Cyclin D1, a Novel Molecular Marker of Minimal Residual Disease, in Metastatic Neuroblastoma (original) (raw)
Abstract
Accurate monitoring of minimal residual disease (MRD) is critical for the management of metastatic neuroblastoma (NB). We evaluated cyclin D1 (CCND1), a cell-cycle control gene, as a novel MRD marker of NB. Using quantitative reverse transcriptase-polymerase chain reaction, we studied CCND1 expression in 133 solid tumors of different histological types, including 39 NB tumors, and examined its potential clinical utility as an early response marker in the bone marrows before and after treatment of 118 stage 4 patients enrolled after induction chemotherapy in an immunotherapy protocol. Based on 40 normal marrow and peripheral blood samples, a CCND1 transcript value greater than the mean + 2 SD was defined as positive. Sensitivity of this assay was one NB cell in 106 normal mononuclear cells. CCND1 transcript levels were high in NB, breast cancer, and Ewing family tumors. Among the NB patients evaluated, early (2.5 months from protocol entry) marrow response was strongly associated with both progression-free (P = 0.0001) and overall survival (P = 0.0006). CCND1 response remained predictive of survival among a subset of 66 patients who had no histological evidence of marrow disease before immunotherapy. We conclude that CCND1 has potential clinical utility as a novel molecular marker of MRD in the bone marrow of patients with metastatic NB.
Sophisticated use of chemotherapy, surgery, and/or radiation therapy can reduce cancers to near complete remission. However, cancer cure remains elusive, the major hurdle being minimal residual disease (MRD), which is typically below the detection limit of conventional clinicopathological tools. Because the current eligibility criteria of most clinical trials require evidence of gross disease, a tumor will not be treated until it is measurable and symptomatic. This killing paradigm may be undesirable for several reasons.1 First, the Goldie-Coldman hypothesis predicts that bigger tumors have higher likelihoods of mutations and resistance.2 Second, visible tumors acquire additional barriers to drug delivery (eg, suboptimal tumor pressure, vasculature, and oxygenation).3,4 Third, a patient with large or extensive disease is in general physically and/or mentally compromised and is less likely to tolerate treatment side effects. An alternative strategy of regulatory control has been proposed.1 This treatment paradigm is particularly relevant to today’s cancer therapeutics. Although novel agents such as angiogenesis inhibitors, growth modulators, or vaccines may not achieve rapid tumor shrinkage, they may nevertheless be effective in controlling MRD, such that patients can live with cancer.
Targeting subclinical disease is especially relevant to metastatic neuroblastoma (NB), a pediatric cancer that poses enormous clinical challenges because of its bulky primaries and widespread metastases. Although most patients with metastatic disease can now achieve near complete remission, they typically relapse because of MRD, and cure after clinical relapse is rare. Adjuvant therapies such as stem cell transplantation and immunotherapy are used, but the ability to measure MRD accurately is crucial to evaluate their anti-tumor effect, to identify the optimal timing for stem cell collection, and to provide early indications of treatment failure. Moreover, there are many potential agents and strategies that can be effective in the MRD setting but are unable to be tested because of the absence of sensitive markers. These include monoclonal antibodies, antiangiogenesis agents, tyrosine kinase inhibitors, demethylators, and their novel combinations. Testing these treatment strategies with the aid of MRD markers will likely hold the key to eradicating metastatic NB.
The repertoire of known markers for NB is very small, with tyrosine hydroxylase being one of most widely used molecular targets. Tyrosine hydroxylase is the first and the rate-limiting enzyme in the biosynthesis of catecholamine, which is secreted by most NBs.5,6,7,8,9 Because NB is inherently heterogeneous, additional markers are necessary to enhance both the sensitivity and specificity of MRD detection.10 In this report, cyclin D1 (CCND1) was evaluated as a novel MRD marker of NB because of its known pivotal role in controlling cyclin-dependent kinases during cell cycle progression11 and its overexpression and adverse prognostic impact in human cancers,11,12,13,14 including NB,12 rhabdomyosarcoma and Ewing’s sarcoma,13 high-grade extremity soft-tissue sarcomas,15 breast cancers,11,14 endometrial adenocarcinoma,16 some lung cancers,17 ovarian cancer,18,19 and squamous cell carcinoma of the oropharynx.20 The molecular-based technique used was quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR), which was shown to be highly specific and sensitive to detect and measure gene transcripts in bone marrow (BM) and blood.21,22,23,24
Materials and Methods
Solid Tumors of Different Histological Types
Tumor samples obtained at diagnosis and relapse at Memorial Sloan-Kettering Cancer Center were snap-frozen in liquid nitrogen. In addition to 39 NBs (four stage 1, five stage 2, five stage 3, five stage 4, and 20 stage 4), there were 12 brain tumors, 22 breast cancers, seven desmoplastic small round cell tumors, 12 Ewing family tumors, five osteogenic sarcomas, 12 prostate cancers, seven rhabdomyosarcomas, five Wilm’s tumor, and 12 other soft tissue sarcomas. Written informed consent was obtained from the patients and/or their guardians in accordance to the guidelines of the institutional review board of Memorial Sloan-Kettering Cancer Center.
