Dynamin 2 is required for the enhancement of HIV-1 infectivity by Nef (original) (raw)
Abstract
Nef is a virulence factor of HIV-1 and other primate lentiviruses that is crucial for rapid progression to AIDS. In cell culture, Nef increases the infectivity of HIV-1 progeny virions by an unknown mechanism. We now show that dynamin 2 (Dyn2), a key regulator of vesicular trafficking, is a binding partner of Nef that is required for its ability to increase viral infectivity. Dominant-negative Dyn2 or the depletion of Dyn2 by small interfering RNA potently inhibited the effect of Nef on HIV-1 infectivity. Furthermore, in Dyn2-depleted cells, this function of Nef could be rescued by ectopically expressed Dyn2 but not by Dyn1, a closely related isoform that does not bind Nef. The infectivity enhancement by Nef also depended on clathrin, because it was diminished in clathrin-depleted cells and profoundly inhibited in cells expressing the clathrin-binding domain of AP180, which blocks clathrin-coated pit formation but not clathrin-independent endocytosis. Together, these findings imply that the infectivity enhancement activity of Nef depends on Dyn2- and clathrin-mediated membrane invagination events.
Keywords: HIV accessory protein, host factor, virion infectivity
Nef is an accessory protein of HIV-1 and other primate lentiviruses that constitutes a crucial determinant of viral pathogenicity (1). Nef induces the clathrin-mediated endocytosis and degradation of cell surface CD4 (2–5), and also triggers the down-regulation of other cell surface molecules, including MHC class I antigens (6), which may facilitate the evasion of an antiviral immune response (7). Nef also modulates T cell function and apoptosis by engaging components of cellular signaling pathways, including Vav, Pak2, ASK1, and the DOCK2–ELMO1 complex (8–12).
Although not essential for virus spreading in cultured cells, Nef significantly enhances virus replication in primary cells, particularly if these are exposed to HIV-1 before activation with mitogens (13, 14). Nef also enhances the intrinsic infectivity of progeny virions (15–17). This effect is independent of CD4 down-regulation and requires the presence of Nef in virus-producing cells (16–18). Because Nef does not affect virus assembly or maturation, the precise nature of its effect on viral infectivity has remained elusive. Nef is N-terminally myristylated and associates with cellular membranes, and small quantities of membrane-bound Nef are taken up into assembling virions (19, 20). It has also been reported that Nef delivers cholesterol to assembling virions, thereby increasing viral infectivity (21). However, a recent study suggests that the uptake of Nef into virions is not required for its ability to enhance viral infectivity (22).
The effect of Nef on viral infectivity may depend on the route of entry, because Nef does not enhance the infectivity of HIV cores pseudotyped with vesicular stomatitis virus glycoprotein (VSV-G), which mediates virus entry through an endocytic compartment rather than directly via the plasma membrane (23). In the absence of Nef, the synthesis of proviral DNA in newly infected target cells is impaired, indicating that Nef affects an early step of the replication cycle (17). Consistent with this notion, Nef has been shown to increase the cytoplasmic delivery of viral cores, possibly by facilitating the movement of the core past the cortical actin network (24).
Because the infectivity of _nef_-negative HIV-1 can be rescued by Nef expressed in trans in the producer but not in the target cells (17), we searched for cellular effectors that mediate this effect of Nef in producer cells. This led to the identification of Dyn2 as a novel binding partner of Nef that is crucial for its infectivity enhancement function.
Results
Nef Specifically Interacts with Dyn2.
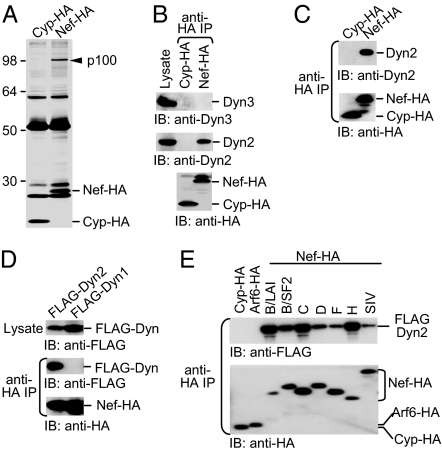
The infectivity enhancement activity of Nef can be readily demonstrated in CD4-negative cells, for instance, in 293T cells (16, 17, 25). To identify effectors that mediate this activity, a HA-tagged version of HIV-1LAI Nef or of an irrelevant control protein was transiently expressed in 293T cells, and the cells were lysed in a buffer containing the mild detergent octyl glucoside. A SDS/PAGE analysis of anti-HA immunoprecipitates revealed a prominent protein band of ≈100 kDa that coprecipitated with Nef-HA but not with the cyclophilin A (Cyp)-HA control (Fig. 1A). The 100-kDa protein could be unambiguously identified by liquid chromatography-coupled tandem mass spectrometry, with 21 tryptic peptides matching the sequence of human Dyn2, a key player in the endocytic fission reaction (26).
Fig. 1.
Nef binds Dyn2 but not other Dyn isoforms. (A) A 100-kDa protein coprecipitates with Nef. Anti-HA immunoprecipitates from 293T cells transiently expressing Cyp-HA or NefLAI-HA were analyzed by SDS/PAGE and silver staining. (B) Nef binds endogenous Dyn2 but not Dyn3. Anti-HA immunoprecipitates (IP) prepared from 293T cells transiently expressing Cyp-HA or NefLAI-HA were analyzed by immunoblotting (IB) as indicated. (C) HIV-1 Nef binds endogenous Dyn2 in a T cell line (Jurkat TAg). (D) Nef does not interact with Dyn1. Lysates and anti-HA immunoprecipitates from 293T cells coexpressing NefLAI-HA and FLAG-Dyn1 or FLAG-Dyn2 were immunoblotted as indicated. (E) Dyn2 binds to the Nef proteins of different HIV-1 subtypes (B, C, D, F, and H) and of SIVmac239. Cyp-HA and Arf6-HA are controls.
