry 1990). This likely reflects the fact that aspartic acid is over 1.0 kcal/mole less stabilizing than is glutamic acid in the ‘g’ position (Krylov et al. 1998).
Of the 108 possible g↔e‘ interactions in the first four heptads, 25% are attractive and only 6% are repulsive. Attractive g↔e‘ interactions show a bias in the orientation of the amino acids. In the first heptad, all attractive g↔e‘ interactions have the same polarity, the ‘g’ position contains a basic amino acid, and the ‘e’ position contains an acidic amino acid (e.g., R↔E or K↔E). In the second heptad, the orientation of the g↔e‘ interaction is reversed (e.g., E↔R, E↔K or D↔K). The PAR family proteins exemplify this observation. In the third and fourth heptad, both orientations of attractive g↔e‘ interactions are observed. Only one B-ZIP protein, CG17836, has both attractive and repulsive g↔e‘ pairs.
Forty-eight percent of the g↔e‘ interactions contain only a single charged amino acid. Leucine zippers with incomplete g↔e‘ interactions will have more promiscuous dimerization activity. They do not contribute to the stability of the homodimer. However, in a heterodimer, they can form complete attractive g↔e‘ interactions and contribute to stability.
Polar or Charged Amino Acids in the ‘a’ and ‘d’ Hydrophobic Interior
All of the Drosophila B-ZIP proteins contain either a polar or charged amino acid in at least one ‘a’ or ‘d’ position in the first four heptads of the leucine zipper. The frequency of polar or charged amino acids in the ‘a’ and ‘d’ position is shown in Table 2. Nineteen proteins contain a polar (N, T, H) and 3 contain a basic (K, R) amino acid in the ‘a’ position of the second heptad, as is frequently observed in mammalian B-ZIP proteins. However, 13 Drosophila B-ZIP proteins (e.g., Pdp1 and CG3136) contain polar amino acids in the ‘a’ position of the first, third, and fourth heptads. These may help prevent heterodimerization with proteins that do not contain a polar amino acid in this position. Charged amino acids are found at the ‘a’ and ‘d’ positions of the leucine zipper in nine B-ZIP proteins, being more frequent in the ‘a’ position. These amino acids should discourage homodimerization. There are more basic amino acids than acidic amino acids in the ‘a’ and ‘d’ positions.
The Energetics of the Valine–Asparagine Interaction in the ‘a’ Position
The large number of polar amino acids in the ‘a’ position of Drosophila B-ZIP leucine zippers prompted us to examine whether these amino acids can affect dimerization specificity. Because ‘a’ position amino acids interact interhelically with the same ‘a’ position of the opposite monomer of the dimer, we needed to use a heterodimerizing system to address dimerization specificity. We previously generated a heterodimerizing system that forces dimerization of leucine zippers (Krylov et al. 1994). This system contains one monomer in which homodimerization is inhibited by repulsive acidic g↔e‘ interactions containing glutamic acid in the ‘g’ and ‘e’ positions (E↔E) of the third and fourth heptads (previously named EE34). We refer to this protein as B-EE34(V): the B represents the basic region and the V highlights the valine in the third ‘a’ position, the amino acid that is changed in this study. The second monomer in the heterodimerizing system contains arginine in the ‘g’ and ‘e’ positions of the third and fourth heptads, resulting in repulsive basic g↔e‘ interactions (R↔R) in the potential homodimer (previously named RR34). We refer to this protein as RR34(V). We replaced the basic region of RR34(V) with a synthetic acidic amphipathic extension (Krylov et al. 1995; Olive et al. 1997; Moll et al. 2000) to produce A-RR34. The acidic amphipathic extension of A-RR34 heterodimerizes with the basic region of EE34, increasing the stability of the EE34‖A-RR34 heterodimer by 2.5 kcal/mole (Moll et al. 2000).