BM Samples before and after Treatment
BM (pooled marrow aspirates from four iliac crest sites) from all stage 4 patients with follow up samples (n = 118) treated on protocol IRB 9418 at Memorial Sloan-Kettering Cancer Center25 was analyzed for CCND1 expression. This immunotherapy protocol used anti-GD2 monoclonal antibody 3F8 plus GM-CSF in patients who had already completed their induction chemotherapy. In brief, treatment cycles were repeated approximately every 1 to 2 months until 24 months from the 1st day of protocol enrollment. Treatment continued until patients either developed progressive disease or had a total of four cycles plus elevated human anti-mouse antibody titer.26 Progressive disease was defined in accordance with International Neuroblastoma Staging Sytem response criteria.27 At the time of protocol entry, these patients had the following disease status: 52 complete remission/very good partial remission (CR/VGPR) (with normal marrow histology, computed tomography/magnetic resonance imaging (CT/MRI), metaiodobenzylguanidine (MIBG) scan, and urinary catecholamine metabolites), 44 primary refractory (with histologic or radiographic evidence of disease after induction therapy), 13 secondary refractory (with evaluable disease after salvage therapy), and nine progressive disease. BMs after treatment were sampled after the patients had completed two treatment cycles at a median of 2.5 months from protocol entry. Patients (109 of 118) were diagnosed at >18 months of age, generally regarded as the highest risk age group. The median age at diagnosis was 56 months. Twenty-seven patients had amplified MYCN with >10 copies per diploid human genome.
Real-Time qRT-PCR
Real-time qRT-PCR was performed on cryopreserved BM as previously described.22,28 CCND1 and endogenous reference glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were quantified from their respective standard curves using serially diluted cDNA from NB cell line NMB7. Dividing the CCND1 level by the GAPDH level resulted in a normalized CCND1 transcript value. The primers and probe for CCND1 were based on the GenBank sequence (NM_053056) and designed using Primer Express (Applied Biosystems, Foster City, CA). CCND1 sense primer was 5′-CCGAGAAGCTGTGCATCTACAC-3′ and anti-sense primer was 5′-AGGTTCCACTTGAGCTTGTTCAC-3′. CCND1 probe was FAM-5′-AGGAGCAGCTCCATTTGCAGCAGCTC-3′-TAMRA. The amplicon was 94 bp. Predeveloped TaqMan assay reagent for human endogenous control glyceraldehyde-3-phosphate dehydrogenase GAPDH (VIC, NM_002046) was also purchased from Applied Biosystems. Samples were assayed at least twice on separate days; overall concordance was >90%, and arithmetic means were used for outcome analyses. They were performed blind with respect to patient outcome.
Statistical Analysis
BM was classified as positive if CCND1 transcript level was greater than the upper limits of normal, as defined as mean + 2 SD of 40 normal marrow and blood samples (7.1 U). Molecular response was defined as follows: patients with BMs that were marker-positive in their pretreatment sample and negative after treatment were scored as CR, marker-positive before and after treatment as refractory, negative pretreatment turning positive after treatment as progressing, and negative marker before and after treatment as uninformative. Proportional hazards Cox models were used to determine associations between CCND1 and progression-free and overall survival, measured from the beginning of immunotherapy. Molecular response was analyzed as a time-dependent covariate.
Results
CCND1 Expression in NB Tumors and Other Human Solid Tumors
Sensitivity of CCND1 mRNA by qRT-PCR was established by spiking NMB7 cells at ratios ranging from 1 to 10,000 tumor cells per 106 normal mononuclear cells. The level of CCND1 transcript for a tumor content of 1/106 was 9.7 U. Among a panel of 133 human solid tumors tested, CCND1 expression was high in 39 NB tumors of all clinical stages (median, 2157 U), in 22 breast cancers (median, 2218 U), and in 12 Ewing family tumors (median, 1987 U) (Table 1).