Dyn2 is a ubiquitously expressed member of a family of large GTPases that also includes Dyn1 and Dyn3, which are expressed in a limited number of tissues (26). Interestingly, although 293T cells express Dyn3 in addition to Dyn2, Western blotting of Nef-HA immunoprecipiates with isoform-specific antisera revealed that Nef interacts with endogenous Dyn2 but not with Dyn3 (Fig. 1B). Endogenous Dyn2 also specifically coprecipitated with Nef-HA expressed in the T lymphoid Jurkat cell line (Fig. 1C), demonstrating that Nef and Dyn2 can interact in different cell types. To determine whether Nef binds to the neuron-specific Dyn1, we coexpressed FLAG-tagged Dyn1 or Dyn2 along with Nef-HA in 293T cells and found that only FLAG-Dyn2 associated with Nef (Fig. 1D). We conclude that Nef discriminates between the three closely related Dyn isoforms.
We also examined whether the ability to interact with Dyn2 is a conserved property of Nef. As shown in Fig. 1E, FLAG-Dyn2 bound to the HA-tagged Nef proteins of various HIV-1 subtypes and of SIVmac239, which belongs to a separate lineage of primate lentiviruses. Arf6-HA was included as a control to rule out that FLAG-Dyn2 bound nonspecifically to myristylated proteins in our assay. We conclude that the ability to interact with Dyn2 is conserved among different Nef alleles.
Nef-HA immobilized on beads specifically interacted with purified FLAG-Dyn2 in an in vitro pull-down assay [see supporting information (SI) Fig. 7]. It is unlikely that the in vitro interaction was mediated by a cellular protein that contaminated immobilized Nef-HA, because the Cyp-HA control showed an essentially identical background pattern, yet only Nef-HA pulled down Dyn2-FLAG. We thus consider it likely that the interaction between Nef and Dyn2 is direct. This notion is further supported by the observation that Nef and Dyn2 interact in a yeast two-hybrid assay (S. Richter and G.P., unpublished work), which indicates that no mammalian cofactor is required for the interaction.
The Middle and GTPase Effector Domains (GEDs) of Dyn2 Are Critical for the Interaction with Nef.
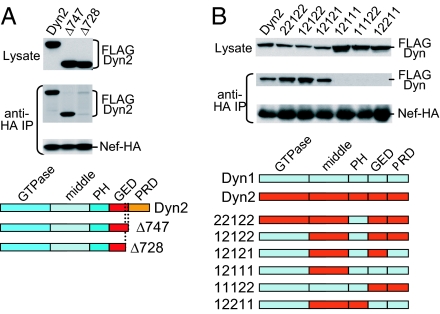
The interaction with Nef does not depend on the C-terminal proline-rich domain (PRD) of Dyn2 but requires an intact GED (Fig. 2A). To map the determinants that allow Nef to distinguish between different isoforms, we swapped entire domains between Dyn1 and Dyn2 and observed that the GTPase and pleckstrin homology domains of Dyn2 do not contribute to the specificity of the interaction (Fig. 2B). In contrast, the middle domain and the GED of Dyn2 were both necessary and together conferred full Nef binding in an otherwise Dyn1 background (Fig. 2B and data not shown). Interestingly, the middle domain and the GED also interact with each other and are both thought to be important for dynamin oligomerization (27).
Fig. 2.
Mapping of the Dyn2 domains required for the interaction with Nef. (A) The PRD of Dyn2 is dispensable for the interaction with Nef, whereas the GED is required. (B) Analysis of Dyn1/Dyn2 chimeras showing that the GTPase and pleckstrin homology domains do not contribute to the specificity of the interaction, whereas the middle and GED domains of Dyn2 together confer full Nef binding in an otherwise Dyn1 background.
Nef Mutants Defective for Dyn2 Binding Lack Infectivity Enhancement Activity.
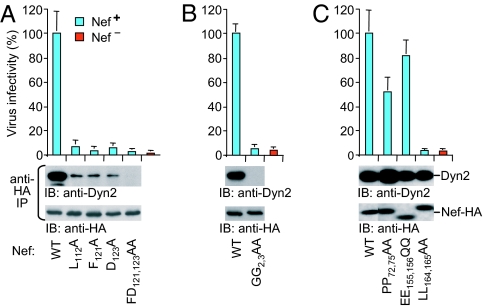
We analyzed a panel of Nef mutants and found that surface-exposed core domain residues (L112, F121, and D123) previously implicated in the interaction with a thioesterase (28, 29) are critical for binding to Dyn2 in vivo (Fig. 3A). Additionally, a nonmyristylated form of Nef (GG2,3AA) failed to bind Dyn2 in vivo (Fig. 3B). Moreover, the GG2,3AA mutant did not interact with purified FLAG-Dyn2 in vitro, confirming that the myristyl moiety of Nef is critical for the interaction with Dyn2 (SI Fig. 7). In contrast, mutations known to affect the binding of Nef to Src kinase SH3 domains (30) (PP72,75AA), to β-Cop (31) (EE155,156QQ), or to clathrin adaptor proteins (32, 33) (LL164,165AA) did not reduce binding to Dyn2 (Fig. 3C).
Fig. 3.