We compared the thermal stability of three heterodimers with either valine or asparagine in the third ‘a’ position. The first heterodimer had valine in both third heptad ‘a’ positions, the second had asparagine in both third heptad ‘a’ positions, and the third had a valine in the third ‘a’ position of one monomer and an asparagine in the second monomer of the dimer. The monomers with asparagine in the third ‘a’ position are B-EE34(N) and A-RR34(N). Comparing the stability of a B-EE34(V) and A-RR34(N) mixture with the stabilities of B-EE34(V)‖A-RR34(V) and B-EE34(N)‖A-RR34(N) allowed us to determine whether stability of the valine–asparagine interaction contributes to dimerization specificity. Table 3 presents the thermodynamic parameters derived from thermal denaturations, as assayed by circular dichroism spectroscopy at 222 nm, of these heterodimers, assuming this is a two-state denaturation process. For the four homodimer denaturations, we find that the valine is more stabilizing than asparagine, as has been observed in another coiled-coil system (Wagschal et al. 1999). Analytical ultracentrifugation of the three heterodimer samples in Table 3 (EE34(V)‖A-RR34(V), EE34(N)‖A-RR34(N), and EE34(V)‖A-RR34(N)) indicate they are dimers (data not shown). The three mixtures are more stable than the four single proteins, indicating that the mixtures form heterodimers. The EE34(V)‖A-RR34(V) heterodimer that produces a valine–valine interaction is 3.0 kcal/mole more stable than the EE34(N)‖A-RR34(N) heterodimer that produces the asparagine–asparagine interaction. This value is consistent with the 2.8 kcal/mole observed in another guest–host leucine zipper system comparing valine–valine interactions with asparagine–asparagine interactions in the ‘a’ position (Wagschal et al. 1999). The EE34(V)‖A-RR34(N) heterodimer that produces an asparagine–valine interaction is 2.3 kcal/mole less stable than an asparagine–asparagine interaction and 5.3 kcal/mole less stable than a valine–valine interaction. These data show that the presence of an asparagine will disfavor heterodimerization with valine and instead drive homodimerization.
Table 3.
Thermodynamic Parameters Derived from Thermal Denaturations of Mixtures of Proteins
| Protein (2.0 μM) | Tm (°C) | ΔG37 (kcal/mole) | ΔH (kcal/mole) |
|---|---|---|---|
| B-EE34(V)‖A-RR34(V) | 57.6 | −14.7 | −111.2 |
| B-EE34(N)‖A-RR34(N) | 50.1 | −11.7 | −99.3 |
| B-EE34(V)‖A-RR34(N) | 43.4 | −9.4 | −81 |
Predicted Dimerization Partners
To predict the dimerization partners for the 27 B-ZIP proteins from Drosophila, we used a two-step approach. In step 1, we examined the number of attractive and repulsive interhelical g↔e‘ interactions in the leucine zipper of each homodimer and the 26 possible heterodimers. In step 2, we examined the amino acid composition of the ‘a’ and ‘d’ positions. The presence of polar or charged amino acids in these positions caused us to modify our predictions of dimerization specificity based on the g↔e‘ interactions determined in step 1.
In Figure 3, the B-ZIP proteins are clustered by the number of attractive minus the number of repulsive g↔e‘ interactions in the homodimer. These values range from three pairs of attractive g↔e‘ interactions for the PAR-like proteins to −2 pairs for the FOS-like protein.
Table 4 lists the predicted dimerization partners for representatives of each cluster. The number of attractive interactions for each predicted pair is shown (column 4) and the basis for each prediction is summarized (column 5).
Table 4.
Predicted B-ZIP Dimerization Partners
| Sum and attractive and repulsive interactions | Protein | B-ZIP family | Predicted dimerization partners (# attractive interactions) | Comments |
|---|---|---|---|---|
| +3 | CG7786 | PAR | Homo (6) CREB (5) | Mammalian PAR proteins homodimerize and heterodimerize within the family; in contrast, Drosophila PAR proteins are predicted to homodimerize. |
| +3 | CG17888 (Pdp1) | PAR | Homo (6) | N in the 4th heptad “a” position will inhibit heterodimerization with other PAR proteins. |
| +3 | CG3136 | ATF6 | Homo (6) | N in the 3rd heptad “a” position will inhibit heterodimerization with other PAR proteins. |
| +2 | CG4575 | PAR | Homo (4) | Reversal of the 3rd heptad salt bridge from E ↔ R to R ↔ E will inhibit heterodimerization with other PAR proteins. |
| +2 | Creb | CREB | Homo (4) Giant (3) | Short zipper and attractive 1st and 3rd g ↔ e′ interactions characteristic of mammalian CREB.Presence of three Q's in the “g” positions of giant may stabilize a CREB/giant heterodimer. |
| +2 | CG8669 | ATF4 | 18619 (4) Sis A (−2) | Charged interface with an E in the 1st heptad “a” position and an R in the 3rd heptad “d” position. May heterodimerize via a salt bridge in the hydrophobic interior with CG18619, an R in the 3rd “d” position with an E in the 3rd “a” position of SisA. An interaction with SisA has been observed in a yeast interaction screen (Jim Erickson, unpublished observations). |
| +2 | CrebA | Oasis | Homo (4) | An N in the 4th heptad “a” position is predicted to promote homodimerization and prevent heterodimerization. |