Table 1.
CCND1 Expression (in Transcript Units) in 133 Human Solid Tumors
| Tumor type | Sample size | Median | 25th Centile | 75th Centile |
|---|---|---|---|---|
| Neuroblastoma | 39 | 2157 | 1053 | 3547 |
| EFT | 12 | 1987 | 1125 | 3695 |
| Breast cancer | 22 | 2218 | 1031 | 3453 |
| DSRCT | 7 | 856 | 747 | 1141 |
| Prostate cancer | 12 | 719 | 509 | 962 |
| Soft tissue sarcoma | 12 | 371 | 247 | 738 |
| Brain tumor | 12 | 137 | 44 | 244 |
| RMS | 7 | 106 | 39 | 514 |
| Wilms’ tumor | 5 | 68 | 17 | 99 |
| OS | 5 | 8 | 7 | 76 |
CCND1 Transcript Levels of BM before and after Treatment of 118 Stage 4 Patients Treated with Immunotherapy Protocol 9418
This immunotherapy protocol used anti-GD2 monoclonal antibody 3F8 plus GM-CSF in patients who had already completed their induction chemotherapy. Marrows after treatment for this analysis were sampled after the completion of two treatment cycles at a median of 2.5 months from protocol entry. This represented generally the first follow-up for these patients so that early molecular response could be evaluated. Forty-eight percent (57 of 118) of pretreatment BMs were CCND1-positive with median transcript level of 15.7 U and an interquartile range of 10.6 to 32.1. Fifty percent of BMs after treatment were positive (median was 18.8, interquartile range of 10.5 to 60.8). Of these 118 stage 4 patients, 66 patients had pretreatment BMs that were histology-negative; ie, their marrows had no detectable NB by biopsy (two sites), by aspirates (four sites), or both. Among these patients with marrow MRD, 53% (35 of 66) were CCND1-positive.
Correlation between CCND1 Molecular Response and Patient Survival
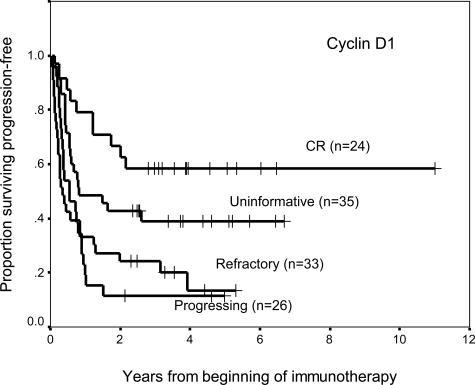
There were 81 progression events and 65 deaths in this cohort of 118 stage 4 NB patients. Median follow-up for survivors was 45 months and 47 months for progression-free survival and overall survival, respectively. According to molecular response, there were 24 CR, 33 refractory, 26 progressing, and 35 uninformative (progression-free survival, Figure 1). CCND1 response was a statistically significant predictor of progression-free survival and overall survival (Table 2). A molecular response status of refractory or progressing influenced patient outcome adversely. There was also evidence that patients who were CCND1-negative before and after treatment (ie, the uninformative group) were likely to have NB that was below detection.
Figure 1.
Kaplan-Meier plot of progression-free survival with respect to cyclin D1 molecular response (P = 0.0001) among 118 patients with stage 4 NB treated with an immunotherapy protocol using anti-GD2 antibody 3F8 plus GM-CSF. Marrow response was defined in the Materials and Methods section. Follow-up marrows were evaluated at 2.5 months from protocol entry.
Table 2.