Ability of Nef mutants to interact with endogenous Dyn2 in human cells and to enhance HIV-1 infectivity in a proviral context. (A) Residues crucial for Dyn2 binding map to a surface patch on the core domain of Nef. (B) The myristyl attachment site of Nef is essential for the interaction with Dyn2. (C) Nef residues required for binding to SH3 domains, β-COP, or clathrin adaptor proteins are dispensable for the interaction with Dyn2. The error bars represent standard deviations from the mean calculated from triplicate determinations.
To examine the effects of these mutations on viral infectivity, each was introduced into the nef gene of the full-length provirus HXB/nef+ without changing any other viral sequences. In the proviral context, all mutations that impaired binding to Dyn2 markedly reduced the ability of Nef to enhance viral infectivity (Fig. 3 A and B). Among the Nef mutants that bound Dyn2 with wild-type efficiency, only the LL164,165AA mutant exhibited a severe defect in infectivity enhancement (Fig. 3C). The PP72,75AA mutation significantly impaired infectivity enhancement by Nef in one study (25), but had only a 2-fold effect in another study (34), in agreement with our results (Fig. 3C). The crucial role of the dileucine motif (LL164,165), which was described earlier (32), implies that the interaction with Dyn2 is not sufficient for the enhancement of infectivity by Nef. However, we note that the dileucine motif has been shown to connect Nef to the clathrin-mediated endocytic pathway (33, 35), which depends on dynamin.
Dominant-Negative Dyn2 Specifically Inhibits the Effect of Nef on HIV-1 Infectivity.
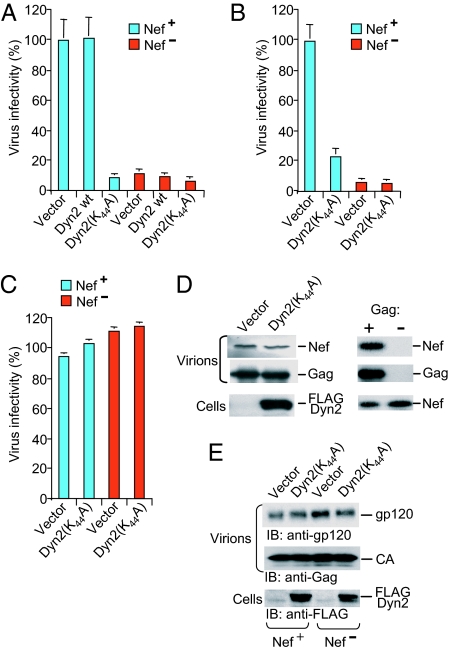
The K44A mutants of Dyn1 and Dyn2 inhibit clathrin-mediated endocytosis (36). Dyn1(K44A) also inhibits the Nef-induced down-regulation of CD4 but not of MHC class I molecules (37, 38). To examine whether Dyn2 plays a role in the infectivity enhancement function of Nef, we produced HIV-1 virions capable of only a single round of replication in transiently transfected Jurkat TAg cells coexpressing exogenous wild-type or dominant-negative Dyn2. Although exogenous Dyn2 had no significant effect, Dyn2(K44A) markedly reduced the infectivity of the _nef_-positive NL4-3 virus to a level close to that obtained in the absence of nef (Fig. 4A). However, Dyn2(K44A) consistently had little or no effect on the residual infectivity of NL4-3 virus lacking nef (Fig. 4A). Similar results were obtained when we used replication-competent rather than _env_-deficient HIV-1 proviruses that depended on Env expressed in trans, confirming that the coexpression of Dyn2(K44A) during virus production inhibited the transmission of _nef_-positive but not of _nef_-negative HIV-1 to target cells (SI Fig. 8).
Fig. 4.
Dominant-negative Dyn2 inhibits the effect of Nef on viral infectivity. (A and B) Dyn2(K44A) selectively decreases the single-round infectivity of _nef_-positive HIV-1 produced by Jurkat TAg lymphoid cells (A) or CD4-negative COS-7 cells (B), as assayed on P4–2 indicator cells. (C) Dyn2(K44A) does not affect the infectivity of VSV-G-pseudotyped HIV-1 particles. Infectivities are the means plus standard deviation from triplicate determinations. (D and E) Dyn2(K44A) does not alter the incorporation of Nef (D) or of the Env glycoprotein gp120 (E) into HIV-1 virions. D demonstrates that the appearance of Nef depended on Gag, confirming its association with viral particles.
Although the Jurkat TAg cells we used for virus production have negligible levels of surface CD4 (data not shown), we also examined the effect of Dyn2(K44A) on the single round infectivity of virus produced in COS-7 cells, which lack CD4. This experiment demonstrated that Dyn2(K44A) can specifically inhibit the effect of Nef on virus infectivity in the absence of CD4 (Fig. 4B).
It has been shown that HIV-1 particles pseudotyped with VSV-G no longer require Nef for optimal infectivity (23). We therefore tested the effect of Dyn2(K44A) on chimeric viral particles composed of the HIV-1 core and VSV-G. As reported previously (23), the infectivity of VSV-G-pseudotyped HIV-1 particles was not enhanced by Nef (Fig. 4C). Importantly, Dyn2(K44A) had no effect on the infectivity of VSV-G-pseudotyped HIV-1 particles, independent of the presence or absence of Nef (Fig. 4C). These results confirm that Dyn2(K44A) counteracts the effect of Nef and does not inhibit viral infectivity per se.
Because a small amount of Nef is incorporated into HIV-1 particles (20, 39), we examined whether Dyn2(K44A) affects the virion-association of Nef. We did not detect any significant difference in the amount of Nef present in virions produced in cells expressing Dyn2(K44A) or a control vector (Fig. 4D). Similarly, Dyn2(K44A) did not affect the levels of virion-associated Env (Fig. 4E). Furthermore, Dyn2(K44A) did not appear to alter the extent of colocalization of Gag and Env (SI Fig. 9_A_) or the subcellular localization of Nef-GFP (SI Fig. 9_B_), which was predominantly perinuclear as previously reported (40). These data indicate that Dyn2(K44A) inhibits the ability of Nef to increase HIV-1 infectivity without affecting the localization of Nef or Env, or their incorporation into virions.