| +2 | CG12850 | ??? | Homo (4) | Three polar “a” and “d” positions are predicted to cause homodimerization. |
| +2 | CG9954 | S-MAF | Homo (4) CG10034 (MAF) | Presence of an aliphatic rather than N in the 2nd heptad “a” position may allow heterodimerization with CG10034, which also has an aliphatic in the 2nd “a” position. Lysine in the 1st heptad “a” position and N in the 3rd heptad “a” position may cause other homodimerization. |
| +1 | Giant | PAR | Homo (2) CG7786 (2) or CREB (3) | |
| +1 | Slbo | C/EBP | A3-3 (2) | Resembles mammalian fos in having lysine in the 2nd “a” position. Two incomplete g ↔ e′ interactions in the 1st and 2nd heptads give the potential for promiscuous dimerization. |
| +1 | CG10034 | L-MAF | MAF (2) | Atypical interface with an L and R in the 2nd and 4th “a” positions, respectively. Predicted to heterodimerize with MAF, which also has L in the 2nd “a” position. |
| +1 | CG13624 | ?? | 4th heptad “d” position K may drive heterodimerization. | |
| +1 | Vri | C/EBP | Homo (2) A3-3 (2); cnc (0) or kay (3) | Has canonical hydrophobic interface with three incomplete g ↔ e′ interactions that are all basic, suggesting heterodimerization with any of the acidic zippers (A3-3, cnc, kay). |
| 0 | CG9415 | ATF6 | Homo (6) | Polar residues in the “a” positions of the 2nd, 4th, and 5th heptads that may drive homodimerization. Attractive interactions in the 5th, 6th, and 7th heptads will also encourage homodimerization. |
| 0 | SisA | ATF4 (1) | The R in the 2nd “a” position and E in the 3rd “a” position is likely to prevent homodimerization. ATF4 is one possible partner. | |
| −1 | Jra | JUN | A3-3 (4); cnc (3); kay (4) | Canonical interface. Presence of a basic repulsive pair in heptad 1 and partial g ↔ e′ interactions in heptads 2–4 indicate likely heterodimerization with acid zipper proteins that can form g ↔ e′ pairs. |
| −1 | CG6272 | C/EBP | A3-3 (4); cnc (3); kay (0) | Like Jra, has a repulsive basic pair in heptad 1, suggesting acidic zipper dimerization partners. Charged residues in “e” positions may promote promiscuous heterodimerization. |
| −1 | Cnc | CNC | Jra (3) CG6272 (1) | Acidic zippers predicted to interact with basic zippers Jra and C6272. |
| −2 | Kay | FOS | Jra (4) | Repulsive interaction in the 1st interaction and a histidine in the 5th “d” position, as seen in mammalian FOS. Salt bridge pattern indicates likely heterodimerization. Kay has been biochemically purified as a heterodimer with Jra. |
Eight B-ZIP proteins have no attractive or repulsive g↔e‘ interactions. The lack of g↔e‘ interactions and the presence of a large number of polar or charged amino acids in the ‘a’ and ‘d’ positions make prediction of dimerization partners for this set difficult. Only SisA and CG9415 have been listed.
DISCUSSION
Previously, a computational annotation of the Drosophila melanogaster genome identified 29 genes containing the B-ZIP motif (Rubin et al. 2000). We have reexamined these data and identified 27 members, including 7 members not identified by the automated InterPro Motif Identification Resource. Twenty-one Drosophila B-ZIP proteins have B-ZIP regions that are highly related to mammalian B-ZIP proteins, including the homodimerizing CREB, C/EBP, and PAR proteins, and the heterodimerizing FOS, JUN, MAF, and NRE2 proteins. Searches between Drosophila and vertebrate B-ZIP proteins identified six that are conserved in both the B-ZIP domain and the rest of the protein. These B-ZIP proteins are putative orthologs and are likely to perform evolutionarily ancient functions.
Automated Versus Manual B-ZIP Protein Identification
Automated annotation by the InterPro Motif Identification Resource (http://www.ebi.ac.uk/proteome) (Apweiler et al. 2001) identified 29 genes containing the B-ZIP motif (Rubin et al. 2000). We have reexamined these data and the Drosophila genome sequence and identified 27 B-ZIP genes, including 7 new members not previously annotated as B-ZIP proteins.
Eight proteins were identified as B-ZIP proteins by the InterPro Motif Identification Resource that do not pass our criteria of what constitutes a B-ZIP protein. One, CG17894 (gi10726715), is identical to the cnc protein (gi73000970) but has an additional 275 amino acids at the N terminus. The remaining seven proteins in the Interpro listing (CG6129, CG18266, CG9274, CG2848, CG18553, CG11774, and CG11745) are not canonical members of the B-ZIP family based on the following criteria. BLAST analysis using each of these proteins as queries failed to identify any known B-ZIP proteins in the protein database (the search was restricted to Drosophila proteins). A search of each protein against the Conserved Domain Database (CDD, National Center for Biotechnology Information [NCBI]) failed to identify the B-ZIP domain, although other domains were identified. And finally, five of the seven Interpro hits fail to meet the significance thresholds set by the databases for “true” hits with B-ZIP signatures and are therefore “false”. Thus, manual query methods for identification of B-ZIP proteins identified six bona fide proteins not found by automated domain identification methods. Furthermore, automated methods identified several putative “false” positives.