Association Between CCND1 Molecular Response and Patient Survival Among 118 Stage 4 Neuroblastoma Patients
| Molecular response | Hazard ratio | 95% CI* | P |
|---|---|---|---|
| Progression-free survival | |||
| CR | Reference* | ||
| Progressing | 4.35 | 1.95, 9.71 | <0.0005 |
| Refractory | 2.76 | 1.27, 5.97 | 0.01 |
| Uninformative | 1.67 | 0.77, 3.62 | 0.2 |
| Overall survival | |||
| CR | Reference* | ||
| Progressing | 5.44 | 2.18, 13.56 | <0.0005 |
| Refractory | 3.78 | 1.52, 9.42 | 0.004 |
| Uninformative | 2.30 | 0.90, 5.84 | 0.08 |
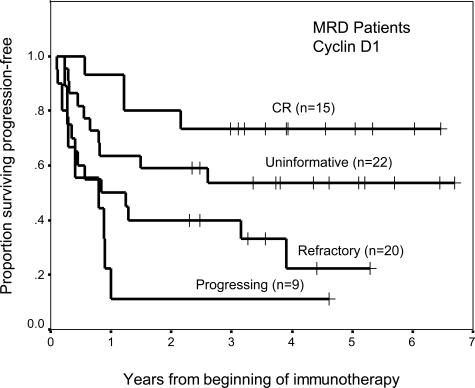
Of particular interest was the molecular response of stage 4 patients with no histological evidence of marrow disease before treatment, ie, patients with marrow MRD (n = 66). Response assessed by CCND1 (P = 0.002) was highly predictive of progression-free survival (Figure 2). Patients who were in molecular remission had greater likelihood of remaining relapse-free. In contrast, the marrows of most patients (61 of 66) after treatment continued to have negative histology, even though 15 of them had CCND1 response of CR, seven were refractory, and 17 were progressing. This finding reflected the relative insensitivity of using histology to assess MRD.
Figure 2.
Prognostic importance of molecular response using cyclin D1 (P = 0.002) among 66 stage 4 NB patients with histologically negative marrows before treatment.
Discussion
Targeting MRD in metastatic NB is critical for cure. At present, there is a paucity of established molecular markers to detect subclinical disease. In addition to tyrosine hydroxylase, GD2 synthase (β1,4-_N_-acetylgalactosaminyltransferase, GalNacT), the key enzyme for biosynthesis of GD2, the antigen highly and homogeneously expressed in NB, has been demonstrated to be a clinically relevant MRD marker.22,23,24 However, because of inherent tumor heterogeneity, multiple markers are needed for the detection of occult NB cells. The advent of real-time quantitative RT-PCR further facilitates the highly sensitive and specific measurement of gene transcripts. Nevertheless, a useful MRD marker must also demonstrate clinical relevance in a well-defined cohort of patients accrued on a clinical trial. The detection of a marker should be associated with poor clinical outcome. The cohort of patient samples reported here was chosen because 1) they were all enrolled on a single phase II protocol using monoclonal antibody to treat MRD after chemotherapy; 2) accrual was completed in 2003, and patients had at least 24 months of follow-up; 3) all patients underwent comprehensive disease workups before, and periodically during, treatment and at follow-up; and 4) BM samples were obtained before and after two cycles of antibody treatment for MRD measurements. Thus, we were able to evaluate CCND1 as a novel early response marker of immunotherapy29 and found it to be highly correlated with patient outcome.
At the time of the follow-up marrow studies for this report, most of the patients (82%) had not shown any signs of progressive disease, even though the disease status at protocol entry (CR/VGPR, primary refractory, secondary refractory, and progressive disease) may be expected to predict outcome of the follow-up marrow. In contrast, CCND1 level identified the different molecular response groups. Among the patients who finally progressed at a median of 45 months from the beginning of immunotherapy, disease progression was evident by either BM histology, MIBG scan, or by CT/MRI. CCND1 molecular response evaluated much earlier in time (ie, 2.5 months after treatment) highly correlated with clinical progression. Moreover, CCND1 had potentials as an MRD marker among stage 4 patients who had histologically negative marrows before protocol entry. Thus, subclinical marrow disease detectable by CCND1 but not by histological examination can have prognostic importance.
Up-regulation of CCND1 has been implicated in many human cancers. Our laboratory identified this gene by means of a marker discovery strategy using genome-wide expression arrays with Affymetrix (Santa Clara, CA) U95 chips A to E to identify genes differentially expressed in tumors from 48 stage 4 patients over nine remission BM, the site where MRD marker is important. Son and colleagues30 also identified CCND1 as a legitimate target for NB based on gene expression profiles of 158 normal human samples from 19 different organs. Besides NB, our results found high CCND1 expression in breast cancer and Ewing family tumors, suggesting this gene may also have potential clinical utility for these cancers.
Acknowledgments
We thank Drs. B. Kushner, K. Kramer, S. Modak, and the nursing staff for their clinical management; and K. Danis for her data management.
Footnotes
Supported in part by the National Institutes of Health (grants CA106450 and CA118845), Robert Steel Foundation, Hope Street Kids, and Pediatric Cancer Foundation.