Dyn2 Is Required for the Infectivity Enhancement Function of Nef.
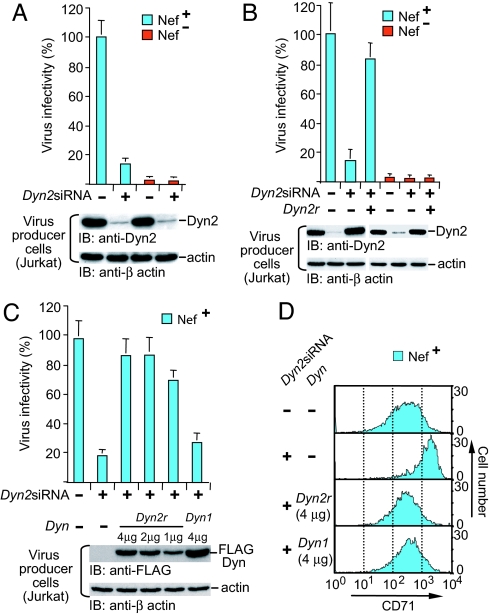
To directly determine whether Dyn2 is necessary for the effect of Nef on HIV-1 infectivity, Jurkat TAg cells were transfected with siRNA directed against Dyn2 (_Dyn2_siRNA) or a control siRNA targeting firefly luciferase, along with _env_-defective HIV-1 proviruses and expression vectors for HIV-1 Env and GFP. Immunoblotting of cell lysates from GFP-expressing cells 96 h after transfection indicated a more than 90% reduction of Dyn2 protein levels by _Dyn2_siRNA (Fig. 5A). _Dyn2_siRNA also markedly reduced the single cycle infectivity of _nef_-positive progeny virus, but did not further diminish the already low infectivity of HIV-1 lacking nef (Fig. 5A).
Fig. 5.
Dyn2 is essential for the infectivity enhancement function of Nef. (A) The siRNA-mediated depletion of Dyn2 in virus-producing Jurkat TAg lymphoid cells decreases the infectivity of _nef_-positive but not _nef_-negative HIV-1NL4-3 progeny virus. (B) Exogenous siRNA-resistant Dyn2 (Dyn2r) rescues the infectivity of _nef_-positive HIV-1NL4-3 produced by siRNA-treated Jurkat TAg cells. (C) Comparison of the ability of FLAG-Dyn2r and FLAG-Dyn1 to rescue the infectivity of _nef_-positive HIV-1NL4-3 produced by siRNA-treated Jurkat TAg cells. Infectivities are the means plus standard deviation from triplicate determinations. (D) Effect of _Dyn2_-siRNA on transferrin receptor (CD71) surface expression levels. The siRNA-mediated depletion of Dyn2 in the virus-producing Jurkat TAg cells increased the surface expression of CD71 as expected, and exogenous siRNA-resistant Dyn2 (Dyn2r) and Dyn1 both restored the original CD71 surface expression levels.
Dyn2 protein levels in cells transfected with _Dyn2_siRNA could be restored by a vector encoding wild-type Dyn2 with four silent mutations at the siRNA target site (denoted Dyn2r), indicating that the silent mutations conferred resistance to RNA interference as expected (Fig. 5B). The reintroduction of Dyn2r also restored the infectivity of _nef_-positive progeny virus to near-normal levels, but had no effect on the infectivity of _nef_-negative NL4-3 virus (Fig. 5B). These results formally prove that the inhibitory effect of _Dyn2_siRNA on the infectivity enhancement function of Nef was caused by the depletion of Dyn2.
Because Nef interacts with Dyn2 but not Dyn1, we examined whether exogenous Dyn1 restores the effect of Nef on viral infectivity in virus producer cells depleted of endogenous Dyn2. To facilitate a comparison of the effects of Dyn1 and Dyn2r, FLAG-tagged versions of both proteins were used in this experiment. As shown in Fig. 5C, the infectivity of _nef_-positive HIV-1 produced by _Dyn2_siRNA-transfected cells could be rescued by FLAG-tagged Dyn2r, whereas in multiple experiments FLAG-Dyn1 had only a minimal effect, even at expression levels up to 5-fold higher than the levels at which FLAG-Dyn2r showed maximal rescue.
As a control for FLAG-Dyn1 activity, we examined the transferrin receptor (CD71) surface levels, which are reported to increase 2-fold if clathrin-mediated endocytosis is inhibited by Dyn1(K44A) (41). _Dyn2_siRNA induced a nearly 5-fold increase of CD71 surface levels, and FLAG-Dyn2r and FLAG-Dyn1 counteracted this effect with comparable potency (Fig. 5D), indicating that both were similarly effective in restoring clathrin-mediated endocytosis. Taken together, these results indicate that Nef requires Dyn2 and cannot use Dyn1 for its infectivity enhancement function.
The Infectivity Enhancement Activity of Nef Depends on Clathrin.