Noncanonical B-ZIP Proteins
Both the manual and automated methods that are used to identify the complete set of B-ZIP proteins in a genome are constrained by our current lack of understanding of these proteins. The most efficient method of identifying B-ZIP proteins that are similar to the well-characterized mammalian B-ZIP proteins is to identify a canonical basic region and then subsequently identify an amphipathic α-helix placed at an invariant distance in the C-terminal direction from the basic region. This approach is flawed by its failure to recognize the possible existence of a class of B-ZIP-like transcription factors in which dimerization is mediated by a leucine zipper but in which DNA binding is mediated by a novel or less-conserved motif. For example, mammalian CHOP-10 (Gadd 153) contains a C/EBP-like leucine zipper but a divergent basic region containing two prolines, and it was initially thought not to bind DNA (Ron and Habener 1992). C/EBP‖CHOP-10 heterodimers, however, are able to bind novel DNA elements (Ubeda et al. 1996). These types of B-ZIP proteins are difficult to identify because there are so many amphipathic α-helices in the genome. Another example may be CG11774 (gi7299089), one of the proteins identified by InterPro but not by our manual analysis. This sequence, and others not discussed, possesses a canonical zipper with good g↔e‘ salt bridge interactions, but lacks a convincing basic region.
Other proteins have an obvious basic region but an ambiguous amphipathic α-helix. For example, there are putative monomeric proteins containing the basic region that bind to DNA. The skn-1 gene in C. elegans (Bowerman et al. 1992) has no dimerization motif but does have a C-terminal four helix bundle to hold the extended α-helical basic region (Rupert et al. 1998). Additionally, the skn basic region has an N-terminal extension of the basic region that helps to stabilize DNA binding (Carroll et al. 1997). Only experiments will determine whether these noncanonical sequences define novel X-ZIP or B-X variants of the B-ZIP transcription factor family or whether they are simply a subset of coiled-coil or basic region-containing proteins.
Structural Features of the B-ZIP Motif
There are several structural features that appear general to the leucine zipper domain of most B-ZIP motifs in the Drosophila melanogaster genome. The leucine zipper is generally four heptads long. In Drosophila, attractive g↔e‘ pairs in the first heptad are always basic↔acidic, whereas in the second heptad, attractive g↔e‘ pairs are reversed to acidic↔basic. Both orientations are observed in the third heptad, whereas the fourth heptad g↔e‘ pairs are acidic↔basic. Arginine is twice as common as lysine in the ‘g’ and ‘e’ positions. A double-mutant thermodynamic cycle analysis of g↔e‘ interactions measures a coupling energy, indicative of amino acid interactions, of −0.5 kcal/mole for the E↔R interaction compared with −0.3 kcal/mole for the E↔K interaction. This indicates that R confers more specific dimerization than does K (Krylov et al. 1998). The preference of R over K in the ‘g’ and ‘e’ positions indicates that this position is used to increase dimerization specificity instead of stability.
All Drosophila B-ZIP leucine zippers contain either a polar or a charged amino acid in an ‘a’ or ‘d’ position. A nonaliphatic amino acid in the second heptad ‘a’ position is observed in 25 of the 27 Drosophila melanogaster B-ZIP proteins, with asparagine occurring 13 times. Asparagine in the ‘a’ position has been shown to limit higher-order oligomerization in the yeast B-ZIP protein GCN4 (Harbury et al. 1993), but it remains obscure why the second heptad ‘a’ position is so often used for this function. Eight B-ZIP proteins with asparagines in the first, third, or fourth heptad ‘a’ position prompted us to determine the energetics of asparagine to dimerization specificity using a heterodimerizing leucine zipper system. The data indicate that asparagine prevents heterodimerization with valine. This, it appears that in the Drosophila melanogaster genome, asparagine has also been used in the first, third, and fourth heptad ‘a’ position to create leucine zippers that prefer to homodimerize and not interact with other leucine zippers that contain aliphatic amino acids in the ‘a’ position, such as Pdp1, CG3136, CrebA, CG954, and CG9415.
Based on our knowledge of the effects of amino acids in the ‘g’, ‘a’, ‘d’, and ‘e’ positions of the leucine zipper on dimerization stability and specificity, we have predicted the potential dimerization partners for the Drosophila melanogaster B-ZIP proteins. In vertebrates, the CREB, C/EBP, or PAR families have multiple members that can homodimerize and heterodimerize within the subfamily. In contrast, in Drosophila melanogaster, each of these families consist either of a single member or multiple members that we predict will only homodimerize and not heterodimerize, even within the subfamily.