References
- Schipper H, Goh CR, Wang TL. Shifting the cancer paradigm: must we kill to cure? J Clin Oncol. 1995;13:801–807. doi: 10.1200/JCO.1995.13.4.801. [DOI] [PubMed] [Google Scholar]
- Goldie JH, Coldman AJ. The somatic mutation theory of drug resistance: the “Goldie-Coldman” hypothesis revisited. Devita HR, editor. Philadelphia: JP Lippincott; Cancer. Principles and Practice of Oncology. 1989:1–12. [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Jain RK. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- Naito H, Kuzumaki N, Uchino J, Kobayashi R, Shikano T, Ishikawa Y, Matsumoto S. Detection of tyrosine hydroxylase mRNA and minimal neuroblastoma cells by the reverse transcription-polymerase chain reaction. Eur J Cancer. 1991;27:762–765. doi: 10.1016/0277-5379(91)90184-f. [DOI] [PubMed] [Google Scholar]
- Miyajima Y, Kato K, Numata SI, Kudo K, Horibe K. Detection of neuroblastoma cells in bone marrow and peripheral blood at diagnosis by the reverse transcriptase-polymerase chain reaction for tyrosine hydroxylase mRNA. Cancer. 1995;75:2757–2761. doi: 10.1002/1097-0142(19950601)75:11<2757::aid-cncr2820751120>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Burchill SA, Lewis IJ, Abrams KR, Riley R, Imeson J, Pearson ADJ, Pinkerton R, Selby P. Circulating neuroblastoma cells detected by reverse transcriptase polymerase chain reaction for tyrosine hydroxylase mRNA are an independent poor prognostic indicator in stage 4 neuroblastoma in children over 1 year. J Clin Oncol. 2001;19:1795–1801. doi: 10.1200/JCO.2001.19.6.1795. [DOI] [PubMed] [Google Scholar]
- Träger C, Kogner P, Lindskog M, Ponthan F, Kullman A, Kagedal B. Quantitative analysis of tyrosine hydroxylase mRNA for sensitive detection of neuroblastoma cells in blood and bone marrow. Clin Chem. 2003;49:104–112. doi: 10.1373/49.1.104. [DOI] [PubMed] [Google Scholar]
- Tchirkov A, Paillard C, Halle P, Bernard F, Bordigoni P, Vago P, Demeocq F, Kanold J. Significance of molecular quantification of minimal residual disease in metastatic neuroblastoma. J Hematother Stem Cell Res. 2003;12:435–442. doi: 10.1089/152581603322286060. [DOI] [PubMed] [Google Scholar]
- Cheung IY, Barber D, Cheung NK. Detection of microscopic neuroblastoma in marrow by histology, immunocytology, and reverse transcription-PCR of multiple molecular markers. Clin Cancer Res. 1998;4:2801–2805. [PubMed] [Google Scholar]
- Donnellan R, Chetty R. Cyclin D1 and human neoplasia. Mol Pathol. 1998;51:1–7. doi: 10.1136/mp.51.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar JJ, van Sluis P, Boon K, Versteeg R, Caron HN. Rearrangements and increased expression of cyclin D1 (CCND1) in neuroblastoma. Genes Chromosom Cancer. 2003;36:242–249. doi: 10.1002/gcc.10166. [DOI] [PubMed] [Google Scholar]
- Baer C, Nees M, Breit S, Selle B, Kulozik AE, Schaefer KL, Braun Y, Wai D, Poremba C. Profiling and functional annotation of mRNA gene expression in pediatric rhabdomyosarcoma and Ewing’s sarcoma. Int J Cancer. 2004;110:687–694. doi: 10.1002/ijc.20171. [DOI] [PubMed] [Google Scholar]
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- Kim SH, Lewis JJ, Brennan MF, Woodruff JM, Dudas M, Cordon-Cardo C. Overexpression of cyclin D1 is associated with poor prognosis in extremity soft-tissue sarcomas. Clin Cancer Res. 1998;4:2377–2382. [PubMed] [Google Scholar]
- Nishimura Y, Watanabe J, Jobo T, Kato N, Fujisawa T, Kamata Y, Kuramoto H. Cyclin D1 expression in endometrioid-type endometrial adenocarcinoma is correlated with histological grade and proliferative activity, but not with prognosis. Anticancer Res. 2004;24:2185–2191. [PubMed] [Google Scholar]