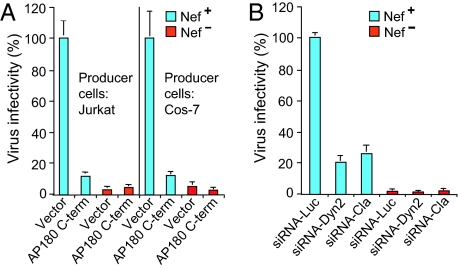
Because Dyn2 functions in endocytosis, we tested whether clathrin is involved in the infectivity enhancement by Nef. First, we examined the effect of AP180-C, a dominant-negative version of the clathrin assembly protein AP180. It has been previously shown that AP180-C inhibits clathrin-mediated but not clathrin-independent endocytosis by blocking clathrin-coated pit formation (42). Both in T lymphoid Jurkat cells and in CD4-negative COS-7 cells, the presence of AP180-C during virus production profoundly reduced the infectivity of _nef_-positive progeny virus but not of virus lacking nef (Fig. 6A), mimicking the effect of Dyn2(K44A). To directly determine whether clathrin is required for the effect of Nef on infectivity, we used a previously described siRNA that has been shown to effectively deplete the clathrin heavy chain (43). As shown in Fig. 6B, the siRNA targeting clathrin led to a comparable reduction in the infectivity of _nef_-positive progeny virions as the _Dyn2_siRNA, but did not further reduce the low infectivity of the _nef_-negative virus. Collectively, these results indicate that clathrin and its ability to form coated vesicles are required for the activity of Nef on infectivity.
Fig. 6.
Clathrin is required for the infectivity enhancement function of Nef. (A) The expression of AP180-C in virus-producing Jurkat TAg lymphoid cells or CD4-negative COS-7 cells selectively decreases the single round infectivity of _nef_-positive HIV-1. (B) The siRNA-mediated depletion of clathrin in virus-producing Jurkat TAg lymphoid cells decreases the infectivity of _nef_-positive but not of _nef_-negative HIV-1NL4-3 progeny virus. Infectivities are the means plus standard deviation from triplicate determinations.
Dyn2 Is Not Specifically Required for the Down-Regulation of CD4 or MHC Class I Molecules by Nef.
Dominant-negative Dyn1 does not affect the Nef-mediated down-modulation of MHC class I molecules (37, 38). In agreement with these reports, we observed that _Dyn2_siRNA did not significantly affect the activity of Nef on MHC class I molecules (SI Fig. 10_A_). As expected, Dyn2siRNA blocked the Nef-induced CD4 down-regulation (SI Fig. 10_B_), which occurs via clathrin-mediated endocytosis (35). However, this Nef activity could be rescued with comparable efficiency by FLAG-Dyn2r and FLAG-Dyn1 (SI Fig. 10_B_), demonstrating that Dyn2 is not specifically required.
Discussion
In this study, we have identified Dyn2 as a Nef-associated protein required for the enhancement of the infectivity of progeny virions by Nef. Dyn2 is a ubiquitously expressed GTPase that is essential for clathrin-mediated endocytosis, and is thought to function as a scission factor that mediates the separation of coated vesicles from the plasma membrane (26). Interestingly, Nef has been shown to trigger the de novo formation of clathrin-coated pits when tethered to the plasma membrane (44), and may thus act as a connector between cargo such as CD4 and the endocytic machinery.
The ability to interact with Dyn2 is conserved among different allelic forms of Nef, suggesting evolutionary pressure to maintain the binding site. The interaction depends on a well conserved surface-exposed hydrophobic patch, reported to form an interface between Nef molecules in crystals of the Nef core domain (45). However, myristylated Nef is predominantly monomeric in solution (46, 47), and the binding site for Dyn2 would thus be expected to be accessible on native Nef. Mutations that inhibited Dyn2 binding also impaired the infectivity enhancement function of Nef, as did the siRNA-mediated depletion of Dyn2. Nevertheless, we cannot formally exclude that the effects of the Nef mutations and of knocking down Dyn2 are unrelated, and that the involvement of Dyn2 in the function of Nef is indirect.
In addition to its essential roles in both clathrin-mediated and clathrin-independent endocytic events, Dyn2 has been implicated in the regulation of actin dynamics (48, 49), in signaling (50), and in centrosome cohesion (51). In particular, Dyn2 controls cytoskeletal remodeling by Rac (52), a small GTPase that has been implicated in the function of Nef (12, 53). However, the modulation of Rac function by Dyn2 appears unrelated to clathrin-mediated endocytosis (52), whereas our results imply that Dyn2 and clathrin are both required for the enhancement of infectivity by Nef, suggesting that this activity of Nef depends on the function of Dyn2 in the budding of clathrin-coated vesicles.
Even though dominant-negative Dyn2 did not affect the incorporation of Env, our results suggest that Env determines the requirement for Dyn2 and clathrin for optimal infectivity, because the infectivity of VSV-G-pseudotyped HIV-1 cores was unaffected by dominant-negative Dyn2. It has been shown that high surface levels of CD4 interfere with the function of HIV-1 Env, and that the Nef-induced endocytosis of CD4 relieves this effect (54). Based on this precedent, it is conceivable that Env is also targeted by another host protein that is subject to down-regulation by Nef. The role of Dyn2 in the down-regulation of CD4 is unlikely to explain its requirement for optimal infectivity in the presence of Nef, because dominant-negative Dyn2 inhibited the effect of Nef on viral infectivity even in CD4-negative cells. Furthermore, although Dyn1 supports the Nef-induced endocytosis of CD4, it cannot substitute for Dyn2 in the enhancement of infectivity by Nef. Of note, the latter observation does not exclude that the endocytosis function of Dyn2 is involved in the infectivity enhancement activity of Nef, because there is evidence that Dyn1 and Dyn2 preferentially function at distinct plasma membrane domains (36).