Homodimerizing Proteins: The PAR Proteins
Four Drosophila proteins share the structural features of PAR proteins. PAR appears to represent the prototypical leucine zipper sequence found throughout metazoans. In the three known vertebrate PAR family proteins, the first four heptads of the leucine zipper have identical attractive g↔e‘ interhelical interactions, R↔E, E↔R, E↔R, E↔R. They also have similar hydrophobic interfaces with an asparagine in the second heptad ‘a’ position. These mammalian proteins are known to form homodimers and to heterodimerize within the family (Hunger et al. 1992; Inaba et al. 1992).
An examination of the PAR-related B-ZIP proteins in Drosophila indicates that two structural strategies have been used to generate new leucine zippers that homodimerize but do not heterodimerize with other PAR family members. One strategy is illustrated by Pdp1 that contains an asparagine in the ‘a’ position of the fourth heptad in addition to the asparagine in the ‘a’ position of the second heptad. We have shown in this study that an asparagine in the ‘a’ position prevents heterodimerization with valine. An asparagine–valine interaction in the ‘a’ position is 2.3 or 5.3 kcal/mole less stable than an asparagine–asparagine or a valine–valine interaction, respectively. Thus, Pdp1 will not interact with the other PAR-like proteins that contain an aliphatic amino acid in this position. The large number of polar amino acids in the ‘a’ position of leucine zippers of Drosophila melanogaster indicates that this mechanism has been used to generate new homodimerizing leucine zippers by changing a single amino acid.
A second strategy to produce new homodimerizing leucine zippers is seen in CG4575 in which the third g↔e‘ pair is reversed from E↔R to R↔E. We calculate this would destabilize heterodimerization with PAR proteins containing an E↔R salt bridge in the third position by 2.9 kcal/mole (Krylov et al. 1998). Reversal of a single salt bridge in a vertebrate PAR family protein has been shown experimentally to prevent heterodimerization (Moll et al. 2000). Interestingly, in a heterodimer, the energetic cost of combining leucine zippers with an E↔R and an R↔E salt bridge is similar to the cost of forming an asparagine–valine pair, indicating that either strategy is capable of producing a new homodimerizing leucine zipper.
Conservation of B-ZIP Proteins Between Drosophila and Humans
Comparisons between Drosophila and vertebrate B-ZIP proteins identified six proteins that are conserved in both the B-ZIP domain and the rest of the protein. These B-ZIP proteins are putative orthologs and are likely to perform evolutionarily ancient functions. Twenty-one Drosophila B-ZIP proteins have B-ZIP regions that are highly related to mammalian B-ZIP proteins, including the homodimerizing CREB, C/EBP, and PAR proteins and the heterodimerizing FOS, JUN, MAF, and NRE2 proteins. We have evaluated whether amino acids that we predict are critical for regulating dimerization specificity in the Drosophila B-ZIP proteins are conserved in the human homolog. The four positions critical for regulating dimerization specificity (‘g’, ‘a’, ‘d’, and‘e’) are more conserved than the entire heptad, indicating that dimerization specificity is actively selected for during evolution. For example, the fourth heptad ‘a’ position asparagine and the third heptad basic–acidic g↔e‘ pair are conserved throughout evolution in CrebA, the Drosophila homolog of the Oasis family in humans. The histidine found in the fifth heptad of Jra, A3–3, and kay are conserved in their human homologs JUN, ATF3, and FOS.
Six putative Drosophila B-ZIP proteins do not have human counterparts. They also do not have any attractive or repulsive g↔e‘ pairs as is observed for canonical leucine zippers. This indicates either that they are not real B-ZIP proteins or are a group of new B-ZIP proteins that have evolved in the insects. The observation that sisA, an insoluble protein, interacts in a yeast two-hybrid screen with two proteins, CG16813 and CG16815 (J. Erickson, pers. comm.), which we have independently identified as putative Drosophila B-ZIP proteins without human homologs, indicates that these proteins heterodimerize, as would be expected for B-ZIP proteins. The function of these proteins in Drosophila sex determination my represent a new function for B-ZIP proteins in the insects.
Dimerization Partner Predictions
Based on our knowledge of the effects of the ‘g’, ‘a’, ‘d’, and ‘e’ positions of the leucine zipper on dimerization stability and specificity, we have predicted the potential dimerization partners for the Drosophila melanogaster B-ZIP proteins. It is likely that dimerization specificity is influenced by other factors in addition to the two simple criteria we have taken into account. For example, the prediction of dimerization partners is complicated by the fact that the DNA sequence bound by B-ZIP dimers can alter dimerization preference (Hai and Curran 1991). Nonetheless, it seems reasonable to explore the idea that these criteria may have some predictive value. For example, our simple rules lead to predicted interactions between Vri, a member of the C/EBP family, and kay, a FOS family member. C/EBP–FOS heterodimers have been observed (Hsu et al. 1994; Ubeda et al. 1996). Likewise, the well-known interaction between JUN and FOS is also predicted by these rules. As our understanding of the energetics of leucine zipper dimerization increases, more valuable predictions will be possible. In the absence of other predictive information, these rules may be a practical starting place in formulating a hypothesis for experimental analysis of possible dimerization partners for the Drosophila B-ZIP proteins.