- Oshita F, Ito H, Ikehara M, Ohgane N, Hamanaka N, Nakayama H, Saito H, Yamada K, Noda K, Mitsuda A, Kameda Y. Prognostic impact of survivin, cyclin D1, integrin beta1, and VEGF in patients with small adenocarcinoma of stage I lung cancer. Am J Clin Oncol. 2004;27:425–428. doi: 10.1097/01.coc.0000128864.15609.5b. [DOI] [PubMed] [Google Scholar]
- Barbieri F, Lorenzi P, Ragni N, Schettini G, Bruzzo C, Pedulla F, Alama A. Overexpression of cyclin D1 is associated with poor survival in epithelial ovarian cancer. Oncology. 2004;66:310–315. doi: 10.1159/000078332. [DOI] [PubMed] [Google Scholar]
- Bali A, O’Brien PM, Edwards LS, Sutherland RL, Hacker NF, Henshall SM. Cyclin D1, p53, and p21Waf1/Cip1 expression is predictive of poor clinical outcome in serous epithelial ovarian cancer. Clin Cancer Res. 2004;10:5168–5177. doi: 10.1158/1078-0432.CCR-03-0751. [DOI] [PubMed] [Google Scholar]
- Yu Z, Weinberger PM, Haffty BG, Sasaki C, Zerillo C, Joe J, Kowalski D, Dziura J, Camp RL, Rimm DL, Psyrri A. Cyclin D1 is a valuable prognostic marker in oropharyngeal squamous cell carcinoma. Clin Cancer Res. 2005;11:1160–1166. [PubMed] [Google Scholar]
- Sidransky D. Nucleic acid-based methods for the detection of cancer. Science. 1997;278:1054–1059. doi: 10.1126/science.278.5340.1054. [DOI] [PubMed] [Google Scholar]
- Cheung IY, Cheung NK. Quantitation of marrow disease in neuroblastoma by real-time reverse transcription-PCR. Clin Cancer Res. 2001;7:1698–1705. [PubMed] [Google Scholar]
- Cheung IY, Lo Piccolo MS, Kushner BH, Kramer K, Cheung NK. Quantitation of GD2 synthase mRNA by real-time reverse transcriptase polymerase chain reaction: clinical utility in evaluating adjuvant therapy in neuroblastoma. J Clin Oncol. 2003;21:1087–1093. doi: 10.1200/JCO.2003.02.055. [DOI] [PubMed] [Google Scholar]
- Cheung IY, Sahota A, Cheung NK. Measuring circulating neuroblastoma cells by quantitative reverse transcriptase-polymerase chain reaction analysis. Cancer. 2004;101:2303–2308. doi: 10.1002/cncr.20660. [DOI] [PubMed] [Google Scholar]
- Kushner BH, Kramer K, Cheung NKV. Phase II trial of the anti-G(D2) monoclonal antibody 3F8 and granulocyte-macrophage colony-stimulating factor for neuroblastoma. J Clin Oncol. 2001;19:4189–4194. doi: 10.1200/JCO.2001.19.22.4189. [DOI] [PubMed] [Google Scholar]
- Cheung NK, Cheung IY, Canete A, Yeh SJ, Kushner B, Bonilla MA, Heller G, Larson SM. Antibody response to murine anti-GD2 monoclonal antibodies: correlation with patient survival. Cancer Res. 1994;54:2228–2233. [PubMed] [Google Scholar]
- Brodeur G, Pritchard J, Berthold F, Carlen NLT, Castel V, Castleberry RP, DeBernardi B, Evans AE, Favrot M, Hedborg F, Kaneko M, Kemshead J, Lampert F, Lee REJ, Look T, Pearson ADJ, Philip T, Roald B, Sawada T, Seeger RC, Tsuchida Y, Voute PA. Revisions of the international criteria for neuroblastoma diagnosis, staging and response to treatment. J Clin Oncol. 1993;11:1466–1477. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- Cheung IY, Cheung NKV. Molecular detection of GAGE expression in peripheral blood and bone marrow: utility as a tumor marker for neuroblastoma. Clin Cancer Res. 1997;3:821–826. [PubMed] [Google Scholar]
- Cheung IY, Lo Piccolo MS, Kushner BH, Cheung NK. Early molecular response of marrow disease to biologic therapy is highly prognostic in neuroblastoma. J Clin Oncol. 2003;21:3853–3858. doi: 10.1200/JCO.2003.11.077. [DOI] [PubMed] [Google Scholar]
- Son CG, Bilke S, Davis S, Greer BT, Wei JS, Whiteford CC, Chen QR, Cenacchi N, Khan J. Database of mRNA gene expression profiles of multiple human organs. Genome Res. 2005;15:443–450. doi: 10.1101/gr.3124505. [DOI] [PMC free article] [PubMed] [Google Scholar]