It has been suggested that Nef enhances HIV-1 infectivity by increasing the cholesterol content of progeny virions, based on the observation that Nef impaired the efflux of cholesterol from macrophages by down-regulating the ATP-binding cassette transporter A1 (55). However, our results argue against the possibility that Dyn2 is required for a Nef-mediated increase in virion cholesterol, because Dyn2(K44A) did not counteract the effect of Nef but rather appeared to moderately increase virion cholesterol levels (SI Fig. 11). Nevertheless, it remains possible that the Dyn2-dependent down-regulation of a host factor by Nef alters the lipid content of the virion envelope. For instance, the plasma-membrane association of certain P-type ATPases that generate transbilayer phospholipid asymmetry is regulated via endocytosis (56). Also, there is evidence that clathrin-dependent endocytosis preferentially internalizes nonraft membrane (57), which may provide a mechanism for the enrichment of plasma membrane microdomains by Nef. Consistent with this scenario, Nef was shown to increase HIV-1 infectivity by enhancing budding from specialized lipid domains (58). In conclusion, although further investigation is needed to determine exactly how Nef modifies progeny virions in a Dyn2- and clathrin-dependent manner, the evidence presented here identifies a cellular pathway exploited by Nef to enhance HIV infectivity.
Materials and Methods
Mammalian Expression Plasmids.
The nef genes of SIVmac239 and of HIV-1 LAI (subtype B), SF2 (subtype B), 97ZA012 (subtype C), 94UG114 (subtype D), 93BR020 (subtype F), and 90CF056 (subtype H) were amplified with a C-terminal HA tag and inserted into pBJ5. A _nef_-deficient variant called pNefFS of the HIV-1LAI Nef expression vector (denoted pNefLAI) harbors a frameshift after nef codon 35. DNAs encoding Dyn2 or Dyn1 with or without an N-terminal FLAG tag were amplified from a human fetus cDNA library (Clontech, Mountain View, CA) or from human EST clone AL538642 (Invitrogen, San Diego, CA) and inserted into pBJ5. Point mutations were introduced by PCR-based mutagenesis. In the vectors encoding Dyn2r and FLAG-Dyn2r, silent mutations at the _Dyn2_siRNA target site changed the wild-type sequence from 472GACATGATCCTGCAGTTCA489 to 471GATATGATTCTCCAGTTTA489 (nucleotides in bold represent positions that were mutated).
Proviral Constructs.
NL4-3/nef− is a _nef_-deficient variant of HIV-1NL4-3 with a frameshift at a unique XhoI site in nef. The _env_-deficient NL4-3/env− and NL4-3/env−/nef− variants harbor a frameshift mutation at the unique NheI site of NL4-3. HXB/nef+, which harbors the HIV-1LAI nef gene, and _nef_- and _env_-deficient variants of HXB/nef+ have been described (59). Mutations into the nef gene of HXB/nef+ were introduced by inserting mutant nef sequences derived from pBJ5-based expression vectors.
Coimmunoprecipitations.
293T and Jurkat TAg cells were transfected with vectors encoding HA- and FLAG-tagged proteins by using Lipofectamine 2000 (Invitrogen) and lysed after 48 h in buffer containing 0.5% _n_-octyl glucoside. The lysates were centrifuged at 16,000 or alternatively at 100,000 × g to rule out artifacts caused by incomplete solubilization, and the supernatants were immunoprecipitated with anti-HA antibody HA.11 (Covance, Princeton, NJ). Immunoprecipitates were analyzed by silver staining or immunoblotting with antisera against the PRD of Dyn2 or Dyn3 (Affinity BioReagents, Neshanic Station, NJ), anti-FLAG antibody M2 (Sigma, St. Louis, MO), or anti-HA antibody. Protein identification was achieved by liquid chromatography coupled-tandem mass spectrometry of tryptic peptides.
Virus Production and Infectivity.
Virions capable of a single round of replication were produced by transfecting Jurkat TAg cells with Lipofectamine 2000 or COS-7 cells with calcium phosphate by using 2 μg of _env_-deficient HIV proviral DNA, vectors encoding HIV-1 Env or VSV-G (0.5 μg), and empty pBJ5 or variants encoding wild-type or mutant Dyn2 (5 μg).
Lipofectamine 2000 was also used to transfect Jurkat TAg and 293T cells with siRNA (80 pmol) along with pEGFP-N1 (Clontech), _env_-deficient HIV proviral DNA, and a vector providing HIV-1 Env. siRNAs targeting Dyn2 (GACAUGAUCCUGCAGUUCAUU), clathrin heavy chain (AAUGGAUCUCUUUGAAUACGG), and a control siRNA (Luciferase GL3 complex) were purchased from Dharmacon (Lafayette, CO). For siRNA rescue assays, Jurkat TAg cells were transfected as above, except that a vector encoding Dyn2r or empty pBJ5 (4 μg) was included. Alternatively, vectors encoding FLAG-tagged versions of Dyn2r or Dyn1 were cotransfected in the amounts indicated, and the total amount of transfected DNA was kept constant with empty pBJ5 vector. At 96 h after transfection, surface CD71 expression levels on cells gated for GFP expression were determined by two-color flow cytometry. Additionally, GFP-positive cells were sorted by flow cytometry and analyzed by immunoblotting with anti-β-actin (Sigma) and anti-Dyn2 or anti-FLAG antibody.
Virus-containing supernatants were harvested 48 h after transfection, or from 84 to 96 h after transfection if siRNA was cotransfected. Single-cycle infectivities were determined in triplicate by challenging HeLa P4–2 indicator cells with viruses normalized for reverse transcriptase activity. Infectivities were evaluated by measuring the HIV-1 Tat-mediated induction of β-galactosidase activity in the target cell lysates, or by staining infected cells with X-Gal and by counting blue-stained foci under a light microscope. Target cells were stained 48 h after infection with trans-complemented viruses, or 18 h after infection with _env_-positive viruses.