METHODS
Pattern Matching
A database of the translated Drosophila melanogaster genome sequence was created from the data released by Celera (dros_na) and deposited into GenBank. This database consists of 14,100 open reading frames. Two types of regular expressions were used to query the database using the gref utility of the SEALS package (NCBI). The “B-ZIP” regular expression ([RKFS]XXXNXX[ASYK][AVK][RKASQNE][SCFYL] R[RKAIFDNQ]XXXXXXXX[LIVTS]XXX[VRATSC] XX[LYVM]XXX[NKVR]XX[LIY]) was generated from a multiple alignment of representative B-ZIP proteins that included VBP, C/EBPα, FOS, P45, CREB, ATF3, GCN4, P18, Sis-A, Cnc, ATF2, and ATF4. Every residue represented at a given position was included in the regular expression. The “BASIC” regular expression ([RQK][NLE][TRK]X[ASY][ASQ]XX[CSFYG][RDL] X[RK][RKL]) was modified from Fernandes et al. (1997). This expression corresponds to the basic region of 30 B-ZIP proteins from various organisms. It was modified at position 2 to include the L found in Yap8p from S. cerevisiae, as well as the more commonly found N. Positions 2, 3, 6, 9, 10, and 13 were modified to include additional residues based on an alignment of CREB sequence from Drosophila melanogaster.
BLAST and PSI-BLAST Analyses
BLAST analysis (Altschul et al. 1990) was performed on the NCBI web server using short B-ZIP sequences as queries. The ungapped parameter was used to force global alignments. Filtering was turned off. Other options were left at their default settings. PSI-BLAST analysis (Altschul et al. 1997) was done using the SPLAT routine of the SEALS (Walker and Koonin 1997). The analysis was performed to convergence with an include threshold (-h) of 0.001.
Multiple Alignment and Phylogenetic Tree Analysis
Multiple alignments were performed with Clustal W (Higgins et al. 1996) [Thompson et al. 1994] using the default options. The pairwise ordering mode was set to fast and approximate. Phylogenetic analysis was performed using TreeView (v. 1.6.1; http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Proteins
The sequence of the 96 amino acid EE34 (Krylov et al. 1994), also named EE34(V) in this manuscript, is ASMTG GQQMGRDP-LEE-KVFVPDEQKDEKYWTRRKKNNVAAKRSRDARRLKENQTI RAAFLEK ENTALRT E(V)AELEK EVGRCEN IVSKYETRYGPL. The leucine zipper is separated into heptads, as presented in Figure 3. The valine in parentheses was changed to N to create EE34(N). The first 13 amino acids are from φ10, the next three amino acids are a cloning linker, and the remaining 80 amino acids comprise the basic region followed by a leucine zipper of EE34. The protein sequence of A-RR34(V) is ASMTGGQQMGRDP-LEE- LEQRAEELARE NEELLEKEAEELEQENAELE RAAFLEK ENTALRT R(V)AELRK RVGRCRN IVSKYKYETRYGPL. The valine in parentheses was changed to N to create A-RR34(N). The LE in bold is the Xho I site that is the border between the leucine zipper and the N-terminal acidic extension. Proteins were expressed in E. coli using the T7 IPTG-inducible system and purified as described previously (Olive et al. 1997).
Circular Dichroism
Circular dichroism (CD) studies were performed using a Jasco J-720 spectropolarimeter. All protein stock solutions were in 12.5 mM potassium phosphate (pH 7.4), 150 mM KCl, and 0.25 mM ethylenediamine tetraacetic acid. One millimolar dithiothreitol and 2 μM of protein sample in 1 ml stock buffer was heated to 65°C for 20 min, cooled to room temperature for 5 min, and added to a 5-mm rectangular CD cell.
Thermodynamic Calculations
Melting temperature (Tm) and enthalpy (ΔH) values were determined from denaturation curves, assuming a two-state equilibrium dissociation of α-helical dimers into unfolded monomers using ΔCp of −2.04 kcal/mole/°C, as described previously (Krylov et al. 1997). ΔG values are reported at 37°C.
WEB SITE REFERENCES
http://taxonomy.zoology.gla.ac.uk/rod/treeview.html; tree-drawing software by Rod Page (University of Glasglow) for displaying phylogenies. Programs can be downloaded.
http://www.ebi.ac.uk/proteome; Proteome Analysis database for comprehensive statistical and comparative analyses of the predicted proteomes of fully sequenced organisms.