Supplementary Material
Supporting Information
Acknowledgments
We thank S. Gygi for LC-MS/MS, S. Khurana and M. Thali for immunofluorescence analyses, H. McMahon (Medical Research Council Laboratory of Molecular Biology, Cambridge, U.K.) for AP180-C, D. Ott (National Cancer Institute, Frederick, MD) for anti-p6 antiserum, B. Strack and S. Craig for research support, and M. O. McClure for helpful suggestions and support (to M.P.). Primary HIV-1 isolates were obtained from the HIV Vaccine Initiative of the World Health Organization–Joint United Nations Programme on HIV/AIDS through the National Institutes of Health AIDS Research and Reference Reagent Program. This study was supported by National Institutes of Health Grants AI54261 and AI29873 (to H.G.G.) and by a grant from Rome AIDS Project N.40F.57 (to G.P.).
Abbreviations
VSV-G
vesicular stomatitis virus glycoprotein
Cyp
cyclophilin A
PRD
proline-rich domain
GED
GTPase effector domain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Kestler HW, III, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 2.Garcia JV, Miller AD. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 3.Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 4.Benson RE, Sanfridson A, Ottinger JS, Doyle C, Cullen BR. J Exp Med. 1993;177:1561–1566. doi: 10.1084/jem.177.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mariani R, Skowronski J. Proc Natl Acad Sci USA. 1993;90:5549–5553. doi: 10.1073/pnas.90.12.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 7.Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 8.Renkema GH, Manninen A, Mann DA, Harris M, Saksela K. Curr Biol. 1999;9:1407–1410. doi: 10.1016/s0960-9822(00)80086-x. [DOI] [PubMed] [Google Scholar]
- 9.Geleziunas R, Xu W, Takeda K, Ichijo H, Greene WC. Nature. 2001;410:834–838. doi: 10.1038/35071111. [DOI] [PubMed] [Google Scholar]
- 10.Fackler OT, Luo W, Geyer M, Alberts AS, Peterlin BM. Mol Cell. 1999;3:729–739. doi: 10.1016/s1097-2765(01)80005-8. [DOI] [PubMed] [Google Scholar]
- 11.Fackler OT, Lu X, Frost JA, Geyer M, Jiang B, Luo W, Abo A, Alberts AS, Peterlin BM. Mol Cell Biol. 2000;20:2619–2627. doi: 10.1128/mcb.20.7.2619-2627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janardhan A, Swigut T, Hill B, Myers MP, Skowronski J. PLoS Biol. 2004;2:E6. doi: 10.1371/journal.pbio.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spina CA, Kwoh TJ, Chowers MY, Guatelli JC, Richman DD. J Exp Med. 1994;179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller MD, Warmerdam MT, Gaston I, Greene WC, Feinberg MB. J Exp Med. 1994;179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowers MY, Spina CA, Kwoh TJ, Fitch NJ, Richman DD, Guatelli JC. J Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowers MY, Pandori MW, Spina CA, Richman DD, Guatelli JC. Virology. 1995;212:451–457. doi: 10.1006/viro.1995.1502. [DOI] [PubMed] [Google Scholar]
- 17.Aiken C, Trono D. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller MD, Warmerdam MT, Page KA, Feinberg MB, Greene WC. J Virol. 1995;69:579–584. doi: 10.1128/jvi.69.1.579-584.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welker R, Harris M, Cardel B, Krausslich HG. J Virol. 1998;72:8833–8840. doi: 10.1128/jvi.72.11.8833-8840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandori MW, Fitch NJ, Craig HM, Richman DD, Spina CA, Guatelli JC. J Virol. 1996;70:4283–4290. doi: 10.1128/jvi.70.7.4283-4290.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng YH, Plemenitas A, Fielding CJ, Peterlin BM. Proc Natl Acad Sci USA. 2003;100:8460–8465. doi: 10.1073/pnas.1437453100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fackler OT, Moris A, Tibroni N, Giese SI, Glass B, Schwartz O, Krausslich HG. Virology. 2006;351:322–339. doi: 10.1016/j.virol.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 23.Aiken C. J Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell EM, Nunez R, Hope TJ. J Virol. 2004;78:5745–5755. doi: 10.1128/JVI.78.11.5745-5755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldsmith MA, Warmerdam MT, Atchison RE, Miller MD, Greene WC. J Virol. 1995;69:4112–4121. doi: 10.1128/jvi.69.7.4112-4121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinshaw JE. Annu Rev Cell Dev Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smirnova E, Shurland DL, Newman-Smith ED, Pishvaee B, van der Bliek AM. J Biol Chem. 1999;274:14942–14947. doi: 10.1074/jbc.274.21.14942. [DOI] [PubMed] [Google Scholar]
- 28.Cohen GB, Rangan VS, Chen BK, Smith S, Baltimore D. J Biol Chem. 2000;275:23097–23105. doi: 10.1074/jbc.M000536200. [DOI] [PubMed] [Google Scholar]