Acknowledgments
We thank Jim Erickson for communicating unpublished observations.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL: vinsonc@dc37a.nci.nih.gov; FAX (301) 496-8419.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.67902.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Alber T. Structure of the leucine zipper. Curr Opin Genet Dev. 1992;2:205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Biswas M, Fleischmann W, Kanapin A, Karavidopoulou Y, Kersey P, Kriventseva EV, Mittard V, Mulder N, Phan I, et al. Proteome Analysis Database: Online application of InterPro and CluSTr for the functional classification of proteins in whole genomes. Nucleic Acids Res. 2001;29:44–48. doi: 10.1093/nar/29.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Capovilla M, Eldon ED, Pirrotta V. The giant gene of Drosophila encodes a b-ZIP DNA-binding protein that regulates the expression of other segmentation gap genes. Development. 1992;114:99–112. doi: 10.1242/dev.114.1.99. [DOI] [PubMed] [Google Scholar]
- Carroll AS, Gilbert DE, Liu X, Cheung JW, Michnowicz JE, Wagner G, Ellenberger TE, Blackwell TK. SKN-1 domain folding and basic region monomer stabilization upon DNA binding. Genes & Dev. 1997;11:2227–2238. doi: 10.1101/gad.11.17.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- Cohen C, Parry D. α-helical coiled coils and bundles: How to design an α-helical protein. Proteins. 1990;7:1–14. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- Day CL, Alber T. Crystal structure of the amino-terminal coiled-coil domain of the APC tumor suppressor. J Mol Biol. 2000;301:147–156. doi: 10.1006/jmbi.2000.3895. [DOI] [PubMed] [Google Scholar]
- Ellenberger T, Brandl C, Struhl K, Harrison S. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: Crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Erickson JW, Cline TW. A bZIP protein, sisterless-a, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes & Dev. 1993;7:1688–1702. doi: 10.1101/gad.7.9.1688. [DOI] [PubMed] [Google Scholar]
- Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George H, Terracol R. The vrille gene of Drosophila is a maternal enhancer of decapentaplegic and encodes a new member of the bZIP family of transcription factors. Genetics. 1997;146:1345–1363. doi: 10.1093/genetics/146.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four -stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Heitzeberg The ATF-3 like bZIP transcription factor of Drosophila melanogaster is involved in development and female fertility. Europ Dros Res Conf. 1999;16:272. [Google Scholar]
- Hewes RS, Schaefer AM, Taghert PH. The cryptocephal gene (ATF4) encodes multiple basic-leucine zipper proteins controlling molting and metamorphosis in Drosophila. Genetics. 2000;155:1711–1723. doi: 10.1093/genetics/155.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D, Thompson J, Gibson T. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Hsu W, Kerppola TK, Chen PL, Curran T, Chen-Kiang S. Fos and Jun repress transcription activation by NF-IL6 through association at the basic zipper region. Mol Cell Biol. 1994;14:268–276. doi: 10.1128/mcb.14.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger SP, Ohyashiki K, Toyama K, Cleary ML. Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes & Dev. 1992;6:1608–1620. doi: 10.1101/gad.6.9.1608. [DOI] [PubMed] [Google Scholar]
- Hurst HC. Transcription factors 1: bZIP proteins. Protein Profile. 1995;2:101–168. [PubMed] [Google Scholar]
- Inaba T, Roberts WM, Shapiro LH, Jolly KW, Raimondi SC, Smith SD, Look AT. Fusion of the leucine zipper gene HLF to the E2A gene in human acute B- lineage leukemia. Science. 1992;257:531–534. doi: 10.1126/science.1386162. [DOI] [PubMed] [Google Scholar]
- Krylov D, Mikhailenko I, Vinson C. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. EMBO J. 1994;13:2849–2861. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov D, Olive M, Vinson C. Extending dimerization interfaces: The bZIP basic region can form a coiled coil. EMBO J. 1995;14:5329–5337. doi: 10.1002/j.1460-2075.1995.tb00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov D, Kasai K, Echlin DR, Taparowsky EJ, Arnheiter H, Vinson C. A general method to design dominant negatives to B-HLHZip proteins that abolish DNA binding. Proc Natl Acad Sci. 1997;94:1227–1229. doi: 10.1073/pnas.94.23.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov D, Barchi J, Vinson C. Inter-helical interactions in the leucine zipper coiled-coil dimer: pH and salt dependence of coupling energy between charged amino acids. J Mol Biol. 1998;279:959–972. doi: 10.1006/jmbi.1998.1762. [DOI] [PubMed] [Google Scholar]
- Landschultz W, Johnson P, McKnight S. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989;243:1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- Lavigne P, Crump MP, Gagne SM, Hodges RS, Kay CM, Sykes BD. Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper. J Mol Biol. 1998;281:165–181. doi: 10.1006/jmbi.1998.1914. [DOI] [PubMed] [Google Scholar]