- 29.Liu LX, Heveker N, Fackler OT, Arold S, Le Gall S, Janvier K, Peterlin BM, Dumas C, Schwartz O, Benichou S, et al. J Virol. 2000;74:5310–5319. doi: 10.1128/jvi.74.11.5310-5319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saksela K, Cheng G, Baltimore D. EMBO J. 1995;14:484–491. doi: 10.1002/j.1460-2075.1995.tb07024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piguet V, Gu F, Foti M, Demaurex N, Gruenberg J, Carpentier JL, Trono D. Cell. 1999;97:63–73. doi: 10.1016/s0092-8674(00)80715-1. [DOI] [PubMed] [Google Scholar]
- 32.Craig HM, Pandori MW, Guatelli JC. Proc Natl Acad Sci USA. 1998;95:11229–11234. doi: 10.1073/pnas.95.19.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bresnahan PA, Yonemoto W, Ferrell S, Williams-Herman D, Geleziunas R, Greene WC. Curr Biol. 1998;8:1235–1238. doi: 10.1016/s0960-9822(07)00517-9. [DOI] [PubMed] [Google Scholar]
- 34.Craig HM, Pandori MW, Riggs NL, Richman DD, Guatelli JC. Virology. 1999;262:55–63. doi: 10.1006/viro.1999.9897. [DOI] [PubMed] [Google Scholar]
- 35.Greenberg M, DeTulleo L, Rapoport I, Skowronski J, Kirchhausen T. Curr Biol. 1998;8:1239–1242. doi: 10.1016/s0960-9822(07)00518-0. [DOI] [PubMed] [Google Scholar]
- 36.Altschuler Y, Barbas SM, Terlecky LJ, Tang K, Hardy S, Mostov KE, Schmid SL. J Cell Biol. 1998;143:1871–1881. doi: 10.1083/jcb.143.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Gall S, Buseyne F, Trocha A, Walker BD, Heard JM, Schwartz O. J Virol. 2000;74:9256–9266. doi: 10.1128/jvi.74.19.9256-9266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swann SA, Williams M, Story CM, Bobbitt KR, Fleis R, Collins KL. Virology. 2001;282:267–277. doi: 10.1006/viro.2000.0816. [DOI] [PubMed] [Google Scholar]
- 39.Welker R, Kottler H, Kalbitzer HR, Krausslich HG. Virology. 1996;219:228–236. doi: 10.1006/viro.1996.0240. [DOI] [PubMed] [Google Scholar]
- 40.Burtey A, Rappoport JZ, Bouchet J, Basmaciogullari S, Guatelli J, Simon SM, Benichou S, Benmerah A. Traffic. 2007;8:61–76. doi: 10.1111/j.1600-0854.2006.00512.x. [DOI] [PubMed] [Google Scholar]
- 41.Damke H, Baba T, Warnock DE, Schmid SL. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 43.Hinrichsen L, Harborth J, Andrees L, Weber K, Ungewickell EJ. J Biol Chem. 2003;278:45160–45170. doi: 10.1074/jbc.M307290200. [DOI] [PubMed] [Google Scholar]
- 44.Foti M, Mangasarian A, Piguet V, Lew DP, Krause KH, Trono D, Carpentier JL. J Cell Biol. 1997;139:37–47. doi: 10.1083/jcb.139.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arold S, Franken P, Strub MP, Hoh F, Benichou S, Benarous R, Dumas C. Structure (London) 1997;5:1361–1372. doi: 10.1016/s0969-2126(97)00286-4. [DOI] [PubMed] [Google Scholar]
- 46.Breuer S, Gerlach H, Kolaric B, Urbanke C, Opitz N, Geyer M. Biochemistry. 2006;45:2339–2349. doi: 10.1021/bi052052c. [DOI] [PubMed] [Google Scholar]
- 47.Dennis CA, Baron A, Grossmann JG, Mazaleyrat S, Harris M, Jaeger J. Proteins. 2005;60:658–669. doi: 10.1002/prot.20544. [DOI] [PubMed] [Google Scholar]
- 48.Lee E, De Camilli P. Proc Natl Acad Sci USA. 2002;99:161–166. doi: 10.1073/pnas.012607799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orth JD, Krueger EW, Cao H, McNiven MA. Proc Natl Acad Sci USA. 2002;99:167–172. doi: 10.1073/pnas.012607899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fish KN, Schmid SL, Damke H. J Cell Biol. 2000;150:145–154. doi: 10.1083/jcb.150.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson HM, Cao H, Chen J, Euteneuer U, McNiven MA. Nat Cell Biol. 2004;6:335–342. doi: 10.1038/ncb1112. [DOI] [PubMed] [Google Scholar]
- 52.Schlunck G, Damke H, Kiosses WB, Rusk N, Symons MH, Waterman-Storer CM, Schmid SL, Schwartz MA. Mol Biol Cell. 2004;15:256–267. doi: 10.1091/mbc.E03-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu X, Wu X, Plemenitas A, Yu H, Sawai ET, Abo A, Peterlin BM. Curr Biol. 1996;6:1677–1684. doi: 10.1016/s0960-9822(02)70792-6. [DOI] [PubMed] [Google Scholar]
- 54.Lama J, Mangasarian A, Trono D. Curr Biol. 1999;9:622–631. doi: 10.1016/s0960-9822(99)80284-x. [DOI] [PubMed] [Google Scholar]
- 55.Mujawar Z, Rose H, Morrow MP, Pushkarsky T, Dubrovsky L, Mukhamedova N, Fu Y, Dart A, Orenstein JM, Bobryshev YV, et al. PLoS Biol. 2006;4:e365. doi: 10.1371/journal.pbio.0040365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, Tanaka K. Mol Biol Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nichols B. J Cell Sci. 2003;116:4707–4714. doi: 10.1242/jcs.00840. [DOI] [PubMed] [Google Scholar]
- 58.Zheng Y, Plemenitas A, Linnemann T, Fackler OT, Peterlin BM. Curr Biol. 2001;11:875–879. doi: 10.1016/s0960-9822(01)00237-8. [DOI] [PubMed] [Google Scholar]
- 59.Dorfman T, Popova E, Pizzato M, Gottlinger HG. J Virol. 2002;76:6857–6862. doi: 10.1128/JVI.76.13.6857-6862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information