- McLachlan A, Stewart M. Tropomyosin coiled-coil interactions: Evidence for an unstaggered structure. J Mol Biol. 1975;98:293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- Mohler J, Vani K, Leung S, Epstein A. Segmentally restricted, cephalic expression of a leucine zipper gene during Drosophila embryogenesis. Mech Dev. 1991;34:3–9. doi: 10.1016/0925-4773(91)90086-l. [DOI] [PubMed] [Google Scholar]
- Moitra J, Szilak L, Krylov D, Vinson C. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry. 1997;36:12567–12573. doi: 10.1021/bi971424h. [DOI] [PubMed] [Google Scholar]
- Moll JR, Olive M, Vinson C. Attractive interhelical electrostatic interactions in the proline- and acidic-rich region (PAR) leucine zipper subfamily preclude heterodimerization with other basic leucine zipper subfamilies. J Biol Chem. 2000;275:34826–34832. doi: 10.1074/jbc.M004545200. [DOI] [PubMed] [Google Scholar]
- Nicklin M, Casari G. A single site mutation in a truncated Fos protein allows it to interact with the TRE in vitro. Oncogene. 1991;6:173–179. [PubMed] [Google Scholar]
- Olive M, Krylov D, Echlin DR, Gardner K, Taparowsky E, Vinson C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- O'Shea E, Klemm J, Kim P, Abler T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel, coiled-coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- O'Shea E, Rutkowski R, Kim P. Mechanism of specificity in the fos-jun oncoprotein heterodimer. Cell. 1992;68:699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- Perkins KK, Dailey GM, Tjian R. Novel Jun- and Fos-related proteins in Drosophila are functionally homologous to enhancer factor AP-1. EMBO J. 1988;7:4265–4273. doi: 10.1002/j.1460-2075.1988.tb03324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins KK, Admon A, Patel N, Tjian R. The Drosophila Fos-related AP-1 protein is a developmentally regulated transcription factor. Genes & Dev. 1990;4:822–834. doi: 10.1101/gad.4.5.822. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes & Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- Rorth P, Montell DJ. Drosophila C/EBP: A tissue-specific DNA-binding protein required for embryonic development. Genes & Dev. 1992;6:2299–2311. doi: 10.1101/gad.6.12a.2299. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert PB, Daughdrill GW, Bowerman B, Matthews BW. A new DNA-binding motif in the Skn-1 binding domain-DNA complex. Nat Struct Biol. 1998;5:484–491. doi: 10.1038/nsb0698-484. [DOI] [PubMed] [Google Scholar]
- Smolik SM, Rose RE, Goodman RH. A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol Cell Biol. 1992;12:4123–4131. doi: 10.1128/mcb.12.9.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- Ubeda M, Wang XZ, Zinszner H, Wu I, Habener JF, Ron D. Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol Cell Biol. 1996;16:1479–1489. doi: 10.1128/mcb.16.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Smolik SM, Goodman RH. Isolation of Drosophila CREB-B: A novel CRE-binding protein. DNA Cell Biol. 1993;12:589–595. doi: 10.1089/dna.1993.12.589. [DOI] [PubMed] [Google Scholar]
- Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Vinson CR, Hai T, Boyd SM. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: Prediction and rational design. Genes & Dev. 1993;7:1047–1058. doi: 10.1101/gad.7.6.1047. [DOI] [PubMed] [Google Scholar]
- Vinson, C., Myakishev, M., Acharya, A., Mir, A., Moll, J., and , B.M. 2002. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell. Biol.22(18): (in press). [DOI] [PMC free article] [PubMed]
- Wagschal K, Tripet B, Lavigne P, Mant C, Hodges RS. The role of position a in determining the stability and oligomerization state of α-helical coiled coils: 20 amino acid stability coefficients in the hydrophobic core of proteins. Protein Sci. 1999;8:2312–2329. doi: 10.1110/ps.8.11.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DR, Koonin EV. SEALS: A system for easy analysis of lots of sequences. Proc Int Conf Intell Syst Mol Biol. 1997;5:333–339. [PubMed] [Google Scholar]
- Zeng X, Herndon AM, Hu JC. Buried asparagines determine the dimerization specificities of leucine zipper mutants. Proc Natl Acad Sci. 1997;94:3673–3678. doi: 10.1073/pnas.94.8.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Chaillet JR, Perkins LA, Halazonetis TD, Perrimon N. Drosophila homolog of the mammalian jun oncogene is expressed during embryonic development and activates transcription in mammalian cells. Proc Natl Acad Sci. 1990;87:6281–6285. doi: 10.1073/pnas.87.16.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Kay C, Hodges R. The net energetic contribution of interhelical electrostatic attractions to coiled-coil stability. Protein Eng. 1994;7:1365–1372. doi: 10.1093/protein/7.11.1365. [DOI] [PubMed] [Google Scholar]